Abstract
Dexamethasone is widely used as an immunosuppressive therapy and recently as COVID-19 treatment. Here, we demonstrate that dexamethasone sensitizes to ferroptosis, a form of iron-catalyzed necrosis, previously suggested to contribute to diseases such as acute kidney injury, myocardial infarction, and stroke, all of which are triggered by glutathione (GSH) depletion. GSH levels were significantly decreased by dexamethasone. Mechanistically, we identified that dexamethasone up-regulated the GSH metabolism regulating protein dipeptidase-1 (DPEP1) in a glucocorticoid receptor (GR)–dependent manner. DPEP1 knockdown reversed the phenotype of dexamethasone-induced ferroptosis sensitization. Ferroptosis inhibitors, the DPEP1 inhibitor cilastatin, or genetic DPEP1 inactivation reversed the dexamethasone-induced increase in tubular necrosis in freshly isolated renal tubules. Our data indicate that dexamethasone sensitizes to ferroptosis by a GR-mediated increase in DPEP1 expression and GSH depletion. Together, we identified a previously unknown mechanism of glucocorticoid-mediated sensitization to ferroptosis bearing clinical and therapeutic implications.
Dexamethasone leads to GR-mediated increased DPEP1 expression and GSH depletion, resulting in higher ferroptosis sensitivity.
INTRODUCTION
Glucocorticoids are used in clinical routine to treat leukemias (1, 2) and COVID-19 (3), for immunosuppression during solid organ transplantation (4), and in conjunction with cancer radiation therapy or, e.g., cataract surgery to reduce edema in critical areas (5). Dexamethasone exhibits a favorably 25-fold higher anti-inflammatory potency compared with cortisol (6), but the use of dexamethasone is limited by several dose-dependent side effects, including but not limited to osteoporosis and osteonecrosis (7), myopathy (8), peptic ulcers (9), and growth retardation (10).
We and others (11, 12) recently reported exquisite sensitivity of steroid-producing adrenocortical carcinomas to ferroptosis induction. Ferroptosis was first reported to be induced by erastin, an inhibitor of system Xc−, a plasma membrane antiporter involved in the regulation of glutathione (GSH) concentrations (13). Ferroptosis is an iron-catalyzed form of regulated necrosis (13, 14) that is critically involved in the pathophysiology of myocardial infarction (15–17), acute stroke (18–20), neurodegeneration (21), kidney injury (22–24), and in the context of transplantation (25–27).
Given the potential role of necrotic cell death in untoward effects of steroids, we hypothesized that dexamethasone might control signaling pathways of regulated necrosis. We found dexamethasone and other glucocorticoids to lower the threshold to ferroptosis by decreasing GSH content in a glucocorticoid receptor (GR)–dependent manner. By means of RNA sequencing (RNA-seq), we identified the glutathione regulator dipeptidase-1 (DPEP1) to mediate the ferroptosis-sensitizing effect by dexamethasone.
RESULTS
Dexamethasone sensitizes to erastin-induced ferroptosis but not to RSL3-induced ferroptosis
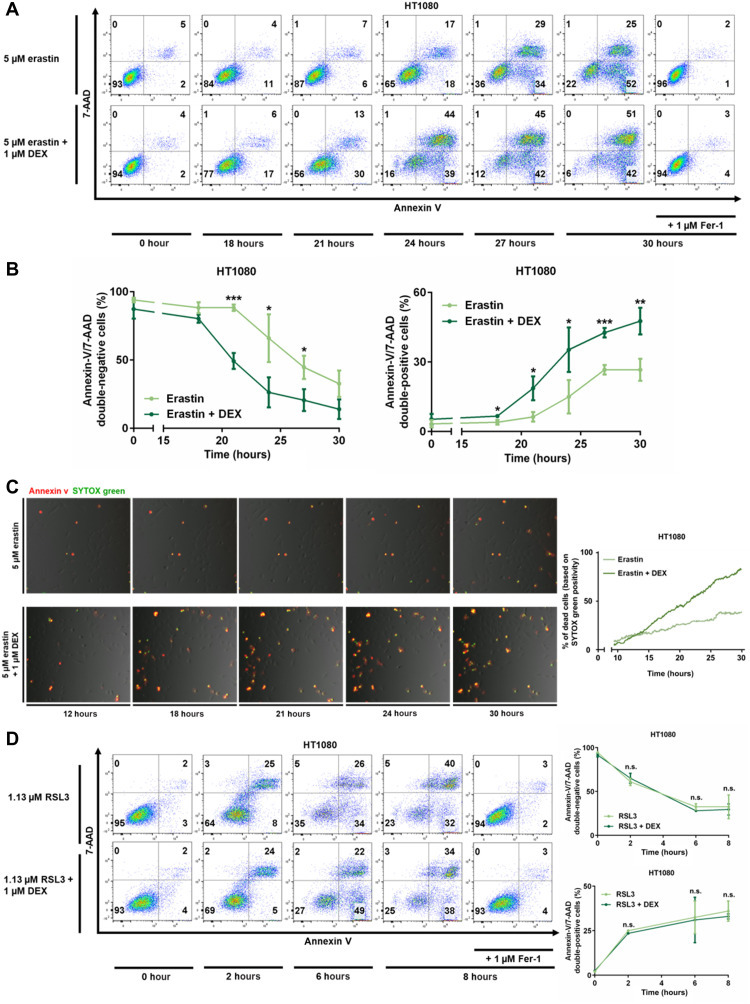
Dexamethasone was previously demonstrated to induce intrinsic apoptosis in selected cells, such as primary murine thymocytes and RS4;11 cells. In our hands, dexamethasone caused fractions of thymocytes to become annexin V positive, and this effect was reversed by the caspase inhibitor emricasan (fig. S1A). Similarly, RS4;11 cells externalized phosphatidylserine to the outer leaflet of the plasma membrane upon dexamethasone stimulation, and again, this effect was reversed by caspase inhibition (fig. S1B). No regulatory role of dexamethasone was detected in anti–Fas-induced extrinsic apoptosis of Jurkat T cells (fig. S1C). With respect to ferroptosis, research has been typically performed in HT1080 cells, but an effect of dexamethasone on ferroptosis progression has not been investigated to the best of our knowledge. We first examined whether dexamethasone alone induces cell death in HT1080 cells. Treatment of these cells for 20 to 40 hours with 20 μM dexamethasone (fig. S2A), and even ultrahigh concentrations of 100 μM for 50 hours (fig. S2B), did not cause any detectable cell death when compared to untreated controls. Next, we induced ferroptosis by inhibition of system Xc− through 5 μM erastin. By flow cytometry, we detected significantly less annexin V/7AAD double-negative cells (living cells) and reciprocally significantly higher percentage of annexin V/7AAD double-positive cells in the erastin + dexamethasone–treated group as compared with the erastin only–treated group within the first 30 hours (Fig. 1, A and B). To directly assess the effect of dexamethasone on ferroptosis, we generated a three-dimensionally (3D) printed incubation chamber that contains two sides of one well separated by a glass slide (see fig. S3 and Materials and Methods for details). This chamber allowed us to perform time-lapse imaging with a single camera for the entire observation period of 30 hours following preincubation with vehicle or 1 μM dexamethasone. We added SYTOX green and annexin V to both sides of the chamber and stimulated the cells with 5 μM erastin. An earlier and more pronounced positivity for SYTOX green was detected in the dexamethasone-treated cells (Fig. 1C). Upon quantification, as many as 85% of cells exhibited a SYTOX-positive signal at 30 hours following 5 μM erastin + 1 μM dexamethasone treatment, while approximately only 40% of the cells were positive in the erastin-stimulated controls (Fig. 1C). Collectively, these data suggested that 1 μM dexamethasone significantly sensitizes HT1080 cells to erastin-induced ferroptosis.
Fig. 1. Dexamethasone sensitizes to erastin-induced, but not to RSL3-induced, ferroptosis.
(A) HT1080 cells were treated for indicated times with 5 μM erastin and 1 μM Fer-1 with or without pretreatment of 1 μM dexamethasone for 12 hours. 7-AAD and annexin V were read out by fluorescence-activated cell sorting (FACS). (B) Quantification of data presented in (A) and repetitions of this experiment. (C) Still images of indicated times of a double-chamber time-lapse imaging and quantification of SYTOX-positive cells over time in HT1080 cells pretreated with dexamethasone as indicated. (D) HT1080 cells were induced to undergo ferroptosis by the GSH peroxidase 4 (GPX4) inhibitor RSL3 and assessed as in (A) after indicated times. Experiments were repeated at least three times, and representative images and FACS plots are shown. The graphs show means ± SD. Statistical analysis was performed using Student’s t test for each time point. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, n.s., nonsignificant.
We next set out to induce ferroptosis by the small-molecule RSL3, a well-characterized inhibitor of the glutathione peroxidase 4 (GPX4). In contrast to ferroptosis induction by erastin, dexamethasone pretreatment did not change the percentage of annexin V/7AAD–negative cells upon RSL3-induced ferroptosis induction (Fig. 1E). The difference between erastin- and RSL3-induced ferroptosis induction suggests that the dexamethasone-induced acceleration of ferroptosis is limited to the GSH regulatory part of the ferroptosis pathway. Theoretically, lower GSH levels might still influence GPX4 activity. To test this possibility in more detail, we investigated 36 hours of pretreatment with dexamethasone before RSL3 treatment (fig. S4A) and tested different concentrations of RSL3 (fig. S4B), especially sublethal concentrations up to 50 nM (fig. S4C). In conclusion, these data suggest that dexamethasone does not affect RSL3-induced ferroptosis.
Glucocorticoids, but not aldosterone or dehydroepiandrostendione, sensitize to erastin-induced ferroptosis
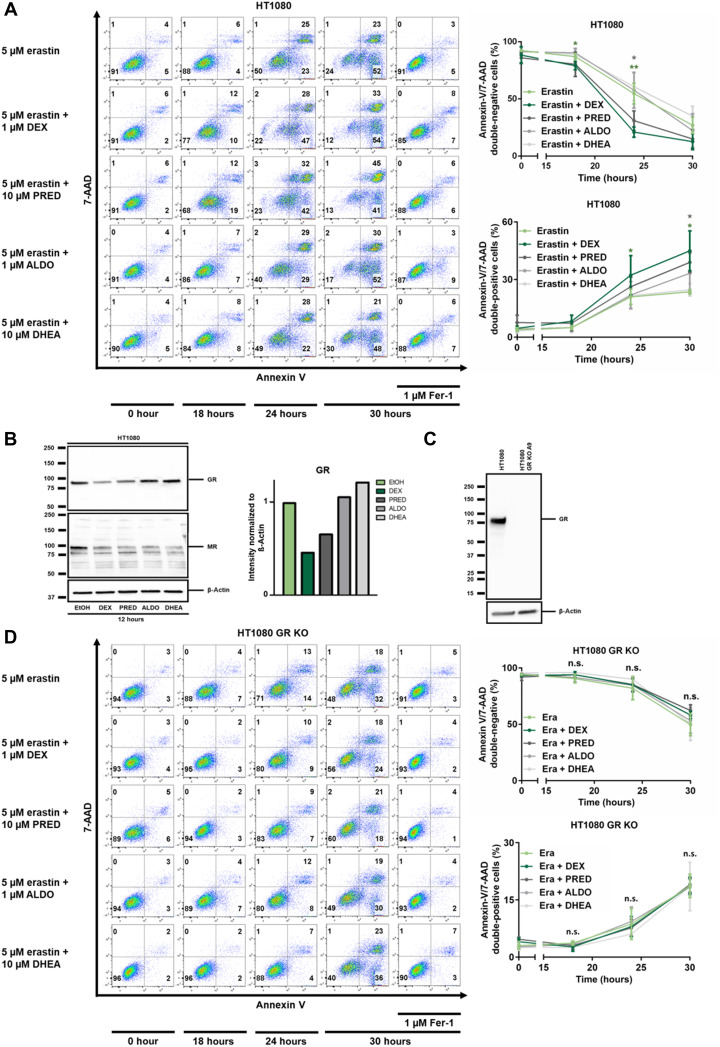
Given the potential redundancy between steroid hormones, we next asked whether other steroid hormones might phenocopy the dexamethasone effect on ferroptosis. While dexamethasone exhibited the strongest effect, prednisolone and, if at all only to a very minor extent, also aldosterone exhibited sensitization in this assay, while the coincubation with dehydroepiandrostendione (DHEA) had no effect (Fig. 2A). It is well known that treatment with glucocorticoids results in receptor down-regulation of the GR, and we confirmed this effect in ferroptosis-sensitive cells following stimulation with dexamethasone and prednisolone. In contrast, aldosterone and DHEA did not result in a similarly significant down-regulation of the GR. As an additional control, we investigated the expression levels of the mineralocorticoid receptor (MR), which is known to be down-regulated by aldosterone and found that also dexamethasone and prednisolone down-regulated the MR (Fig. 2B). In summary, these data suggested that the dexamethasone-induced sensitization to ferroptosis might be mediated through the GR.
Fig. 2. Steroid-induced sensitization to erastin-induced ferroptosis requires the GR 1.
(A) HT1080 cells were treated with erastin, Fer-1, 1 μM dexamethasone (DEX), 10 μM prednisolone (PRED), 1 μM aldosterone (ALDO), and 10 μM dehydroepiandrostendione (DHEA) as indicated. Primary FACS plots and respective quantifications of indicated populations are demonstrated. Note the sensitization to ferroptosis induced by DEX and PRED, but not by ALDO and DHEA. (B) Assessment of GR and mineralocorticoid receptor (MR) down-regulation in response to indicated steroid hormones. β-Actin serves as a loading control. (C) CRISPR-Cas9–mediated knockout of GR from HT1080 cells, confirmed by Western blotting. (D) Experiment performed as described for (A), but using GR-knockout cells. Note the loss of sensitization to ferroptosis. All experiments shown are representative of at least three independent complete repetitions performed. The graphs show means ± SD. Statistical analysis was performed using Student’s t test for each time point. *P ≤ 0.05, **P ≤ 0.01, n.s., nonsignificant.
Dexamethasone-induced sensitization to ferroptosis requires the GR
Dexamethasone and other glucocorticoids were described to control cellular processes either transcriptionally through the GR or in a GR-independent manner. To test the role of the GR in our system, we stably transfected HT1080 cells with Cas9 and added guide RNAs to generate an HT1080 crKO (CRISPR knockout) of the GR (Fig. 2C). In contrast to control cells, deletion of the GR entirely reversed the sensitization to erastin-induced ferroptosis mediated by dexamethasone or prednisolone (Fig. 2D). These results demonstrate that glucocorticoid-induced sensitization to erastin-induced ferroptosis requires the GR.
Dexamethasone treatment reduces GSH in HT1080 cells
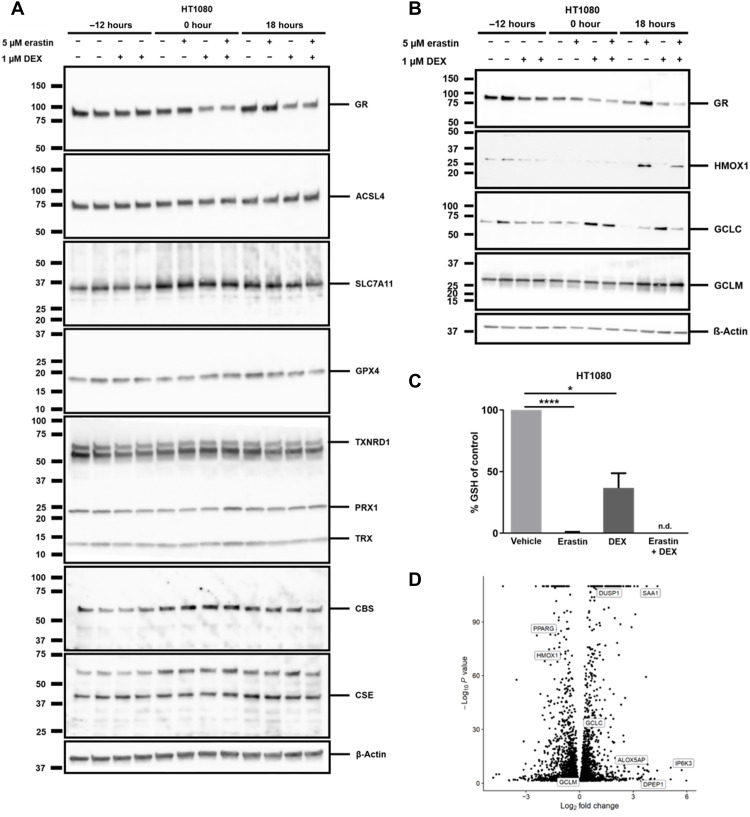
To understand how GR stimulation by dexamethasone influences erastin-induced but not RSL3-induced ferroptosis, we first looked at protein expression levels of known key players of the ferroptosis pathway. As demonstrated in Fig. 3A, no significant changes in protein expression of acyl-CoA synthetase long chain family member 4 (ACSL4), solute carrier family 7 member 11 (SLC7A11), GPX4, thioredoxin reductase 1 (TXNRD1), peroxiredoxin 1 (PRX1), thioredoxin (TRX), cystathionine beta-synthase (CBS), or cystathionine gamma lyase (CSE) were detected following dexamethasone stimulation. In addition, we investigated the protein expression levels of heme oxygenase 1 (HMOX1), glutamate-cysteine ligase catalytic subunit (GCLC), and glutamate-cysteine ligase modifier subunit (GCLM) (Fig. 3B). As expected, HOMX1 was up-regulated by erastin (28, 29), while GCLC was up-regulated by dexamethasone. To gain further insight into the cellular redox state, we used liquid chromatography electrospray ionization quadrupole-time-of-flight (LC-ESI-QToF) mass spectrometry (MS) to accurately assess the level of GSH upon treatment with 1 μM dexamethasone (fig. S5A). As a GSH-depletion control, erastin was added as indicated in Fig. 3C. While erastin entirely depleted the GSH pool, dexamethasone resulted in a greater than 50% reduction of GSH content without significant numbers of cells undergoing actual ferroptosis (plasma membrane rupture; fig. S5B). This experiment explains why pretreatment with dexamethasone sensitizes to erastin-induced ferroptosis, while RSL3-induced ferroptosis remains unaltered, as the latter bypasses the need for GSH depletion upon ferroptosis induction. In line with this conclusion, β-mercaptoethanol also prevented cell death and GSH depletion in comparable settings (fig. S6, A and B).
Fig. 3. An unbiased bulk RNA-seq of parental HT1080 and GR-crKO cells reveals dexamethasone-induced genes involved in ferroptosis.
(A) Protein expression of key players in ferroptosis 12 hours after treatment with or without 1 μM dexamethasone with subsequent treatment of induction of ferroptosis for 18 hours with erastin. β-Actin serves as a loading control. (B) Protein expression of HMOX1, GCLC, and GCLM 12 hours after treatment with or without 1 μM dexamethasone with subsequent treatment of induction of ferroptosis for 18 hours with erastin. β-Actin serves as a loading control. (C) GSH content in HT1080 cells treated with or without 1 μm dexamethasone before inducing ferroptosis with 5 μM erastin for 14 hours. (D) Volcano plot of differently regulated genes upon dexamethasone treatment in HT1080 cells. The bar graph shows means ± SD. Statistical analysis was performed using Student’s t test. *P ≤ 0.05, ****P ≤ 0.0001 (GR: glucocorticoid receptor, ACSL4: acyl-CoA synthetase long chain family member 4, SCL7A11: solute carrier family 7 member 11, GPX4: GSH peroxidase 4, TXNRD1: thioredoxin reductase 1, PRX1: peroxiredoxin 1, TRX: thioredoxin, CBS: cystathionine beta-synthase, CSE: cystathionine gamma lyase, HMOX1: heme oxygenase 1, GCLC: glutamate-cysteine ligase catalytic subunit, GCLM: glutamate-cysteine ligase modifier subunit, GSH: glutathione, n.d.: not detectable).
Dexamethasone induces DPEP1 expression
To identify potentially unknown regulators of ferroptosis sensitivity downstream of GR activation, we performed an unbiased bulk RNA-seq (fig. S7A). We confirmed accuracy of the investigated groups by principal component 1 (PC1)/PC2 variance (fig. S7B) and quality control assessments for RNA-seq accuracy (fig. S8, A and B) and assessed a hierarchical cluster of RNA-seq results (fig. S8C). In addition, we performed alignment statistics for RNA-seq analysis (fig. S8D). Moreover, we validated the GR knockdown in the RNA-seq analysis (fig. S8E). Among the most abundantly up- and down-regulated genes (data S1 and S2), several genes encoding for proteins previously associated with redox homeostasis appeared as demonstrated in a volcano plot in Fig. 3D. Among the top hits of up-regulated proteins, we decided to further validate DPEP1, also known as dehydropeptidase-I (DHP-I), microsomal dipeptidase (MDP), or renal dipeptidase (30). DPEP1 is a zinc-dependent metalloproteinase that has been shown to process antibiotics and hydrolyze a variety of peptides, including GSH breakdown products [cysteinylglycine (Cys-Gly)] (31). GSH was previously suggested to be involved in acute kidney injury (32).
Knockdown of DPEP1 reverts dexamethasone-licensed sensitization to erastin-induced ferroptosis
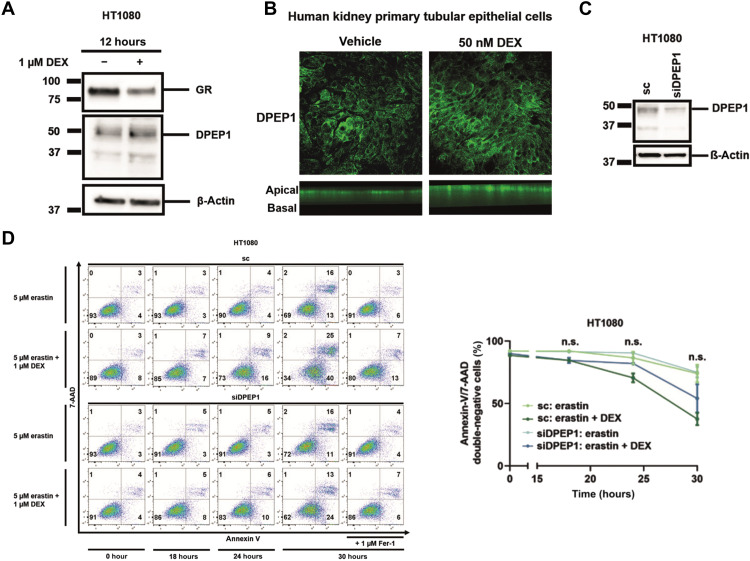
DPEP1 is up-regulated by treatment with 1 μM dexamethasone for 12 hours at the protein level (Fig. 4A). In keeping with this observation, DPEP1-labeled immunofluorescence in human primary kidney tubular epithelial cells cultured in Transwells exhibited an increase upon dexamethasone treatment while maintaining apical expression/polarity (Fig. 4B). In summary, these results confirm that dexamethasone indeed up-regulates protein expression of DPEP1. To test a functional role of DPEP1, we performed knockdown experiments in HT1080 cells and confirmed knockdown efficiency (Fig. 4C). As demonstrated in Fig. 4D, the knockdown of DPEP1 reverses dexamethasone sensitization to erastin-induced ferroptosis. However, as observed for the knockdown of DPEP1, the DPEP1 inhibitor cilastatin alone does not protect from erastin-induced ferroptosis (fig. S9, A to C).
Fig. 4. Dexamethasone-mediated sensitization to erastin-induced ferroptosis is mediated by DPEP1.
(A) HT1080 cells were treated for 12 hours with dexamethasone before investigating protein expression levels of the GR and DPEP1. (B) Human kidney primary tubular epithelial cells (hTEC) were grown on Transwells and treated for 24 hours with dexamethasone before fluorescently labeling DPEP1. (C) HT1080 cells were treated 48 hours with siRNA against DPEP1, and knockdown efficacy was confirmed by Western blot. (D) HT1080 cells were treated with erastin and dexamethasone as indicated following the 48-hour pretreatment with an siRNA against DPEP1. Primary FACS plots and respective quantifications of indicated populations are demonstrated. The graphs show means ± SD. Statistical analysis was performed using Student’s t test for each time point. sc, scrambled; n.s., nonsignificant.
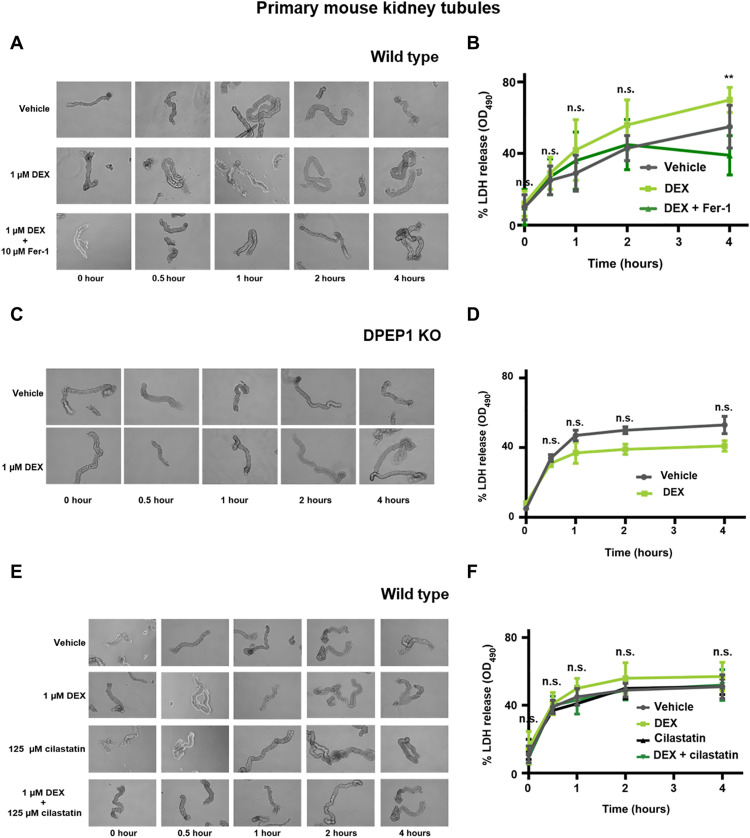
Dexamethasone accelerates the spontaneous lactate dehydrogenase release from freshly isolated murine renal tubules
Ferroptosis drives spontaneous acute tubular necrosis in freshly isolated renal tubules (33). We used this ex vivo model (fig. S10) to assess the effect of dexamethasone on lactate dehydrogenase (LDH) release from renal tubules. Indeed, dexamethasone cotreatment led to significantly increased LDH release compared with vehicle-treated tubules, and this effect was reversed by the addition of ferrostatin-1 (Fer-1) (Fig. 5, A and B) or β-mercaptoethanol (fig. S11, A and B). Dexamethasone failed to accelerate LDH release from tubules isolated from DPEP1-deficient mice (Fig. 5, C and D). In keeping with a role of DPEP1 during the dexamethasone-induced sensitization, dexamethasone/cilastatin cotreated tubules did not exhibit higher LDH release levels as compared with vehicle-treated tubules (Fig. 5, E and F). Intriguingly, and in line with the data obtained from cell culture (fig. S9, B and C), cilastatin alone did not protect kidney tubules from LDH release. In summary, these data suggest a permissive pro-ferroptotic effect of dexamethasone mediated by the GR and DPEP1 (fig. S12).
Fig. 5. Ferroptosis in freshly isolated renal tubules is accelerated by dexamethasone in wild-type but not in DPEP1 knockout mice.
(A) Representative images of freshly isolated murine kidney tubules undergoing spontaneous cell death in the presence of either vehicle or 1 μM dexamethasone in the presence or absence of 10 μM Fer-1. (B) LDH release of respective time points. (C) Representative images of freshly isolated murine kidney tubules from DPEP1 knockout mice undergoing spontaneous cell death in the presence of vehicle or 1 μM dexamethasone. (D) LDH release of respective time points. (E) Representative images of freshly isolated murine kidney tubules undergoing spontaneous cell death in the presence of either vehicle, 1 μM dexamethasone, 125 μm cilastatin, or dexamethasone and cilastatin. (F) LDH release of respective time points. The graphs show means ± SD. Statistical analysis was performed using Student’s t test for each time point. **P ≤ 0.01.
DISCUSSION
A growing body of evidence suggests the importance of ferroptosis in acute kidney injury and tubular necrosis (22, 24, 33–39). The molecular trigger of ferroptosis, however, still remains unclear, but intracellular lactate acidosis and a shift toward lower pH might facilitate intracellular Fenton reactions. Our data urge caution on the use of dexamethasone and, potentially, other steroids, as they may sensitize to tubular necrosis by ferroptosis. Trials using DPEP1 inhibitors to protect from acute kidney injury may appear far at the horizon given this first preclinical set of data. However, the clinical use of DPEP1 inhibitors upon treatment with carbapenems, such as imipenem-cilastatin, suggests sufficient safety at least in the class of DPEP1 inhibitors.
The DPEP1 inhibitor cilastatin is in clinical use as a combined therapeutic with β-lactam antibiotics to prevent DPEP1-mediated hydrolysis of drugs such as imipenem. Dipeptidases are well-established Cys-Gly cleaving enzymes that were demonstrated to affect intra- and extracellular concentrations of GSH by regulating GSH degradation (40). Dipeptidase mutant Saccharomyces cerevisiae cannot grow with GSH as the sole sulfur source (41). Likewise, human DPEP1, initially identified as a disulfide-linked homodimer that is glycosylphosphatidylinositol anchored to the renal brush border of the kidney (42), functions as a plasma membrane–bound zinc peptidase, which regulates renal GSH metabolism (43). DPEP1 is reported to have a subcellular localization on the brush border (luminal face) of the proximal tubular epithelium (44). As ferroptosis occurs in a non–cell autonomous manner (24, 45), osmotic conditions outside of the plasma membrane, e.g., calcium gradients (46), may affect ferroptosis propagation.
DPEP1 and ferroptosis were identified in the pathogenesis of experimental contrast-induced acute kidney injury (CI-AKI) (36). We demonstrated a protective effect of the molecule necrostatin-1 (Nec-1) in a similar model (47). As Nec-1 is known to also inhibit ferroptosis (22), in retrospect, we consider it possible that the beneficial effects of Nec-1 on the CI-AKI model may be due to ferroptosis modulation rather than inhibition of receptor-interacting protein kinase 1 (RIPK1). Furthermore, a recent study identified single-nucleotide polymorphisms (SNPs) in close proximity to the DPEP1 gene to be frequently associated with AKI to chronic kidney disease progression in a large patient cohort (48). In addition, carnosine dipeptidase II (CNDP2) was demonstrated to protect cells under cysteine insufficiency by hydroliyzing GSH-related peptides (49). Further work is warranted to investigate the details of other dipeptidases in murine models of AKI and ferroptosis-related disorders.
Since the SARS-CoV-2 crisis, high-dose dexamethasone is applied to severely sick patients suffering from COVID-19. Given the millions of patients treated with this regimen, it is of paramount importance to better understand dexamethasone-associated side effects. The insight that dexamethasone lowers the threshold to ferroptosis through up-regulation of DPEP1 might be considered in future studies on the pathogenesis of steroid-induced untoward effects. In conclusion, we found the mechanism of a previously unrecognized ferroptosis-regulating function of dexamethasone.
MATERIALS AND METHODS
Reagents
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Antibodies | ||
| GR (D6H2L) XP | Cell Signaling Technology |
12041 |
| Anti-FACL4 antibody (EPR8640) | Abcam | ab155282 |
| PRX pathway (TRX, TXNRD1, and PRX1) WB cocktail |
Abcam | ab184868 |
| NR3C2 polyclonal antibody | Thermo Fisher Scientific |
PA5-79761 |
| CBS monoclonal antibody (mAB) (GT519) | Thermo Fisher Scientific |
MA5-17273 |
| Anti-gamma cystationase mouse mAB | Proteintech | 60234-1-Ig |
| Anti-GPX4 antibody (EPNCIR144) | Abcam | ab125066 |
| xCT/SLC7A11 (D2M7A) rabbit mAB | Cell Signaling Technology |
12691 |
| Recombinant anti–heme oxygenase 1 antibody (EPR1390Y) |
Abcam | ab68477 |
| Recombinant anti-GCLM antibody (EPR6667) | Abcam | ab126704 |
| Recombinant anti-GCLC antibody (EP13475) | Abcam | ab190685 |
| DPEP1 polyclonal antibody | Proteintech | 12222-1-AP |
| β-Actin cell signaling | Cell Signaling Technology |
3700S |
| Anti-mouse immunoglobulin (IgG); horseradish peroxidase (HRP)–linked antibody |
Cell Signaling Technology |
7076S |
| Anti-rabbit IgG; HRP-linked antibody | Cell Signaling Technology |
7074S |
| Alexa Fluor 488 anti-rabbit IgG | Invitrogen | A21206 |
| Compounds and chemicals | ||
| Erastin [type 1 ferroptosis inducer (FIN)] | Sigma-Aldrich | E7781 |
| RSL3 (type 2 FIN) | Selleck Chemicals |
S8155 |
| αFASL human, CH11 | Merck Millipore |
05-201 |
| Fer-1 | Merck Millipore |
341494 |
| 2-Mercaptoethanol | Thermo Fisher Scientific |
31350010 |
| zVAD fmk | BD Biosciences |
550377 |
| Emricasan | Sigma-Aldrich | SML2227 |
| Dexamethasone | Sigma-Aldrich | D4902 |
| Prednisolone | Sigma-Aldrich | P6004 |
| Aldosterone | Sigma-Aldrich | A9477 |
| DHEA | Avanti/ Sigma-Aldrich |
SKU 7000087P |
| Cilastatin | Sigma-Aldrich | C5743 |
| Octyl-β-d-glucopyranosid | Sigma-Aldrich | O8001 |
| Annexin V, Alexa Fluor 647 conjugate | Thermo Fisher Scientific |
A23204 |
| SYTOX Green | Life Technologies |
S7020 |
| 7-AAD | BD Biosciences |
559925 |
| Annexin V–fluorescein isothiocyanate (FITC) | BD Biosciences |
556420 |
| Annexin V binding buffer | BD Biosciences |
556454 |
| ProLong Gold antifade | Invitrogen | P36391 |
| LDH release assay | Promega | G1780 |
| Bradford assay | Thermo Fisher Scientific |
23225 |
| ECL Prime Western Blotting System | Thermo Fisher Scientific |
GERPN2232 |
| Opti-MEM I Medium | Thermo Fisher Scientific |
31985062 |
| Lipofectamine RNAiMAX | Thermo Fisher Scientific |
13778075 |
| DPEP1 small interfering RNA (siRNA) (s4246) | Thermo Fisher Scientific |
4392420 |
| Silencer Select Negative Control #1 siRNA | Thermo Fisher Scientific |
4390843 |
Cell lines and cell culture
Human HT1080 cells were purchased from the American Type Culture Collection. HT1080 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific) supplemented with 10% (v/v) fetal bovine serum (FBS; Thermo Fisher Scientific), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Thermo Fisher Scientific). RS4;11 cells were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ) and cultured in α-MEM with ribo- and deoxyribonucleosides (Thermo Fisher Scientific) supplemented with 10% (v/v) FBS (Thermo Fisher Scientific), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Thermo Fisher Scientific). Jurkat T cells were provided by A. Rösen-Wolff and cultured in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% (v/v) FBS (Thermo Fisher Scientific), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Thermo Fisher Scientific). Murine thymocytes were isolated as described below and cultured in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% (v/v) FBS (Thermo Fisher Scientific), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Thermo Fisher Scientific). All cells were cultured in a humidified 5% CO2 atmosphere.
Cell culture of human proximal tubular epithelial cells
Nondiseased residual kidney tissue was obtained from medically indicated human nephrectomies. Kidney tissues were collected within 2 hours of surgery in Hanks’ balanced salt solution (HBSS) supplemented with penicillin-streptomycin (5 mg/ml) on ice. Human nephrectomy sample collection was approved by the Conjoint Health Research Ethics Board at the University of Calgary and Alberta Health Services. Following removal of the renal capsule, cortical tissue was carefully cut away and minced using aseptic techniques. After digestion in collagenase (1.5 mg/ml in HBSS) at 37°C for 60 min, samples were passed through serial filters from 200 to 45 μm to remove intact glomeruli and large cellular debris. Samples were then plated on plastic cell culture plates at 37°C for 90 min in K1 culture medium DMEM/F12 containing 10% FBS, 1% penicillin-streptomycin, prostaglandin E1 (125 ng/ml), l-thyroxine (1.8 μg/ml), hydrocortisone (3.38 ng/ml), insulin-transferrin-sodium selenite supplement (2.5 mg/ml), and epithelial growth factor (10 ng/ml; Sigma-Aldrich). Human proximal tubular epithelial cells (HPTC) were collected and plated onto collagen IV–coated cell culture plates. To maintain the epithelial phenotype, HPTC were used less than two passages.
Generation of GR-deficient HT1080 cells
We used a two-vector CRISPR-Cas9 approach. In the first step, HT1080 cells were transduced with viral supernatant containing the lentiCas9-Blast vector and packing plasmids (pMDLg/pRRE, pRSV-Rev, and pMD2.G). Cells were selected using Blasiticidin (5 μg/ml; InvivoGen). After outgrowth of single cells, Cas9 expression was verified by Western blotting. Guide RNAs targeting the GR (GAACACTGGTCGACCTATTG) were cloned into the lentiGuide-Puro vector. Subsequently, Cas9-expressing HT1080 cells were transduced with viral supernatant containing the lentiGuide-Puro vector and packing plasmids. After selection with puromycin (1 μg/ml; InvivoGen), knockout efficacy was determined on protein level in single cells.
siRNA-mediated knockdown of DPEP1
HT1080 cells were plated in a petri dish in 15 ml of antibiotic-free medium. After 24 hours, 180 pmol RNAi and 24 μl of Lipofectamine (Thermo Fisher Scientific) were each mixed in 1.5 ml of Opti-MEM I Medium (Thermo Fisher Scientific) without serum, combined, and incubated at room temperature for 20 min before dropping the mixture on the cells. The following day, cells were harvested, plated into six-well plates, and cell death assays were performed as described below. Knockdown efficacy was determined on protein level.
Western blotting
Cells were lysed in ice-cold 50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 5 mM EDTA supplemented with PhosSTOP (Merck), cOmplete (Merck), and 1 mM phenylmethylsulfonyl fluoride (PMSF) for 30 min on ice. To determine DPEP1 protein levels, cells were lysed in ice-cold 50 mM tris-HCl (pH 8), 150 mM NaCl, 0.5% sodium desoxychelat, 0.1% SDS supplemented with PhosSTOP (Merck), cOmplete (Merck), 1 mM PMSF, and 100 mM Octyl-β-d-glucopyranosid for 30 min on ice. Insoluble material was removed by centrifugation (14,000g, 30 min, 4°C). Protein concentration was determined using a commercial bicinchoninic acid assay kit according to the manufacturer’s instructions (Thermo Fisher Scientific). Equal amounts of protein (typically 25 μg per lane) were resolved on a 4 to 15% gradient SDS–polyacrylamide gel electrophoresis gel and transferred to a polyvinylidene difluoride membrane (Bio-Rad). After blocking for 1 hour at room temperature, primary antibody incubation was performed at 4°C overnight. Secondary antibodies (anti-mouse, HRP-linked antibody, and anti-rabbit, HRP-linked antibody, Cell Signaling Technology) were applied at concentrations of 1:5000. Proteins were then visualized by enhanced chemiluminescence (Amersham Biosciences).
Cell death assays
Ferroptosis was induced using established FINs—type 1 FIN: erastin (Sigma-Aldrich), or type 2 FIN: RSL3 (Selleckchem). Cells were seeded in six-well plates or petri dishes and pretreated for 12 hours with 1 μM dexamethasone before induction of ferroptosis. Unless otherwise indicated, we used 5 μM erastin and 1.13 μM RSL3. After indicated time points, cells were collected and prepared for flow cytometry, immunoblotting, or live-cell imaging.
Flow cytometry
Cells were harvested, and the pellets were washed twice in phosphate-buffered saline (PBS) and stained with 5 μl of 7-AAD (BD Biosciences) and 5 μl of annexin V–FITC (BD Biosciences) added to 100 μl of annexin V–binding buffer (BD Biosciences). After 15 min, cells were recorded on the LSR II with the FACS Diva 6.1.1 software (BD Biosciences) and subsequently analyzed with the FlowJo v10 software (Tree Star). The flow cytometry procedure was supported by the Flow Cytometry Core Facility of the Center for Molecular and Cellular Bioengineering (CMCB) Technology Platform at Technische Universität (TU) Dresden.
LDH release assay
The LDH release of cells or of freshly isolated kidney tubules was measured according to the manufacturers’ instructions at indicated time points. In brief, an aliquot of the supernatant was taken, and lysis solution was added for 45 min to induce maximal LDH release before another aliquot of the supernatant was taken. Subsequently, the supernatants were incubated with CytoTox 96 reagent for 15 min, protected from light at room temperature before adding stop solution. Absorbance was measured at 490 nm.
Mass Spectrometry (LC-ESI-QToF)
Cells were placed on ice and washed with 0.9% NaCl before covering them with −80°C cold quenching buffer (80% LC-MS grade methanol) followed by a 20-min incubation time at −80°C. Subsequently, cells were scraped and centrifuged at 14,000g, 4°C for 10 min. The supernatant was centrifuged at 14,000g, 4°C for 10 min again before freezing it at −80°C until further analysis. For the MS-based analysis using an Aquity I-class ultra performance LC system (Waters) coupled to a high-resolution QToF-MS (Vion IMS QToF, Waters), 200-μl aliquots of the aforementioned cell lysates were dried down in a vacuum-assisted centrifuge for at least 2.5 hours and afterward reconstituted in 100 μl of initial mobile phase (10/90% water/acetonitrile containing 0.1% formic acid). For chromatographic separation, a Waters ACQUITY UPLC BEH column (2.1 × 100 mm, 1.7 μm) at 45°C and a gradient of mobile phases A (water/0.1% formic acid) and B (acetonitrile/0.1% formic acid) was used. Calibrators and samples (7.5 μl), kept at 4°C in the autosampler, were injected into the LC-QToF-MS system at a flow rate of 0.4 ml/min. Directly after injection at 10% mobile phase A, proportions of A linearily increased to 20% at 2.0 min to 48% at 4.0 min and further to 90% at 5.0 min. After a hold for 1 min at 90% mobile phase A, the gradient returned to initial conditions at 6.3 min, followed by another 1.2 min for column reequilibration. For identification and quantification of GSH, a single standard of GSH was purchased from Sigma-Aldrich (Munich, Germany). After disolvation and respective dilution in mobile phase, the GSH standard was injected into the LC-QToF-MS to determine analyte-specific characteristics, such as LC retention time (3.30 min), accurate mass [(M + H+) = 308.0915 Da], and the collision cross section (CCS) value (168.06 Å2), a metabolite-specific measure of ion mobility. GSH in calibrators and samples was detected by using the high-definition MSE data acquisition mode that includes ion mobility separation and determination of CCS data, and accurate precursor and respective fragment ion masses of all ions. Positive electrospray ionization source parameters were set to 1.0 kV for capillary voltage, to 120° and 550°C for source and desolvation temperatures, respectively, as well as to respective cone- and desolvation gas flows at 50 and 900 liter/hour. Data were evaluated by using Waters with UNIFI Software package (version 1.9.4.053).
Immunofluorescence microscopy
HPTC (see above) were cultured on collagen-coated coverslips and treated with dexamethasone as indicated. Cells were fixed with 4% paraformaldehyde and incubated with NH4Cl (50 μM) to quench free aldehyde groups. Cells were blocked in 3% bovine serum albumin (BSA) before incubation with a primary DPEP1 antibody overnight at 4°C. Cells were washed in PBS before incubation in secondary fluorescent antibody (Alexa Fluor 488 anti-rabbit IgG, A21206, Invitrogen). Coverslips were mounted onto slide in ProLong Gold antifade (P36391, Invitrogen) containing 4′, 6-diamidino-2-phenylindole. Images were acquired with a confocal fluorescence microscope (Olympus IX-70) using the Fluoview1000 software (Olympus, Tokyo, Japan).
3D printing of a double chamber for simultaneous live-cell imaging
For improved comparability of dexamethasone-treated and untreated cells, we aimed to design a two-well system. This allows for direct comparison of differentially treated cells using the same fluorescence camera. The used chambers consist of a border including a retainer for a glass wall and a glass wall, which is separating the two chambers. The border was printed out of a silicone elastomer (SE 1700, Dow Corning) with the 3DDiscovery bioprinter from RegenHU using a conical nozzle with an inner diameter of 250 μm. Print layouts were developed in the BIOCAD software (RegenHU). The printing speed and extrusion pressure were adjusted to get the thickness of the printed line around 500 μm (these parameters depend on the age of silicone). The height of the border was 8 mm.
The chambers’ border was printed on top of a silanized microscope slide and cured at 100°C for 30 min. A line of SE1700 was printed in the center of the chambers using higher printing speed, thus making it much thinner than the border of the chambers (around 200 μm). SE1700 was applied to the retainer manually (via a syringe with a conical needle) to bond the glass wall. For the glass wall, a coverslip (20 mm by 20 mm; thickness, 0.12 mm) was cut to a size of 8 mm by 20mm, cleaned with ethanol, and treated with air plasma for 10 s using a cold-plasma generator Piezobrush PZ2-i equipped with a nearfield nozzle from Relyon Plasma. Then, the glass wall was inserted into the retainer and pushed down until it was in direct contact with the thin line of SE1700. This structure was again cured at 100°C for 30 min. The structure was removed from the microscope slide, and its bottom side as well as the inside of a six-well plate was treated with air plasma for around 10 s (using Piezobrush PZ2-i). Then, the structure was attached to a well of the six-well plate and kept at 60°C for 2 hours for bonding. In some cases, certain places of the border were poorly bonded to the surface. These places were sealed with SE1700 manually, and then the structure was placed again to 60°C for 4 hours.
Time-lapse imaging
Time-lapse imaging was performed using a 2.5×/0.12 Fluar objective for the custom-made 3D-printed well plated with HT1080 cells on an Axiovert 200M equipped with a large incubation chamber (37°C), 5% CO2, and humidity control. Transmitted light and fluorescent images (LED 475-nm Spectra X light source, emission filter BP 525 to 550 and LED 631-nm Spectra X light source, emission filter BP 663 to 738) were acquired using an Orca flash 4.0 camera. The protocol used for staining was adapted from Wallberg et al. (50). HT1080 cells were plated in the above-described double chamber in DMEM without phenol red (Thermo Fisher Scientific) and pretreated for 12 hours with 1 μM dexamethasone or vehicle. Subsequently, 5 μM erastin was added, as well as 0.5 μM SYTOX Green (Life Technologies), annexin V (5 μg/ml), Alexa Fluor 647 conjugate (Thermo Fisher Scientific), and 2.5 mM CaCl2. The live-imaging procedure was supported by the Light Microscopy Facility, a core facility of the CMCB Technology Platform at TU Dresden.
RNA-seq analysis
RNA was isolated using the RNeasy Mini Kit (QIAGEN). For quantification of gene abundances of the processed paired-end RNA-seq reads, a STAR (51) (version 2.7.6a) index was created on the basis of the human hg38 transcriptome. Gene abundances were quantified with RSEM (52) (version 1.3.1) using default settings. The estimated gene abundances were imported to R with tximport (53), and genes that had summed counts below 10 across all samples were removed from the downstream analysis. For outlier detection, gene counts were normalized with varianceStabilizingTransformation (blind = TRUE) from the DESeq2 (54) package (version 1.28.1), and a principal components analysis (PCA) was performed with RunPCA (ntop = 2000). Hierarchical clustering based on Euclidean distances was performed on the variance stabilized data and visualized with heatmap.2. Differential gene expression analysis was performed with DESeq using a log2 fold change threshold of 1 and false discovery rate <0.05. Significantly differentially expressed genes were visualized in volcano plots.
Mice
Male mice (8 to 12 weeks old) were cohoused with two to five mice per cage in IVCs (individually ventilated cages) in our facility at the Medizinisch-Theoretisches Zentrum at the Medical Faculty of the Technical University of Dresden (TU Dresden). All wild-type mice (C57Bl/6N) were initially provided by Charles River, Sulzfeld, Germany, at the age of 6 to 7 weeks. DPEP1 knockout mice were described previously (55). All experiments were performed according to German animal protection laws and were approved by ethics committees and local authorities of Dresden (Germany).
Isolation of murine thymocytes
After removal of thymi, organs were placed on a 100 μm cell strainer. Using a plunger of a 3-ml syringe, thymi were pressed through the cell strainer and rinsed with RPMI 1640 medium (Thermo Fisher Scientific). Thymocytes were washed twice before dead cells were removed by Ficoll-Paque PLUS (VWR). Subsequently, cells were counted, plated in 24-well plates, and treated with vehicle, 1 μM dexamethasone with or without 5 μM emricasan for the indicated time points.
Isolation of primary murine renal tubules
Primary murine renal tubules were isolated following a modified previously published protocol (33). In detail (demonstrated in fig. S7), murine kidneys were removed, washed with PBS, and decapsulized and sliced in four to five slices. Kidney slices of each kidney were transferred in 2-ml reaction tube containing collagenase type II (2 mg/ml) in incubation solution [trypsin inhibitor (48 μg/ml), DNAse I (25 μg/ml), 140 mM NaCl, 0.4 mM KH2PO4, 1.6 mM K2HPO4 · 3 H2O, 1 mM MgSO4 · 7 H2O, 10 mM CH3COONa · 3 H2O, 1 mM α-ketoglytarate, and 1.3 mM Ca-gluconate] and digested for 5 min at 37°C, 850 rpm. Because of the presence of damaged tubules, the first resulting supernatant was discarded, and 1 ml of collagenase type II (2 mg/ml) in incubation solution was added to the kidney slices and digested for 5 min at 37°C, 850 rpm. The supernatant was collected and transferred in a 2-ml reaction tube containing 1 ml of ice-cold sorting solution (0.5 mg/ml BSA in incubation solution). The reaction tubes were left on ice for the tubules to precipitate. The supernatant was removed, and the tubules were washed twice with ice-cold incubation solution. Once the tubules precipitated, the supernatant was removed, and ice-cold sorting solution was added (the volume was adjusted depending on the number of samples needed for the experiment). Tubules were distributed in a 24-well plate containing DMEM F-12 nutrient mixture without phenol red (DMEM/F12, Thermo Fisher Scientific), supplemented with recombinant human insulin (0.01 mg/ml), human transferrin (5.5 μg/ml), sodium selenite (Na2SeO3, 0.005 μg/ml) (ITS without linoleic acid, Sigma-Aldrich), 50 nM hydrocortisone, penicillin (100 U/ml), and streptomycin (100 μg/ml) (Thermo Fisher Scientific).
Statistical analysis
Statistical analyses were performed with Prism 8 (GraphPad software, San Diego, CA, USA) using Student’s t test. Data were considered significant when *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, or ****P ≤ 0.0001.
Acknowledgments
We would like to thank R. Opitz, C. Laqua, and A. Pioch for expert technical assistance, M. Langner for expert help with mass spectrometry, and the Light Microscopy Facility at Dresden BIOTEC, in particular H. Hartmann and E. Geibelt, for the expert help. We thank the Microstructure Facility of the BIOTEC at Technische Universitat Dresden [partly funded by the State of Saxony and the European Fund for Regional Development—EFRE (100344812)].
Funding: Work in the Linkermann laboratory is funded by the SFB-TRR 205, SFB-TRR 127, and the international research training group (IRTG) 2251. This work was additionally supported by the German Research Foundation (DFG), priority program on ferroptosis (SPP2306) to A.v.M. and A.L., the Heisenberg-Professorship to A.L. (project number 324141047), and an instrument grant support to M.P. (INST 515/28-1 FUGG). We further thank the Else Kröner-Fresenius Stiftung and the Sander-Stiftung for supporting our laboratory.
Author contributions: A.v.M., N.Z.G., F.M., A.B., W.T., C.M., K.B., A.La., M.P., P.H., and S.L. performed the experiments. M.T.H. performed the bulk sequencing analysis codesigned by A.v.M., A.Li., and R.K. S.R.B. provided critical reagents and conceptual input. A.v.M., T.C., R.K., D.A.M., C.H., S.R.B., and A.Li. designed the experiments. A.v.M. and A.Li. wrote the paper.
Competing interests: A.La. and D.A.M. have equity positions in Arch Biopartners Inc. and have patents issued or pending focused on DPEP1 inhibition: (i) DPEP1 binding compositions and methods of use. Inventors: S. M. Robbins; (Calgary, CA), D. L. Senger (Calgary, CA), J. J. Rahn (Calgary, CA), A. W. S. Lau (Calgary, CA), D. A. Muruve (Calgary, CA), S. R. Choudhury (Calgary, CA), L. Babes (Calgary, CA), and P. Kubes (Calgary, CA) [patent application: #15/234521 (57983413), filed 11 August 2016; international application: #PCT/IB2016/001244, filed August 11, 2016; U.S. patent application: #16/504909, filed 8 July 2019; U.S. patent #10,493,127, issued 3 December 2019]. (ii) DPEP1 binding compositions and methods of use. Inventors: S. M. Robbins (Calgary, CA), D. L. Senger (Calgary, CA), J. J. Rahn (Calgary, CA), A. W. S. Lau (Calgary, CA), D. A. Muruve (Calgary, CA), S. R. Choudhury (Calgary, CA), L. Babes (Calgary, CA), and P. Kubes (Calgary, CA) [U.S. patent application: #16/700901 (70853301), filed 2 December 2019; international application: #PCT/IB2019/001289, filed 2 December 2019]. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Legends for data S1 and S2
Legends for movies S1 and S2
Other Supplementary Material for this manuscript includes the following:
Data S1 and S2
Movies S1 and S2
REFERENCES AND NOTES
- 1.Jackson R. K., Irving J. A., Veal G. J., Personalization of dexamethasone therapy in childhood acute lymphoblastic leukaemia. Br. J. Haematol. 173, 13–24 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Teachey D. T., Pui C. H., Comparative features and outcomes between paediatric T-cell and B-cell acute lymphoblastic leukaemia. Lancet Oncol. 20, e142–e154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The RECOVERY Collaborative , Dexamethasone in hospitalized patients with Covid-19—Preliminary report. N. Eng. J. Med. covidwho-650223, (2020). [Google Scholar]
- 4.Thomusch O., Wiesener M., Opgenoorth M., Pascher A., Woitas R. P., Witzke O., Jaenigen B., Rentsch M., Wolters H., Rath T., Cingöz T., Benck U., Banas B., Hugo C., Rabbit-ATG or basiliximab induction for rapid steroid withdrawal after renal transplantation (Harmony): An open-label, multicentre, randomised controlled trial. Lancet 388, 3006–3016 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Moshirfar M., Somani A. N., Motlagh M. N., Ronquillo Y. C., Management of cataract in the setting of uveitis: A review of the current literature. Curr. Opin. Ophthalmol. 31, 3–9 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Kauffman S. L., Acceleration of canalicular development in lungs of fetal mice exposed transplacentally to dexamethasone. Lab. Invest. 36, 395–401 (1977). [PubMed] [Google Scholar]
- 7.Weinstein R. S., Glucocorticoid-induced osteonecrosis. Endocrine 41, 183–190 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pereira R. M., Freire de Carvalho J., Glucocorticoid-induced myopathy. Joint Bone Spine 78, 41–44 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Bandyopadhyay U., Biswas K., Bandyopadhyay D., Ganguly C. K., Banerjee R. K., Dexamethasone makes the gastric mucosa susceptible to ulceration by inhibiting prostaglandin synthetase and peroxidase—Two important gastroprotective enzymes. Mol. Cell. Biochem. 202, 31–36 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Gibson A. T., Pearse R. G., Wales J. K., Growth retardation after dexamethasone administration: Assessment by knemometry. Arch. Dis. Child. 69, 505–509 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belavgeni A., Bornstein S. R., von Massenhausen A., Tonnus W., Stumpf J., Meyer C., Othmar E., Latk M., Kanczkowski W., Kroiss M., Hantel C., Hugo C., Fassnacht M., Ziegler C. G., Schally A. V., Krone N. P., Linkermann A., Exquisite sensitivity of adrenocortical carcinomas to induction of ferroptosis. Proc. Natl. Acad. Sci. U.S.A. 116, 22269–22274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weigand I., Schreiner J., Röhrig F., Sun N., Landwehr L. S., Urlaub H., Kendl S., Kiseljak-Vassiliades K., Wierman M. E., Angeli J. P. F., Walch A., Sbiera S., Fassnacht M., Kroiss M., Active steroid hormone synthesis renders adrenocortical cells highly susceptible to type II ferroptosis induction. Cell Death Dis. 11, 192 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon S. J., Lemberg K. M., Lamprecht M. R., Skouta R., Zaitsev E. M., Gleason C. E., Patel D. N., Bauer A. J., Cantley A. M., Yang W. S., Morrison B. III, Stockwell B. R., Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conrad M., Lorenz S. M., Proneth B., Targeting Ferroptosis: New hope for as-yet-incurable diseases. Trends Mol. Med. 27, 113–122 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Del Re D. P., Amgalan D., Linkermann A., Liu Q., Kitsis R. N., Fundamental mechanisms of regulated cell death and implications for heart disease. Physiol. Rev. 99, 1765–1817 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang X., Wang H., Han D., Xie E., Yang X., Wei J., Gu S., Gao F., Zhu N., Yin X., Cheng Q., Zhang P., Dai W., Chen J., Yang F., Yang H. T., Linkermann A., Gu W., Min J., Wang F., Ferroptosis as a target for protection against cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 116, 2672–2680 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao M., Monian P., Quadri N., Ramasamy R., Jiang X., Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell 59, 298–308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alim I., Caulfield J. T., Chen Y., Swarup V., Geschwind D. H., Ivanova E., Seravalli J., Ai Y., Sansing L. H., Ste Marie E. J., Hondal R. J., Mukherjee S., Cave J. W., Sagdullaev B. T., Karuppagounder S. S., Ratan R. R., Selenium drives a transcriptional adaptive program to block ferroptosis and treat stroke. Cell 177, 1262–1279.e25 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Tuo Q.-Z., Lei P., Jackman K. A., Li X. L., Xiong H., Li X.-L., Liuyang Z.-Y., Roisman L., Zhang S. T., Ayton S., Wang Q., Crouch P. J., Ganio K., Wang X. C., Pei L., Adlard P. A., Lu Y. M., Cappai R., Wang J. Z., Liu R., Bush A. I., Tau-mediated iron export prevents ferroptotic damage after ischemic stroke. Mol. Psychiatry 22, 1520–1530 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Zille M., Karuppagounder S. S., Chen Y., Gough P. J., Bertin J., Finger J., Milner T. A., Jonas E. A., Ratan R. R., Neuronal death after hemorrhagic stroke in vitro and in vivo shares features of ferroptosis and necroptosis. Stroke 48, 1033–1043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guiney S. J., Adlard P. A., Bush A. I., Finkelstein D. I., Ayton S., Ferroptosis and cell death mechanisms in Parkinson’s disease. Neurochem. Int. 104, 34–48 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Angeli J. P. F., Schneider M., Proneth B., Tyurina Y. Y., Tyurin V. A., Hammond V. J., Herbach N., Aichler M., Walch A., Eggenhofer E., Basavarajappa D., Radmark O., Kobayashi S., Seibt T., Beck H., Neff F., Esposito I., Wanke R., Forster H., Yefremova O., Heinrichmeyer M., Bornkamm G. W., Geissler E. K., Thomas S. B., Stockwell B. R., O’Donnell V. B., Kagan V. E., Schick J. A., Conrad M., Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 16, 1180–1191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L. L., Liao X. H., Sun H., Jiang X., Liu Q., Zhang L., Augmenter of liver regeneration protects the kidney from ischaemia-reperfusion injury in ferroptosis. J. Cell. Mol. Med. 23, 4153–4164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linkermann A., Skouta R., Himmerkus N., Mulay S. R., Dewitz C., De Zen F., Prokai A., Zuchtriegel G., Krombach F., Welz P. S., Weinlich R., Vanden Berghe T., Vandenabeele P., Pasparakis M., Bleich M., Weinberg J. M., Reichel C. A., Brasen J. H., Kunzendorf U., Anders H. J., Stockwell B. R., Green D. R., Krautwald S., Synchronized renal tubular cell death involves ferroptosis. Proc. Natl. Acad. Sci. U.S.A. 111, 16836–16841 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruni A., Pepper A. R., Pawlick R. L., Gala-Lopez B., Gamble A. F., Kin T., Seeberger K., Korbutt G. S., Bornstein S. R., Linkermann A., Shapiro A. M. J., Ferroptosis-inducing agents compromise in vitro human islet viability and function. Cell Death Dis. 9, 595 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W., Feng G., Gauthier J. M., Lokshina I., Higashikubo R., Evans S., Liu X., Hassan A., Tanaka S., Cicka M., Hsiao H. M., Ruiz-Perez D., Bredemeyer A., Gross R. W., Mann D. L., Tyurina Y. Y., Gelman A. E., Kagan V. E., Linkermann A., Lavine K. J., Kreisel D., Ferroptotic cell death and TLR4/Trif signaling initiate neutrophil recruitment after heart transplantation. J. Clin. Invest. 129, 2293–2304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schumann-Bischoff A., Schmitz J., Scheffner I., Schmitt R., Broecker V., Haller H., Brasen J. H., Gwinner W., Distinct morphological features of acute tubular injury in renal allografts correlate with clinical outcome. Am. J. Physiol. Renal Physiol. 315, F701–F710 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Kwon M. Y., Park E., Lee S. J., Chung S. W., Heme oxygenase-1 accelerates erastin-induced ferroptotic cell death. Oncotarget 6, 24393–24403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adedoyin O., Boddu R., Traylor A., Lever J. M., Bolisetty S., George J. F., Agarwal A., Heme oxygenase-1 mitigates ferroptosis in renal proximal tubule cells. Am. J. Physiol. Renal Physiol. 314, F702–f714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakagawa H., Inazawa J., Inoue K., Misawa S., Kashima K., Adachi H., Nakazato H., Abe T., Assignment of the human renal dipeptidase gene (DPEP1) to band q24 of chromosome 16. Cytogenet. Cell Genet. 59, 258–260 (1992). [DOI] [PubMed] [Google Scholar]
- 31.Song D., Lin Z., Yuan Y., Qian G., Li C., Bao Y., DPEP1 Balance GSH involve in cadmium stress response in blood clam tegillarca granosa. Front. Physiol. 9, 964 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paller M. S., Renal work, glutathione and susceptibility to free radical-mediated postischemic injury. Kidney Int. 33, 843–849 (1988). [DOI] [PubMed] [Google Scholar]
- 33.Tonnus W., Meyer C., Steinebach C., Belavgeni A., von Mässenhausen A., Gonzalez N. Z., Maremonti F., Gembardt F., Himmerkus N., Latk M., Locke S., Marschner J., Li W., Short S., Doll S., Ingold I., Proneth B., Daniel C., Kabgani N., Kramann R., Motika S., Hergenrother P. J., Bornstein S. R., Hugo C., Becker J. U., Amann K., Anders H. J., Kreisel D., Pratt D., Gütschow M., Conrad M., Linkermann A., Dysfunction of the key ferroptosis-surveilling systems hypersensitizes mice to tubular necrosis during acute kidney injury. Nat. Commun. 12, 4402 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolisetty S., Traylor A., Joseph R., Zarjou A., Agarwal A., Proximal tubule-targeted heme oxygenase-1 in cisplatin-induced acute kidney injury. Am. J. Physiol. Renal Physiol. 310, F385–F394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng F., Sharma I., Dai Y., Yang M., Kanwar Y. S., Myo-inositol oxygenase expression profile modulates pathogenic ferroptosis in the renal proximal tubule. J. Clin. Invest. 129, 5033–5049 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lau A., Chung H., Komada T., Platnich J. M., Sandall C. F., Choudhury S. R., Chun J., Naumenko V., Surewaard B. G. J., Nelson M. C., Ulke-Lemée A., Beck P. L., Benediktsson H., Jevnikar A. M., Snelgrove S. L., Hickey M. J., Senger D. L., James M. T., Macdonald J. A., Kubes P., Jenne C. N., Muruve D. A., Renal immune surveillance and dipeptidase-1 contribute to contrast-induced acute kidney injury. J. Clin. Invest. 128, 2894–2913 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin-Sanchez D., Ruiz-Andres O., Poveda J., Carrasco S., Cannata-Ortiz P., Sanchez-Niño M. D., Ruiz Ortega M., Egido J., Linkermann A., Ortiz A., Sanz A. B., Ferroptosis, but not necroptosis, is important in nephrotoxic folic acid-induced AKI. J. Am. Soc. Nephrol. 28, 218–229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mishima E., Sato E., Ito J., Yamada K. I., Suzuki C., Oikawa Y., Matsuhashi T., Kikuchi K., Toyohara T., Suzuki T., Ito S., Nakagawa K., Abe T., Drugs repurposed as antiferroptosis agents suppress organ damage, including AKI, by functioning as lipid peroxyl radical scavengers. J. Am. Soc. Nephrol. 31, 280–296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zarjou A., Bolisetty S., Joseph R., Traylor A., Apostolov E. O., Arosio P., Balla J., Verlander J., Darshan D., Kuhn L. C., Agarwal A., Proximal tubule H-ferritin mediates iron trafficking in acute kidney injury. J.Clin.Invest 123, 4423–4434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baudouin-Cornu P., Lagniel G., Kumar C., Huang M. E., Labarre J., Glutathione degradation is a key determinant of glutathione homeostasis. J. Biol. Chem. 287, 4552–4561 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ganguli D., Kumar C., Bachhawat A. K., The alternative pathway of glutathione degradation is mediated by a novel protein complex involving three new genes in Saccharomyces cerevisiae. Genetics 175, 1137–1151 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keynan S., Habgood N. T., Hooper N. M., Turner A. J., Site-directed mutagenesis of conserved cysteine residues in porcine membrane dipeptidase. Cys 361 alone is involved in disulfide-linked dimerization. Biochemistry 35, 12511–12517 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Keynan S., Hooper N. M., Turner A. J., Identification by site-directed mutagenesis of three essential histidine residues in membrane dipeptidase, a novel mammalian zinc peptidase. Biochem. J. 326 ( Pt. 1), 47–51 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kropp H., Sundelof J. G., Hajdu R., Kahan F. M., Metabolism of thienamycin and related carbapenem antibiotics by the renal dipeptidase, dehydropeptidase. Antimicrob. Agents Chemother. 22, 62–70 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S. E., Zhang L., Ma K., Riegman M., Chen F., Ingold I., Conrad M., Turker M. Z., Gao M., Jiang X., Monette S., Pauliah M., Gonen M., Zanzonico P., Quinn T., Wiesner U., Bradbury M. S., Overholtzer M., Ultrasmall nanoparticles induce ferroptosis in nutrient-deprived cancer cells and suppress tumour growth. Nat. Nanotechnol. 11, 977–985 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riegman M., Sagie L., Galed C., Levin T., Steinberg N., Dixon S. J., Wiesner U., Bradbury M. S., Niethammer P., Zaritsky A., Overholtzer M., Ferroptosis occurs through an osmotic mechanism and propagates independently of cell rupture. Nat. Cell Biol. 22, 1042–1048 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Linkermann A., Heller J. O., Prokai A., Weinberg J. M., De Zen F., Himmerkus N., Szabo A. J., Brasen J. H., Kunzendorf U., Krautwald S., The RIP1-kinase inhibitor necrostatin-1 prevents osmotic nephrosis and contrast-induced AKI in mice. J. Am. Soc. Nephrol. 24, 1545–1557 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guan Y., Liang X., Ma Z., Hu H., Liu H., Miao Z., Linkermann A., Hellwege J. N., Voight B. F., Susztak K., A single genetic locus controls both expression of DPEP1/CHMP1A and kidney disease development via ferroptosis. Nat. Commun. 12, 5078 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi S., Homma T., Okumura N., Han J., Nagaoka K., Sato H., Konno H., Yamada S., Takao T., Fujii J., Carnosine dipeptidase II (CNDP2) protects cells under cysteine insufficiency by hydrolyzing glutathione-related peptides. Free Radic. Biol. Med. 174, 12–27 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Wallberg F., Tenev T., Meier P., Time-lapse imaging of cell death. Cold Spring Harb. Protoc. 2016, pdb.prot087395 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soneson C., Love M. I., Robinson M. D., Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choudhury S. R., Babes L., Rahn J. J., Ahn B. Y., Goring K. R., King J. C., Lau A., Petri B., Hao X., Chojnacki A. K., Thanabalasuriar A., McAvoy E. F., Tabariès S., Schraeder C., Patel K. D., Siegel P. M., Kopciuk K. A., Schriemer D. C., Muruve D. A., Kelly M. M., Yipp B. G., Kubes P., Robbins S. M., Senger D. L., Dipeptidase-1 is an adhesion receptor for neutrophil recruitment in lungs and liver. Cell 178, 1205–1221.e17 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Legends for data S1 and S2
Legends for movies S1 and S2
Data S1 and S2
Movies S1 and S2