Abstract
Translational regulation plays an important role in gene expression and function. Although the transcriptional dynamics of mouse preimplantation embryos have been well characterized, the global mRNA translation landscape and the master regulators of zygotic genome activation (ZGA) remain unknown. Here, by developing and applying a low-input ribosome profiling (LiRibo-seq) technique, we profiled the mRNA translation landscape in mouse preimplantation embryos and revealed the translational dynamics during mouse preimplantation development. We identified a marked translational transition from MII oocytes to zygotes and demonstrated that active translation of maternal mRNAs is essential for maternal-to-zygotic transition (MZT). We further showed that two maternal factors, Smarcd2 and Cyclin T2, whose translation is activated in zygotes, are required for chromatin reprogramming and ZGA, respectively. Our study thus not only filled in a knowledge gap on translational regulation during mammalian preimplantation development but also revealed insights into the critical function of maternal mRNA translation in MZT.
LiRibo-seq reveals translational dynamics and key regulators of chromatin reprogramming and ZGA in mouse preimplantation embryos.
INTRODUCTION
Mammalian life starts with the fusion of two terminally differentiated gametes, sperm and oocyte, resulting in a totipotent zygote. After going through preimplantation development, the zygote reaches blastocyst before implantation. The two most important events taking place during preimplantation development are zygotic genome activation (ZGA) and the first cell lineage differentiation to generate inner cell mass cells and trophoblast cells (1, 2). For these events to occur successfully, gene expression has to be tightly regulated at different levels. One of the most studied gene expression regulation is at the transcriptional level. In addition, recent advances in low-input profiling techniques have made the characterization of various epigenetic and chromatin changes during preimplantation development possible (3, 4). However, the mechanism underlying mammalian ZGA is still not fully understood (5–7).
Most biological processes are carried out by proteins. The steady-state protein levels are determined by the overall gene expression activity regulated at multiple levels. Consequently, proteomic analysis of mouse preimplantation embryos has been attempted (8, 9). However, because of the limitation of obtaining a large quantity of preimplantation embryos, the proteomic analyses have provided limited coverage and information. In addition, proteomics data can only provide an overall readout of gene expression, but not the dynamics of mRNA translation. During the time window from fully grown oocytes to early zygotes, transcription is completely silenced. Thus, mRNA translation serves as a major regulatory step by which maternally stored mRNAs are translated in a spatially and temporally regulated manner (10), indicating that translational and posttranslational regulation may play a critical role during early embryogenesis. Therefore, understanding the translational dynamics during mouse preimplantation development may shed light on gene expression regulation during embryogenesis. Ribosome profiling is a well-known method for quantifying ribosome occupancy and mRNA translation (11, 12). However, conventional ribosome profiling methods require millions of cells, which preclude their use in mouse early embryos. Thus, the mRNA translational dynamics in mouse preimplantation embryos are largely unknown.
In contrast to mammals, studies in other model organisms with readily accessible early embryos have revealed a clear role of translational regulation in early embryogenesis (10). For example, polysome profiling and ribosome footprinting have revealed a dynamic change in translation between mature eggs and activated eggs, and identified Pan Gu kinase as a major translational regulator in Drosophila (13). In addition, the polyadenylic acid [poly(A)] tail length of mRNAs was found to positively correlate with translation efficiency (TE) during Drosophila egg activation (14). Ribosome profiling in early zebrafish embryos [2 hours post-fertilization (hpf)] has revealed that the pluripotent factors Pou5f3, SoxB, and Nanog are highly translated, and their translation is required for subsequent ZGA (15). Because mouse ZGA takes place at one-cell (minor ZGA) and two-cell (major ZGA) stages when SoxB and Nanog are not detectable, while maternal depletion of the mouse Pou5f1, the zebrafish Pou5f3 homolog, does not affect mouse ZGA (16), it is not clear whether translation of other factors is needed for mouse maternal-to-zygotic transition (MZT).
A study performed 17 years ago suggested that translation might be important for mouse MZT, as treatment of fertilized eggs with a translation inhibitor could lead to one-cell arrest (17). However, it is not clear which newly translated factors are essential for mouse MZT. The development and utilization of the polysome profiling technique for oocytes and zygotes have made the identification of actively translating mRNAs during MZT possible (18). Recently, an oocyte-specific (Zp3-Cre) RiboTag strategy has been used to characterize the dynamics of mRNA translation during oocyte maturation, which revealed CPEB1 as an important factor in maintaining constitutive translation (19). Although these studies have provided valuable information about mRNA translation during MZT and oocyte maturation, ribosome profiling can provide more detailed translational regulatory features at the single-codon level (20). Thus, the development and application of low-input ribosome profiling techniques can help our understanding of mRNA translation during embryonic development.
By improving the recently developed RiboLace technique (21), we performed low-input ribosome profiling of mouse preimplantation embryos, which revealed the translational dynamics during preimplantation development and a translational switch from oocytes to zygotes. In addition, we found that maternal proteins are insufficient to support mouse MZT and that active translation of some maternal mRNAs, including the nucleosome remodeling factor Smarcd2 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily D, member 2) and the transcription elongation regulator Cyclin T2, is required for chromatin remodeling and ZGA, respectively, during mouse MZT.
RESULTS
Development of low-input ribosome sequencing method applicable to mouse preimplantation embryos
Ribosome profiling is a well-known technique for analyzing mRNAs actively engaged in translation (12). However, conventional ribosome profiling method requires millions of cells to enrich the ribosome-protected fragments (RPFs), preventing its application to low-input samples, such as mammalian preimplantation embryos. A recent study reported the use of RiboLace beads to capture RPFs (21), raising the possibility that this method might be further improved for low-input samples. To this end, we made several modifications, including the use of a ligation-free protocol (22) to avoid gel purification in library preparation steps (see details in Materials and Methods). We have named our improved method low-input ribosome footprint sequencing (LiRibo-seq). To determine whether LiRibo-seq works for low-input ribosome profiling, we first applied LiRibo-seq to 5000 mouse embryonic stem cells (mESCs) (table S1) and compared the results with the ribosome profiling data generated using millions of mESCs (11). We found that our LiRibo-seq data correlated well with each other (fig. S1A) and also highly correlated with the published data (fig. S1A). Furthermore, we observed similar P-site enrichment in coding sequence regions (CDS) between LiRibo-seq and that generated using millions of cells (fig. S1B). Occupancy meta-profiles showed the typical trinucleotide periodicity of P-site along CDS (fig. S1C), confirming that the transcripts we captured were from ribosome footprints. These results indicate that LiRibo-seq can generate reliable profiles using as few as 5000 mESCs.
Next, we applied LiRibo-seq to preimplantation embryos (Fig. 1A). In parallel, we also performed total RNA sequencing (RNA-seq) to quantify RNA expression (tables S1 and S2). For each developmental stage, we generated two biological replicates, which showed high reproducibility (figs. S2A and S3), and all LiRibo-seq samples showed P-site enrichment in CDS (fig. S2B) and trinucleotide periodicity (fig. S2C). In total, we detected at least 12,479 genes actively engaging in translation at any of the developmental stage, which is more than three times that of the proteins detected by proteomics analysis (Fig. 1B) (8), indicating the powerfulness of LiRibo-seq.
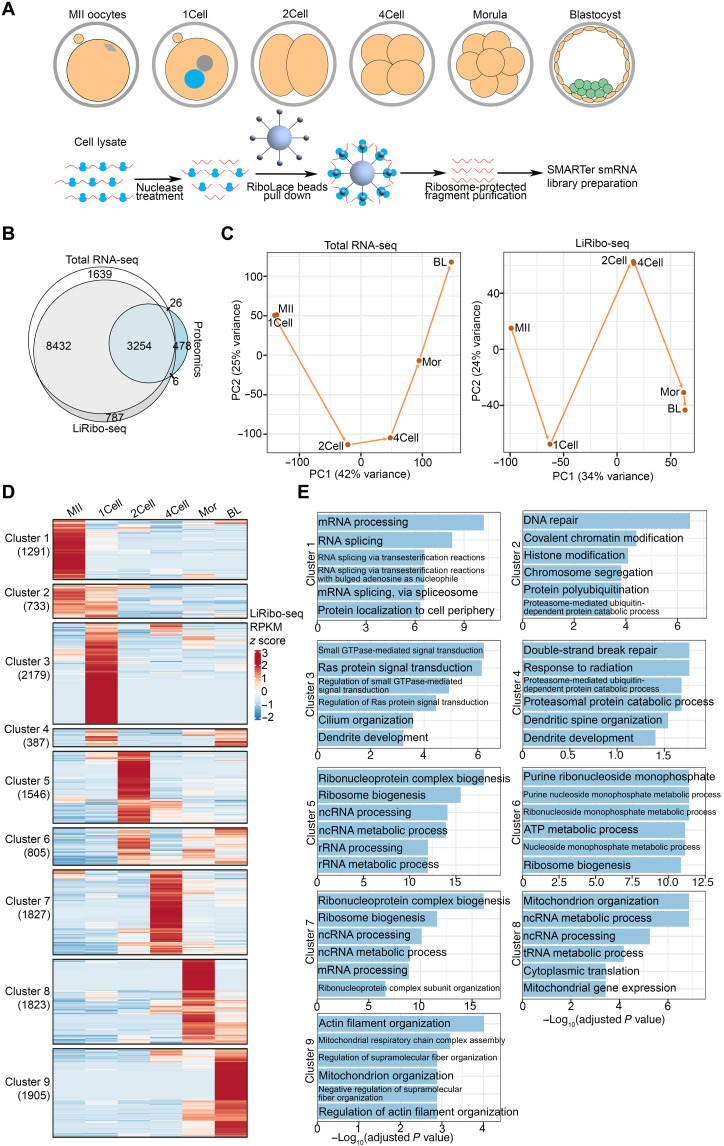
Fig. 1. LiRibo-seq reveals translational dynamics in mouse preimplantation embryos.
(A) Schematic of LiRibo-seq for capturing mRNAs engaged in translation in mouse preimplantation embryos. One-cell (1Cell), two-cell (2Cell), four-cell (4Cell), morula (Mor), and blastocyst (BL) embryos were collected at 12, 30, 48, 72, and 96 hpf, respectively. (B) Venn diagram of detected gene product by total RNA-seq [RPKM (reads per kilobase of transcript per million reads mapped) ≥ 1 at any stage], LiRibo-seq (RPKM ≥ 1 at any stage), and proteomic identified genes in oocytes and preimplantation embryos. (C) Principal components analysis (PCA) of total RNA-seq (left) and LiRibo-seq (right) in mouse MII oocytes and preimplantation embryos. (D) Heatmap of the mRNA clusters exhibiting stage-specific translation patterns in mouse MII oocytes and preimplantation embryos. (E) Gene ontology enrichment analysis of different mRNA clusters in (D). The enriched terms are ranked by −log10(adjusted P value).
LiRibo-seq reveals translational dynamics in mouse preimplantation embryos
With the LiRibo-seq data available, we analyzed the translational dynamics during mouse preimplantation development. Principal components analysis (PCA) showed that MII oocytes and zygotes were clustered together on the basis of their total RNA-seq data, consistent with the lack of global transcription from MII oocytes to zygotes (Fig. 1C). In contrast, LiRibo-seq revealed a marked difference between MII oocytes and zygotes (Fig. 1C). Consistent with the notion that major ZGA and mid-preimplantation gene activation occur at late two-cell stage and around morula stage, respectively (17), we observed transcriptional changes from one-cell to two-cell, and four-cell to morula, and even from morula to blastocyst (Fig. 1C). In LiRibo-seq data, we also observed obvious separation from one-cell to two-cell, and from four-cell to morula, while two-cell and four-cell, morula, and blastocyst were clustered together (Fig. 1C). These data suggest that translational changes can occur with or without transcriptional changes during preimplantation development.
To better characterize the translational dynamics during mouse preimplantation development, we divided the LiRibo-seq–detected genes into nine different clusters based on their translation activity at different stages (Fig. 1D) and performed gene ontology (GO) enrichment analysis on each gene cluster. Cluster 1 genes showed the highest translation activity in MII oocytes and were enriched for mRNA processing (Fig. 1E). Cluster 2 genes were highly translated in both MII oocytes and zygotes (Fig. 1D) and were enriched for chromosome segregation and chromatin modification (Fig. 1E), consistent with the meiotic-to-mitotic transition taking place during this time window. Cluster 3 genes showed specific high translation activity in zygotes (Fig. 1D) and were enriched for small GTPase (guanosine triphosphatase)–mediated signal transduction (Fig. 1E), consistent with their function in spindle assembly after fertilization (23). Cluster 4 genes, which were highly translated in zygotes and blastocysts (Fig. 1D), showed enrichment of DNA double-strand break repair and protein degradation (Fig. 1E). Cluster 5 genes showed a high translation activity in two-cell embryos (Fig. 1D) and were enriched for noncoding RNA (ncRNA) processing (Fig. 1E), consistent with recent findings that Neat1 and LincGet ncRNAs play important roles in cell fate determination as early as two-cell-stage embryos (24, 25). Cluster 5 to 8 genes showed high translation activity from and after two-cell stages (Fig. 1D) and were mostly enriched for ribonucleoprotein complex, ribosome biogenesis, or transfer RNA (tRNA) and ncRNA metabolism (Fig. 1E), indicating that zygotic ribosomes may be used to prepare the embryos for subsequent development. Last, cluster 9 genes showed the highest translation only in blastocyst stage (Fig. 1D) and were enriched for cofactors and coenzymes involved in metabolism and actin filament organization (Fig. 1E), which is consistent with the metabolic preparation for the transition to post-implantation development (26) and the function of actin filaments in the hatching process of mouse blastocyst (27). Collectively, our LiRibo-seq analyses revealed the translational dynamics during preimplantation development, which prepare the embryos for subsequent development.
TE change in preimplantation embryos
To understand the relationship between translation and transcription, we integrated the LiRibo-seq dataset with the transcriptome of mouse preimplantation embryos. First, we analyzed the correlation between LiRibo-seq and RNA-seq data. In general, LiRibo-seq showed good correlation with RNA-seq (Fig. 2A), which is consistent with previous studies (28). We observed a clear decrease in the correlation in two-cell embryos (Fig. 2A). On the basis of the correlation between transcription and translation, MII and zygotes were clustered together because maternal transcripts dominated the transcriptome and translatome in these two stages. However, after ZGA takes place at the two-cell stage, newly generated transcripts gradually dominate the transcriptome. Consistent with this maternal-to-zygotic transcriptome shift, mRNA translation was also shifted from maternal to zygotic, which is reflected in the clear separation of the two domains (Fig. 2A).
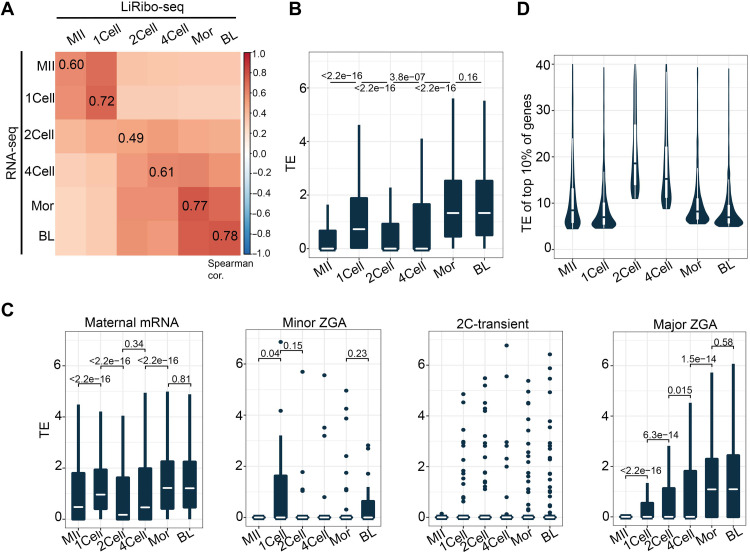
Fig. 2. TE change in preimplantation embryos is functionally relevant.
(A) Heatmap depicting the Spearman correlation coefficient between total RNA-seq and LiRibo-seq at different stages during preimplantation development. (B) Box plot showing the TE of all expressed genes (RNA-seq RPKM ≥ 1) in each stage during preimplantation development. The white line in each box indicates the median value. Box hinges indicate the 25th and 75th percentiles. The whiskers indicate the hinge ± 1.5 × interquartile range. P values were calculated with two tailed Mann-Whitney U test. (C) Box plot showing the TE of maternal, minor ZGA, 2C-transient, and major ZGA genes in each stage during preimplantation development. TE was assigned as 0 when both LiRibo-seq RPKM and RNA-seq RPKM were 0 for a gene. The specifications of the box plot are the same as that described in (B). Dots in minor ZGA and 2C-transient groups indicate mRNAs with higher TE. P values were calculated with two-tailed Mann-Whitney U test. (D) Violin plot showing the TE of the top 10% high-TE mRNAs at different stages during preimplantation development. The whiskers indicate the hinge ± 1.5 × interquartile range.
TE (TE = normalized RPF counts/normalized mRNA counts) serves as a sensitive and quantitative measurement of ribosome occupancy in each mRNA molecule and represents a genome-wide measurement of translation landscape. Consistent with the marked transition in translation activity from MII oocytes to zygotes (Fig. 1C), the maternal mRNA TE showed a significant increase (P < 2.2 × 10−16) (Fig. 2B). In addition, the median of zygotic mRNA TE also gradually increased from two-cell to morula embryos after ZGA (Fig. 2B). To reveal the details of translational dynamics, we analyzed the TE of different mRNA groups, including maternal, minor ZGA, 2C-transient, and major ZGA genes. We found that the TE of maternal mRNAs was significantly increased from MII oocytes to zygotes (Fig. 2C). The TE of minor ZGA genes was specifically high in zygotes, consistent with their activation in zygotes (Fig. 2C). For the two-cell transient genes, although the overall TE was very low in early embryos, translation did occur for some two-cell transient genes (Fig. 2C). In addition, the well-known two-cell transient genes, such as Zfp352, Zscan4f, Zscan4d, and Usp17lc, also showed two-cell–specific translation (fig. S4A). For the major ZGA genes, their TEs were gradually increased from two-cell to blastocyst stage (Fig. 2C), indicating that ZGA genes are not translated simultaneously (fig. S4B). Collectively, the above analyses revealed the translational dynamics of different gene groups during preimplantation development.
To gain insight into the potential function of TE regulation, we attempted to analyze TE distribution during preimplantation development. To this end, we focused the analysis on the top 10% high-TE (high-TE) mRNAs. Although MII oocytes, two-cell embryos, and four-cell embryos showed low TE for all mRNAs (Fig. 2B), the TE of high-TE mRNAs was relatively high (Fig. 2D), suggesting a dispersed distribution of TE at these stages. To identify the biological processes subjected to translational regulation, we performed GO analysis on high-TE mRNAs at each stage. We found that the top enriched GO terms in oocyte high-TE mRNAs included RNA processing (fig. S4C), which is required for oocyte maturation (29). Top enriched GO terms in zygotes were involved in nucleosome organization (fig. S4C), indicating that the high-TE mRNAs in zygotes may function in chromatin reprogramming. In addition, high-TE mRNAs after the two-cell stage were enriched with GO term of metabolic process (fig. S4C). These results indicate that regulation of TE is functionally relevant to the changes taking place during preimplantation development.
Translational switch occurs during MII-to-zygote transition
The TE is determined by both translation activity and mRNA abundance. To determine the relative contribution of mRNA abundance and translation activity to the changes in TE regulation at the different developmental stages, we performed deltaTE analysis (30), which can divide genes into differentially transcribed genes (DTGs) for genes regulated by mRNA abundance and differential TE genes (DTEGs) for genes regulated by translation (fig. S5A). This analysis revealed that, from MII to zygotes, 6.4% of the total regulatory changes were due to changes in mRNA abundance (DTGs), while 84.6% of the total regulatory changes can be ascribed to translational regulation (DTEGs) (fig. S5B). In contrast, about 50% of the total regulatory changes were due to changes in mRNA abundance (DTGs) and only about 15 to 30% were caused by translational regulation (DTEGs) during preimplantation development (fig. S5C). Collectively, these data support that translational regulation from MII oocytes to one-cell embryo may play an important role for MZT.
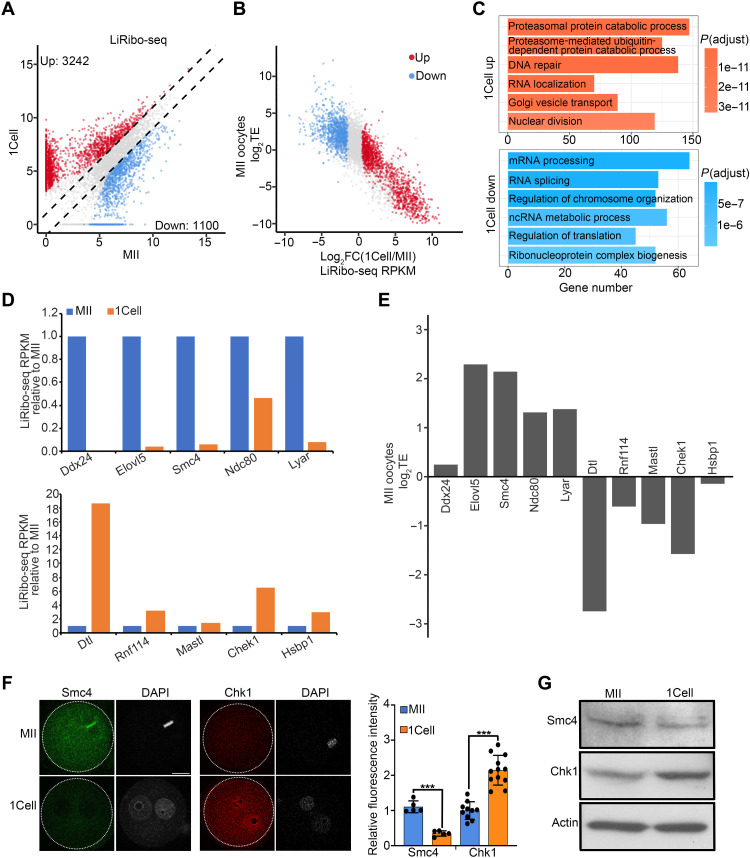
To further characterize the translational regulation from MII oocytes to one-cell embryos, we compared their LiRibo-seq data. We found that 3242 mRNAs were translationally up-regulated, while 1100 were down-regulated from MII oocytes to zygotes (Fig. 3A and table S3). Consistently, with the Click-iT O-propargyl-puromycin (OPP) staining as a measurement of protein synthesis, we observed a significant increase in the OPP signals in zygotes, indicating more protein synthesis in zygotes compared with that in MII oocytes (fig. S5D). Further analyses showed that mRNAs with lower TE in MII oocytes became translationally up-regulated in zygotes, while mRNAs with higher TE tended to be translationally down-regulated (Fig. 3B), indicating a translational switch upon fertilization (fig. S5E). In addition, GO analysis revealed that translationally down-regulated mRNAs were enriched for RNA splicing and processing, which has an essential role in the regulation and establishment of the maternal transcriptome in oocytes (31), while translational up-regulated mRNAs were enriched for protein degradation, RNA localization, and cell division (Fig. 3C), consistent with the MZT induced by fertilization. For example, genes with function in oocyte maturation, such as Ndc80 (32), was translationally down-regulated from MII oocytes to zygotes (Fig. 3, D and E), while genes involved in MZT, such as Rnf114 (33) and Mastl (34), were translationally up-regulated in zygotes (Fig. 3, D and E). To confirm these observations, we performed immunostaining and Western blotting analysis, which confirmed the increased protein levels for Chek1, Igf2bp2, Yap1, Hsf1, Mastl, Ybx1, and Dtl in zygotes (Fig. 3, F and G, and fig. S6) and decreased level for Smc4 (Fig. 3, F and G). Collectively, the above differential translational analyses revealed a translational switch upon fertilization, and this switch may play an important role in regulating MZT.
Fig. 3. A translational switch during MII to zygote transition.
(A) Scatterplot showing the differentially translated genes between MII oocytes and zygotes. Fold change ≥ 2, false discovery rate (FDR) < 0.01. Red and blue dots indicate mRNAs whose translation from MII oocytes to zygotes was up- and down-regulated, respectively. The x and y axes are log2-transformed normalized read counts. (B) Scatterplot showing the relationship between TE in MII oocytes and translational fold change of differentially translated genes from MII oocytes to zygotes. Red and blue dots indicate mRNAs whose translation from MII oocytes to zygotes were up- and down-regulated in (A), respectively. (C) Gene ontology enrichment analysis of the maternal mRNAs that was translationally up- and down-regulated during MII oocyte–to–zygote transition. The enriched terms are ranked by adjusted P value. (D) Bar graph showing the relative translational levels of some maternal mRNAs down-regulated (top) and up-regulated (bottom) in zygotes. (E) Bar graph showing the log2TE of genes in (D) at the MII stage. (F) Left panels showing representative images of immunostaining of Smc4 and Chk1 in MII oocytes and zygotes. Scale bar, 20 μm. Right panel showing the quantification of relative fluorescence intensity. ***P < 0.001. (G) Western blot showing Smc4 and Chk1 protein levels in MII oocytes and zygotes.
Active translation is essential for MZT
To understand the biological significance of the translational switch, particularly the increased translation in zygotes, we next attempted to determine whether this translational switch has a role in MZT. Previous studies using the translation inhibitor cycloheximide (CHX) showed that translation of maternal mRNAs is required for minor ZGA and maternal mRNA decay (17). However, in that study, embryos treated with CHX were blocked at one-cell stage, but the samples were not collected until 43 hours after human chorionic gonadotropin (hCG) injections for microarray analysis. At this time point, the untreated control embryos are at an early two-cell stage, making the comparison inappropriate because of their different developmental stages. To reveal the true effect of CHX treatment on mouse MZT, we first determined the exact time when embryonic development is affected by CHX treatment. To this end, we treated embryos with CHX at fertilization and analyzed its effect on pronuclei (PN) formation and cell cycle progression (fig. S7A). We found that CHX treatment did not affect PN formation as indicated by the successful TH2B and H2A deposition to the parental PN (PPN) (fig. S7B). Analysis of rH2AX level, an indicator of DNA damage, revealed a great increase in PPN at 6 hpf after CHX treatment (fig. S7C), indicating that CHX treatment impaired the repair of DNA lesions caused by the rapid DNA demethylation occurring in the PPN (35). However, a detailed cell cycle progression analysis indicated that CHX-treated embryos had successfully gone through S phase [indicated by the bromodeoxyuridine (BrdU) staining] and G2 phase (indicated by phospho-histone 3 staining) but were blocked to enter M phase (fig. S7D), indicating that translation is required for the mitotic cell cycle progression.
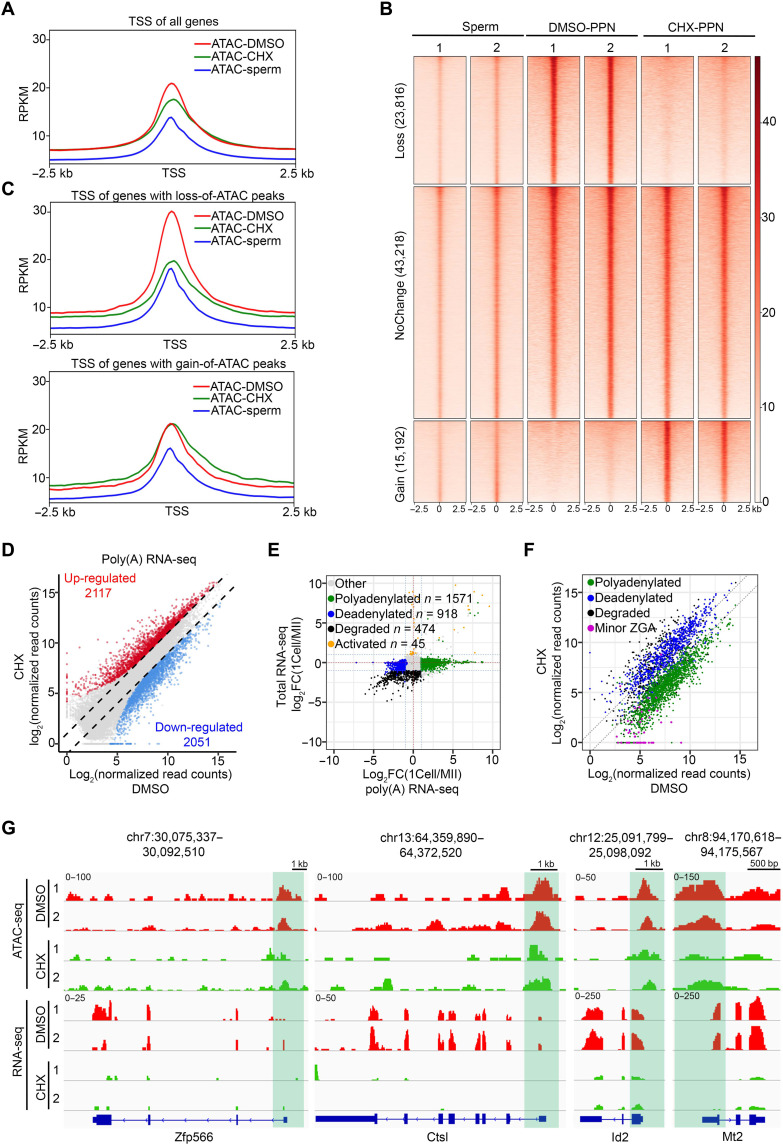
Given that CHX treatment did not affect cell cycle progression in the first 12 hours after fertilization (fig. S7D), our previous studies have demonstrated that, at 12 hours after fertilization, chromatin accessibility has been globally reprogrammed (36), raising the question whether maternal mRNA translation is required for chromatin accessibility reprogramming. To answer this question, we performed ATAC-seq (assay for transposase-accessible chromatin using sequencing) analysis on PPN at 12 hours after fertilization from embryos treated or untreated with CHX (fig. S8A and table S4). Compared to sperms (37), a notable increase in chromatin accessibility was observed in the PPN, which was impeded by CHX treatment (Fig. 4A). Consistently, PCA revealed that the chromatin accessibility landscape of CHX-treated embryos was between sperms and dimethyl sulfoxide (DMSO) control (fig. S8B), suggesting that CHX treatment compromised chromatin accessibility reprogramming. When compared with the DMSO control, CHX treatment resulted in 29% of loss and 18.5% of gain of ATAC peaks (Fig. 4B). While the gained ATAC peaks were largely outside the transcription start site (TSS) (Fig. 4C, bottom), the lost ATAC peaks were mostly around TSS (Fig. 4C, top) and made the ATAC signal closer to that in sperm. This observation indicates that translation inhibition impaired the accessibility reprogramming from sperms to PPN by blocking chromatin opening at promoter, which may affect the subsequent ZGA.
Fig. 4. Active translation is essential for the MZT.
(A) Metaplot showing the ATAC-seq signals at TSSs in sperms and zygotes treated with DMSO or CHX. (B) Heatmap showing the ATAC peaks classified on the basis of their changes in zygotes treated by CHX versus DMSO. Each row represents a locus (ATAC-seq peak center ± 2.5 kb), and the red gradient color indicates the ATAC-seq signal intensity. The numbers above the column indicate the replicates. (C) Metaplot showing the ATAC-seq signals at the TSS of genes with loss (top) and gain (bottom) of ATAC peaks in CHX-treated zygotes. (D) Scatterplot showing the comparison of RNA-seq profiles of DMSO- and CHX-treated zygotes. Two replicates for each treatment were used for differential gene expression analyses (FC ≥ 2, FDR < 0.01, RPKM ≥ 1). The x and y axes are transformed normalized read counts. (E) Scatterplot showing the classification of polyadenylated, deadenylated, degraded, and activated genes based on comparison of total RNA-seq and poly(A) RNA-seq data. Two replicates for each group were used for differential gene expression analysis (FC ≥ 2, FDR < 0.01, RPKM ≥ 1). (F) Scatterplot showing expression of polyadenylated, deadenylated, degraded, and minor ZGA genes in DMSO- and CHX-treated zygotes. Two replicates for each group were used for differential gene expression analysis (FC ≥ 2, FDR < 0.01, RPKM ≥ 1). The x and y axes are log2-transformed normalized read counts. (G) Examples of the genome browser view of ATAC-seq and RNA-seq results from mouse zygotes treated with DMSO or CHX. Shaded area indicates the ATAC-seq peaks around the TSS region.
Gene regulation during MZT is at multiple levels, including maternal mRNA decay, minor ZGA, and mRNA processing such as polyadenylation and deadenylation (38–40). Before analyzing each of their contribution, we first determined the effect of CHX treatment on the overall steady-state mRNA levels. To this end, transcriptome profiling was performed with 12-hpf zygotes treated with DMSO or CHX (fig. S8C and table S5). We found that the mRNA level of a large number of genes was changed, with 12.1 and 12.5% of total detected genes down- and up-regulated, respectively (Fig. 4D). To determine respective contribution at various levels affected by translation inhibition, we performed an integrative analysis of total RNA-seq and poly(A) RNA-seq of MII oocytes and zygotes. This analysis allowed us to divide genes into activated, degraded, polyadenylated, and deadenylated gene groups (Fig. 4E and table S6). To make the detection of minor ZGA genes more accurate, we used an RNA-seq dataset comparing the transcriptomes in the absence or presence of the transcriptional inhibitor α-amanitin (AMA) (table S7). This analysis revealed that CHX treatment resulted in down-regulation of most polyadenylated and minor ZGA genes (Fig. 4F, green and purple dots), while degraded genes tended to be up-regulated (Fig. 4F, black dots). Although CHX treatment may indirectly affect mRNA stability by inhibiting mRNA decapping (41, 42), it is possible that factors required for maternal mRNA decay are translated after fertilization. The poly(A) length of mRNAs is known to positively correlate with mRNA translation (43); here, our results indicated that mRNA polyadenylation may, in turn, be regulated by active translation of some maternal mRNAs. To determine whether down-regulated minor ZGA genes are associated with loss of chromatin accessibility, we analyzed the ATAC-seq signals for minor ZGA genes and noticed an obvious decrease in the ATAC-seq signals at the TSS region in CHX-treated samples compared to the control (Fig. 4G and fig. S8D), indicating that translation inhibition–caused loss of chromatin accessibility may be responsible for the down-regulation of these genes. Collectively, our analyses demonstrate that translation at zygotes plays a critical function in MZT, including mitotic progression, chromatin accessibility establishment, minor ZGA, maternal mRNA degradation, and polyadenylation.
Smarcd2 is required for chromatin reprogramming after fertilization
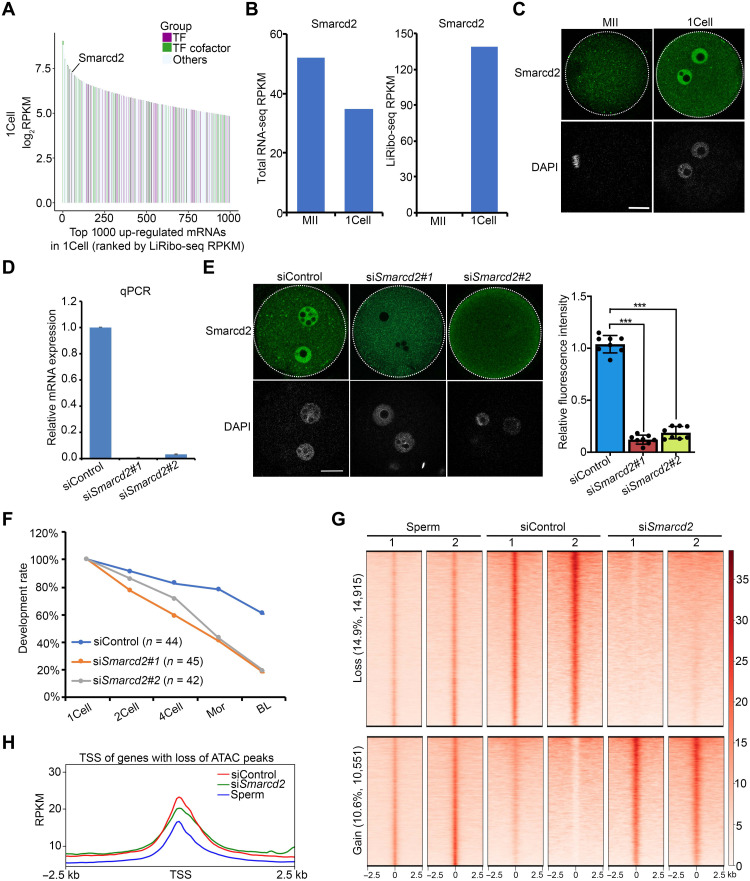
Having established the function of maternal mRNA translation in MZT, we next attempted to identify factors required for MZT, particularly those that regulate chromatin accessibility and ZGA. To this end, we focused on mRNAs whose translation activities were increased after fertilization and ranked them on the basis of their LiRibo-seq RPKM (reads per kilobase of transcript per million reads mapped) value in zygotes (Fig. 5A and table S8). Because a major function of adenosine triphosphate (ATP)–dependent chromatin remodeling factors is to regulate chromatin accessibility (44), we first checked the translation activity of chromatin remodeling factors and identified Smarcd2, also known as Baf60b (BRG1/Brahma-associated factor 60b), as the highest translated chromatin remodeling factor (Fig. 5A), although its mRNA level did not change much from MII oocytes to zygotes (Fig. 5B). Immunostaining confirmed its marked increase in its protein level from MII oocytes to zygotes (Fig. 5C), and this increase was impaired with CHX treatment (fig. S9A), supporting the notion that it is actively translated after fertilization.
Fig. 5. Smarcd2 is required for chromatin reprogramming after fertilization.
(A) Rank plot showing the translation level of the top 1000 translationally up-regulated mRNAs in zygotes. (B) Bar graph showing transcriptional and translational level (RPKM) of Smarcd2 in both MII oocytes and zygotes. (C) Representative images of immunostaining of Smarcd2 in MII oocytes and zygotes. Scale bar, 20 μm. (D) Bar graph showing the relative mRNA level of Smarcd2 in siControl and siSmarcd2 GV oocytes after 24 hours of arrest. (E) Immunostaining showing the reduced Smarcd2 protein level by siRNA in zygotes at 6 hpf. Scale bar, 20 μm. The right panel is the quantification of the Smarcd2 protein level. The average intensity of siControl zygotes were set as 1.0. Each dot represents a single embryo analyzed. ***P < 0.001. (F) Line chart showing the percentage of embryos reaching the indicated stages. The numbers of embryos examined were 44 (siControl), 45 (siSmarcd2#1), and 42 (siSmarcd2#2). (G) Heatmap showing the lost and gained ATAC peaks in siSmarcd2 versus siControl zygotes. Each row represents a locus (ATAC-seq peak center ± 2.5 kb), and the red gradient color indicates the ATAC-seq signal intensity. (H) Metaplot showing the ATAC-seq signals at the TSS of genes with loss of ATAC peaks in siSmarcd2 zygotes.
Smarcd2 is a component of the SWI/SNF complex and has been identified as a key regulator of granulopoiesis and myeloid differentiation (45, 46). Recent studies indicated that the integrity of the SWI/SNF complex is required for maintaining chromatin accessibility in embryonic stem cells (47, 48). To determine whether Smarcd2 has a role in chromatin accessibility during MZT, we attempted to deplete maternal Smarcd2 mRNA by injecting small interfering RNA (siRNA) into germinal vesicle (GV) oocytes. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) and immunostaining analyses confirmed that Smarcd2 mRNA and protein were both efficiently depleted (Fig. 5, D and E). To evaluate the effect of Smarcd2 depletion on preimplantation development, siRNA-injected GV oocytes were in vitro matured and fertilized, and their developmental potential was compared with that of control. We found that Smarcd2 depletion greatly reduced the blastocyst formation rate when compared to control (Fig. 5F), indicating that Smarcd2 has an important role in preimplantation development. To further confirm that the function of Smarcd2 in preimplantation development depends on its translational activity after fertilization, we used Smarcd2 Atg morpholino (MO) to block its translation in zygotes. After confirming the depletion of Smarcd2 protein (fig. S9B), we observed a similar defect in blastocyst formation (fig. S9C), supporting a role of active Smarcd2 translation in preimplantation development.
To assess its role in remodeling chromatin accessibility in zygotes, we isolated the PPN from control and siSmarcd2-injected zygotes and performed ATAC-seq (fig. S9D and table S9). This analysis revealed that about 25% of all protein-coding genes changed their chromatin accessibility in response to Smarcd2 depletion, including 14.7% of genes that lost their ATAC peaks and 10.9% of genes that gained ATAC peaks (Fig. 5, G and H, and fig. S9E). These findings are consistent with previous results demonstrating a role of Brg1 (also known as Smarca4), the catalytic subunit of SWI/SNF-related complexes, in regulating genome reprogramming and the subsequent ZGA (49). Collectively, we conclude that Smarcd2 is an actively translated protein in zygotes, whose depletion affects paternal genome accessibility during MZT.
Cyclin T2 is a regulator for ZGA
To identify factors regulating ZGA, we analyzed translationally up-regulated genes in zygotes (Fig. 3A). Among these genes, 50 have been reported to be embryonic lethal before or at implantation when knocked out (table S10), indicating that they play an important role in preimplantation development. To further narrow down this list, we overlapped the 50 genes with transcription factor/cofactor list and identified 18 genes with DNA binding/activating capability (Fig. 6A). The Ccnt2 gene, which encodes Cyclin T2, caught our attention because it is one component of the positive transcription elongation factor b (P-TEFb). Cyclin T2 recruits cyclin-dependent kinase 9 (CDK9) to phosphorylate the CTD (C-terminal domain) of the largest subunit of RNA polymerase II (RNAP II), thus activating transcription elongation (50). Ccnt2 has a nonredundant function with Ccnt1, as its depletion resulted in preimplantation lethality (51). However, its role in ZGA has not been explored.
Fig. 6. Cyclin T2 is a regulator of ZGA.
(A) Venn diagram showing the overlaps of three gene lists for identifying ZGA candidate regulators. KO, knockout. (B) Bar graph showing the transcriptional and translational level (RPKM) of Ccnt2 in both MII oocytes and zygotes. (C) Representative images of immunostaining of Cyclin T2 in MII oocytes and zygotes. Scale bar, 20 μm. (D) Bar graph showing the relative mRNA level of Ccnt2 in siControl and siCcnt2 GV oocytes after 24 hours of arrest. (E) Immunostaining showing the reduced Cyclin T2 protein level in two independent siRNAs in zygotes at 6 hpf. Scale bar, 20 μm. The right panel is a quantification of Cyclin T2 protein level. The average intensity of siControl zygotes was set as 1.0. Each dot represents a single embryo analyzed. ***P < 0.001. (F) Line chart showing the percentage of embryos reaching the indicated stages. The numbers of embryos examined were 40 (siControl), 36 (siCcnt2 #1), and 38 (siCcnt2 #2). (G) Scatterplots showing the RNA-seq profiles of all genes in siCcnt2 relative to siControl embryos at late two-cell stage. Two replicates for each group were used for differential gene expression analyses (FC ≥ 2, FDR < 0.05, RPKM ≥ 1). The x and y axes are log2-transformed normalized read counts. (H) Scatterplots showing the RNA-seq profile of ZGA genes in siCcnt2 relative to siControl embryos at late two-cell stage. Two replicates for each group were used for differential gene expression analyses (FC ≥ 2, FDR < 0.05, RPKM ≥ 1). The x and y axes are log2-transformed normalized read counts.
RNA-seq and LiRibo-seq revealed that Ccnt2 is a maternal gene, but its translation was markedly increased in zygotes (Fig. 6B). Immunostaining confirmed the marked increase of its protein level in zygotes compared to that in MII oocytes (Fig. 6C), and translation inhibition by CHX greatly reduced Cyclin T2 protein level (fig. S10A), consistent with the notion that it is a maternal factor highly translated in zygotes. To assess its potential role in ZGA, we generated two independent siRNAs and performed Ccnt2 knockdown successfully in GV oocytes (Fig. 6, D and E). Similar to the previously reported preimplantation lethal phenotype (51), Ccnt2 depletion caused many embryo arrests at two- or four-cell stage, and few can reach the morula stage (Fig. 6F). To further confirm that the role of Cyclin T2 in preimplantation development depends on its translational activity, we used Ccnt2 Atg MO to block its translation in zygotes. After confirming its depletion (fig. S10B), we observed embryo arrest at two- or four-cell stage (fig. S10C), consistent with siRNA knockdown results. Collectively, these results support a role of Ccnt2 mRNA translation in preimplantation development.
The two- to four-cell arrest phenotype suggests a possible ZGA defect. To test this notion, we performed RNA-seq with late two-cell embryos when ZGA takes place (fig. S10D and table S11). Comparative transcriptome analysis revealed that siRNA-mediated Ccnt2 knockdown resulted in 370 gene up-regulations and 809 gene down-regulations, respectively (Fig. 6G). Of the 2252 mouse ZGA genes, 289 genes (12.8%) were significantly down-regulated [fold change (FC) ≥ 2, false discovery rate (FDR) < 0.05], while only 3 genes were up-regulated (Fig. 6H and fig. S10E). This result strongly suggests that Cyclin T2 plays an important role in regulating mouse ZGA.
DISCUSSION
Despite great progress in our understanding of the transcriptional regulation in mouse preimplantation development, the role of translational regulation during this process is poorly understood because of the technical limitation in profiling translation activity using limited materials. In this study, we improved the Ribo-seq technique to make this study possible and performed the first LiRibo-seq profiling of mRNAs actively engaged in translation at different stages of mouse preimplantation development. Integrative analyses of LiRibo-seq and RNA-seq allowed us to reveal several new insights into the translational regulation of mouse preimplantation development. We found that translational activity was dynamically regulated during preimplantation and observed a translational switch during MZT from MII oocytes to zygotes. In addition, we provided several pieces of evidence demonstrating that translation of maternal mRNAs is required for MZT. Last, we identified important maternal factors, whose translation is activated by fertilization and is required for chromatin reprogramming and ZGA.
Comparative analysis of LiRibo-seq and RNA-seq datasets from oocytes and preimplantation embryos revealed dynamic changes of TE. From MII oocytes to zygotes transition, mRNA translation contributed to 84.6% of the regulatory changes, which is much more than its contribution (15.8 to 29%) during preimplantation development, indicating that translational regulation may play a more important role in MZT than during preimplantation development. Notably, morula and blastocyst embryos do not show many differences in the level of translation. This finding is different from proteomic analysis (8, 52), which revealed a marked change in protein level from morula to blastocyst embryos. One explanation is that proteomic analysis is a measurement of the steady-state protein level that is also controlled by posttranslational regulation in addition to translation activity. Thus, a direct measurement of translational activity by Ribo-seq is the only way to determine the true translational contribution of mRNA to protein expression.
It is known that maternal proteins are important for preimplantation development (7). However, whether translation of maternal stored mRNAs is absolutely required for fertilization-triggered MZT in mammals is not clear. Our detailed time course analysis of the effect of CHX treatment on cell cycle progression, chromatin accessibility, and transcriptomes demonstrates that translation of maternal mRNA immediately after fertilization is required for chromatin reprogramming and minor ZGA, key events of MZT. In addition, our LiRibo-seq data also indicate that mRNA translation activity is markedly increased after fertilization. With regard to the mechanism of how translation is triggered by fertilization, there are several hypotheses (10, 53), including the “masking model” in which the maternal mRNAs are believed to be masked by some RNA binding proteins (RBPs), preventing them from translation in oocytes. Fertilization may trigger some RBPs to change their modifications and release them from mRNAs, allowing mRNAs to be translated. The second hypothesis is the opposite. It is believed that some maternal mRNAs may need to be bound by RBPs to promote their translation, and fertilization can facilitate binding of RBPs to mRNAs. Besides RBPs, the length of poly(A) has also been reported to regulate mRNA translation (38). For example, in Xenopus and zebrafish, it has been reported that poly(A) lengthening occurs after fertilization and is positively correlated with mRNA translation (43). Whether any of these hypotheses explains the translation transition from MII oocytes to zygotes awaits to be investigated.
ZGA is one of the most important molecular events taking place during preimplantation development. However, the major regulators that control mammalian ZGA still await to be identified. Previous studies have been mainly focused on maternal proteins that may be important for ZGA. Here, we found that translational activation of maternal mRNAs also plays an important role for ZGA. Among these mRNAs, we demonstrated that Cyclin T2, a component of P-TEFb, is highly translated in zygotes. After maternal Ccnt2 knockdown, activation of about 13% of ZGA genes is impaired, supporting a role of Cyclin T2 in ZGA. Because there are still some Cyclin T2 proteins left in knockdown mice, the ZGA defects should be more marked in knockout mice. Previous studies indicated that Cyclin T1, a gene related to Cyclin T2, can cooperate with CDK9 to promote ZGA (54). However, Cyclin T1 protein is not detected in nuclei until late two-cell stage (54). Given their different expression and localization patterns, it is not surprising that Cyclin T1 and Cyclin T2 function in a nonredundant way during mouse embryonic development (50). Consistent with a previous hypothesis (55), here, we showed that Cyclin T2 protein is highly translated after fertilization and is localized in PN in zygotes, which is similar to that of CDK9 (54), indicating that Cyclin T2 may function earlier than Cyclin T1 in ZGA.
In summary, our study not only revealed mRNA translational dynamics during mouse preimplantation development but also identified factors important for MZT by regulating chromatin reprogramming and ZGA. Our work thus has filled in a knowledge gap in the study of translational regulation in mammalian preimplantation development.
MATERIALS AND METHODS
All animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Harvard Medical School.
Collection of mouse oocytes and embryos
Mouse MII oocytes were collected from wild-type (WT) B6D2F1/J strain (BDF1; Jackson stock: 100006) female mice (5). For ovarian hyperstimulation, 6- to 9-week-old female mice were injected intraperitoneally with 7.5 IU of pregnant mare serum gonadotropin (PMSG; Millipore) on day 1 and 7.5 IU of hCG (Millipore) on day 3 (44 to 48 hours after PMSG injection). MII oocytes were collected from oviducts 12 to 16 hours after hCG injection, and cumulus cells were removed from oocytes by briefly incubating in M2 medium with hyaluronidase (Millipore).
For preimplantation embryo collection, MII oocytes from WT B6D2F1/J female mice were fertilized with sperms from WT B6D2F1/J male in vitro. Specifically, spermatozoa collected from the caudal epididymis were first incubated in human tubal fluid (HTF) medium supplemented with bovine serum albumin (BSA) (10 mg/ml) (Sigma-Aldrich) for 1 hour. The capacitated spermatozoa were used to fertilize cumulus-oocyte complexes (COCs) collected from oviducts. Approximately 6 hours after in vitro fertilization (IVF), zygotes with two PN were transferred from HTF medium to KSOM (Millipore) and cultured at 37°C under 5% CO2. The time when sperm was added to COCs was considered as 0 hpf. One-cell, two-cell, four-cell, morula, and blastocyst embryos were collected at 12, 30, 48, 72, and 96 hpf, respectively.
For GV oocyte collection, ovaries were washed in M2 medium (Sigma-Aldrich) with 0.2 mM 3-isobutyl-1-methylxanthine (IBMX) (Sigma-Aldrich) and punctured with a 27-gauge needle to release COCs. The cumulus cells were removed from oocytes by a narrow-bore glass pipette. GV oocytes were then cultured in α-MEM (minimum essential medium) (Life Technologies) supplemented with 5% fetal bovine serum (FBS; Sigma-Aldrich), epidermal growth factor (EGF; 10 ng/ml; Sigma-Aldrich), and 0.2 mM IBMX (Sigma-Aldrich). IBMX was supplemented to inhibit meiotic resumption of GV oocytes. After GV oocytes were incubated in MEM + IBMX medium for 1 hour, oocytes with perivitelline space were collected for siRNA injection. The siRNA concentrations for siControl, siSmarcd2 (Thermo Fisher Scientific, siRNA ID: 175190), and siCcnt2 (Horizon Discovery, J-044000-06-0002 and J-044000-08-0002) were 5 μM. To test knockdown efficiency, injected GV oocytes were cultured in MEM + IBMX medium for 24 hours and then collected for RT-qPCR. Oligonucleotides used in this study were listed in table S12. To test developmental phenotype, injected GV oocytes were cultured in MEM + IBMX medium for 8 hours and then transferred to MEM medium for in vitro maturation (IVM). After 16 to 18 hours of IVM, MII oocytes were collected and used for IVF. Atg MOs targeting Smarcd2 and Ccnt2 (Gene Tools) were injected into zygotes 2 hours after fertilization at a concentration of 0.5 mM.
Chemical treatment
For DMSO, CHX (Sigma-Aldrich), and AMA (Sigma-Aldrich) treatment, MII oocytes were fertilized with sperms in HTF medium with CHX or AMA (20 μg/ml). Approximately 6 hours after IVF, zygotes with two PN were transferred to KSOM (Millipore) with CHX or AMA (20 μg/ml) and cultured at 37°C under 5% CO2. DMSO (0.2%, v/v) was used as a control.
Whole-mount immunostaining
One-cell embryos were fixed in 3.7% paraformaldehyde containing 0.2% Triton X-100 at room temperature for 20 min. After washing four times in 1% BSA/phosphate-buffered saline (PBS), embryos were incubated with primary antibodies at 4°C overnight. The primary antibodies used for immunostaining were rabbit anti-Smc4 (1:200 dilution; Cell Signaling Technology, 5547S), mouse anti-Chk1 (1:200; Cell Signaling Technology, 2360S), rabbit anti-Igf2bp2 (1:100; Thermo Fisher Scientific, PA5-96202), rabbit anti-Yap1 (1:100; Proteintech, 13584-1-AP), rabbit anti-Hsf1 (1:200; Cell Signaling Technology, 4356S), rabbit anti-Mastl (1:200; Cell Signaling Technology, 12069S), rabbit anti-Ybx1 (1:100; Proteintech, 20339-1-AP), rabbit anti-Dtl (1:100; Proteintech, 12896-1-AP), mouse anti-TH2B (1:2000; a gift from S. Ishii), rabbit anti-H2A (1:2000; MBL, D210-3), mouse anti-rH2AX (1:1000; Millipore, 05-636), mouse anti-pH3 (1:1000; Millipore, 06-570), mouse anti-BrdU (1:200; Roche, 11170376001), rabbit anti-Smarcd2 (1:200; Abcam, ab220164), and rabbit anti–Cyclin T2 (1:500; Novus Biologicals, NBP1-87592). After washing three times in 1% BSA/PBS, embryos were incubated with a 1:200 dilution of Alexa Flour 488 donkey anti-mouse immunoglobulin G (IgG) or Alexa Fluor 568 donkey anti-rabbit IgG (Life Technologies) for 1 hour. After washing with 1% BSA/PBS, the embryos were mounted on a glass slide in Vectashield anti-bleaching solution with DAPI (4′,6-diamidino-2-phenylindole) (Vector Laboratories). Fluorescence was detected using a laser scanning confocal microscope (Zeiss, LSM800).
Western blotting
A total of 100 MII oocytes and zygotes were collected, and Western blotting was performed as described previously (56). Primary antibodies against Smc4 (1:1000 dilution; Cell Signaling Technology, 5547S), Chk1 (1:1000 dilution; Cell Signaling Technology, 2360S), and β-actin (1:1000 dilution; Cell Signaling Technology, 4967S) were used in this study.
LiRibo-seq library preparation and sequencing
LiRibo-seq libraries were prepared using Active Ribo-Seq with the RiboLace Kit (IMMAGINA Biotechnology, RL001) and the SMARTer smRNA-Seq Kit (Takara, 635030) following the manufacturer’s instructions. Briefly, MII oocytes and embryos were cultured in KSOM with CHX (10 μg/ml) and cultured at 37°C under 5% CO2 for 15 min. Then, oocytes and embryos were briefly treated with Acidic Tyrode (Millipore) to remove zona pellucida, washed with M2 medium (Sigma-Aldrich), and finally washed with 0.2% BSA/PBS. In total, 100 to 250 oocytes or embryos were collected and lysed with 100 μl of lysis buffer containing 1% sodium deoxycholate, deoxyribonuclease I (5 U/ml), and RiboLock RNase inhibitor (200 U/ml). Oocytes and embryos were triturated with a 30-gauge needle and kept on ice for 15 min. After centrifugation at 20,000g for 5 min, the supernatant was transferred to a new tube and kept on ice for 20 min. Absorbance of lysate was checked at 260 nm with NanoDrop. Ribosome footprinting was generated with nuclease treatment according to the manufacturer’s instructions. RPFs were captured by functionalized RiboLace beads and purified by acid phenol:chloroform following the manufacturer’s instructions. Purified RPFs were dephosphorylated with T4 polynucleotide kinase diphosphatase and then used for library preparation with the SMARTer smRNA-Seq Kit (Takara, 635030). Artificial poly(A) tails were added to RPFs, and complementary DNAs (cDNAs) were synthesized with 3′ smRNA dT primer. After amplification, 175– to 200–base pair (bp) PCR products were purified with polyacrylamide gel electrophoresis gel purification. The prepared LiRibo-seq libraries were sequenced on NextSeq 550 (Illumina) with paired-ended 75-bp reads.
RNA-seq library preparation and sequencing
Total RNA-seq libraries were prepared using the SMART-Seq Stranded Kit (Clontech) following the manufacturer’s instructions. Poly(A) RNA-seq libraries were prepared as previously described (56) using a SMARTer ultralow input RNA cDNA preparation kit (Clontech). A Nextera XT DNA library preparation kit (Illumina) was used for poly(A) RNA-seq cDNA fragmentation, adaptor ligation, and amplification according to the manufacturer’s instructions. The prepared RNA-seq libraries were sequenced on NextSeq 550 (Illumina) with paired-ended 75-bp reads.
ATAC-seq library preparation and sequencing
For ATAC-seq with PPN, one-cell embryos (12 hpf) were cultured in M2 medium containing 10 μM cytochalasin B (Calbiochem) for 15 min. PPN were collected with minimal cytoplasm carryover using a piezo-driven micromanipulator (Eppendorf). After washing with 1% BSA/PBS, 75 to 100 PPN were collected for ATAC-seq following the same protocol as previously described (56).
LiRibo-seq data analysis
The first three nucleotides of each read that were generated from template switch oligo (TSO) from LiRibo-seq were removed, and the poly(A) and adaptor sequences in each read were trimmed using Cutadapt (v2.10) (57) with the following parameters: -m 20 -u 3 -a AAAAAAAA. Trimmed reads that were shorter than 20 bp were discarded. To remove reads from ribosomal RNA (rRNA) and tRNA, we aligned the reads to sequences from SILVA rRNA database (www.arb-silva.de; release 138.1) and Genomic tRNA database (http://gtrnadb.ucsc.edu; mm10-tRNAs.fa) using bowtie2 (v2.4.2) (58) with default parameters. Reads not mapped to rRNA and tRNA were used for further analysis. These reads were aligned to mouse reference genome GRCm38 using STAR (v2.7.8a) (59) in an end-to-end alignment mode and allowing two maximum mismatches per read. The genome alignments were converted to transcriptome alignments by STAR with Gencode M24 mouse gene annotations, and only protein-coding genes were used. To quantify the ribosome footprints for each protein-coding gene, RSEM (v1.3.1) (60) was used with reads mapped to transcriptome as input and parameters as follows: --estimate-rspd --calc-ci --strandedness forward.
To assess the consistency between replicates, Pearson correlation coefficients were calculated using log2-transformed RPKM values generated from RSEM. To evaluate the quality of LiRibo-seq data, P-site distribution in CDS, untranslated region, and P-site metaplot around the start codon and stop codon were generated using riboWaltz (v1.2.0) (61). The public mESC Ribo-seq data (11) were downloaded from Gene Expression Omnibus (GEO) with accession number GSM765302.
TE was calculated as RPKM from LiRibo-seq divided by RPKM from total RNA-seq. DTG and DTEG were identified following the deltaTE (30) method, with read counts generated by RSEM from total RNA-seq and LiRibo-seq as input. DTGs and DTEGs were defined with FDR < 0.05.
The ranking of the one-cell newly translated gene candidates was ordered by the mean RPKM from LiRibo-seq at one-cell stage and was not translated (LiRibo-seq mean RPKM < 1) at MII stage. The mouse TF and TF cofactor gene lists were retrieved from AnimalTFDB3 (62).
RNA-seq data analysis
For paired-end sequencing reads of total RNA-seq, the first three nucleotides of read 2 that were generated from TSO were first removed. For total RNA-seq and poly(A) RNA-seq, adaptors if presented in reads were trimmed with Trimmomatic (v0.39) (63). The cleaned reads were mapped to mouse reference genome GRCm38 using STAR (v2.7.8a) (59) with the following parameters: --outFilterMultimapNmax 20 --outFilterMismatchNmax 999 --outFilterMismatchNoverReadLmax 0.04 --alignIntronMin 20 --alignIntronMax 1000000 --alignMatesGapMax 1000000 --alignSJoverhangMin 8 --alignSJDBoverhangMin 1 --sjdbScore 1 and were converted to transcriptome alignments. RSEM (v1.3.1) (60) was used to quantify the expression level of each gene. For total RNA-seq, the parameters for RSEM were --estimate-rspd --calc-ci --paired-end --strandedness reverse. For poly(A) RNA-seq, the parameters for RSEM were --estimate-rspd --calc-ci --paired-end --strandedness none.
For differentially expressed gene (DEG) analysis, DESeq2 (v1.32.0) (64) package in R was used with read counts generated from RSEM as input. To obtain the genome browser views, bigwig files were generated as input of IGV (v2.7.2) (65) by using deepTools (66) bamCoverage with the following parameters: --binSize 20 --minMappingQuality 30 --scaleFactor 1 --normalizeUsing RPKM.
Minor ZGA genes were identified by comparing RNA-seq of AMA-treated one-cell embryos and control (DMSO treated), with FC ≥ 5, FDR < 0.01, and control mean FPKM (fragments per kilobase of exon per million mapped fragments) ≥ 1. Major ZGA genes were identified as genes up-regulated in two-cell stage, with zygote as control (FC ≥ 5, FDR < 0.01, and two-cell mean RPKM ≥ 1). Two-cell transient genes were defined by comparing RNA-seq of two-cell to one-cell and four-cell embryos with FC ≥ 5, FDR < 0.01, and two-cell mean FPKM ≥ 1. Maternal genes were defined as genes with RNA-seq RPKM ≥ 20 in MII oocytes.
Polyadenylated, deadenylated, degraded, and activated genes were defined by comparing the expression difference between MII oocytes and one-cell embryos of poly(A) RNA-seq data and total RNA-seq data. DEGs were identified with the parameter that mean RPKM ≥ 1, FC ≥ 2, and FDR < 0.01. Polyadenylated genes were defined as genes up-regulated in poly(A) RNA-seq but not changed in total RNA-seq. Deadenylated genes were defined as genes down-regulated in poly(A) RNA-seq but not changed in total RNA-seq. Degraded genes were defined as genes down-regulated or not changed in poly(A) RNA-seq but down-regulated in total RNA-seq. Activated genes were defined as genes up-regulated or not changed in poly(A) RNA-seq but up-regulated in total RNA-seq.
ATAC-seq data analysis
All the ATAC-seq reads were first trimmed of the adaptors with Cutadapt (version 2.10) and then aligned to the GRCm38 genomes using bowtie2 (version 2.2.5) (67) with the following parameters: -X 800 -I 0 --no-mixed --no-discordant --no-unal. All the unmapped reads and duplicated reads were removed. The nonuniquely mapped reads were filtered by “XS:” flag in bam files. Reads with mapping quality (MAPQ) of less than 30 were removed by samtools (version 1.11). The Pearson correlation between ATAC-seq replicates was calculated using log2-transformed RPKM in 10-kb bins with the multiBigwigSummary function of deepTools (66).
To obtain the genome browser views, bigwig files were generated as input of IGV by using deepTools bamCoverage with the following parameters: --binSize 20 -e 250 --minMappingQuality 30 --scaleFactor 1 --normalizeUsing RPKM. All the ATAC-seq peaks were called by macs2 (68) with the following parameters: -g mm --bdg --SPMR --nomodel -q 0.05 --nolambda --shift -100 --extsize 200. Metaplot was generated using computeMatrix and plotProfile function of deepTools (66).
To classify ATAC-seq peaks according to their changes, peaks identified in the two replicates were merged to get all the possible loci. Then, the detectable peaks were filtered with the following standard to remove noise signal: (i) RPKM normalized peak signal in one sample replicate > 1 and (ii) mean of RPKM normalized peak signal in two sample replicates > 2. Last, the peaks with FC > 5 in treated samples over control samples were classified as gain or loss peaks.
Clustering, PCA, and GO analysis
Genes with LiRibo-seq RPKM ≥ 1 in at least one stage were used for clustering. Before clustering analysis, RPKMs from pooled replicates of LiRibo-seq among different developmental stages for each gene were z score–normalized. Hierarchical clustering of genes was performed using the hclust function in R with Pearson correlation as distance measure metric. hcut in factoextra (v1.0.7) package was used to divide the cluster tree into nine clusters. The clustering results were visualized by the ComplexHeatmap (v2.8.0) (69) package.
The PCA analysis was performed with prcomp function in R. For LiRibo-seq and RNA-seq data, log2-transformed RPKM of pooled replicates for each stage were used as input. For ATAC-seq data, log2-transformed RPKM in 10-kb bins were used as input and bins with zero value in all samples were removed. The GO enrichment analysis was performed with the clusterProfiler (v4.0.5) (70) package, and the biological process ontology was used.
Statistical analysis
All the statistical analysis was performed using R programming language. The “cor” function was used to calculate Pearson correlation coefficient. The “t.test” was used to perform two-sample Student’s t tests, and the “wilcox.test” was used to perform two-sample Wilcoxon rank sum and signed-rank tests. The P value was adjusted with FDR or Benjamini-Hochberg correction for multiple comparisons. The ggplot2 package (v3.3.1) was used to generate most of the plots.
Acknowledgments
We thank S. Ishii for the mouse anti-TH2B antibody. We thank Z. Chen for the critical discussion of this project and help of total RNA-seq library preparation.
Funding: This project was supported by the NIH (R01HD092465) and HHMI. Y.Z. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: Y.Z. conceived the project. C.Z. and Y.Z. designed the experiments. C.Z. performed the experiments. M.W. and Y.L. analyzed the high-throughput data. All authors were involved in the interpretation of the data. C.Z. and Y.Z. wrote the manuscript, with some input from M.W. and Y.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All sequencing data have been deposited in the GEO under the accession number GEO: GSE169632. The code used to analyze LiRibo-seq and RNA-seq data is available at https://doi.org/10.5281/zenodo.5748293 and at GitHub: https://github.com/YiZhang-lab/LiRibo-seq.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Legends for tables S1 to S12
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S12
Erratum published 15 August 2025:
There was an error in the originally published version of fig. S10B. The following correction has been implemented:
• In fig. S10B, the “Ccnt2 Atg MO” Cyclin T2 immunofluorescence image was inadvertently a duplicate of the “siCcnt2 #1” Cyclin T2 immunofluorescence image in Fig. 6E. This error occurred during the figure assembly process. Figure S10B has been corrected.
The authors’ conclusions are not affected by this correction. The Supplementary Materials have been updated.
The original version is available here:
REFERENCES AND NOTES
- 1.Lu F., Zhang Y., Cell totipotency: Molecular features, induction, and maintenance. Natl. Sci. Rev. 2, 217–225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiorentino J., Torres-Padilla M. E., Scialdone A., Measuring and modeling single-cell heterogeneity and fate decision in mouse embryos. Annu. Rev. Genet. 54, 167–187 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Xia W., Xie W., Rebooting the epigenomes during mammalian early embryogenesis. Stem Cell Rep. 15, 1158–1175 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu X., Zhang C., Zhang Y., Epigenetic regulation of mouse preimplantation embryo development. Curr. Opin. Genet. Dev. 64, 13–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu F., Liu Y., Inoue A., Suzuki T., Zhao K., Zhang Y., Establishing chromatin regulatory landscape during mouse preimplantation development. Cell 165, 1375–1388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z., Zhang Y., Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat. Genet. 51, 947–951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz K. N., Harrison M. M., Mechanisms regulating zygotic genome activation. Nat. Rev. Genet. 20, 221–234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Y., Liu X., Tang B., Li C., Kou Z., Li L., Liu W., Wu Y., Kou X., Li J., Zhao Y., Yin J., Wang H., Chen S., Liao L., Gao S., Protein expression landscape of mouse embryos during pre-implantation development. Cell Rep. 21, 3957–3969 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Wang S., Kou Z., Jing Z., Zhang Y., Guo X., Dong M., Wilmut I., Gao S., Proteome of mouse oocytes at different developmental stages. Proc. Natl. Acad. Sci. U.S.A. 107, 17639–17644 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teixeira F. K., Lehmann R., Translational control during developmental transitions. Cold Spring Harb. Perspect. Biol. 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingolia N. T., Lareau L. F., Weissman J. S., Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingolia N. T., Ghaemmaghami S., Newman J. R. S., Weissman J. S., Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kronja I., Yuan B., Eichhorn S. W., Dzeyk K., Krijgsveld J., Bartel D. P., Orr-Weaver T. L., Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition. Cell Rep. 7, 1495–1508 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichhorn S. W., Subtelny A. O., Kronja I., Kwasnieski J. C., Orr-Weaver T. L., Bartel D. P., mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. eLife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee M. T., Bonneau A. R., Takacs C. M., Bazzini A. A., DiVito K. R., Fleming E. S., Giraldez A. J., Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu G., Han D., Gong Y., Sebastiano V., Gentile L., Singhal N., Adachi K., Fischedick G., Ortmeier C., Sinn M., Radstaak M., Tomilin A., Schöler H. R., Establishment of totipotency does not depend on Oct4A. Nat. Cell Biol. 15, 1089–1097 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamatani T., Carter M. G., Sharov A. A., Ko M. S., Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell 6, 117–131 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Potireddy S., Vassena R., Patel B. G., Latham K. E., Analysis of polysomal mRNA populations of mouse oocytes and zygotes: Dynamic changes in maternal mRNA utilization and function. Dev. Biol. 298, 155–166 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Luong X. G., Daldello E. M., Rajkovic G., Yang C. R., Conti M., Genome-wide analysis reveals a switch in the translational program upon oocyte meiotic resumption. Nucleic Acids Res. 48, 3257–3276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapeli K., Yeo G. W., Genome-wide approaches to dissect the roles of RNA binding proteins in translational control: Implications for neurological diseases. Front. Neurosci. 6, 144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clamer M., Tebaldi T., Lauria F., Bernabò P., Gómez-Biagi R. F., Marchioretto M., Kandala D. T., Minati L., Perenthaler E., Gubert D., Pasquardini L., Guella G., Groen E. J. N., Gillingwater T. H., Quattrone A., Viero G., Active ribosome profiling with RiboLace. Cell Rep. 25, 1097–1108.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Hornstein N., Torres D., Das Sharma S., Tang G., Canoll P., Sims P. A., Ligation-free ribosome profiling of cell type-specific translation in the brain. Genome Biol. 17, 149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Courtois A., Schuh M., Ellenberg J., Hiiragi T., The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J. Cell Biol. 198, 357–370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hupalowska A., Jedrusik A., Zhu M., Bedford M. T., Glover D. M., Zernicka-Goetz M., CARM1 and paraspeckles regulate pre-implantation mouse embryo development. Cell 175, 1902–1916.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., Wang L., Feng G., Wang Y., Li Y., Li X., Liu C., Jiao G., Huang C., Shi J., Zhou T., Chen Q., Liu Z., Li W., Zhou Q., Asymmetric expression of LincGET biases cell fate in two-cell mouse embryos. Cell 175, 1887–1901.e18 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., Zhao J., Dahan P., Lu V., Zhang C., Li H., Teitell M. A., Metabolism in pluripotent stem cells and early mammalian development. Cell Metab. 27, 332–338 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Cheon Y. P., Gye M. C., Kim C.-H., Kang B. M., Chang Y. S., Kim S. R., Kim M. K., Role of actin filaments in the hatching process of mouse blastocyst. Zygote 7, 123–129 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Atlasi Y., Jafarnejad S. M., Gkogkas C. G., Vermeulen M., Sonenberg N., Stunnenberg H. G., The translational landscape of ground state pluripotency. Nat. Commun. 11, 1617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu X., Gao Z., Qin D., Li L., A maternal functional module in the mammalian oocyte-to-embryo transition. Trends Mol. Med. 23, 1014–1023 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Chothani S., Adami E., Ouyang J. F., Viswanathan S., Hubner N., Cook S. A., Schafer S., Rackham O. J. L., deltaTE: Detection of translationally regulated genes by integrative analysis of Ribo-seq and RNA-seq data. Curr. Protoc. Mol. Biol. 129, e108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Do D. V., Strauss B., Cukuroglu E., Macaulay I., Wee K. B., Hu T. X., Igor R. D. L. M., Lee C., Harrison A., Butler R., Dietmann S., Jernej U., Marioni J., Smith C. W. J., Göke J., Surani M. A., SRSF3 maintains transcriptome integrity in oocytes by regulation of alternative splicing and transposable elements. Cell Discov. 4, 33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gui L., Homer H., Hec1-dependent cyclin B2 stabilization regulates the G2-M transition and early prometaphase in mouse oocytes. Dev. Cell 25, 43–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y., Zhou C., Wang Y., Liu W., Liu C., Wang L., Liu Y., Shang Y., Li M., Zhou S., Wang Y., Zeng W., Zhou J., Huo R., Li W., The E3 ubiquitin ligase RNF114 and TAB1 degradation are required for maternal-to-zygotic transition. EMBO Rep. 18, 205–216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soeda S., Yamada-Nomoto K., Michiue T., Ohsugi M., RSK-MASTL pathway delays meiotic exit in mouse zygotes to ensure paternal chromosome stability. Dev. Cell 47, 363–376.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Ladstatter S., Tachibana-Konwalski K., A surveillance mechanism ensures repair of DNA lesions during zygotic reprogramming. Cell 167, 1774–1787.e13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Djekidel M. N., Inoue A., Matoba S., Suzuki T., Zhang C., Lu F., Jiang L., Zhang Y., Reprogramming of chromatin accessibility in somatic cell nuclear transfer is DNA replication independent. Cell Rep. 23, 1939–1947 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jung Y. H., Kremsky I., Gold H. B., Rowley M. J., Punyawai K., Buonanotte A., Lyu X., Bixler B. J., Chan A. W.-S., Corces V. G., Maintenance of CTCF- and transcription factor-mediated interactions from the gametes to the early mouse embryo. Mol. Cell 75, 154–171.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vastenhouw N. L., Cao W. X., Lipshitz H. D., The maternal-to-zygotic transition revisited. Development 146, (2019). [DOI] [PubMed] [Google Scholar]
- 39.Lee M. T., Bonneau A. R., Giraldez A. J., Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 30, 581–613 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe K. I., Funaya S., Tsukioka D., Kawamura M., Suzuki Y., Suzuki M. G., Schultz R. M., Aoki F., Minor zygotic gene activation is essential for mouse preimplantation development. Proc. Natl. Acad. Sci. U.S.A. 115, E6780–E6788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huch S., Nissan T., Interrelations between translation and general mRNA degradation in yeast. Wiley Interdiscip. Rev. RNA 5, 747–763 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herrick D., Parker R., Jacobson A., Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 2269–2284 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subtelny A. O., Eichhorn S. W., Chen G. R., Sive H., Bartel D. P., Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clapier C. R., Cairns B. R., The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Witzel M., Petersheim D., Fan Y., Bahrami E., Racek T., Rohlfs M., Puchałka J., Mertes C., Gagneur J., Ziegenhain C., Enard W., Stray-Pedersen A., Arkwright P. D., Abboud M. R., Pazhakh V., Lieschke G. J., Krawitz P. M., Dahlhoff M., Schneider M. R., Wolf E., Horny H. P., Schmidt H., Schäffer A. A., Klein C., Chromatin-remodeling factor SMARCD2 regulates transcriptional networks controlling differentiation of neutrophil granulocytes. Nat. Genet. 49, 742–752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Priam P., Krasteva V., Rousseau P., D’Angelo G., Gaboury L., Sauvageau G., Lessard J. A., SMARCD2 subunit of SWI/SNF chromatin-remodeling complexes mediates granulopoiesis through a CEBPɛ dependent mechanism. Nat. Genet. 49, 753–764 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Schick S., Grosche S., Kohl K. E., Drpic D., Jaeger M. G., Marella N. C., Imrichova H., Lin J.-M. G., Hofstätter G., Schuster M., Rendeiro A. F., Koren A., Petronczki M., Bock C., Müller A. C., Winter G. E., Kubicek S., Acute BAF perturbation causes immediate changes in chromatin accessibility. Nat. Genet. 53, 269–278 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iurlaro M., Stadler M. B., Masoni F., Jagani Z., Galli G. G., Schübeler D., Mammalian SWI/SNF continuously restores local accessibility to chromatin. Nat. Genet. 53, 279–287 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Bultman S. J., Gebuhr T. C., Pan H., Svoboda P., Schultz R. M., Magnuson T., Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 20, 1744–1754 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurosu T., Zhang F., Peterlin B. M., Transcriptional activity and substrate recognition of cyclin T2 from P-TEFb. Gene 343, 173–179 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Kohoutek J., Li Q., Blazek D., Luo Z., Jiang H., Peterlin B. M., Cyclin T2 is essential for mouse embryogenesis. Mol. Cell. Biol. 29, 3280–3285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Israel S., Ernst M., Psathaki O. E., Drexler H. C. A., Casser E., Suzuki Y., Makalowski W., Boiani M., Fuellen G., Taher L., An integrated genome-wide multi-omics analysis of gene expression dynamics in the preimplantation mouse embryo. Sci. Rep. 9, 13356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tadros W., Lipshitz H. D., Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev. Dyn. 232, 593–608 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Oqani R. K., Kim H. R., Diao Y. F., Park C. S., Jin D. I., The CDK9/cyclin T1 subunits of P-TEFb in mouse oocytes and preimplantation embryos: A possible role in embryonic genome activation. BMC Dev. Biol. 11, 33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svoboda P., Franke V., Schultz R. M., Sculpting the transcriptome during the oocyte-to-embryo transition in mouse. Curr. Top. Dev. Biol. 113, 305–349 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Zhang C., Chen Z., Yin Q., Fu X., Li Y., Stopka T., Skoultchi A. I., Zhang Y., The chromatin remodeler Snf2h is essential for oocyte meiotic cell cycle progression. Genes Dev. 34, 166–178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 3 (2011). [Google Scholar]
- 58.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lauria F., Tebaldi T., Bernabò P., Groen E. J. N., Gillingwater T. H., Viero G., riboWaltz: Optimization of ribosome P-site positioning in ribosome profiling data. PLOS Comput. Biol. 14, e1006169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu H., Miao Y.-R., Jia L.-H., Yu Q.-Y., Zhang Q., Guo A.-Y., AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 47, D33–D38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thorvaldsdottir H., Robinson J. T., Mesirov J. P., Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gu Z., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Legends for tables S1 to S12
Tables S1 to S12
Erratum published 15 August 2025:
There was an error in the originally published version of fig. S10B. The following correction has been implemented:
• In fig. S10B, the “Ccnt2 Atg MO” Cyclin T2 immunofluorescence image was inadvertently a duplicate of the “siCcnt2 #1” Cyclin T2 immunofluorescence image in Fig. 6E. This error occurred during the figure assembly process. Figure S10B has been corrected.
The authors’ conclusions are not affected by this correction. The Supplementary Materials have been updated.
The original version is available here: