ABSTRACT
In recent researches, circular RNAs (circRNAs) have been shown to exert critical functions in osteosarcoma biology. Nevertheless, the contribution of circRNAs to osteosarcoma remains largely unclear. Results indicated that expression of circATRNL1 was higher in osteosarcoma tissues and cells. The high-expression of circATRNL1 was significantly correlated with aggressive features and acted as an independent risk factor for osteosarcoma patients’ overall survival. Functionally, our findings demonstrate that circATRNL1 promotes the osteosarcoma aerobic glycolysis in vitro. Mechanistically, circATRNL1 up-regulated the expression level of LDHA, which was also targeted by miR-409-3p. Therefore, circATRNL1 exerted the accelerative roles of osteosarcoma aerobic glycolysis through miR-409-3p/LDHA axis. In conclusion, circATRNL1 promoted osteosarcoma progression by enhancing glycolysis via circATRNL1/miR-409-3p/LDHA axis, which may inspire a novel therapeutic target for osteosarcoma.
KEYWORDS: Osteosarcoma, circular rna, circATRNL1, aerobic glycolysis, ldha
1. Introduction
Osteosarcoma is the most common bone cancer among adolescents with rapid progression and high metastasis [1,2]. The 5-year survival of localized osteosarcoma is approximately 70%, which is purely 30% for metastatic osteosarcoma. Worldwide, osteosarcoma contributes to a remarkable share of cancer-related death [3]. At the pathological level, the high rate of metastasis occurrence takes the major charge for the unfavorable prognosis of osteosarcoma [4,5]. Although adjuvant chemotherapy and novel surgeries are widely utilized to cure osteosarcoma, the results remain unsatisfied [6,7]. Therefore, we should pay close attention to the osteosarcoma pathogenesis.
Circular RNA (circRNA), which is a group of transcribed RNA generated by back splicing, plays critical roles in the human tumorigenesis [8,9]. CircRNA is involved in diverse biological processes, including chromatin modification, cellular differentiation, apoptosis and metastasis. For instance, circRNA circTADA2A is highly expressed in osteosarcoma cells and tissues, where silencing attenuates the migration, invasion and proliferation of osteosarcoma [10]. An overexpressed circRNA circ_001621 promotes the osteosarcoma proliferation and migration through inhibiting matrix metallopeptidase 9 (MMP9) and cyclin-dependent kinase 4 (CDK4) via miR-578 [11]. Thus, these data suggest that circRNAs play critical role in the osteosarcoma.
In the present study, by performing circRNA sequencing (circRNA-seq), we found an up-regulated circRNA (circATRNL1, hsa_circ_0092796) in osteosarcoma. CircATRNL1 is derived from the 24–22 exon of ATRNL1 gene, therefore named as circATRNL1. CircATRNL1 acted as a sponge to absorb miR-409-3p, thereby releasing LDHA, forming an axis of circATRNL1/miR-409-3p/LDHA. Our research systematically investigated the role of circATRNL1 in osteosarcoma and discovered its deep going mechanism in the tumorigenesis. Besides, we performed the auxiliary diagnosis, including computed tomography (CT) and Magnetic Resonance Imaging (MRI), to detect the examination efficiency. Overall, the work tries to investigate the critical role of circATRNL1 in the osteosarcoma tumorigenesis, thereby investigating the therapeutic strategy for osteosarcoma.
2. Materials and methods
2.1. Osteosarcoma tissues
The current study totally enrolled 60 pairs of osteosarcoma tissue and adjacent normal specimens at Zibo Central Hospital. All included tissues were histopathologically diagnosed by two independent pathologists. The clinicopathological data of osteosarcoma patients was analyzed based on the circATRNL1 expression (Table 1). All patients were diagnosed by MR examination and CT examination. All patients provided informed consent. This clinical research was approved by the Ethics Committee of Zibo Central Hospital.
Table 1.
The clinicopathological characteristics of OS patients
| N | circATRNL1 |
p | |||
|---|---|---|---|---|---|
| High | Low | ||||
| Gender | Male | 26 | 14 | 12 | 0.497 |
| Female | 34 | 16 | 18 | ||
| Stage | I/II | 41 | 22 | 19 | 0.006* |
| III | 19 | 8 | 11 | ||
| Location | Tibia/femur | 33 | 21 | 12 | 0.395 |
| Elsewhere | 27 | 9 | 18 | ||
| Histological type | Osteoblastic | 7 | 4 | 3 | 0.632 |
| Chondroblastic | 12 | 6 | 6 | ||
| Fibroblastic | 20 | 11 | 9 | ||
| Mixed | 21 | 9 | 12 | ||
| Tumor size (cm) | <5 | 20 | 12 | 8 | 0.011* |
| ≥5 | 40 | 18 | 22 | ||
| CT/MRI auxiliary diagnosis | Checkout | 57 | 30 | 27 | 0.103 |
| No-checkout | 3 | 0 | 3 | ||
*P < 0.05 represents statistical differences.
2.2. Cell lines
Osteosarcoma cell lines (U2OS, Saos-2, MG63) and normal osteoblastic cell line (hFOB) were provided from Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in high-glucose Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Gibco) and 100 g/ml streptomycin, 100 U/ml penicillin at 37°C in humidified air containing 5% CO2.
2.3. Transfection
For the overexpression of circATRNL1, the full length circATRNL1 cDNA was cloned into vector pLCDH-ciR (Ribobio, Guangzhou, China). For the knockdown of circATRNL1, the shRNA oligonucleotides toward circATRNL1 were provided from RiboBio. Then, miRNA mimics and scramble were synthesized by GenePharma (Shanghai, China) and transfected (50 nM) using Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol.
2.4. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated from osteosarcoma cells and tissues by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) as per the manufacturers’ protocols [12]. RNA was reversely transcripted into cDNA by using GoScript Reverse Transcription System with SuperScript First-Stand Synthesis system (Invitrogen, US). RT-PCR was conducted using 7500 FAST Real-Time PCR Detection System (Applied Biosystems, Cambridge, UK). Primers were designed using computer-assisted Primer3 software (Table S1). Relative expression level = 2−ΔΔCt.
2.5. Actinomycin D and RNase R assay
Cells were treated by transcription inhibitor Actinomycin D (2 mg/mL) or control (DMSO) and then collected at indicated time points [12]. The RNA stability was analyzed using RT-qPCR. For RNase R assay, the total RNA (2 mg) was incubated at 37°C for 30 min with RNase R (Epicenter Technologies, 5 U/μg) or Mock. The remaining RNA was purified by RNeasy MinElute Cleaning Kit and then analyzed by RT-qPCR.
2.6. Apoptosis analysis
Apoptosis analysis was calculated using flow cytometry as described previously [13] using FITC Annexin V Apoptosis Detection Kit I (BD Pharmagen, USA). In brief, osteosarcoma cells were washed by cold PBS twice and resuspended in Binding buffer (100 μl). Then, FITC Annexin V (5 μl) and propidium iodide (PI, 5 μl) were administrated to cells for at room temperature 15 min in the dark. After incubation, buffer (400 μl) were administrated to cells and then apoptotic rate was detected by FACS Canto II flow cytometry (BD Biosciences).
2.7. Glucose analysis, lactate analysis and ECAR detection
Glucose and lactate levels were, respectively, measured using Glucose Uptake Colorimetric Assay Kits (Biovision, USA) and Lactate Colorimetric Assay Kits (Biovision, USA) [14]. For extracellular acidification rate (ECAR), the glycolytic capacity and the glycolysis rate of osteosarcoma cells were detected using Seahorse XF 96 Extracellular Flux Analyzer (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions.
2.8. Western blot assay
Protein isolation was extracted using radioimmunoprecipitation assay lysis buffer supplemented with phenylmethanesulfonyl fluoride (Roche Applied Science, USA). The protein concentrations were detected using BCA protein assay kit (Beyotime, China). Proteins were separated to 10–12% gels of SDS-PAGE and then to polyvinylidene fluoride membranes (PVDF, 0.22 nm, Millipore). The PVDF members carried proteins were incubated with primary antibodies (anti-LDHA, ab52488, Abcam, 1:1000) and then blocked with 5% skim milk powder (Boster, USA). Membranes were shown using Image Pro-Plus system (Media Cybernetics, Silver Spring, MD, USA). Bands were visualized and its intensity was carried out by the ImageJ software.
2.9. Subcellular fractionation
For the subcellular fractionation analysis of circATRNL1, the nuclear and cytoplasmic fragments were extracted using PARIS Kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocols.
2.10. Fluorescence In Situ Hybridization (FISH)
FISH assay using osteosarcoma cells was carried out by using the FISH Hybridization Kit (GenePharm, Guangzhou, China) according to the manufacturer’s instructions [15]. In brief, DAPI-labeled U6 probes, FAM-labeled miR-409-3p probes and Cy3-labeled circATRNL1 probes obtained from Genepharma (Shanghai, China). Nucleus was stained by 40,6-diamidino-2-phenylindole (DAPI) and images were obtained with a confocal microscope (Olympus).
2.11. Luciferase reporter assay
The interaction within circATRNL1 and miR-409-3p, or the interaction within miR-409-3p and LDHA was identified by the luciferase reporter assay. In brief, psicheck2 plasmid containing circATRNL1 or LDHA 3ʹ-UTR region was constructed, being named as circATRNL1 wild type and LDHA wild type. Correspondingly, the control mutants were also synthesized, being named as circATRNL1 mutants and LDHA mutants. 293 T cells were co-transfected with the circATRNL1 wild type or LDHA wild-type plasmids with miR-409-3p mimics (or miRNA controls). After transfection, Renilla luciferase activity was detected using Renilla Luciferase Assay System (Promega, Madison, WI).
2.12. Animal in vivo assay
All in vivo animal assays and manipulations were conducted in compliance with the guidelines, which had been approved by ethical committee of Zibo Central Hospital. Firstly, approximately 1 × 106 MG63 cells (circATRNL1 transfected cells) suspended in 100 μl PBS were injected into the flank of male BALB/c nude mice (10 mice, 15–20 g, 5 weeks). The tumor size was detected using vernier caliper every 3 days. Finally, mice were anesthetized and sacrificed after 3 weeks for tumor weight. Tumor size was calculated by width× width× length×0.52.
2.13. Statistical analysis
Statistical analyses were conducted using SPSS 20.0 software (Abbott Laboratories, Chicago, IL, USA) and GraphPad Prism (GraphPad Software, La Jolla, CA, USA). Data was analyzed using unpaired Student’s t-test or one-way analysis of variance (ANOVA). The results are presented as the mean ± SD. p-value less than 0.05 was considered statistically significant.
3. Results
Our research found that high-expression of circATRNL1 acted as an independent risk factor for osteosarcoma. Functionally, our findings demonstrate that circATRNL1 promotes the osteosarcoma aerobic glycolysis in vitro. Mechanistically, circATRNL1 up-regulated the expression level of LDHA, which was also targeted by miR-409-3p. In conclusion, circATRNL1 promoted osteosarcoma progression by enhancing glycolysis via circATRNL1/miR-409-3p/LDHA axis, which may inspire a novel therapeutic target for osteosarcoma.
3.1. Overexpression of circATRNL1 indicated the unfavorable prognosis of osteosarcoma patients
CircATRNL1 was a novel circRNA derived from the 24–22 exon of ATRNL1 gene (Figure 1(a)). In the circRNA microarray on the osteosarcoma tissue, several up-regulated or down-regulated circRNAs were discovered (Figure 1(b)). The sequential verification was tested using the Sanger sequencing. Results indicated that the junction sites’ sequences were found using the Sanger sequencing and the circATRNL1 was indeed a circular transcript (Figure 1(c)). Actinomycin D (Act D) administration indicated that circATRNL1 could resist the degradation as compared to the ATRNL1 mRNA (Figure 1(d)). Moreover, RNase R administration assay found that circATRNL1 was more stable than the ATRNL1 mRNA transcript (Figure 1(e)). In the enrolled osteosarcoma patients’ specimens, circATRNL1 expression dramatically up-regulated compared to the normal controls (figure 1(f)). In the present research, we added an extra auxiliary diagnosis (CT, MRI) for the osteosarcoma patients (Table 1). Clinically, patients with high-expression of circATRNL1 demonstrated a lower prognosis than those with low-expression of circATRNL1 (figure 1(f)). Taken together, the above findings suggested that circATRNL1 overexpression indicated the unfavorable prognosis of osteosarcoma patients.
Figure 1.
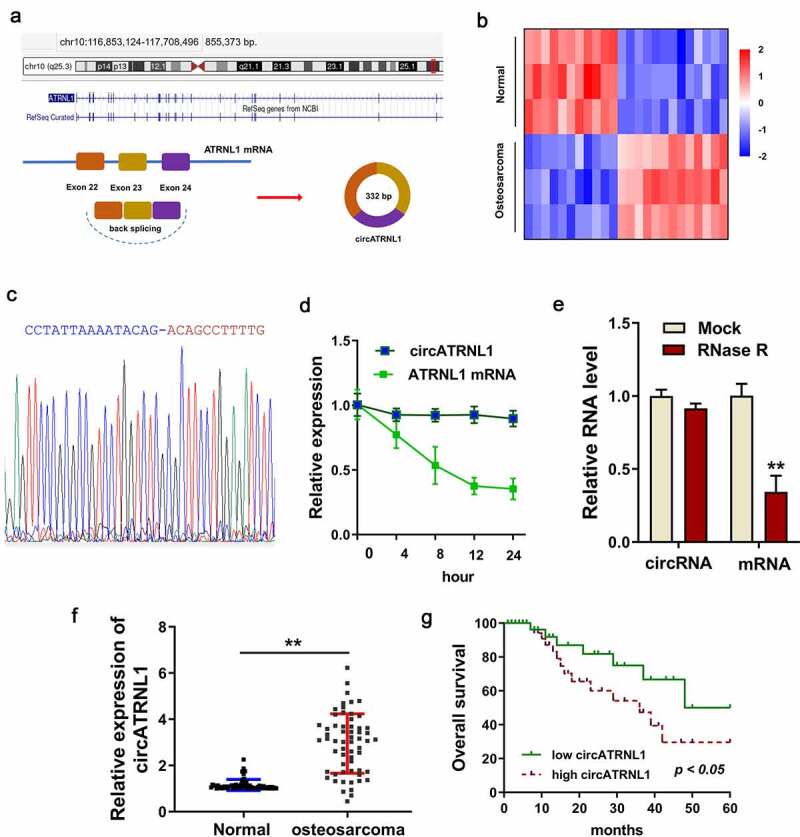
Overexpression of circATRNL1 indicated the unfavorable prognosis of osteosarcoma patients. (a) schematic diagram illustrated the genesis of circATRNL1 from the ATRNL1 gene loci through backsplicing. (b) screening analysis using the RT-PCR discovered the potential up/down-expressed circRNA in the osteosarcoma tissue and normal tissue. (c) sanger sequencing revealed the junction sites of circATRNL1 using the cDNA in the cells. (d) act D administration and (e) RNase R administration were performed to detect the stability of circATRNL1 and ATRNL1 mRNA. the levels of RNA were measured by RT-PCR. (f) in the enrolled osteosarcoma patients’ specimens, circATRNL1 expression was detected using RT-PCR. (g) clinically, the patients with high-expression of circATRNL1 demonstrated a lower prognosis than who with low-expression of circATRNL1. two-group comparison was calculated by student’s t-test. experiments were performed in triplicate. multiple group comparison was calculated by one-way analysis of variance (ANOVA). data are presented as mean ± SD. *p < 0.05, **p < 0.01
3.2. CircATRNL1 promoted the glycolytic capacity of osteosarcoma cells in vitro
Firstly, the expression of circATRNL1 in the osteosarcoma cells (U2OS, Saos-2, MG63) was detected as compared to the normal cells. Results indicated that circATRNL1 level was remarkably up-regulated in the osteosarcoma cells (Figure 2(a)). Moreover, the overexpression of circATRNL1 was constructed in the Saos-2 cells and the silencing of circATRNL1 was constructed in MG63 cells (Figure 2(b)). Apoptosis analysis found that circATRNL1 overexpression reduced the apoptotic rate and circATRNL1 knockdown accelerated the apoptotic rate of osteosarcoma cells (Figure 2(c)). Further, glucose uptake analysis and lactate production analysis revealed that circATRNL1 overexpression promoted glucose uptake and lactate production quantity (Figure 2(d)). Extracellular acidification rate (ECAR) analysis revealed that circATRNL1 overexpression accelerated the acidification rate of osteosarcoma cells (Saos-2) and circATRNL1 silencing weakened the acidification rate (MG63) (Figure 2(e)). In vivo, the MG63 cells transfected with circATRNL1 knockdown could remarkably reduce the tumor growth in mice subcutaneous injection (figure 2(f), Supplementary Figure S1). Taken together, these results showed that circATRNL1 promoted the glycolytic capacity of osteosarcoma cells in vitro, and knockdown of circATRNL1 reduced the tumor growth.
Figure 2.
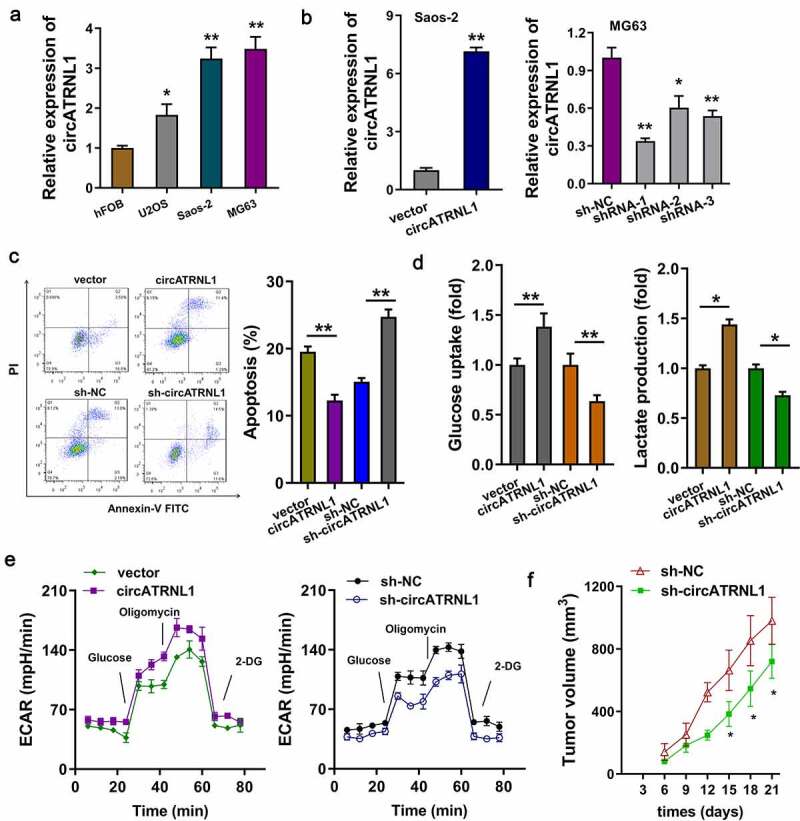
CircATRNL1 promoted the glycolytic capacity of osteosarcoma cells in vitro. (a) RT-PCR illustrated that expression of circATRNL1 in the osteosarcoma cells (U2OS, saos-2, MG63) as compared to the normal cell (hFOB). (b) cellular transfections were constructed by plasmids stable transfection for up-regulation (saos-2) and silencing (MG63). (c) apoptosis analysis by flow cytometry detected the apoptotic rate in saos-2 cells with circATRNL1 up-regulation and in MG63 with circATRNL1 silencing. (d) glucose uptake analysis and lactate production analysis uncovered glucose consumption and lactate generation with circATRNL1 up-regulation (saos-2) or silencing (MG63). (e) extracellular acidification rate (ECAR) analysis reflected the glycolytic capacity with circATRNL1 up-regulation (saos-2) or silencing (MG63). (f) in vivo mice assay indicated that the MG63 cells transfected with circATRNL1 knockdown could remarkably reduce the tumor growth in subcutaneous injection. tumor volume was calculated by detecting length and width. two-group comparison was calculated by student’s t-test. experiments were performed in triplicate. data are presented as mean ± SD. *p < 0.05, **p < 0.01
3.3. CircATRNL1 targeted miR-409-3p by miRNA sponge in osteosarcoma cells
In the initial stage, subcellular fractionation location analysis found that circATRNL1 is significantly located in the cytoplasm portion of osteosarcoma cells (Saos-2, MG63) (Figure 3(a)). Moreover, in order to discover the downstream of circATRNL1, we found that miR-409-3p might act as the downstream effector of circATRNL1. The complementary binding sites on the circATRNL1, which was combined by miR-409-3p, was marked in schematic diagram (Figure 3(b)). The co-location analysis by fluorescence in situ hybridization (FISH) illustrated that circATRNL1 and miR-409-3p were both located in the cytoplasm of osteosarcoma cells (Saos-2) (Figure 3(c)). Mutation and wild type of circATRNL1 were established for the luciferase assay. Luciferase gene reporter assay indicated that miR-409-3p could interact with circATRNL1 wild type in the co-transfection, indicating the molecular binding within miR-409-3p and circATRNL1 (Figure 3(d)). RT-qPCR analysis found that circATRNL1 overexpression reduced that level of miR-409-3p and the circATRNL1 silencing promoted the miR-409-3p expression (Figure 3(e)). Taken together, these results showed that circATRNL1 targeted miR-409-3p by miRNA sponge in osteosarcoma cells.
Figure 3.
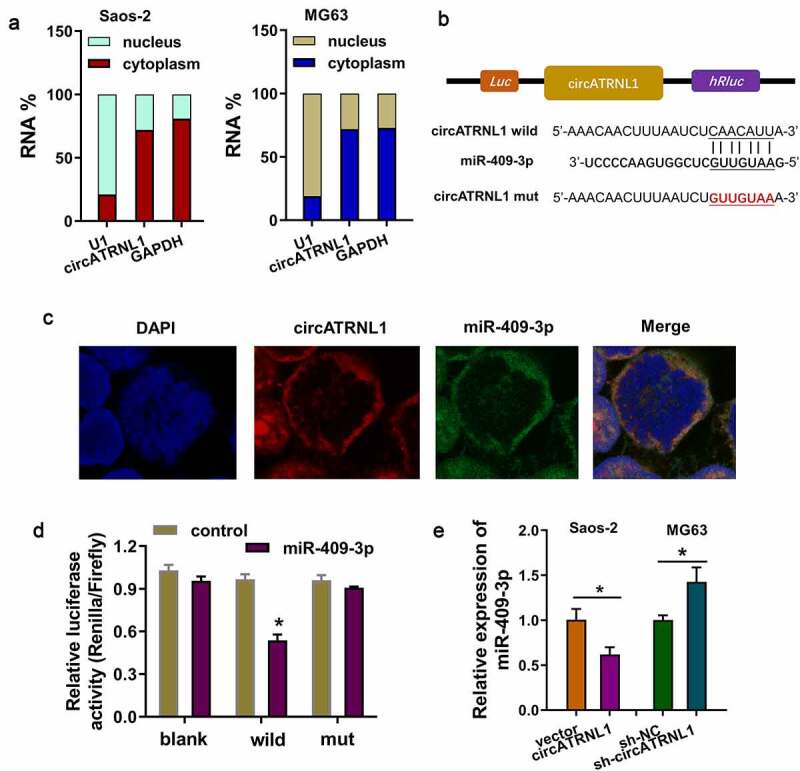
CircATRNL1 targeted miR-409-3p by miRNA sponge in osteosarcoma cells. (a) subcellular fractionation location analysis using RT-PCR revealed the circATRNL1 expression in the cytoplasm/nucleus portion of osteosarcoma cells (saos-2, MG63). (b) there were several complementary binding sites within miR-409-3p and circATRNL1. wild type and mutants were both constructed. (c) the co-location analysis by fluorescence in situ hybridization (FISH) illustrated the location of circATRNL1 and miR-409-3p in osteosarcoma cells (Saos-2). DAPI indicated the nucleus. (d) luciferase gene reporter assay indicated the activity in the co-transfection of miR-409-3p and circATRNL1 wild type/mutant. (e) RT-qPCR analysis detected the miR-409-3p expression level in circATRNL1 overexpression/silencing group. two-group comparison was calculated by student’s t-test. experiments were performed in triplicate. data are presented as mean ± SD. *p < 0.05, **p < 0.01
3.4. MiR-409-3p targeted LDHA axis in the osteosarcoma cells
Using the bioinformatics predictive manners (TargetScan, miRanda, PITA), LDHA was found to be the most promising target of miR-409-3p (Figure 4(a)). There were several complementary binding sites within miR-409-3p and LDHA (Figure 4(b)). Mutation and wild type of LDHA were established for the luciferase assay. Luciferase gene reporter assay indicated that miR-409-3p could interact with LDHA wild type in the co-transfection, indicating the molecular binding within miR-409-3p and LDHA (Figure 3(c)). RT-PCR illustrated that circATRNL1 overexpression enhanced the LDHA mRNA (Figure 3(d)) and protein (Figure 3(e)) expression, and the miR-409-3p mimics transfection reduced the LDHA mRNA and protein expression. Moreover, co-transfection of miR-409-3p and circATRNL1 recovered the LDHA mRNA and protein expression. Taken together, miR-409-3p targeted LDHA axis in the osteosarcoma cells. Forming circATRNL1/miR-409-3p/LDHA axis.
Figure 4.
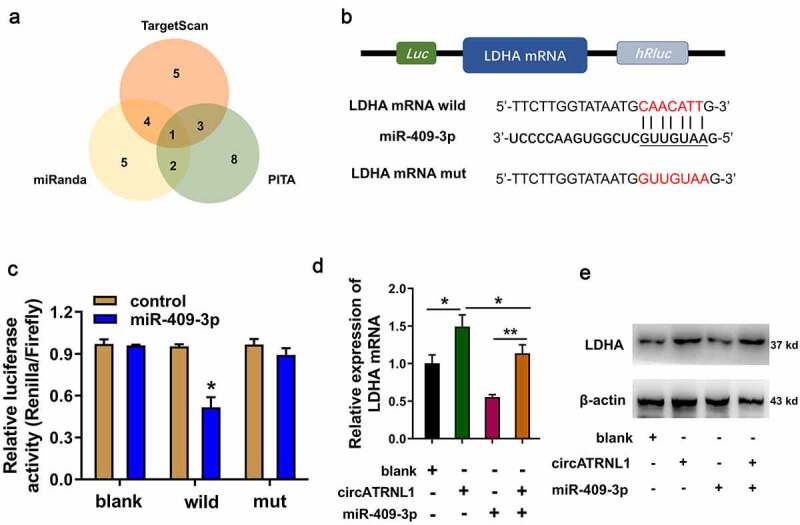
MiR-409-3p targeted LDHA axis in the osteosarcoma cells. (a) screening found that LDHA acted as the most promising target of miR-409-3p using the bioinformatics predictive manners (TARGETSCAN, miRanda, PITA). (b) mutation and wild type of LDHA were established for the luciferase assay. (c) luciferase gene reporter assay indicated the activity in the co-transfection of miR-409-3p and LDHA wild type. (d) RT-PCR illustrated the LDHA mRNA in Saos-2 cells transfected with circATRNL1 or/and miR-409-3p. (e) western blot analysis detected the LDHA protein levels in the saos-2 cells transfected with circATRNL1 or/and miR-409-3p. data are presented as mean ± SD. two-group comparison was calculated by student’s t-test. experiments were performed in triplicate. *p < 0.05, **p < 0.01
4. Discussion
In the constant exploration for osteosarcoma pathogenesis, increasing number of circRNAs have been discovered and their functions attract more and more attention [16,17]. In terms of biogenesis, circRNAs were generated from certain gene via back-splicing, which is a special splicing manner [18,19]. Circular RNAs are a novel class of non-coding RNAs that are regulating more and more pathophysiological process in the regulation of osteosarcoma.
In the present study, circATRNL1 is a novel circRNA derived from the ATRNL1 locus. In the osteosarcoma tissue and cells, circATRNL1 expression significantly up-regulated as compared to the normal controls. Clinically, the high-expression of circATRNL1 closely correlated to the poor prognosis of osteosarcoma patients. CT and MRI have auxiliary diagnostic value for osteosarcoma, and the checkout rate of CT examination is higher than that of MRI examination [20–22]. In the present results, we found that CT/MRI combined auxiliary examination in the preoperative diagnosis had high positive checkout rate. Therefore, we analyzed the correlation within checkout rate and circATRNL1 expression (high/low) in the enrolled patients’ specimens. Results showed that high circATRNL1 expression could be entirely identified by the CT/MRI auxiliary examination. However, the statistical significance was not remarkable due to the sample size. In spite of this, the CT/MRI auxiliary examination still demonstrated a conspicuous diagnostic value.
In the functional cellular experiments, our team demonstrated that circATRNL1 promoted the glycolytic capacity of osteosarcoma cells in vitro and the knockdown of circATRNL1 reduced the tumor growth. These findings illustrated that circATRNL1 might act as an oncogene in the osteosarcoma. More and more circRNAs have been reported as the oncogenic elements in the osteosarcoma tumorigenesis. For instance, circMYO10 is significantly upregulated in osteosarcoma cells and tissue samples and circMYO10 silencing inhibits the cellular proliferation and endothelial-mesenchymal transition (EMT) through complexing with chromatin remodeling and histone-modifying factor TIP60 and lymphoid enhancer factor-1 (LEF1), thus promoting histone H4K16 acetylation (H4K16Ac) in the promoter region of gene c-myc [23]. Moreover, circCAMSAP1 effectively reduces the osteosarcoma cellular growth, migration and invasion through circCAMSAP1/miR-145-5p/FLI1 (friend leukemia virus integration 1) axis [24]. Taken together, these findings suggest critical regulation of circRNAs on the osteosarcoma progression.
The aberrant energy metabolism in cancer was first described by Otto Warburg in the 1920s, thus termed as Warburg effect (aerobic glycolysis). It is widely reconsidered that abnormal metabolism acts as an essential promoting factor. There are several critical elements in the aerobic glycolysis, including rate-limiting enzymes HK2, GLUT1, LDHA at al. In present research, we found that circRNA circATRNL1 functioned as miR-409-3p sponge to reduce the expression of miR-409-3p. Due to the negative targeting of miR-409-3p toward LDHA, the repression of miR-409-3p, which was induced by circATRNL1, could recover the LDHA expression. Overall, the circATRNL1/miR-409-3p/LDHA axis promoted the osteosarcoma aerobic glycolysis (Figure 5).
Figure 5.

circATRNL1/miR-409-3p/LDHA axis promoted the osteosarcoma aerobic glycolysis
Existing evidence has indicated that circRNA regulates the osteosarcoma aerobic glycolysis. For instance, a novel circRNA circECE1 is highly expressed in osteosarcoma cells and tissues. Mechanistically, circECE1 silencing inhibits both tumor metastasis and proliferation in vitro/vivo through interacting with c-Myc to restraining c-Myc ubiquitination and degradation [25]. Overall, these findings, including ours and other scholars’ work, both suggest the role of circRNA on osteosarcoma tumorigenesis.
5. Conclusion
In conclusion, our findings in this research provide novel insights for the role of circRNAs in osteosarcoma tumorigenesis through aerobic glycolysis. The function of circATRNL1/miR-409-3p/LDHA axis presents supporting base for molecular mechanisms by which circATRNL1 accelerates the Warburg effect. Our results might help develop novel diagnostic and therapeutic targets for osteosarcoma.
Supplementary Material
Funding Statement
The authors reported that there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
No research data shared.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Garcia MB, Ness KK, Schadler KL.. Exercise and physical activity in patients with osteosarcoma and survivors. Adv Exp Med Biol. 2020;1257:193–207. [DOI] [PubMed] [Google Scholar]
- [2].Mujtaba B, Nassar SM, Aslam R, et al. Primary osteosarcoma of the breast: pathophysiology and imaging review. Curr Probl Diagn Radiol. 2020;49(2):116–123. [DOI] [PubMed] [Google Scholar]
- [3].Wu CC, Livingston JA.. Genomics and the immune landscape of osteosarcoma. Adv Exp Med Biol. 2020;1258:21–36. [DOI] [PubMed] [Google Scholar]
- [4].Egea-Gámez RM, Ponz-Lueza V, Cendrero-Torrado A, et al. Spinal osteosarcoma in the paediatric age group: case series and literature review, revista espanola de cirugia ortopedica y traumatologia. 2019;63(2):122–131. [DOI] [PubMed] [Google Scholar]
- [5].Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy, nature reviews. Endocrinology. 2017;13:480–491. [DOI] [PubMed] [Google Scholar]
- [6].Roberts RD, Lizardo MM, Reed DR, et al. Provocative questions in osteosarcoma basic and translational biology: a report from the children’s oncology group. Cancer. 2019;125(20):3514–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yang C, Tian Y, Zhao F, et al. Bone microenvironment and osteosarcoma metastasis. Int J Mol Sci. 2020;21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shi X, Wang B, Feng X, et al. circRNAs and exosomes: a mysterious frontier for human cancer, molecular therapy. Nucleic acids. 2020;19:384–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wu Y, Xie Z, Chen J, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol Cancer. 2019;18(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ji X, Shan L, Shen P, et al. Circular RNA circ_001621 promotes osteosarcoma cells proliferation and migration by sponging miR-578 and regulating VEGF expression. Cell Death Dis. 2020;11(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [12].Guo Y, Guo Y, Chen C, et al. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol Cancer. 2021;20(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shang A, Gu C, Wang W, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 2020;19:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liao M, Liao W, Xu N, et al. LncRNA EPB41L4A-AS1 regulates glycolysis and glutaminolysis by mediating nucleolar translocation of HDAC2. EBioMedicine. 2019;41:200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shao T, Pan YH, Xiong XD. Circular RNA: an important player with multiple facets to regulate its parental gene expression. Mol Ther Nucleic Acids. 2021;23:369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Saikishore R, Velmurugan P, Ranjithkumar D, et al. The circular RNA-miRNA axis: a special RNA signature regulatory transcriptome as a potential biomarker for OSCC. Mol Ther Nucleic Acids. 2020;22:352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mao X, Cao Y, Guo Z, et al. Biological roles and therapeutic potential of circular RNAs in osteoarthritis. Mol Ther Nucleic Acids. 2021;24:856–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang C, Ding R, Sun Y, et al. Circular RNA in tumor metastasis. Mol Ther Nucleic Acids. 2021;23:1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Luo Z, Chen W, Shen X, et al. CT and MRI features of calvarium and skull base osteosarcoma (CSBO). Br J Radiol. 2020;93(1105):20190653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sue M, Oda T, Sasaki Y, et al., Osteosarcoma of the Mandible: a case report with CT, MRI and scintigraphy, the Chinese journal of dental research: the official journal of the scientific section of the Chinese Stomatological Association (CSA). (2017);20:169–172. [DOI] [PubMed] [Google Scholar]
- [22].Zhang X, Guan Z. PET/CT in the diagnosis and prognosis of osteosarcoma. Front Biosci (Landmark Ed). 2018;23(11):2157–2165. [DOI] [PubMed] [Google Scholar]
- [23].Chen J, Liu G, Wu Y, et al. CircMYO10 promotes osteosarcoma progression by regulating miR-370-3p/RUVBL1 axis to enhance the transcriptional activity of β-catenin/LEF1 complex via effects on chromatin remodeling. Mol Cancer. 2019;18(1):150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chen Z, Xu W, Zhang D, et al. circCAMSAP1 promotes osteosarcoma progression and metastasis by sponging miR-145-5p and regulating FLI1 expression, molecular therapy. Nucleic acids. 2021;23:1120–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shen S, Yao T, Xu Y, et al. CircECE1 activates energy metabolism in osteosarcoma by stabilizing c-Myc. Mol Cancer. 2020;19(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No research data shared.


