Abstract
Animal genomes show networks of deeply conserved gene linkages whose phylogenetic scope and chromosomal context remain unclear. Here, we report chromosome-scale conservation of synteny among bilaterians, cnidarians, and sponges and use comparative analysis to reconstruct ancestral chromosomes across major animal groups. Comparisons among diverse metazoans reveal the processes of chromosome evolution that produced contemporary karyotypes from their Precambrian progenitors. On the basis of these findings, we introduce a simple algebraic representation of chromosomal change and use it to establish a unified systematic framework for metazoan chromosome evolution. We find that fusion-with-mixing, a previously unappreciated mode of chromosome change, has played a central role. We find that relicts of several metazoan chromosomal units are preserved in unicellular eukaryotes. These conserved pre-metazoan linkages include the chromosomal unit that encodes the most diverse set of metazoan homeobox genes, suggesting a candidate genomic context for the early diversification of this key gene family.
Conserved chromosomes reveal ancestral animal linkage groups, establishing a phylogenetic system for karyotype evolution.
INTRODUCTION
The great variability of chromosome number across animals naively suggests that interchromosomal rearrangements have occurred frequently over the course of metazoan evolution (1). Unexpectedly, comparative analysis of subchromosomal draft genome sequences of diverse bilaterians, cnidarians, sponges, and placozoans (2–5) reveals gene linkages that are conserved across more than half a billion years of evolution. These linkages are conserved without regard to gene order, a property referred to as conserved synteny [“synteny” meaning “same ribbon” as introduced by Renwick (6)]. Since chromosomal breaks disrupt synteny, the discovery of anciently conserved syntenies between diverse animals suggests that such breaks may be rare, at least in some animal lineages. If this is the case, then it may be possible to use conservation of synteny to infer ancestral metazoan linkage groups.
Chromosome-scale genome sequences, which have only recently become practical for nonmodel systems, are required for testing the hypothesis of conserved chromosome-scale synteny and for exploring the possibility of inferring ancestral metazoan linkage groups. With respect to bilaterians, the chromosomes of sea scallop (7) and amphioxus (8) were shown to be simply related to each other, confirming predictions based on subchromosomal sequences (3). Relicts of these ancient bilaterian syntenies can also be detected in vertebrate chromosomes after accounting for the effects of ancient vertebrate whole-genome duplications and vertebrate-specific rearrangements (2, 5, 8, 9).
The phylogenetic extent of chromosome-scale conserved synteny, however, remains poorly understood. Even among bilaterians, conservation of chromosome-scale synteny appears to be robust in some lineages but subject to disruption by interchromosomal rearrangement in others. For example, while patterns of chromosome-scale conserved synteny are easily detectable between scallop and amphioxus, they are absent or only weakly evident between these bilaterians and fruit flies (10) or the nematode Caenorhabditis elegans (7); nevertheless, flies (11–13) and rhabditid nematodes (14, 15) exhibit extensive within-group conserved synteny. A comprehensive framework for metazoan chromosome evolution is needed.
Here, we compare the genomes of diverse animals to (i) document patterns of chromosome-scale conservation; (ii) reconstruct the ancestral chromosomes of bilaterians, cnidarians, and sponges; and (iii) analyze subsequent genome evolution across these groups. To aid this effort, we report the first chromosome-scale assembly of hydra, a venerable cnidarian model for cell and developmental biology, and an improved assembly of the cephalochordate amphioxus. We find that the syntenies (i.e., chromosomal linkages without regard to gene order) of many diverse animals are remarkably similar even after half a billion years of independent evolution, often differing by a handful of discrete events superimposed on a background rate of small-scale gene transfers between chromosomes. The nature of these genome rearrangements sheds light on the mechanisms of “genome tectonics,” that is, the processes by which contemporary chromosomes were built up from ancestral elements. We identify three basic kinds of chromosomal change that have occurred repeatedly during metazoan evolution and explore chromosome evolution within spiralians, cnidarians, and deuterostomes in the context of this picture, leading to predictions that will be tested by future genome sequencing. Several metazoan syntenic units are also conserved in the unicellular relatives of animals, implying their existence before the emergence of animals.
RESULTS
Finding ancestral metazoan linkage groups
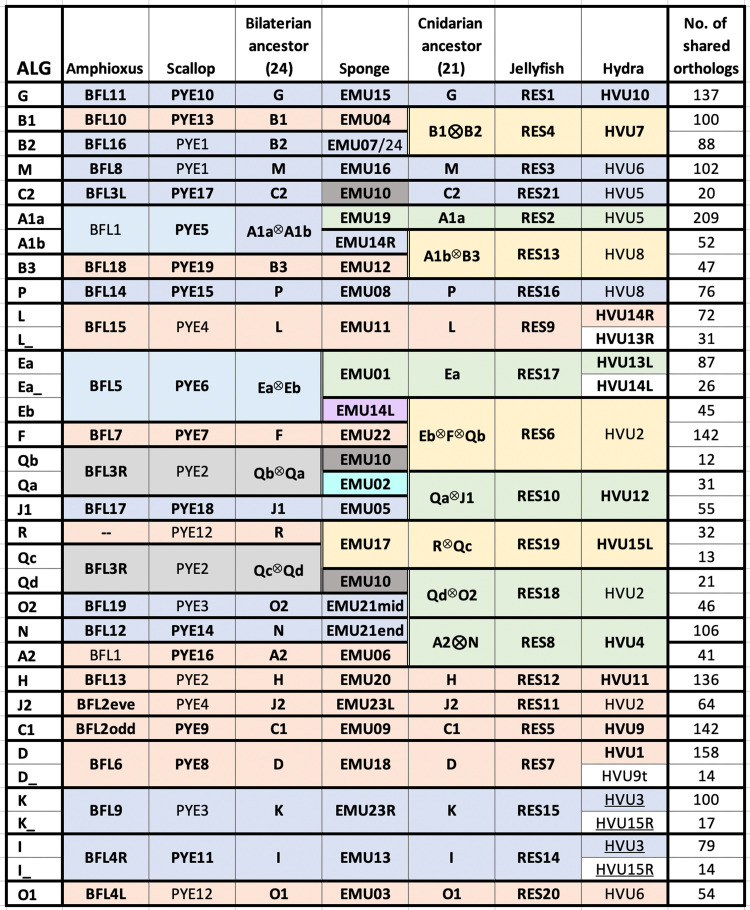
By analyzing a small but diverse set of chromosome-scale genome sequences, we find 29 groups of genes whose chromosomal linkages (without regard to gene order) are conserved across bilaterians, cnidarians, and sponges (Figs. 1 to 3 and Materials and Methods). For simplicity, we refer to the crown group that includes these three lineages as the “BCS” clade (for bilaterians, cnidarians, and sponges), which is either synonymous with metazoans (i.e., comprises all animals) or represents the sister clade to ctenophores (16, 17). To expand the diversity of chromosome-scale animal genome sequences, we report a high-quality chromosome-scale genome sequence for the model hydrozoan cnidarian Hydra vulgaris (three-letter acronym HVU) and an improved chromosomal assembly of the Florida lancelet Branchiostoma floridae (BFL) (Materials and Methods, Supplementary Materials, and fig. S1). Our analysis takes advantage of published chromosome-scale genomes of the sea scallop Patinopecten yessoensis (PYE) (7), the fire jellyfish Rhopilema esculentum (RES) (18), and the freshwater sponge Ephydatia muelleri (EMU) (19). For simplicity, we generally refer to these five focal species by their common names and use three-letter acronyms to refer to their chromosomes (Fig. 1 and Materials and Methods). Extensive conservation among this limited sampling of the BCS clade allows us to infer chromosome-scale linkages at multiple ancestral nodes. The validity and usefulness of these inferred ancestral linkage groups (ALGs) can be demonstrated and tested using other metazoan draft and chromosome-scale genomes (table S1).
Fig. 1. Anciently conserved synteny across bilaterians, sponge, and cnidarians.
(Right) Numbered horizontal bars represent the chromosomes of five species. (Left) Phylogenetic tree is shown with the root marked by a black circle. Sponge is shown in a central location to display conserved syntenies with both bilaterians (top) and cnidarians (bottom). Common names and three-letter acronyms for each species are shown (see text for details). On the right, colored vertical lines connect orthologous genes across the five species. Only connections between chromosome pairs with significantly enriched conservation of synteny are shown. Each color represents a distinct ALG, as listed in Fig. 3. Two or more colors converging on a chromosome (e.g., amphioxus BFL5 and hydra HVU6) indicate fusion-with-mixing of ancestral units. See also Fig. 4.
Chromosome-scale conserved synteny among bilaterians, cnidarians, and sponges can be demonstrated in complementary ways using pairwise dot plots and significance grids (Fig. 2; see also figs. S2 and S3). In pairwise dot plots, each dot represents the ordinal positions of a pair of orthologous genes in the two genomes so that a rectangular cluster of dots represents a group of genes that are consistently linked in both species (Materials and Methods). In most cases, these clusters span either entire chromosomes or discrete subchromosomal segments (see below). In pairwise comparisons, each chromosome typically has a significant conserved linkage to only a few chromosomes in the other species (Bonferroni-corrected P < 0.01; see Materials and Methods, Fig. 2, and fig. S4), often just one.
Fig. 2. Pairwise dot plots and chromosome-chromosome significance.
(Top) Dot plots showing the ordinal location of orthologous genes between jellyfish and amphioxus (left) and sponge (right). Genes with orthologs are numbered consecutively without regard to distance. (Bottom) Sponge versus amphioxus dot plot (left) and chromosome-chromosome significance by Fisher’s exact test (right). In dot plots, colored dots represent genes with consistent statistically significant conserved chromosomal synteny as shown in Figs. 1 and 3; genes of variable synteny are shown in gray. n.s., not significant.
The mutual consistency of these pairwise chromosome associations across bilaterians, cnidarians, and sponge is shown in Figs. 1 and 3, which organizes the network of significant chromosomal associations among amphioxus, sea scallop, sponge, jellyfish, and hydra according to 29 “ancestral (BCS) linkage groups.” By “ALG,” we mean a collection of genes whose synteny (that is, chromosomal linkages without regard to gene order) has persisted in multiple lineages since their most recent common (BCS) ancestor (Fig. 3). These findings (i) extend to chromosome scale the subchromosomal networks of conserved synteny that were originally found in the draft genome sequences of various invertebrates (2–4, 9, 20, 21), (ii) expand the phylogenetic scope of chromosomal relationships beyond bilaterians to include cnidarians (18) and sponges (19), and (iii) organize these observations in a phylogenetic framework that reveals the nature of transitions between chromosomal units in the distant past.
Fig. 3. Correspondence between ALGs and chromosomes of contemporary organisms.
Bold indicates direct association with ALG. Note that BFL3R (shaded gray) represents a single unit split to better indicate relationships with other species.
Although chromosome-scale syntenies (sometimes “macrosyntenies”) are highly conserved among these deeply divergent genomes, we emphasize that gene order within each ALG is largely scrambled among the deeply divergent BCS lineages (2, 8, 9, 21). This is evident from the uniform distribution of dots between homologous pairs of chromosomes seen in Fig. 2 (see also figs. S2 and S3). Quantitative comparisons between our five focal species show that conserved runs of consecutive orthologs (colinearity or sometimes “microsynteny”) are rare in comparisons between phyla (table S2 and Materials and Methods), consistent with near-complete loss of gene adjacency, although specific residual short-range conserved linkages with possible function have been described (3, 4, 22, 23).
Scrambling of gene order within chromosomes (i.e., loss of colinearity) over long time spans is the cumulative effect of numerous overlapping inversions and other intrachromosomal rearrangements, as notably documented for the Muller elements of Drosophila (12), which represent stable chromosome arms across drosophilids (11, 13). Since, even among Drosophila genomes, only short blocks of colinearity are found (24), it is not surprising to find near-complete loss of colinearity among deeply divergent clades of bilaterians (8), cnidarians (2), and sponges (21).
The dot plots make clear that while most genes participate in deeply conserved syntenies, some genes have moved between chromosomes (indicated in gray in Fig. 2 and figs. S2 and S3; see Materials and Methods). These genes of variable synteny, which account for between 29% (scallop-jellyfish) and 43% (amphioxus-sponge) of recognized orthologs (table S3), are distributed across the remaining chromosomes and are omitted for clarity in Fig. 1. Genes of variable synteny are enriched for specific functional categories including transmembrane receptors, transporters, and peptidases, although the biological significance of this is unclear. The dispersal of genes of variable synteny is the cumulative effect of numerous small-scale interchromosomal translocations over time (25). We estimate the rate of gene translocation to be ~1% per ~40 million years, which corresponds to a handful of genes translocated per million years. This is approximately 10-fold slower than the typical rate of gene duplication (26).
Syntenic equivalence
We observe numerous 1:1 chromosome-to-chromosome relationships across our five focal species, which enables powerful evolutionary inferences. We refer to pairwise 1:1 associations between chromosomes (without regard to gene order) as “syntenic equivalence” and represent it by the symbol “≡,” using parentheses as needed to convey phylogenetic relationships (Fig. 4A). For example, at the far right of Fig. 1 (and at the top of Fig. 3), we observe a chain of equivalences (PYE10 ≡ BFL11) ≡ EMU15 ≡ (RES1 ≡ HVU10) among chromosomes of two bilaterians, one sponge, and two cnidarians. These equivalences imply that these five chromosomes are all directly descended (with only internal rearrangement) from an ALG that must have been present in the most recent BCS common ancestor. We refer to this linkage group as ALG_G. The alternative to direct descent would be convergent gathering of these same genes onto the same chromosome in multiple lineages. The same logic implies that ALG_G existed as a syntenic unit in the most recent common ancestors of bilaterians, cnidarians, and sponges.
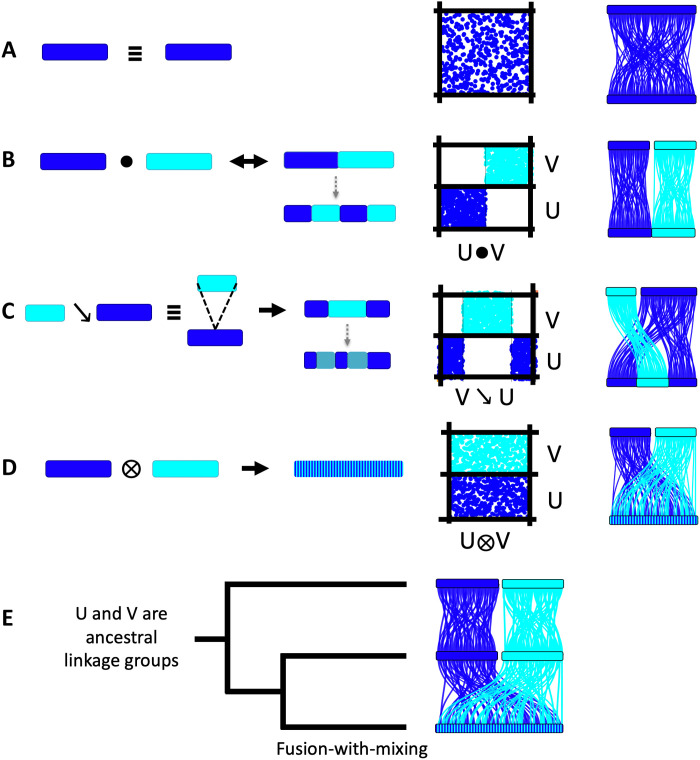
Fig. 4. Elementary algebraic operations underlying metazoan chromosome dynamics.
Left: Cartoon of chromosomal translocations. Right: The resulting dot plot. (A) Syntenic equivalence, i.e., conserved synteny without regard to colinearity, denoted symbolically by ≡. (B) Robertsonian translocation in which two chromosomes “fuse” without mixing, with stable boundary denoted symbolically by ●. Robertsonian fusions are reversible if no mixing has occurred. This diagram may also describe end-to-end fusions. (C) Centric insertion in which one chromosome is inserted into another, denoted symbolically by ↘. (D) Fusion-with-mixing in which the genes of two chromosomes are brought together (by either Robertsonian or end-to-end translocation or centric insertion) and, through a series of intrachromosomal rearrangements, become interspersed, denoted symbolically by ⊗. Both (C) and (D) are irreversible changes. Dotted gray arrows in (B) and (C) show the result of large-scale inversions that cross fusion boundaries. Note that in (B), the distal segments have different ancestry, while in (C), they have the same ancestry. (E) Phylogenetic inference using chromosomal characters. When chromosomes of two lineages are in 1:1 correspondence (e.g., the top two lineages in this rooted three-taxon tree), their shared syntenic configuration must be ancestral to the entire clade. Internal branches (bottom lineage) that show genes of these ALGs interspersed on the same chromosome are said to be fused-and-mixed.
To infer the existence of ALGs using this logic, it is sufficient to find syntenically equivalent chromosomes in just one pair of species that span the phylogenetic node of interest (e.g., Fig. 4E). For example, the chain of equivalences BFL8 ≡ RES3 ≡ EMU16 between chromosomes of one bilaterian, one cnidarian, and one sponge is sufficient to infer the conservation of ALG_M in the most recent BCS ancestor (as well as the most recent ancestors of bilaterians, cnidarians, and medusozoans) and implies independent rearrangement in scallop and hydra. Similarly, the chromosomal relationship (BFL14 ≡ PYE15) ≡ RES16 ≡ EMU08 between two bilaterians, jellyfish, and sponge implies that these chromosomes represent an ancestral BCS unit (ALG_P), with rearrangement on the hydra lineage after its divergence from jellyfish (Fig. 1; see below).
The broad patterns of chromosome-scale conservation of ancient linkage groups across our five focal species (two bilaterians, two cnidarians, and one sponge) are sufficient to (i) robustly infer the ALGs at deep phylogenetic nodes including bilaterians, cnidarians, and BCS; (ii) characterize three types of evolutionary change that combine pairs of ALGs to produce new linkages; and (iii) map these changes onto metazoan phylogeny, which allows gene linkages of as yet unsequenced genomes to be predicted. We use additional chromosome-scale and subchromosomal genome sequences to test these inferences below (notes S2 to S7). Although it may seem surprising that comparisons among such a small number of animal genomes (including only a single sponge) can lead to unambiguous reconstructions of ancestral chromosomes, this is a consequence of the widespread chromosome-scale conservation across diverse invertebrate lineages and the logic of cladistics, which allows ancestral characters to be inferred on the basis of patterns of conservation across phylogeny.
Nomenclature
We find that the 29 BCS ALGs reported in Fig. 3 are nearly all subsets of the 17 previously described “chordate” linkage groups labeled CLGA-Q (8). For consistency, we therefore denote each ALG by the same A-Q code and add suffixes as needed to designate consistently found subgroupings. Numerical suffixes are used to represent subdivisions in bilaterians, and alphabetical suffixes are used to represent subdivisions in cnidarians. The sole exception is a new linkage group (ALG_R) that was not recognized previously because it is dispersed across multiple chromosomes in chordates (i.e., amphioxus, ascidians, and vertebrates) but is recognizable in comparisons among mollusks, cnidarians, and sponge and is found as an intact chromosome in the sea urchin Lytechinus variegatus (LVA; fig. S12) (27). The combinatorial dynamics of these ancient units are explored below.
Tectonic process that built metazoan chromosomes
With the ancestral BCS units in hand, we can consider how the chromosomes of contemporary species are built from them. We observe three elementary processes of chromosomal change: (i) Robertsonian or end-end fusion/translocation, (ii) centric insertion, and (iii) fusion-with-mixing [diagrammed in Fig. 4 (B to D)]. These three processes are visible in Fig. 1 and in dot plots and are described further below. The three processes are conveniently expressed using a concise algebraic notation that describes the ALG ancestry of a chromosome (Fig. 4). We refer to these basic mechanisms and the systematic construction of contemporary chromosomes from ancestral units as genome tectonics (from “tectonic” meaning “related to building or construction”).
Centric and end-end translocations
The first type of combination is revealed by cases in which ALGs occur as discrete subchromosomal segments rather than entire chromosomes. For example, the left arm of BFL4 (denoted BFL4L) is syntenically equivalent to the ancestral BCS linkage group ALG_O1 (i.e., PYE12 ≡ RES20 ≡ EMU03) (Fig. 3). It follows from the transitivity of syntenic equivalence that BFL4L ≡ ALG_O1, which remains intact on the amphioxus lineage as a chromosome arm. Similarly, the right arm of BFL4 is BFL4R ≡ ALG_I (i.e., PYE11 ≡ RES14 ≡ EMU13). We symbolically represent the juxtaposition of two ALGs in this amphioxus chromosome by BFL4 ≡ ALG_O1●ALG_I, where “●” indicates a fusion boundary without mixing of the left and right arms (Fig. 4B). Similarly, BFL3 ≡ ALG_C2●ALG_Q. Such fusions are also found in EMU23 ≡ ALG_J2●ALG_K and EMU14 ≡ ALG_Eb●ALG_A1b. Side-by-side juxtapositions of ALGs are the expected outcome of Robertsonian translocations [with the boundaries between ALGs corresponding to ancient centromeres (28)] or end-to-end fusions [as seen, for example, in human chromosome 2 (29)].
Such exchanges of entire chromosome arms with stable boundaries at centromeres are famously observed in drosophilids, leading to the identification of six Muller elements (11, 13). Muller elements represent ancestral drosophilid linkage groups that correspond to ancestral acrocentric chromosomes. While these elements can join by centric (Robertsonian) translocation to form a biarmed chromosome, subsequent mixing of the two arms is blocked by the centromere. This process is reversible, and a biarmed chromosome can dissociate into two acrocentric chromosomes (28). Since centric fusion/fission is reversible, we cannot determine whether our ALGs were chromosomes or chromosome arms in the BCS ancestor. Last, we note that BFL4 also exhibits what appears to be a large inversion across a fusion boundary. This is analogous to the pericentric inversion after fusion of Muller elements B and C in the common Drosophila yakuba–Drosophila erecta ancestor (12, 24).
Centric insertion
A second type of combining pattern is found in EMU21, which comprises an internal segment (EMU21mid in Fig. 3) corresponding to ALG_O2 sandwiched between two distal segments (EMU21end) that together correspond to ALG_N. This is the signature of a centric insertion (Fig. 4C), in which one chromosome is inserted into the centromeric region of another, a type of chromosomal translocation that has been described in several plant genomes (28, 30–32) but that, to our knowledge, has not been previously described in animals. We represent this centric insertion symbolically as EMU21 ≡ ALG_O2 ↘ ALG_N. An alternative scenario, in which ALG_O2 and ALG_N are first juxtaposed side by side and the ALG_N segment is then split by an inversion containing the entire ALG_O2 segment, is possible but is less likely because the presence of ALG_N genes at both ends of EMU21 would require that the inversion breakpoint be very close to a telomere.
Similarly, BFL2 in amphioxus shows an alternating pattern of ancient synteny, with odd-numbered segments (BFL2odd in Fig. 3) having ALG_C1 ancestry and the even-numbered elements (BFL2eve) having ALG_J2 ancestry. Notably, the odd segments include both telomere-adjacent regions. The most parsimonious explanation of this pattern is that BFL2 arose by a centric insertion of ALG_J2 ≡ RES11 ≡ EMU23L into ALG_C1 ≡ PYE9 ≡ (RES5 ≡ HVU9) ≡ EMU09, followed by an inversion that spanned the boundary of the insertion (Fig. 4B). The alternative hypothesis of Robertsonian fusion followed by two inversions (with one breakpoint required to be precisely positioned close to the telomere) invokes more tectonic events and is therefore less parsimonious.
Fusion-with-mixing
Last, our analysis reveals a third previously undescribed process in karyotype evolution in which chromosomes corresponding to two different ALGs fuse into a single chromosome and the genes from these ALGs become interspersed along the fusion chromosome by subsequent intrachromosomal rearrangement. We refer to this process as “fusion-with-mixing” and represent it symbolically by “⊗” (Fig. 4D). For example, HVU8 is a fusion-with-mixing of two jellyfish-like chromosomes RES13⊗RES16, and BFL1 is a fusion-with-mixing of two scallop-like chromosomes PYE5⊗PYE16 (the direction or polarity of these changes in time is determined by our knowledge of the ancestral state, as shown in Fig. 4E). Fusion-with-mixing is (a) “commutative” in the sense that the ancestral units that become combined can be written in either order and (b) irreversible, in the sense that, once two ALGs have fused and mixed, they will not spontaneously unmix. This is in contrast to Robertsonian fusion (which occurs without mixing), which can be reversed by pericentromeric double-strand breaks (28).
Mixing may occur over an extended period of time, presumably by the same intrachromosomal processes that scramble gene order within Muller elements as documented in drosophilids (12) and as described above for syntenically equivalent chromosomes between deeply divergent groups. Since proper segregation of a chromosome requires exactly one functional centromere, the initial “fusion” may occur by an end-to-end process (29) with centromeric loss/nonfunctionalization (33). Since centromeres typically serve as barriers to mixing due to the meiotic deficits that accompany pericentric inversion (11), mixing after Robertsonian fusion probably requires the relocation of a centromere or the emergence of a neocentromere.
To estimate the degree of mixing after fusion, we computed the length of uninterrupted runs of genes from the ALG that contribute to fused-and-mixed chromosomes (Materials and Methods). With complete mixing, these runs are expected to be short and geometrically distributed. Simulations show that random inversions effectively intersperse genes from initially separate ALGs and rapidly generate complete mixing (Materials and Methods and fig. S5). Scallop and jellyfish chromosomes derived by fusion-with-mixing show the expected geometric distributions of run lengths. In contrast, chromosomes that arise from end-to-end fusion or centric insertion (such as BFL2, BFL3, and BFL4) have long uninterrupted runs of adjacent genes from the same ALG, indicating that extensive mixing has not occurred (BFL1, which is a fusion-with-mixing of ALG_A1 and ALG_A2, shows longer-than-expected residual adjacent blocks of ancestry, consistent with a more recent fusion-with-mixing in the amphioxus lineage). Similar to the mixing of ink and water, the irreversibility of fusion-with-mixing can be quantified by entropy [in this case, Shannon entropy (34)], and spontaneous unmixing as a consequence of intrachromosomal rearrangement cannot occur over any practical time frame (Materials and Methods). The rarity of fusion-with-mixing coupled with its irreversibility means that fusion-with-mixing can be used as a strong character for phylogenetic inference as discussed below.
Early evolution of bilaterian, cnidarian, and sponge karyotypes
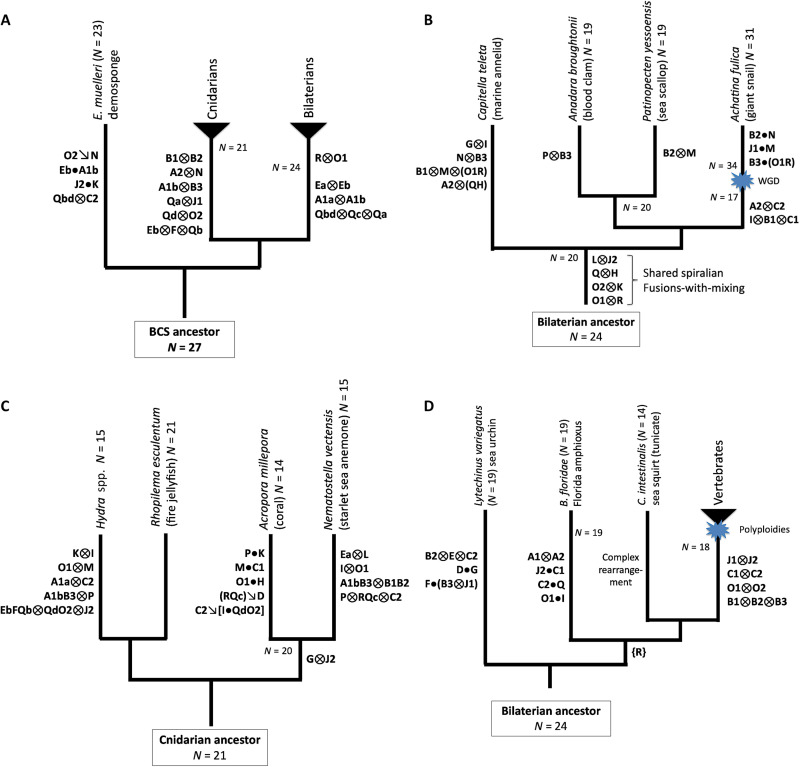
With the ALGs in hand, we can use cladistic logic (e.g., Fig. 4E) and the three elementary combining processes to establish the ancestral chromosome organization of bilaterians, cnidarians, and sponges and to describe the early chromosome evolution leading to these three ancient metazoan lineages (Fig. 5A, fig. S6, and note S2). Four ALGs (B1, B2, A2, and N) are each represented in both sponges and bilaterians by syntenically equivalent chromosomes (or discrete segments) and must therefore correspond to the ancestral BCS state. In medusozoans, these ALGs participate in fusion-and-mixing rearrangements (RES4 ≡ HVU7 ≡ ALG_B1⊗ALG_B2 and RES8 ≡ HVU4 ≡ ALG_A2⊗ALG_N), which represent derived states. We show below that these same mixing events are found in anthozoan cnidarians, which implies that they occurred on the cnidarian stem. A somewhat more complicated situation involves the three ALGs A1a, A1b, and B3, which are (i) distinct in sponge, (ii) fused as ALG_A1a⊗ALG_A1b ≡ PYE5 in bilaterians, with ALG_B3 ≡ BFL18 ≡ PYE19 remaining distinct, and (iii) fused as ALG_A1b⊗ALG_B3 ≡ RES13 in medusozoans, with ALG_A1a ≡ RES2 remaining distinct (the fusions-with-mixing shared by jellyfish and hydra are also found in anthozoans and are therefore pan-cnidarian; see below). Here, again, we can infer that the sponge E. muelleri retains the primitive state, and bilaterian and cnidarian chromosomes arose by distinct fusion-with-mixing processes. In this manner, we can polarize most early karyotype changes even without an outgroup and define 24 ancestral bilaterian linkage groups (BLGs) and 21 ancestral cnidarian synteny groups (note S2). The most complex cases involve the ancestral state of the chromosomes derived from the four ALGs Qa to Qd and R, which are the smallest and apparently most labile units (fig. S6). The only ambiguity of the ancestral BCS state involves Qc and R (note S2).
Fig. 5. Chromosome evolution in metazoan lineages.
Phylogenetic distribution of chromosome changes in (A) early bilaterian-cnidarian-sponge divergence, (B) spiralians, (C) cnidarians, and (D) chordates. All ancestral states and changes are inferred from genome comparisons and reconstructions as described in the text. ALGs not shown on branches retain ancestral state. Algebraic symbols are defined in Fig. 4 and in the text. Blue starbursts represent whole-genome duplications. Note that prior rearrangements in the indicated ancestor are denoted in shorthand, e.g., A1bB3 represents the cnidarian ancestral state A1b⊗B3 and B1B2 represents B1⊗B2.
Fusion-with-mixing and the evolution of spiralian karyotypes
The importance of fusion-with-mixing in karyotype evolution and the utility of our chromosomal algebra for describing and timing specific tectonic events are demonstrated by comparative analysis of spiralian karyotypes (Fig. 5B and note S3). As evident from Figs. 1 and 3, fusion-with-mixing occurred five times in the scallop lineage: PYE1 ≡ ALG_M⊗ALG_B2, PYE2 ≡ ALG_H⊗ALG_Q, PYE3 ≡ ALG_K⊗ALG_O2, PYE4 ≡ ALG_J2⊗ALG_L, and PYE12 ≡ ALG_R⊗ALG_O1. By comparing the chromosomes of scallop to the chromosomes of two other mollusks [the blood clam Anadara broughtonii (ABR) (35) and giant snail Achatina fulica (AFU) (36)], and using amphioxus and the sea urchin L. variegatus (LVA) (27) as outgroups, we see that four of these five fusions-with-mixing (ALG_H⊗ALG_Q, ALG_K⊗ALG_O2, ALG_J2⊗ALG_L, and ALG_R⊗ALG_O1) are shared by all three mollusks, which implies that they occurred before the bivalve-gastropod split (fig. S7 and note S3). The alternative is that these fusions-with-mixing occurred convergently in bivalves and gastropods. It follows from the irreversibility of fusion-with-mixing that all descendants of the most recent bivalve-gastropod ancestor must share these four fusions-with-mixing. This is a prediction of our approach that can be tested by future genome sequencing efforts. For example, analysis of the recently published subchromosomal Nautilus pompilius scaffolds (37) shows that, as anticipated, these four fusions-with-mixing are likely present in a cephalopod (fig. S7).
Comparisons among mollusks also show that equal chromosome numbers do not necessarily imply 1:1 chromosomal homology. In particular, although the blood clam and sea scallop both have haploid chromosome number N = 19, they derive from their common N = 20 ancestor by different lineage-specific fusion-with-mixing events. Last, although the giant snail is a paleotetraploid (38), its karyotype evolution can also be described by our algebra, further demonstrating its utility (note S3). In the paleotetraploid giant snail lineage, the ancestral N = 20 bivalve-gastropod number is first (i) reduced to 17 by three fusion-with-mixing events, then (ii) doubled to 34 by a genome duplication (38), and lastly (iii) reduced to 31 by three Robertsonian fusions that occurred after the genome doubling (Fig. 5B).
Although no chromosome-scale annelid genomes are available to date, we can nevertheless use the draft genome of the marine annelid Capitella teleta (CTE) (3) and our framework for metazoan chromosome evolution to make predictions about annelid and other spiralian genomes. Analysis of the subchromosomal sequences of C. teleta suggests that multiple bilaterian ALGs [bilaterian linkage groups (BLGs)] are present in unmixed form in this genome, which implies that these syntenic groups will be found as chromosomes or chromosomal arms/blocks (fig. S8 and note S3). Multiple scaffolds are enriched for genes from pairs of ALGs, indicating fusion-with-mixing. Since the same ALG pairs are fused-and-mixed in bivalve and gastropod mollusks and since fusions-with-mixings are irreversible, we can make another testable prediction: These fusions-with-mixings must be shared by all spiralians (which descend from the most recent annelid-molluscan common ancestor). Additional possibly lineage-specific fusions-with-mixing are also detected in C. teleta (Fig. 5B and note S3).
Evolution of cnidarian karyotypes
We find that the fire jellyfish preserves the N = 21 ancestral cnidarian chromosome number (Fig. 5C). While N ~ 14 to 15 karyotypes are common among cnidarians (e.g., Nematostella vectensis, Acropora millepora, and H. vulgaris), we find that this chromosome number has been independently derived in multiple lineages (Fig. 5C). Satisfyingly, the chromosomes of the coral A. millepora (the only available chromosome-scale anthozoan genome to date) (39) can be straightforwardly represented in terms of the ancestral RES-like cnidarian units using our chromosome algebra, although they were not included in the analysis that defined the ALGs (fig. S9 and note S4). We also updated the analyses of Putnam et al. (2) and Srivastava et al. (21) by grouping their subchromosomal scaffolds of the starlet sea anemone N. vectensis (NVE) into 15 clusters based on conserved syntenies with jellyfish and coral (fig. S9 and note S4). These 15 clusters are presumed to be in one-to-one correspondence with the N = 15 N. vectensis chromosomes, consistent with (40) and validating a long series of predictions based on conserved synteny (2, 3, 8, 21). Comparing the 14 A. millepora chromosomes with the 15 N. vectensis clusters, however, we infer that the two anthozoans reached similar chromosome numbers by parallel reductions from their N = 20 anthozoan ancestor. Both coral and sea anemone have a chromosome that combines ALG_G and ALG_J2, as a likely Robertsonian fusion in A. millepora and as a fusion-with-mixing in N. vectensis. This suggests that a Robertsonian fusion on the anthozoan stem broke down by mixing in the N. vectensis lineage, although we cannot rule out convergent fusions.
The N = 15 chromosomes of H. vulgaris are derived from those of the N = 21 cnidarian ancestor by a different set of six fusion-with-mixing rearrangements from that found in anthozoans (Fig. 3 and fig. S3). These fusions-with-mixing are shared with the distantly related green hydra Hydra viridissima based on an analysis of the subchromosomal scaffolds of this species (fig. S9) (41), consistent with the stability of the N = 15 karyotype across the genus Hydra despite variation in genome size (42). In contrast with other focal species in our analysis, however, H. vulgaris chromosomes also display balanced translocations that break ALGs (Fig. 3). For example, HVU9 comprises ALG_C1 ≡ RES5 joined to a small portion of ALG_D ≡ RES7. Since ALG_D genes are only found at the tip of HVU9, it is likely that HVU9 was formed by the translocation of a short (presumably distal) portion of an ALG_D-like chromosome onto an ALG_C1-like ancestor. HVU13 and HVU14 arose by reciprocal translocation of a pair of ancestral chromosomes resembling ALG_L ≡ RES9 and ALG_Ea ≡ RES17, possibly through arm exchange. Sharp breakpoint boundaries suggest recent rearrangements. Somewhat similarly, hydra chromosomes HVU3 and HVU15R display the signature of a reciprocal translocation of ALG_I ≡ RES14 and ALG_K ≡ RES15. Unlike the ALG_L/ALG_Ea case, however, the reciprocal translocation of ALG_I/ALG_K was followed by mixing. We note that the sequenced 105 strain of H. vulgaris has been maintained asexually in the laboratory for nearly 50 years, and these (and other smaller translocations) may be the result of relaxed constraint on or, possibly, selection for chromosome rearrangements under these conditions.
Nematodes, flies, and ecdysozoans
Although the small chromosome numbers of rhabditid nematodes and drosophilids are evidently highly reduced relative to the 24 ancestral BLGs, we can nevertheless detect relicts of these ancient linkages. The N = 6 chromosomes of C. elegans (CEL) can be simply expressed in terms of the ancestral BLGs. In particular, CEL_V corresponds directly with BLG_A1 (≡ ALG_A1a⊗ALG_A1b), and the other five C. elegans chromosomes are fusions-with-mixing of between two and six BLGs (fig. S10 and note S5). These correspondences expand upon and mechanistically explain the conserved synteny observed by Wang et al. (7) between C. elegans and the computationally inferred 17 ALGs of chordates (5). Within nematodes, Tandonnet et al. (15) recently showed that chromosomes of diverse rhabditid species can be described in terms of seven “Nigon elements,” the rhabditid analogs of the six Muller elements in drosophilids. This is wholly consistent with our phylogenetically broader scheme since the rhabditid Nigon elements correspond to fusions-with-mixing of the ancient bilaterian linkage units (note S5). Although our focus is on conserved macrosynteny, we note that chromosome fusions in the rhabditid lineage were also accompanied by a higher rate of small-scale interchromosome translocations than seen in other bilaterians, producing a higher number of genes of variable synteny (table S3).
Searches for large-scale conserved synteny between Drosophila and other phyla have been unsuccessful (7, 10). Within insects, however, macrosynteny is conserved, as previously noted between fruit flies and mosquitos (43) and shown in fig. S10 between fruit fly and the flour beetle Tribolium castaneum (TCA) (44). We find several weak but significant macrosyntenic associations between the drosophilid Muller elements and BLGs (fig. S11 and note S5). In contrast, scaffolds of the centipede Strigamia maritima (SMA) (45)are consistent with the preservation of intact BLGs in the chilopod lineage. These findings (and the formation of rhabditid nematode genomes by fusion-with-mixing of BLGs) imply a scenario in which early ecdysozoans preserved many ancestral bilaterian syntenies, with large-scale rearrangements occurring on the insect/fly lineage. These predictions will be tested by future ecdysozoan genome sequencing.
Deuterostomes
With the reconstructed ancestral bilaterian chromosomes in hand, we can refine the reconstruction of the protovertebrate genome before early genome-wide duplications (8) and, by comparison with the chromosome-scale sea urchin genome (LVA) (27), explore early deuterostome chromosome evolution. We find that (i) fusions-with-mixing produced 18 syntenic units (chromosomes and/or chromosome arms) on the protovertebrate lineage and (ii) the syntenies of the most recent chordate ancestor resembled those of the bilaterian ancestor (Fig. 5D and note S6). The discrepancy between the 18 syntenic units reported here and the 17 “chordate linkage groups” (CLGs) of (8) is accounted for by our new finding that the amphioxus chromosome BFL1 is a fused-and-mixed combination of two ancestral bilaterian units, BLG_A1 and BLG_A2. These BLGs are separate in sea urchin (fig. S12), scallop, sponge, and cnidarians as well as in lamprey (46) and tunicate (47) chromosomes. The BLG_A1⊗BLG_A2 fusion-with-mixing must therefore have occurred on the amphioxus lineage, not the ancestral chordate lineage, as assumed in (8). Residual short blocks of BLG_A1 and BLG_A2 ancestry are consistent with relatively recent fusion-with-mixing on the amphioxus lineage.
Several other fusions-with-mixing are shared by vertebrates but not amphioxus and other bilaterians and therefore occurred on the vertebrate stem. For example, ALG_B1, B2, and B3 are each preserved as separate linkage units in sponge and amphioxus (and, therefore, the most recent chordate ancestor) but were combined on the vertebrate lineage (since they are found in fused-and-mixed form in jawed vertebrate genomes as CLG_B). Similarly, the ALG pairs ALG_C1/C2, J1/J2, and O1/O2 are distinct units in bilaterians but always occur in fused-and-mixed form in jawed vertebrates (8). From the broader phylogenetic perspective described here, we note that the “CLGs” defined by Simakov et al. (8) should properly be referred to as “protovertebrate linkage groups” since they include ALG fusions on the preduplication vertebrate stem that are not found in other chordates. While the chromosomes of Ciona intestinalis (type A) (47) retain some ancestral bilaterian syntenies, these are highly disrupted, with several ALGs split across two chromosomes (fig. S12), indicating the occurrence of fissions and/or partial translocations in the more divergent tunicates (48), similar to the disruptions seen in H. vulgaris. Last, ALG_R, which we identified as an ancestral unit that is (i) fused-and-mixed with ALG_O1 in scallop and (ii) fused-and-mixed with ALG_Qc in sponge and cnidarians, is present as an intact chromosome in sea urchin (LVA6; see fig. S12), confirming ALG_R as an ancestral bilaterian and deuterostome chromosome (or arm). The genes of ALG_R are dispersed across several chromosomes in chordates (amphioxus, C. intestinalis, and vertebrates), suggesting a possible fourth mode of chromosome evolution.
Placozoans, ctenophores, and sponges
Although the genome of the placozoan Trichoplax adhaerens (TAD) is not yet assembled to chromosome scale, we find that its subchromosomal scaffolds (9, 49) generally preserve BCS ALGs, supporting earlier findings of conserved synteny between Trichoplax and human (3, 9). We can also use the patterns of ALG mixture as phylogenetic characters to test hypotheses about the nature of placozoans (9, 49–51). These arguments critically rely on (i) the irreversibility of fusion-with-mixing, which implies that all descendants of an ancestrally fused-and-mixed chromosome must also show that same intermixing of genes from two ALGs, and (ii) the rarity of these events, which suggests that convergent fusions-with-mixing in unrelated lineages are unlikely. These principles imply that the placozoans cannot be crown group cnidarians or bilaterians since Trichoplax lacks both the ALG_A2⊗ALG_N and ALG_A1b⊗ALG_B3 mixings shared by all cnidarians and the ALG_A1a⊗ALG_A1b mixing shared by all bilaterians (fig. S13). However, Trichoplax does exhibit the ALG_Eb⊗ALG_F fusion-with-mixing shared by all cnidarians (note S7). Thus, ALG_Eb⊗ALG_F is a synapomorphy (shared derived character) that unites placozoans and cnidarians as sister groups, consistent with some gene-based phylogenies (51).
Among sponges, we find that the Great Barrier Reef demosponge Amphimedon queenslandica (AQU) (21) shows extensive conserved synteny with BCS ALGs and the freshwater demosponge E. muelleri (19), consistent with prior findings of deeply conserved synteny between subchromosomal assemblies of A. queenslandica and bilaterians (21), now extended to chromosome scale. We note in passing that although the sponge E. muelleri has n = 23 chromosomes, the assembly of (19) has 24 major scaffolds. Conservation of synteny shows that the 24th largest scaffold has the same pattern of synteny as EMU07, corresponding to ALG_B2 (Fig. 3). This fact suggests that the two scaffolds are linked in the sponge genome. The Hi-C contact map reported in (19) supports this linkage, affirming the power of our approach. Unfortunately, the currently available subchromosomal assemblies of ctenophores (17, 52) are too fragmented to apply this logic to infer the relationships among ctenophores, sponges, and other animals; such analysis awaits chromosome-scale genome assemblies.
Conserved synteny between animals and their unicellular relatives
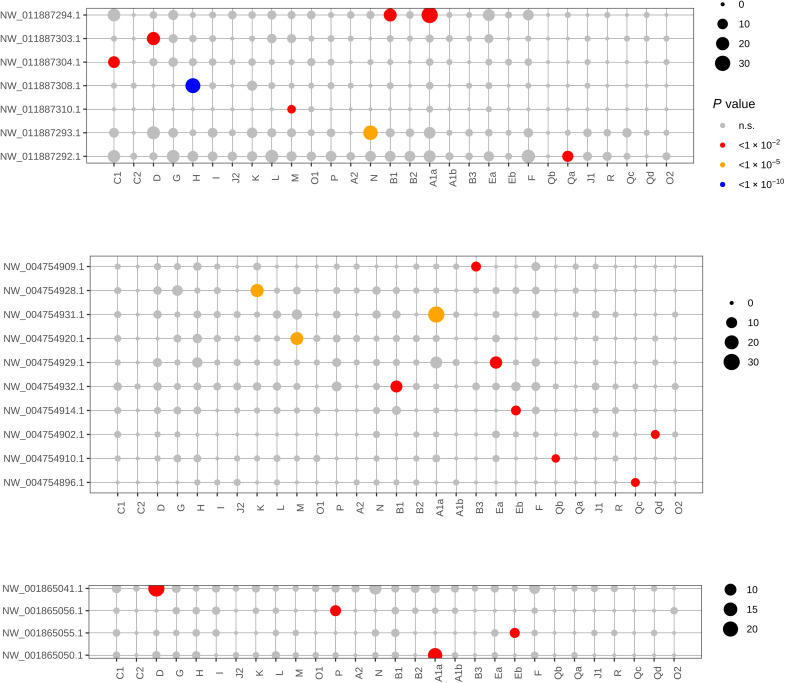
We also find relicts of ancient linkages shared between animals and their unicellular relatives. While the available draft genomes of choanoflagellates Monosiga brevicollis (53) and Salpingoeca rosetta (54) and the filasterean Capsaspora owczarzaki (55) are not yet assembled into chromosomes, we tested their longest genomic scaffolds for significant conserved synteny with ALGs (Materials and Methods). We find that 16 ALGs show conserved synteny with one or more choanoflagellate and/or filasterean genome scaffolds, revealing gene linkages that have persisted across more than 800 million years of independent evolution (Fig. 6). These extraordinarily ancient conserved linkages include one ALG (ALG_A1a) that is at least partially preserved in all three unicellular genomes and seven whose preservation is detected in two of the three genomes.
Fig. 6. Significant associations between BCS ALG and scaffolds of three unicellular holozoans imply deep ancestry of BCS linkage groups.
(Top) Filasterean amoeba C. owczarzaki. (Middle) Colony-forming choanoflagellate S. rosetta. (Bottom) Solitary choanoflagellate M. brevicollis. For each species, P values are Bonferroni-corrected for the number of scaffolds tested against the BCS ALGs for conserved synteny (Materials and Methods). Circle size corresponds to the number of shared genes.
We emphasize that the ancient conserved syntenies between animals and their unicellular relatives shown in Fig. 6 are underestimates since the fragmentation of the current genome assemblies of unicellular relatives of animals reduces the statistical power to detect conserved synteny by (i) reducing the number of genes involved in each scaffold-ALG test and (ii) increasing the multiple testing burden due to the large number of scaffolds. Furthermore, these scaffold-based tests can only confirm partial preservation of ALGs in unicellular genomes; it remains possible that some ALGs are (possibly ancestrally) “split” in these lineages. Chromosome-scale assemblies for these unicellular organisms will be necessary for more complete tests.
DISCUSSION
As the physical carriers of genetic information, chromosomes are heritable and mutable entities that evolve over time. Here, we show that the chromosomes of diverse bilaterians, cnidarians, and sponges are often in simple correspondence, implying preservation of groups of anciently linked genes in diverse lineages (Fig. 1). This finding validates and extends earlier predictions based on subchromosomal draft genomes (2–5, 9, 20, 21, 45). Notably, we find that chromosome-scale gene linkages are generally conserved without conservation of gene order, that is, conserved synteny in its general sense as introduced by Renwick (6). As described for drosophilids (12, 13, 24), scrambling of gene order is due to the cumulative effect of intrachromosomal rearrangements. The loss of gene order among divergent lineages suggests that any selection favoring the preservation of gene order must be weak over long time scales.
The observed patterns of conserved synteny imply that the chromosome sets of diverse species can be interpreted as simple combinations of 29 ALGs. These ALGs can be inferred by cladistic principles and the known phylogeny of bilaterians, cnidarians, and sponges (Fig. 3). As an extreme example of deep conservation of synteny, we find that the n = 21 chromosomes of fire jellyfish represent the ancestral state of cnidarians, although the ordering of genes along these ancestral chromosomes cannot be determined. On top of the general patterns of chromosome-scale conserved linkage, we find that a handful of genes change chromosomes every million years, presumably through small-scale translocations (25).
Across animal phyla, we find that, typically, only a handful of chromosome-scale fusions have occurred on each lineage since the Cambrian era. It is this slow rate of change that allows us to infer the ALGs from only a small number of diverse genomes. We find that there are three types of fusions: Robertsonian or end-to-end translocations, insertional centric fusion, and the newly described process of fusion-with-mixing (Fig. 4). Fusion-with-mixing presumably occurs by fusion [or, more properly, whole-chromosome translocation (28)] followed by numerous subsequent intrachromosomal rearrangements that eventually obscure fusion boundaries. Fusion-with-mixing is a rare and irreversible process, which makes it a useful phylogenetic character (56). Fusion-with-mixing events therefore (i) generate testable predictions of our model of chromosome evolution (since shared fusion-with-mixing between two genomes requires this character to be shared across an entire clade) and (ii) provide shared-derived genomic characters that may be useful in phylogenetic analysis.
Why have chromosome-scale syntenic units been stable over such long periods of time? There are two broad, nonexclusive types of selective constraints on chromosome evolution. First, chromosome rearrangements may be constrained by gene regulation since structural variations that disrupt cis-regulatory interactions will generally be selected against. For example, gene regulation in vertebrates has been shown to involve long-range chromatin interactions (22, 57), and overlapping networks of such intrachromosomal interactions could, in theory, stabilize arrays of syntenic linkages while still allowing for local changes in gene order. A recent study in fruit flies with highly rearranged chromosomes, however, suggests lack of coupling between genome topology and gene expression (58). The widespread lack of gene order conservation described above among bilaterians, cnidarians, and sponges suggests that long-range cis-regulatory constraints on chromosome-scale evolution, if they exist, must be subtle. This is consistent with our inability to detect shared molecular function of genes in each ALG.
A second general constraint on chromosome evolution arises from the requirement that new chromosome-scale mutations must successfully navigate meiosis as heterozygotes on their way to fixation in a sexual population (59). In general, even balanced chromosomal rearrangements can be deleterious to the fertility of heterozygotes because of the disruption of meiotic pairing (11) and the production of unbalanced gametes with altered gene dosage (25, 59). While these effects are expected to reduce the fertility of fusion heterozygotes, reversible centric (Robertsonian) fusions and fissions can evidently evade these constraints, and the balance between them may be controlled by meiotic drive (60, 61).
The effects of both regulatory and meiotic constraints are enhanced in large panmictic populations, which are generally resistant to the spread of “underdominant” mutations (i.e., mutations that are deleterious as heterozygotes) and deleterious mutations in general (59). The spread of such variants is thought to require special conditions such as subdivision of a population into small demes, which allows such variations to become established by genetic drift and dispersal through colonization (59, 62), or variation in nonrandom segregation of centromeres in female meiosis (61). These conditions may account for the prevalence of chromosomal rearrangements in mammals (63). Broadcast spawning invertebrates, however, typically form large panmictic populations. Their large population sizes may also stabilize their genomes against chromosomal change, contributing to the remarkable persistence of ancient metazoan linkage groups through evolution.
Although slow chromosome evolution is widespread among invertebrates (Fig. 5 and notes S2 to S7), it is not absolute. Exceptions to the general metazoan rule that ALGs are combined but not broken include disruptive change on the lineages leading from the chordate ancestor to ascidians [known for their rapid genome evolution (48)], from the protostome ancestor to insects, and possibly from the molluscan ancestor to coleoid cephalopods [which show stable but much larger chromosome numbers than other invertebrates (64)]. While, in these cases, disruptive chromosome rearrangements that break ALGs have occurred, they appear to be restricted to a specific time interval followed by a return to slow evolution (as seen, for example, in flies). These intervals of rapid chromosomal change could correspond to bottlenecks when population size drops, and unusual chromosome rearrangements that break ALGs can more readily become fixed. Substantial morphological evolution can occur (e.g., between bilaterians, cnidarians, and sponges and within these groups) with only relatively conservative chromosomal change following the mechanisms of Fig. 4.
Unexpectedly, we also find evidence for at least partial conservation of the ancestral metazoan syntenic units in unicellular relatives of animals, testifying to their deep antiquity. These ancient linkage units provide a framework for understanding gene and genome evolution in animals. For example, in metazoans, ALG_A1 carries the most diverse collection of homeobox genes, with members of ANTP, PRD, PROS, and HNF homeobox gene classes as well as the more ancient TALE class that is also found in fungi and plants (65). Although these homeobox genes (except TALE) are not found in unicellular eukaryotes, ALG_A1 is conserved with unicellular relatives of animals and may represent the chromosomal context within which metazoan homeoboxes arose and diversified. Our observation is consistent with the proposal of a homeobox-rich protovertebrate linkage group (66) and discussions of ancient homeobox giga-clusters (67, 68).
MATERIALS AND METHODS
Chromosomal assemblies of H. vulgaris and amphioxus
We generated a de novo chromosomal assembly for H. vulgaris (HVU) by combining 29-fold coverage in PacBio circular consensus (“HiFi”) shotgun sequencing with in vivo (69) chromatin conformational capture datasets (note S1). To avoid challenges arising from heterozygosity, PacBio sequencing was performed on genomic DNA from an F1 hybrid of the Japanese H. vulgaris (HVU) strain 105 [the substrate for previous subchromosomal assemblies of Hydra (20)] crossed with the North American AEP strain. These two H. vulgaris strains differ by approximately 1% sequence differences so that a haplotype-resolved assembly could be generated with Flye (70). Contigs derived from the strain 105 parent were identified by mapping in vitro chromatin conformation capture (“Hi-C”) Illumina data (Dovetail Genomics, Scotts Valley, CA) from strain 105 to the haplotype-resolved assembly. Hi-C scaffolding was performed using Juicebox (71) with manual curation as described in note S1. Centromeric tandem repeat loci were identified, and chromosomes were reoriented p-to-q. The final Hi-C contact map is shown in fig. S1. Chromatin contact data suggest that the genome of this asexual strain of H. vulgaris, which has been propagated asexually by budding for almost 50 years, is heterozygous for a reciprocal translocation. Since one of the two alternative conformations shows chromosome-scale conserved synteny with jellyfish, amphioxus, and sea scallop, we report this most parsimonious structure. Our overall results are not sensitive to this choice since the alternative form would simply represent a recent rearrangement in the Hydra lineage (note S1). For amphioxus, we updated the existing chromosome-scale assembly (8) by manual curation with Juicebox (71). This process incorporated 6 million base pairs of a previously unplaced sequence into chromosomes and corrected 11 inversion errors. BFL chromosomes were numbered according to Simakov et al. 2020 (8). Annotations for both hydra and amphioxus were mapped forward from previous assemblies by nucleotide BLAST of transcripts (72).
Other genomic datasets
To define the ancestral BCS linkage groups, we also used several published genome datasets, including the yesso sea scallop P. yessoensis (PYE) (7), fire jellyfish R. esculentum (RES) (18), and freshwater sponge E. muelleri (EMU) (19). Table S1 lists the source of all genome data used in this study. For uniformity, we labeled each chromosome by the species’ three-letter species acronym and the scaffold or chromosome number assigned by the original authors. For E. muelleri, as noted by the original authors, the assembly recovers 22 of the n = 23 chromosomes as single sequences, with the remaining chromosome “represented by two sequences likely split at the centromere.” On the basis of our synteny analyses, the scaffold pair EMU07 and EMU24 together corresponds to ALG_B2 (Fig. 3). The Hi-C data of (19) support the physical association (although this is not noted there). We conclude that these scaffolds together represent the split chromosome.
Identification of ALGs
We identified 29 ancient conserved linkage groups (ALGs) across BCSs by two complementary methods for assessing chromosome homology. Both approaches produced the same results as shown in Fig. 3.
In the first approach, we inferred the anchor genes on the basis of multiway mutual best hit (MBH) clusters of orthologs. To identify homology between chromosomes, we used orthology information from “three-way” MBH (synonymous with reciprocal blast hit) relationships between scallop, amphioxus, and jellyfish, requiring consistent MBH pairs across all three species. To define the best hits, we used the hit with the highest score using BLASTP version 2.11.0+ (73). This resulted in 6766 MBH gene families. To add more species to this core set, MBHs were first inferred in pairwise comparisons between each species and the three focal species and then merged into “multiway” MBHs by requiring consistent 1-1 matches in MBH gene pairs among scallop, amphioxus, and jellyfish from the same MBH core family to ensure stringent orthology assignment. ALGs were then defined by chromosomal combinations (chromosomal location of the orthologous genes from a given MBH family) that had the highest number of supporting MBH families. Using this approach, 2361 MBH families could be assigned to one of the ALGs (data S1), with the remaining MBHs comprising very rare chromosomal combinations. Groupings of chromosomes into ALGs were insensitive to the method of grouping genes into orthologous clusters. The 2361 MBH families comprise the anchor gene families. The number of such gene families contributing to each group is shown in the last column of Fig. 3.
In the second complementary approach, we considered pairwise ortholog dot plots (Fig. 2 and figs. S2 and S3) among the five core species in Fig. 3. The significance of chromosome-chromosome associations (Fig. 2 and fig. S4) between species was assessed with Fisher’s exact test against a null model of random gene permutation; for each species pair, we applied a Bonferroni correction for the number of chromosome-chromosome tests. Pairwise associations between the five focal species (amphioxus, scallop, jellyfish, hydra, and sponge) were nearly all commutative: If chromosome A from species 1 and chromosome B from species 2 were associated and chromosome B from species 2 and chromosome C from species 3 were associated, then, generally, chromosome A from species 1 and chromosome C from species 3 were also associated. The resulting network of chromosome-chromosome associations between pairs of species recapitulates Fig. 3, obtained using orthologous gene sets across all five species. ALG_R is an exception, as it was not found as a significant association with any amphioxus chromosomes. We note that ALG_R and ALG_O1 occur in fused-and-mixed form in scallop, and in developing Fig. 5, we note the uncertain phylogenetic position of this fusion-with-mixing due to its uncertainty in amphioxus. Although it was defined without reference to sea urchin, ALG_R is in 1:1 correspondence with chromosome LVA6 of the green sea urchin L. variegatus (fig. S12 and note S6).
Conserved syntenic relationships are shown in Fig. 3 as well as in the ribbon diagram of Fig. 1. In Fig. 1, vertical lines connect orthologous genes between chromosomes of adjacent species (74) if their chromosomes show significant conserved synteny, i.e., those that appear in the same row of Fig. 3. Note that genes of variable synteny are not shown in Fig. 1. The 29 ALGs were 10-color-labeled using R’s RColorBrewer “Dark2” palette.
We note that in some cases, an ALG corresponds uniquely to single chromosomes in each species (e.g., ALG_G; top row of Fig. 3). In other cases, two or more ALGs correspond to distinct chromosomes in some species but the same chromosome in others, which we indicate in Fig. 3 by partially merged rows. For example, ALG_B1 and ALG_B2 (of Fig. 3) are distinct in amphioxus, scallop, and sponge but are both associated with RES4 and HVU7. Since ALG_B1 and ALG_B2 genes are found throughout RES4 and HVU7, this configuration implies that ALG_B1 and ALG_B2 have become fused and mixed as described here.
Loss of colinearity between phyla
To measure the degree of colinearity between chromosomes of different species, we identified colinear runs of consecutive orthologs (as defined by pairwise MBHs between those two species). Orthologs were considered colinear if they occurred in the same order in both genomes, in forward or reverse orientation, without intervening genes from the ortholog set (that is, genes without MBHs were not considered). Comparisons between phyla showed that the typical gene does not participate in a colinear run (i.e., its adjacent genes in one species are not adjacent in another). No colinear runs of more than four genes were observed (table S2).
ALG and BLG nomenclature and correspondence with chordate notation
ALGs were named to ensure consistency with the previously defined CLGs derived from scallop-amphioxus-jawed vertebrate comparisons (see note S2) (8). Since amphioxus chromosomes have been partitioned into segments labeled as CLG_A through CLG_Q, we adopted these 17 letter codes and subdivided them as needed by suffixes.
To represent subdivisions of previously defined CLGs that are split in bilaterians, we use numerical suffixes. For example, several amphioxus chromosomes (BFL10, BFL16, and BFL18) were grouped together into CLG_B since homologs of these chromosomal units are combined (fused and mixed) in the vertebrate lineage. These three amphioxus chromosomes, however, are orthologous to distinct chromosomes in scallop and sponge and must therefore represent separate ALGs, and we designate them as ALG_B1, B2, and B3. Similarly, we split CLG_C, J, and O into ALG_C1/C2, J1/J2, and O1/O2, respectively. These represent discrete segments of amphioxus that were previously grouped together based on their fusion in vertebrates.
We found that amphioxus chromosome BFL1, which previously defined CLG_A, is a fusion-with-mixing relative to scallop and lamprey. Thus, we conclude that CLG_A should be split into two (bilaterian) BLGs and use this and other information (e.g., corroboration by jellyfish and sponge) to define BLG_A1 and BLG_A2. As predicted, we find that these are each represented by distinct chromosomes in the sea urchin L. variegatus (LVA7 and LVA8, respectively).
To represent subdivisions of previously defined chordate/bilaterian units that are split in sponge and/or cnidarians, we added a lowercase letter suffix. For example, ALG_E is split into two parts, ALG_Ea and ALG_Eb. ALG_Ea comprises those BLG E genes whose orthologs are found on EMU01 and RES17, while ALG_Eb comprises those genes whose orthologs are found on EMU14L and RES6. Similar considerations apply to ALG_A1a/b and ALG_Qa/b/c/d. When there is no confusion in a bilaterian context we refer to ALGs as BLGs and omit the lowercase letter suffix.
Last, ALG_R is a unit of conserved synteny between scallop, sponge, jellyfish, and hydra that was not detected in amphioxus and so was assigned a new letter code. It is defined by conserved ortholog locations in PYE12, RES19, and HVU15, without regard to its chromosome position in other species. Although the genes of ALG_R are present in amphioxus and vertebrates, they do not show conserved linkage in these genomes, which explains why it was not found previously in (8). ALG_R is found intact as chromosome LVA6 of the green sea urchin L. variegatus, indicating (based on its conservation in PYE12) that it was present in the bilaterian and deuterostome common ancestor.
Analysis of subchromosomal segments
As previously noted by Simakov et al. (8), amphioxus chromosomes BFL2, 3, and 4 are divided into discrete units with distinct syntenic relationships. For example, BFL3 and BFL4 are divided into left and right halves denoted by the suffixes L and R in Fig. 3 (see also Fig. 1, but note that, for clarity, some of these chromosomes have been reverse-complemented, exchanging right and left on the image; L and R are defined relative to the FASTA sequence). BFL3 and BFL4 therefore represent fusion chromosomes relative to the ALGs but, in contrast to RES4 and HVU7, without subsequent mixing. We find several cases where chromosomes are split into odd numbers of segments.
For example, BFL2 is divided into five segments. Since even- and odd-numbered segments have different patterns of conserved synteny across other species, we denote them as BFL2eve and 2odd, respectively, in Fig. 3. Notably, BFL2odd includes both distal (i.e., subtelomeric) regions. Similarly, EMU21 is split into three distinct patterns of conserved synteny, with EMU21end labeling the distal segments and EMU21mid labeling the internal segment. These are the signal of centric insertions as described here.
In some cases, H. vulgaris showed unique rearrangements relative to the other four species. For example, ALG_D, K, L, Ea, and I are split in hydra (with minor component indicated by an underscore in Fig. 3) but not in the other species. The uniqueness of these hydra configurations relative to jellyfish and the other species indicates that these splits are due to rearrangement in the hydra lineage and do not affect the definition of the ALGs.
Genes of variable synteny
We define genes of variable synteny as MBH gene pairs that lie on chromosomes that do not belong to ALG groupings, i.e., gene pairs inconsistent with Fig. 3 and its extension to other species. For maximum resolution, we defined these in pairwise comparisons. These are shown in gray in Fig. 2 and other pairwise dot plots. Table S3 shows the number of mutual hit gene pairs between species and the number of such pairs lying on significantly syntenically conserved chromosomes. Note that these counts only include genes assigned to chromosomes in both species, accounting for the low rate of MBHs for P. yessoensis, which has a substantial unassigned gene complement (identified as PYE0 “chromosome”). For the purposes of assessing potential functional enrichments of genes of variable synteny, we used InterProScan version 5.47-82.0 (75), which provided gene ontology (GO) terms (76). We tested GO term enrichment using clusterProfiler R package (77).
Simulation of fusion-with-mixing
In fusion-with-mixing, the genes of two distinct ALGs become interspersed along the same fused-and-mixed chromosome. As a simple model for this process, we simulated a chromosome of 100 genes beginning from an initial configuration of two juxtaposed ALGs of 50 genes each. We then randomly selected a block of 10 consecutive genes and inverted its order. Snapshots of the simulation are shown in fig. S5, along with the lengths of runs of consecutive genes from the same original ALG. After 1000 inversions, the runs of consecutive genes from the same ALG become geometrically distributed, as expected for a random sequence of ALG identities. Such rapid mixing is expected, independent of the details of intrachromosomal rearrangement, similar to findings for random card shuffling (56, 78). Figure S5 also shows several examples of ALG fusion-with-mixing in scallop and jellyfish chromosomes.
The irreversibility of fusion-with-mixing unmixing can be quantified by the Shannon entropy (34). The pattern of ALG identity along a fused-and-mixed chromosome (Materials and Methods) is consistent with complete mixing among bilaterians, cnidarians, and sponge since ALG identity decays geometrically in these cases, as expected for uncorrelated sequences. For a fully mixed sequence, the Shannon entropy in bits is S = NAlog2(NA/N) + (NB/N)log2(NB/N), where NA and NB are the number of genes from the two participating ALGs and N = NA + NB is the total number of genes. Roughly speaking, 2S is the relative ratio of the number of mixed to unmixed configurations. If rearrangements occur randomly (i.e., without regard to ALG identity of the genes involved), then the waiting time for unmixing is therefore 2S τ, where τ is the typical time between rearrangements. For N ~ 100, and even allowing for a very rapid τ ~ 105 years, the waiting time for spontaneous unmixing is ~1022 years, more than 10 orders of magnitude longer than the age of the universe.
Conserved synteny in draft genomes
To integrate subchromosomal genome datasets into our evolutionary scheme, we adapted the approach pioneered by Putnam and colleagues (2, 3, 5, 21). Briefly, for draft genomes, we (i) found MBHs of protein-coding genes with one or more related chromosome-scale genomes; (ii) made an ortholog grid between the scaffolds of the draft genome and the related chromosome-scale sequences and/or versus the 2361 ALG-defining anchor genes, considering only scaffolds with more than 10 orthologs; and (iii) grouped scaffolds by hierarchical clustering based on their vector of conserved syntenies, using neighbor joining with ward.D distance metric as implemented in R hclust function. Dot plot code is available under https://bitbucket.org/viemet/public/src/master/CLG/.
Scaffolds grouped by similar patterns of conserved synteny are hypothesized to belong to the same chromosome. To infer fusion-with-mixing, we require multiple scaffolds with the same pattern of conserved synteny to two different ALGs or chromosomes of another species. By insisting that two or more scaffolds share the same pattern of conserved synteny, this method is insensitive to individual misassembled scaffolds.
The prediction that scaffolds showing similar patterns of conserved synteny are indeed linked on the same chromosome has been confirmed in amphioxus by comparing the synteny-based groupings found by Putnam et al. (5) to the chromosome-scale assembly of amphioxus (8). Note that we cannot exclude the existence of recent translocations that separate scaffolds with closely related patterns of synteny onto different chromosomes. Similarly, the synteny-based grouping method cannot confidently recognize Robertsonian, end-end, or centric fusions since these produce sharp boundaries in ALG ancestry that cannot be easily differentiated from misassembly.
Supplementary Material
Acknowledgments
We thank N. Putnam, M. Srivastava, A. Dernberg, and G. Karpen for discussion and critical reading of the manuscript and J. Chapman, M. Levine, D. Hockemeyer, S. Haddock, R. Harland, C. Lowe, and J. Gerhart for helpful and encouraging discussions.
Funding: This work was supported by internal funds of the OIST Molecular Genetics Unit, the Chan Zuckerberg Biohub, and NIH RO1 HD080708 to D.S.R. D.T.S. was supported by NSF DEB-1542679. This work was made possible, in part, through access to the Genomics High Throughput Facility Shared Resource of the Cancer Center Support Grant (P30CA-062203) at the University of California, Irvine and NIH shared instrumentation grants 1S10RR025496-01, 1S10OD010794-01, and 1S10OD021718-01. D.S.R. is grateful for the support of the Marthella Foskett Brown Chair in Biology. O.S. was supported by the Austrian Science Fund grant P32190 and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 945026).
Author contributions: D.S.R. and O.S. designed the study and performed analysis of synteny. D.S.R., C.N.D., R.E.S., and O.S. designed hydra genome sequencing and assembly. R.E.S., D.E.M., and P.D. provided materials and resources for Hydra genome analysis. R.E.G., B.L.O., and D.T.S. generated and analyzed Hi-C data. J.B., K.B., and F.M. assembled and annotated hydra and amphioxus genomes. T.M. performed comparative analysis and visualization. D.S.R. and O.S. wrote the paper with input from R.E.S., R.E.G., and C.N.D. All authors read and approved the manuscript.
Competing interests: R.E.G. and D.S.R. are paid consultants and equity holders of Dovetail Genomics. The other authors declare that they have no competing interests.
Data and materials availability: Illumina sequencing reads from Chicago and Hi-C libraries are deposited at NCBI BioProject PRJNA703404. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Correction (22 March 2022): In two instances in the Discussion section “Conserved synteny between animals and their unicellular relatives,” the authors inadvertently referred to “ichthyosporean” instead of the correct “filasterean,” first in describing “Capsaspora owczarzaki,” and then in the phrase “choanoflagellate and/or ichthyosporean genome.” These have now been corrected to “filasterean.”
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S13
Tables S1 to S3
Legends for data files S1 to S5
References
Other Supplementary Material for this manuscript includes the following:
Data files S1 to S5
REFERENCES AND NOTES
- 1.Waddington C. H., Callan H. G., Animal cytology and evolution. Nature 175, 436 (1955). [Google Scholar]
- 2.Putnam N. H., Srivastava M., Hellsten U., Dirks B., Chapman J., Salamov A., Terry A., Shapiro H., Lindquist E., Kapitonov V. V., Jurka J., Genikhovich G., Grigoriev I. V., Lucas S. M., Steele R. E., Finnerty J. R., Technau U., Martindale M. Q., Rokhsar D. S., Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Simakov O., Marletaz F., Cho S.-J., Edsinger-Gonzales E., Havlak P., Hellsten U., Kuo D.-H., Larsson T., Lv J., Arendt D., Savage R., Osoegawa K., de Jong P., Grimwood J., Chapman J. A., Shapiro H., Aerts A., Otillar R. P., Terry A. Y., Boore J. L., Grigoriev I. V., Lindberg D. R., Seaver E. C., Weisblat D. A., Putnam N. H., Rokhsar D. S., Insights into bilaterian evolution from three spiralian genomes. Nature 493, 526–531 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simakov O., Kawashima T., Marlétaz F., Jenkins J., Koyanagi R., Mitros T., Hisata K., Bredeson J., Shoguchi E., Gyoja F., Yue J.-X., Chen Y.-C., Freeman R. M. Jr., Sasaki A., Hikosaka-Katayama T., Sato A., Fujie M., Baughman K. W., Levine J., Gonzalez P., Cameron C., Fritzenwanker J. H., Pani A. M., Goto H., Kanda M., Arakaki N., Yamasaki S., Qu J., Cree A., Ding Y., Dinh H. H., Dugan S., Holder M., Jhangiani S. N., Kovar C. L., Lee S. L., Lewis L. R., Morton D., Nazareth L. V., Okwuonu G., Santibanez J., Chen R., Richards S., Muzny D. M., Gillis A., Peshkin L., Wu M., Humphreys T., Su Y.-H., Putnam N. H., Schmutz J., Fujiyama A., Yu J.-K., Tagawa K., Worley K. C., Gibbs R. A., Kirschner M. W., Lowe C. J., Satoh N., Rokhsar D. S., Gerhart J., Hemichordate genomes and deuterostome origins. Nature 527, 459–465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Putnam N. H., Butts T., Ferrier D. E. K., Furlong R. F., Hellsten U., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu J.-K., Benito-Gutiérrez E. L., Dubchak I., Garcia-Fernàndez J., Gibson-Brown J. J., Grigoriev I. V., Horton A. C., de Jong P. J., Jurka J., Kapitonov V. V., Kohara Y., Kuroki Y., Lindquist E., Lucas S., Osoegawa K., Pennacchio L. A., Salamov A. A., Satou Y., Sauka-Spengler T., Schmutz J., Shin-I T., Toyoda A., Bronner-Fraser M., Fujiyama A., Holland L. Z., Holland P. W. H., Satoh N., Rokhsar D. S., The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Renwick J. H., The mapping of human chromosomes. Annu. Rev. Genet. 5, 81–120 (1971). [DOI] [PubMed] [Google Scholar]
- 7.Wang S., Zhang J., Jiao W., Li J., Xun X., Sun Y., Guo X., Huan P., Dong B., Zhang L., Hu X., Sun X., Wang J., Zhao C., Wang Y., Wang D., Huang X., Wang R., Lv J., Li Y., Zhang Z., Liu B., Lu W., Hui Y., Liang J., Zhou Z., Hou R., Li X., Liu Y., Li H., Ning X., Lin Y., Zhao L., Xing Q., Dou J., Li Y., Mao J., Guo H., Dou H., Li T., Mu C., Jiang W., Fu Q., Fu X., Miao Y., Liu J., Yu Q., Li R., Liao H., Li X., Kong Y., Jiang Z., Chourrout D., Li R., Bao Z., Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 1, 0120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simakov O., Marlétaz F., Yue J. X., O’Connell B., Jenkins J., Brandt A., Calef R., Tung C. H., Huang T. K., Schmutz J., Satoh N., Yu J. K., Putnam N. H., Green R. E., Rokhsar D. S., Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol. 4, 820–830 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srivastava M., Begovic E., Chapman J., Putnam N. H., Hellsten U., Kawashima T., Kuo A., Mitros T., Salamov A., Carpenter M. L., Signorovitch A. Y., Moreno M. A., Kamm K., Grimwood J., Schmutz J., Shapiro H., Grigoriev I. V., Buss L. W., Schierwater B., Dellaporta S. L., Rokhsar D. S., The Trichoplax genome and the nature of placozoans. Nature 454, 955–960 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Zdobnov E. M., Von Mering C., Letunic I., Bork P., Consistency of genome-based methods in measuring Metazoan evolution. FEBS Lett. 579, 3355–3361 (2005). [DOI] [PubMed] [Google Scholar]
- 11.H. J. Muller, Bearing of the Drosophila work on systematics, in The New Systematics (1940), pp. 185–268; https://ci.nii.ac.jp/naid/10004957361/.
- 12.Drosophila 12 Genomes Consortium , Evolution of genes and genomes on the Drosophila phylogeny. Nature 450, 203–218 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Schaeffer S. W., Muller “elements” in Drosophila: How the search for the genetic basis for speciation led to the birth of comparative genomics. Genetics 210, 3–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hillier L. W., Miller R. D., Baird S. E., Chinwalla A., Fulton L. A., Koboldt D. C., Waterston R. H., Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLOS Biol. 5, e167 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tandonnet S., Koutsovoulos G. D., Adams S., Cloarec D., Parihar M., Blaxter M. L., Pires-daSilva A., Chromosome-wide evolution and sex determination in the three-sexed nematode Auanema rhodensis. G3-Genes Genom. Genet. 9, 1211–1230 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn C. W., Hejnol A., Matus D. Q., Pang K., Browne W. E., Smith S. A., Seaver E., Rouse G. W., Obst M., Edgecombe G. D., Sørensen M. V., Haddock S. H. D., Schmidt-Rhaesa A., Okusu A., Kristensen R. M., Wheeler W. C., Martindale M. Q., Giribet G., Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745–749 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Ryan J. F., Pang K., Schnitzler C. E., Nguyen A.-D., Moreland R. T., Simmons D. K., Koch B. J., Francis W. R., Havlak P.; NISC Comparative Sequencing Program, Smith S. A., Putnam N. H., Haddock S. H. D., Dunn C. W., Wolfsberg T. G., Mullikin J. C., Martindale M. Q., Baxevanis A. D., The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342, 1242592 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Gao L., Pan Y., Tian M., Li Y., He C., Dong Y., Sun Y., Zhou Z., Chromosome-level reference genome of the jellyfish Rhopilema esculentum. GigaScience 9, giaa036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenny N. J., Francis W. R., Rivera-Vicéns R. E., Juravel K., de Mendoza A., Díez-Vives C., Lister R., Bezares-Calderón L. A., Grombacher L., Roller M., Barlow L. D., Camilli S., Ryan J. F., Wörheide G., Hill A. L., Riesgo A., Leys S. P., Tracing animal genomic evolution with the chromosomal-level assembly of the freshwater sponge Ephydatia muelleri. Nat. Commun. 11, 3676 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapman J. A., Kirkness E. F., Simakov O., Hampson S. E., Mitros T., Weinmaier T., Rattei T., Balasubramanian P. G., Borman J., Busam D., Disbennett K., Pfannkoch C., Sumin N., Sutton G. G., Viswanathan L. D., Walenz B., Goodstein D. M., Hellsten U., Kawashima T., Prochnik S. E., Putnam N. H., Shu S., Blumberg B., Dana C. E., Gee L., Kibler D. F., Law L., Lindgens D., Martinez D. E., Peng J., Wigge P. A., Bertulat B., Guder C., Nakamura Y., Ozbek S., Watanabe H., Khalturin K., Hemmrich G., Franke A., Augustin R., Fraune S., Hayakawa E., Hayakawa S., Hirose M., Hwang J. S., Ikeo K., Nishimiya-Fujisawa C., Ogura A., Takahashi T., Steinmetz P. R. H., Zhang X., Aufschnaiter R., Eder M.-K., Gorny A.-K., Salvenmoser W., Heimberg A. M., Wheeler B. M., Peterson K. J., Böttger A., Tischler P., Wolf A., Gojobori T., Remington K. A., Strausberg R. L., Venter J. C., Technau U., Hobmayer B., Bosch T. C. G., Holstein T. W., Fujisawa T., Bode H. R., David C. N., Rokhsar D. S., Steele R. E., The dynamic genome of Hydra. Nature 464, 592–596 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srivastava M., Simakov O., Chapman J., Fahey B., Gauthier M. E. A., Mitros T., Richards G. S., Conaco C., Dacre M., Hellsten U., Larroux C., Putnam N. H., Stanke M., Adamska M., Darling A., Degnan S. M., Oakley T. H., Plachetzki D. C., Zhai Y., Adamski M., Calcino A., Cummins S. F., Goodstein D. M., Harris C., Jackson D. J., Leys S. P., Shu S., Woodcroft B. J., Vervoort M., Kosik K. S., Manning G., Degnan B. M., Rokhsar D. S., The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irimia M., Tena J. J., Alexis M. S., Fernandez-Miñan A., Maeso I., Bogdanović O., De La Calle-Mustienes E., Roy S. W., Gómez-Skarmeta J. L., Fraser H. B., Extensive conservation of ancient microsynteny across metazoans due to cis-regulatory constraints. Genome Res. 22, 2356–2367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmermann B., Robert N. S. M., Technau U., Simakov O., Ancient animal genome architecture reflects cell type identities. Nat. Ecol. Evol. 3, 1289–1293 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Bhutkar A., Schaeffer S. W., Russo S. M., Xu M., Smith T. F., Gelbart W. M., Chromosomal rearrangement inferred from comparisons of 12 drosophila genomes. Genetics 179, 1657–1680 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lv J., Havlak P., Putnam N. H., Constraints on genes shape long-term conservation of macro-synteny in metazoan genomes. BMC Bioinformatics. 12, S11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch M., Conery J. S., The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Davidson P. L., Guo H., Wang L., Berrio A., Zhang H., Chang Y., Soborowski A. L., McClay D. R., Fan G., Wray G. A., Chromosomal-level genome assembly of the sea urchin Lytechinus variegatus substantially improves functional genomic analyses. Genome Biol. Evol. 12, 1080–1086 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schubert I., Lysak M. A., Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 27, 207–216 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Ijdo J. W., Baldini A., Ward D. C., Reeders S. T., Wells R. A., Origin of human chromosome 2: An ancestral telomere-telomere fusion. Proc. Natl. Acad. Sci. U.S.A. 88, 9051–9055 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The International Brachypodium Initiative , Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Wang X., Jin D., Wang Z., Guo H., Zhang L., Wang L., Li J., Paterson A. H., Telomere-centric genome repatterning determines recurring chromosome number reductions during the evolution of eukaryotes. New Phytol. 205, 378–389 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Luo M. C., Deal K. R., Akhunov E. D., Akhunova A. R., Anderson O. D., Anderson J. A., Blake N., Clegg M. T., Coleman-Derr D., Conley E. J., Crossman C. C., Dubcovsky J., Gill B. S., Gu Y. Q., Hadam J., Heo H. Y., Huo N., Lazo G., Ma Y., Matthews D. E., McGuire P. E., Morrell P. L., Qualset C. O., Renfro J., Tabanao D., Talbert L. E., Tian C., Toleno D. M., Warburton M. L., You F. M., Zhang W., Dvorak J., Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc. Natl. Acad. Sci. 106, 15780–15785 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiatante G., Giannuzzi G., Calabrese F. M., Eichler E. E., Ventura M., Centromere destiny in dicentric chromosomes: New insights from the evolution of human chromosome 2 ancestral centromeric region. Mol. Biol. Evol. 34, 1669–1681 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.C. E. Shannon, W. Weaver, The Mathematical Theory of Communication (1998), p. 125.
- 35.Bai C.-M., Xin L.-S., Rosani U., Wu B., Wang Q.-C., Duan X.-K., Liu Z.-H., Wang C.-M., Chromosomal-level assembly of the blood clam, Scapharca (Anadara) broughtonii, using long sequence reads and Hi-C. GigaScience 8, giz067 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Y., Zhang Y., Liu Q., Huang Y., Mao G., Yue Z., Abe E. M., Li J., Wu Z., Li S., Zhou X., Hu W., Xiao N., A chromosomal-level genome assembly for the giant African snail Achatina fulica. GigaScience 8, giz124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Mao F., Mu H., Huang M., Bao Y., Wang L., Wong N. K., Xiao S., Dai H., Xiang Z., Ma M., Xiong Y., Zhang Z., Zhang L., Song X., Wang F., Mu X., Li J., Ma H., Zhang Y., Zheng H., Simakov O., Yu Z., The genome of Nautilus pompilius illuminates eye evolution and biomineralization. Nat. Ecol. Evol. 5, 927–938 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C., Ren Y., Li Z., Hu Q., Yin L., Wang H., Qiao X., Zhang Y., Xing L., Xi Y., Jiang F., Wang S., Huang C., Liu B., Liu H., Wan F., Qian W., Fan W., Giant African snail genomes provide insights into molluscan whole-genome duplication and aquatic–terrestrial transition. Mol. Ecol. Resour. 21, 478–494 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Przeworski Lab, Acropora millepora genome resource (2020); https://przeworskilab.com/acropora-millepora-genome/.
- 40.B. Zimmermann, S. M. C. Robb, G. Genikhovich, W. J. Fropf, L. Weilguny, S. He, S. Chen, J. Lovegrove-Walsh, E. M. Hill, K. Ragkousi, D. Praher, D. Fredman, Y. Moran, M. C. Gibson, U. Technau, Sea anemone genomes reveal ancestral metazoan chromosomal macrosynteny. bioRxiv 10.1101/2020.10.30.359448 [Preprint] (2020). [DOI]
- 41.Hamada M., Satoh N., Khalturin K., A reference genome from the symbiotic hydrozoan, Hydra viridissima. G3-Genes Genom. Genet. 10, 3883–3895 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zacharias H., Anokhin B., Khalturin K., Bosch T. C. G., Genome sizes and chromosomes in the basal metazoan Hydra. Fortschr. Zool. 107, 219–227 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Zdobnov E. M., von Mering C., Letunic I., Torrents D., Suyama M., Copley R. R., Christophides G. K., Thomasova D., Holt R. A., Subramanian G. M., Mueller H.-M., Dimopoulos G., Law J. H., Wells M. A., Birney E., Charlab R., Halpern A. L., Kokoza E., Kraft C. L., Lai Z., Lewis S., Louis C., Barillas-Mury C., Nusskern D., Rubin G. M., Salzberg S. L., Sutton G. G., Topalis P., Wides R., Wincker P., Yandell M., Collins F. H., Ribeiro J., Gelbart W. M., Kafatos F. C., Bork P., Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298, 149–159 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Herndon N., Shelton J., Gerischer L., Ioannidis P., Ninova M., Dönitz J., Waterhouse R. M., Liang C., Damm C., Siemanowski J., Kitzmann P., Ulrich J., Dippel S., Oberhofer G., Hu Y., Schwirz J., Schacht M., Lehmann S., Montino A., Posnien N., Gurska D., Horn T., Seibert J., Jentzsch I. M. V., Panfilio K. A., Li J., Wimmer E. A., Stappert D., Roth S., Schröder R., Park Y., Schoppmeier M., Chung H.-R., Klingler M., Kittelmann S., Friedrich M., Chen R., Altincicek B., Vilcinskas A., Zdobnov E., Griffiths-Jones S., Ronshaugen M., Stanke M., Brown S. J., Bucher G., Enhanced genome assembly and a new official gene set for Tribolium castaneum. BMC Genomics 21, 47 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chipman A. D., Ferrier D. E. K., Brena C., Qu J., Hughes D. S. T., Schröder R., Torres-Oliva M., Znassi N., Jiang H., Almeida F. C., Alonso C. R., Apostolou Z., Aqrawi P., Arthur W., Barna J. C. J., Blankenburg K. P., Brites D., Capella-Gutiérrez S., Coyle M., Dearden P. K., Du Pasquier L., Duncan E. J., Ebert D., Eibner C., Erikson G., Evans P. D., Extavour C. G., Francisco L., Gabaldón T., Gillis W. J., Goodwin-Horn E. A., Green J. E., Griffiths-Jones S., Grimmelikhuijzen C. J. P., Gubbala S., Guigó R., Han Y., Hauser F., Havlak P., Hayden L., Helbing S., Holder M., Hui J. H. L., Hunn J. P., Hunnekuhl V. S., Jackson L., Javaid M., Jhangiani S. N., Jiggins F. M., Jones T. E., Kaiser T. S., Kalra D., Kenny N. J., Korchina V., Kovar C. L., Kraus F. B., Lapraz F., Lee S. L., Lv J., Mandapat C., Manning G., Mariotti M., Mata R., Mathew T., Neumann T., Newsham I., Ngo D. N., Ninova M., Okwuonu G., Ongeri F., Palmer W. J., Patil S., Patraquim P., Pham C., Pu L.-L., Putman N. H., Rabouille C., Ramos O. M., Rhodes A. C., Robertson H. E., Robertson H. M., Ronshaugen M., Rozas J., Saada N., Sánchez-Gracia A., Scherer S. E., Schurko A. M., Siggens K. W., Simmons D., Stief A., Stolle E., Telford M. J., Tessmar-Raible K., Thornton R., van der Zee M., von Haeseler A., Williams J. M., Willis J. H., Wu Y., Zou X., Lawson D., Muzny D. M., Worley K. C., Gibbs R. A., Akam M., Richards S., The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLOS Biol. 12, e1002005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith J. J., Keinath M. C., The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Res. 25, 1081–1090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Satou Y., Nakamura R., Yu D., Yoshida R., Hamada M., Fujie M., Hisata K., Takeda H., Satoh N., O’Neill R., A nearly complete genome of Ciona intestinalis Type A (C. robusta) reveals the contribution of inversion to chromosomal evolution in the genus Ciona. Genome Biol. Evol. 11, 3144–3157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holland L. Z., Gibson-Brown J. J., The Ciona intestinalis genome: When the constraints are off. Bioessays 25, 529–532 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Eitel M., Francis W. R., Varoqueaux F., Daraspe J., Osigus H. J., Krebs S., Vargas S., Blum H., Williams G. A., Schierwater B., Wörheide G., Comparative genomics and the nature of placozoan species. PLOS Biol. 16, e2005359 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whelan N. V., Kocot K. M., Moroz T. P., Mukherjee K., Williams P., Paulay G., Moroz L. L., Halanych K. M., Ctenophore relationships and their placement as the sister group to all other animals. Nat. Ecol. Evol. 1, 1737–1746 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laumer C. E., Fernández R., Lemer S., Combosch D., Kocot K. M., Riesgo A., Andrade S. C. S., Sterrer W., Sørensen M. V., Giribet G., Revisiting metazoan phylogeny with genomic sampling of all phyla. Proc. R. Soc. B Biol. Sci. 286, 20190831 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moroz L. L., Kocot K. M., Citarella M. R., Dosung S., Norekian T. P., Povolotskaya I. S., Grigorenko A. P., Dailey C., Berezikov E., Buckley K. M., Ptitsyn A., Reshetov D., Mukherjee K., Moroz T. P., Bobkova Y., Yu F., Kapitonov V. V., Jurka J., Bobkov Y. V., Swore J. J., Girardo D. O., Fodor A., Gusev F., Sanford R., Bruders R., Kittler E., Mills C. E., Rast J. P., Derelle R., Solovyev V. V., Kondrashov F. A., Swalla B. J., Sweedler J. V., Rogaev E. I., Halanych K. M., Kohn A. B., The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.King N., Westbrook M. J., Young S. L., Kuo A., Abedin M., Chapman J., Fairclough S., Hellsten U., Isogai Y., Letunic I., Marr M., Pincus D., Putnam N., Rokas A., Wright K. J., Zuzow R., Dirks W., Good M., Goodstein D., Lemons D., Li W., Lyons J. B., Morris A., Nichols S., Richter D. J., Salamov A.; JGI Sequencing, Bork P., Lim W. A., Manning G., Miller W. T., McGinnis W., Shapiro H., Tjian R., Grigoriev I. V., Rokhsar D., The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451, 783–788 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fairclough S. R., Chen Z., Kramer E., Zeng Q., Young S., Robertson H. M., Begovic E., Richter D. J., Russ C., Westbrook M. J., Manning G., Lang B. F., Haas B., Nusbaum C., King N., Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 14, R15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suga H., Chen Z., De Mendoza A., Sebé-Pedrós A., Brown M. W., Kramer E., Carr M., Kerner P., Vervoort M., Sánchez-Pons N., Torruella G., Derelle R., Manning G., Lang B. F., Russ C., Haas B. J., Roger A. J., Nusbaum C., Ruiz-Trillo I., The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat. Commun. 4, 2325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hillis D. M., SINEs of the perfect character. Proc. Natl. Acad. Sci. U.S.A. 96, 9979–9981 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naville M., Ishibashi M., Ferg M., Bengani H., Rinkwitz S., Krecsmarik M., Hawkins T. A., Wilson S. W., Manning E., Chilamakuri C. S. R., Wilson D. I., Louis A., Lucy Raymond F., Rastegar S., Strähle U., Lenhard B., Bally-Cuif L., Van Heyningen V., Fitzpatrick D. R., Becker T. S., Roest Crollius H., Long-range evolutionary constraints reveal cis-regulatory interactions on the human X chromosome. Nat. Commun. 6, 6904 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghavi-Helm Y., Jankowski A., Meiers S., Viales R. R., Korbel J. O., Furlong E. E. M., Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet. 51, 1272–1282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright S., On the probability of fixation of reciprocal translocations. Am. Nat. 75, 513–522 (1941). [Google Scholar]
- 60.Hedrick P. W., The establishment of chromosomal variants. Evolution 35, 322–332 (1981). [DOI] [PubMed] [Google Scholar]
- 61.de Villena F. P.-M., Sapienza C., Female meiosis drives karyotypic evolution in mammals. Genetics 159, 1179–1189 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lande R., The fixation of chromosomal rearrangements in a subdivided population with local extinction and colonization. Heredity 54, 323–332 (1985). [DOI] [PubMed] [Google Scholar]
- 63.Bush G. L., Case S. M., Wilson A. C., Patton J. L., Rapid speciation and chromosomal evolution in mammals. Proc. Natl. Acad. Sci. U.S.A. 74, 3942–3946 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J.-H., Zheng X.-D., Comparison of the genetic relationship between nine Cephalopod species based on cluster analysis of karyotype evolutionary distance. Comp. Cytogenet. 11, 477–494 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bürglin T. R., Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25, 4173–4180 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Popovici C., Leveugle M., Birnbaum D., Coulier F., Homeobox gene clusters and the human paralogy map. FEBS Lett. 491, 237–242 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Holland P. W. H., Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol. 2, 31–45 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Ferrier D. E. K., Evolution of homeobox gene clusters in animals: The giga-cluster and primary vs secondary clustering. Front. Ecol. Evol. 4, 36 (2016). [Google Scholar]
- 69.Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B. R., Sabo P. J., Dorschner M. O., Sandstrom R., Bernstein B., Bender M. A., Groudine M., Gnirke A., Stamatoyannopoulos J., Mirny L. A., Lander E. S., Dekker J., Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kolmogorov M., Yuan J., Lin Y., Pevzner P. A., Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Durand N. C., Robinson J. T., Shamim M. S., Machol I., Mesirov J. P., Lander E. S., Aiden E. L., Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Altschul S., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 73.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T. L., BLAST+: Architecture and applications. BMC Bioinformatics. 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y., Tang H., Debarry J. D., Tan X., Li J., Wang X., Lee T.-h., Jin H., Marler B., Guo H., Kissinger J. C., Paterson A. H., MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zdobnov E. M., Apweiler R., InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848 (2001). [DOI] [PubMed] [Google Scholar]
- 76.Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., Heger A., Hetherington K., Holm L., Mistry J., Sonnhammer E. L. L., Tate J., Punta M., Pfam: The protein families database. Nucleic Acids Res. 42, D222–D230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aldous D., Diaconis P., Shuffling cards and stopping times. Am. Math. Mon. 93, 333–348 (2018). [Google Scholar]
- 79.Burton J. N., Adey A., Patwardhan R. P., Qiu R., Kitzman J. O., Shendure J., Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 31, 1119–1125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Durand N. C., Shamim M. S., Machol I., Rao S. S. P., Huntley M. H., Lander E. S., Aiden E. L., Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dudchenko O., Batra S. S., Omer A. D., Nyquist S. K., Hoeger M., Durand N. C., Shamim M. S., Machol I., Lander E. S., Aiden A. P., Aiden E. L., De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O. Dudchenko, M. S. Shamim, S. S. Batra, N. C. Durand, N. T. Musial, R. Mostofa, M. Pham, B. G. St. Hilaire, W. Yao, E. Stamenova, M. Hoeger, S. K. Nyquist, V. Korchina, K. Pletch, J. P. Flanagan, A. Tomaszewicz, D. M. Aloose, C. P. Estrada, B. J. Novak, A. D. Omer, E. L. Aiden, The juicebox assembly tools module facilitates de novo assembly of mammalian genomes with chromosome-length scaffolds for under $1000. bioRxiv 254797 [Preprint] (2018).
- 84.H. Li, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN] (2013); https://arxiv.org/abs/1303.3997v2.
- 85.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bateson Z. W., Hammerly S. C., Johnson J. A., Morrow M. E., Whittingham L. A., Dunn P. O., Specific alleles at immune genes, rather than genome-wide heterozygosity, are related to immunity and survival in the critically endangered Attwater’s prairie-chicken. Mol. Ecol. 25, 4730–4744 (2016). [DOI] [PubMed] [Google Scholar]
- 87.G. Formenti, A. Rhie, B. P. Walenz, F. Thibaud-Nissen, K. Shafin, S. Koren, E. W. Myers, E. D. Jarvis, A. M. Phillippy, Merfin: Improved variant filtering and polishing via k-mer validation. bioRxiv 2021.07.16.452324 [Preprint] (2021). [DOI] [PMC free article] [PubMed]
- 88.Danecek P., Bonfield J. K., Liddle J., Marshall J., Ohan V., Pollard M. O., Whitwham A., Keane T., McCarthy S. A., Davies R. M., Li H., Twelve years of SAMtools and BCFtools. GigaScience 10, giab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Melters D. P., Bradnam K. R., Young H. A., Telis N., May M. R., Ruby J. G., Sebra R., Peluso P., Eid J., Rank D., Garcia J. F., DeRisi J. L., Smith T., Tobias C., Ross-Ibarra J., Korf I., Chan S. W. L., Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14, R10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hallinan N. M., Lindberg D. R., Comparative analysis of chromosome counts infers three paleopolyploidies in the mollusca. Genome Biol. Evol. 3, 1150–1163 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Albertin C. B., Simakov O., Mitros T., Wang Z. Y., Pungor J. R., Edsinger-Gonzales E., Brenner S., Ragsdale C. W., Rokhsar D. S., The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524, 220–224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wong W. Y., Simakov O., Bridge D. M., Cartwright P., Bellantuono A. J., Kuhn A., Holstein T. W., David C. N., Steele R. E., Martínez D. E., Expansion of a single transposable element family is associated with genome-size increase and radiation in the genus Hydra. Proc. Natl. Acad. Sci. U.S.A. 116, 22915–22917 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kamm K., Osigus H. J., Stadler P. F., DeSalle R., Schierwater B., Trichoplax genomes reveal profound admixture and suggest stable wild populations without bisexual reproduction. Sci. Rep. 8, 11168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dieterich C., Clifton S. W., Schuster L. N., Chinwalla A., Delehaunty K., Dinkelacker I., Fulton L., Fulton R., Godfrey J., Minx P., Mitreva M., Roeseler W., Tian H., Witte H., Yang S. P., Wilson R. K., Sommer R. J., The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat. Genet. 40, 1193–1198 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tribolium Genome Sequencing Consortium , The genome of the model beetle and pest Tribolium castaneum. Nature 452, 949–955 (2008). [DOI] [PubMed] [Google Scholar]
- 97.Braasch I., Gehrke A. R., Smith J. J., Kawasaki K., Manousaki T., Pasquier J., Amores A., Desvignes T., Batzel P., Catchen J., Berlin A. M., Campbell M. S., Barrell D., Martin K. J., Mulley J. F., Ravi V., Lee A. P., Nakamura T., Chalopin D., Fan S., Wcisel D., Cañestro C., Sydes J., Beaudry F. E. G., Sun Y., Hertel J., Beam M. J., Fasold M., Ishiyama M., Johnson J., Kehr S., Lara M., Letaw J. H., Litman G. W., Litman R. T., Mikami M., Ota T., Saha N. R., Williams L., Stadler P. F., Wang H., Taylor J. S., Fontenot Q., Ferrara A., Searle S. M. J., Aken B., Yandell M., Schneider I., Yoder J. A., Volff J.-N., Meyer A., Amemiya C. T., Venkatesh B., Holland P. W. H., Guiguen Y., Bobe J., Shubin N. H., Di Palma F., Alföldi J., Lindblad-Toh K., Postlethwait J. H., The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 48, 427–437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S13
Tables S1 to S3
Legends for data files S1 to S5
References
Data files S1 to S5