ABSTRACT
Chemotherapy resistance is identified as an obstacle for breast cancer (BC) therapy, and, besides, increasing evidence indicates that long-noncoding RNAs (lncRNAs) participate in the regulation of BC adriamycin (ADR) resistance. Here, our work shows that lncRNA DLGAP1 antisense RNA 1 (DLGAP1-AS1) is up-regulated in ADR-resistant BC cells (MCF-7/ADR). Clinically, higher DLGAP1-AS1 expression was closely correlated to poorer clinical prognosis. Results showed that DLGAP1-AS1 promoted the ADR IC50 and proliferation of ADR-resistant cells. Moreover, N6-methyladenosine (m6A) methyltransferase WT1 associated protein (WTAP) binds to the m6A modified site of DLGAP1-AS1 and motivates its stability. Mechanistically, DLGAP1-AS1 sponged miR-299-3p through 3ʹ-untranslated region (3ʹ-UTR) binding, which in turn relieved the repression of WTAP and thus upregulated WTAP expression. In conclusion, above findings conclude that lncRNA DLGAP1-AS1 promotes BC ADR-resistance through WTAP/DLGAP1-AS1/miR-299-3p feedback loop.
KEYWORDS: Adriamycin resistance, N6-methyladenosine, breast cancer, DLGAP1-AS1, WTAP
1. Introduction
Breast cancer (BC) is the most common gynecological malignancy, resulting in the critical leading cancer-related death [1,2]. Although great progresses have been achieved for the BC treatment, the prognosis for BC patients remains dismal [3,4]. It has been identified that complicated risk factors contribute to the BC tumorigenesis [5]. Chemotherapy resistance could cause the treatment dilemmas of BC therapy [6,7]. Adriamycin (ADR) acts as a major first-line treatment for BC, which has reduced the mortality for patients with advanced BC and prolonged the survival. However, the resistance to ADR remarkably reduces the treatment effectiveness [8]. Although the chemotherapy resistance has a well-known association with BC recurrence, the molecular mechanism of BC ADR-resistance remains unclear.
An increasing number of literature has reported the essential roles of long noncoding RNAs (lncRNAs) in BC pathogenesis and therapy [9,10]. LncRNAs are a group of RNA transcripts with more than 200 nucleotides without proteins encoding potential [11]. More and more findings have been recognized the functions of lncRNAs in the ADR-resistance. For example, LINC01977 acts as an oncogenic driver and significantly promotes BC progression and doxorubicin chemoresistance by sponging miR-212-3p to prevent GOLM1 gene [12]. LncRNA LOC645166 downregulation promotes BC cells sensitivity to ADR both in vitro and in vivo. Moreover, lncRNA LOC645166 upregulation strengthens the tolerance of BC cells to ADR [13]. The expression of XIST is higher in ADR-resistant MDA-MB-231/ADM cells and XIST overexpression promotes the cell proliferation and inhibits the apoptosis of ADR-treated MDA-MB-231 cells via promoting ANLN expression [14]. This evidence supports the role of lncRNA in BC ADR-resistance.
N6-Methyladenosine (m6A) modification specifically refers to the methylation of N6-position adenosine. This modification is widespread in RNA, including non-coding RNAs (ncRNAs) and messenger RNA (mRNA). With regard to lncRNA, the specific m6A modification could regulate the fate of lncRNA and indirectly modulate the tumor progression. For instance, IGF2BP2 acts as an m6A reader to modify DANCR and stabilizes DANCR. Mechanistically, IGF2BP2 interacts with DANCR at 664 of DANCR to promote cellular stemness-like properties and proliferation [15]. For another example, methylated RNA immunoprecipitation sequencing (MeRIP-Seq) revealed that there is m6A modification ABHD11-AS1 and methyltransferase-like 3 (METTL3) installs the m6A modification to enhance the stability of ABHD11-AS1 [16]. Thus, these increasing findings show the critical function of m6A on lncRNA characteristic
In the current study, we found that DLGAP1-AS1 is an ADR-resistance associated lncRNA that accelerates the malignant phenotypes of BC. Moreover, DLGAP1-AS1 functions as an independent predictor for the BC patients’ survival outcome. Mechanistically, m6A methyltransferase WT1 associated protein (WTAP)-mediated DLGAP1-AS1 upregulation subsequently enhanced the BC progression and ADR-resistance. These findings unveiled the function of m6A-lncRNA, which might bring a novel superiority for BC treatment.
2. Materials and methods
2.1. Human tissue specimens
BC tissues and adjacent normal tissues (40 pairs) were collected from BC patients who underwent surgery at Zibo Central Hospital. This clinical research had been approved by the ethics committee. Informed consent had obtained from the patients. None of the patients received radiotherapy before surgery. These enrolled tissue samples were independently diagnosed by two experienced clinical pathologists. The histological type was determined according to the TNM (Tumor, Node, Metastasis) stage according to the criteria of the American Joint Committee on Cancer (AJCC, 8th edition).
2.2. Cell lines culture and transfection
BC cell lines (MDA-MB-231, BT-549, MCF-7, and MCF-7/ADR) and normal mammary cells (MCF-10A) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS, Gibco) in a humidified atmosphere with 5% CO2 at 37°C. For MCF-7/ADR cells, medium was supplemented with ADR (1 mg/mL) to maintain the drug-resistant phenotype [17]. For the stable knockdown of DLGAP1-AS1, lentiviruses were produced in HEK293T cells by co-transfection with sh-DLGAP1-AS1 and packaging plasmids GAG and VSVG. After transfection, cells were screened for stable cell clones. The sequences were shown in Table S1. Transfection was performed using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions.
2.3. Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from BC cells using RNAiso Plus (TaKaRa Bio Technology, Dalian, China). Prime Script TM RT Master Mix (TaKaRa Bio Technology, Dalian, China) was performed to reversely transcribe RNA into cDNA. The purity of RNA was determined by OD260/OD280 ratio of 1.7–2.1. qPCR was conducted using SYBR Premix Ex Taq II (TaKaRa Bio Technology, Dalian, China) on Applied Biosystems 7500 RealTime PCR System (Applied Biosystems, Foster City, CA, USA). Primers for qRT-PCR were demonstrated in Table S1. The relative expression of target genes was calculated using the 2−ΔΔCt method.
2.4. CCK-8 assay and ADR sensitivity assay
After 24 h transfection, cells were digested and seeded into 96-well plates (3 × 103/well). At 0, 24, 48, 72, and 96 h as indication, culture medium was replaced by 100 μL fresh complete medium, and then 10 μL CCK8 (TransGen Biotech, Beijing, China) was administrated at 37°C with 5% CO2 for 1 h. For the sensitivity assay [18], the absorbance was measured at 450 nm using a Spectra Max i3x Multifunctional microplate detection system (Molecular Devices, San Jose, CA).
2.5. Subcellular isolation of cytoplasmic and nuclear RNA
The nuclear or cytoplasmic fraction of DLGAP1-AS1 was isolated using PARIS Kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol [19]. The separated nuclear and cytoplasmic fractions were identified using qRT-PCR.
2.6. Luciferase activity assay
The wild-type (WT) or mutant (Mut) sequences of DLGAP1-AS1 and WTAP mRNA 3ʹ-UTR binding miR-299-3p were constructed as previously described [20]. Sequences were subcloned into pGL3 Basic vector (Promega). 293 T cells were seeded in 24-well plates for 48 hours and miR-299-3p mimics (10 μg) (RiboBio, Guangzhou, China) were co-transfected with pLuc-WT-DLGAP1-AS1 or pLuc-Mut-DLGAP1-AS1. Luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega, Madison,WI).
2.7. Methylated RNA immunoprecipitation PCR (MeRIP-qPCR)
The quantification of m6A-modified DLGAP1-AS1 was detected using MeRIP-qPCR [21]. First, the total RNA was isolated from BC cell by Trizol. In the IP buffer (140 mM NaCl, 20 mM Tris pH 7.5, 1% NP-40, 2 mM EDTA), anti-m6A antibody (A19841, Abclonal) and anti-IgG were conjugated to the protein A/G magnetic beads at 4°C overnight supplemented with RNase inhibitor and protease inhibitor. Then, the RNA was eluted using an evolution buffer from beads to detect the expression of DLGAP1-AS1 using PCR.
2.8. RNA stability
The BC cells were added with actinomycin D (5 μg/ml Act-D, Sigma, USA). At the indicated times, RNAs were collected and extracted using Trizol for real-time PCR [22].
2.9. Xenograft assay
For xenograft tumor model, 5-week-old male BALB/c nude mice (n = 10) were gained from Beijing Laboratory Animal Center (Beijing, China). BC cells (MCF-7/ADR, 1 × 107 cells/0.2 mL PBS:Matrigel = 1:3, v/v) were transfected with NC or sh-DLGAP1-AS1 and then subcutaneously implanted into the flank of mice. Every 3 days, the length and width were recorded and calculated for the volume: V = (width2× length × 0.52). At the indicated time, nude mice were sacrificed, and the parameters of tumor weight were detected. All the animal assays were approved by the Ethics Committee of Zibo Central Hospital.
2.10. Statistical analysis
Statistical analysis was performed using GraphPad Prism software 7.0 (GraphPad Software, La Jolla, CA, USA). Comparison within two groups was performed using two-tailed Student’s t-test. Pearson’s correlation was performed to detect the correlation within circRNA and other RNA. Kaplan–Meier survival curve and log-rank test were performed to depict the overall survival probability of BC patients with higher or lower DLGAP1-AS1 expression. Data were presented as mean ± standard deviation (SD) and p value <0.05 was considered statistically significant.
3. Results
Our research found that high-expression of DLGAP1-AS1 promoted the ADR IC50 and proliferation of ADR-resistant BC cells. Regarding the genesis of DLGAP1-AS1, mechanistical results illustrated that N6-methyladenosine (m6A) methyltransferase WTAP binds the m6A site of DLGAP1-AS1 and motivated its stability. Mechanistically, DLGAP1-AS1 acted as a sponge binding miR-299-3p through 3ʹ-UTR, which in turn relieved the repression of WTAP expression and thus upregulated WTAP expression. In conclusion, above findings conclude that lncRNA DLGAP1-AS1 promotes BC ADR-resistance through WTAP/DLGAP1-AS1/miR-299-3p feedback loop.
3.1. WTAP-induced DLGAP1-AS1 was overexpressed in ADR-resistant BC cells and correlated to unfavorable prognosis for BC patients
In the present study, ADR-resistant BC cells (MCF-7/ADR) were transfected with WTAP overexpression plasmids and corresponding controls. Then, screening analysis using RT-PCR revealed that numerous lncRNAs were up-regulated or down-regulated in WTAP overexpression as compared to the blank control transfection (Figure 1(a)). In the BC cell lines, RT-qPCR demonstrated that DLGAP1-AS1 expression was increased, especially in the MCF-7/ADR cells (Figure 1(b)). In the BC patients’ population, DLGAP1-AS1 expression was found to be up-regulated as compared to the normal control group (Figure 1(c), Table 1). Kaplan–Meier survival curve and log-rank test showed that the BC patients with higher DLGAP1-AS1 expression had a lower survival compared to the lower group (Figure 1(d)). In conclusion, these findings suggested that DLGAP1-AS1 was overexpressed in ADR-resistant BC cells and correlated with the unfavorable prognosis for BC patients.
Figure 1.
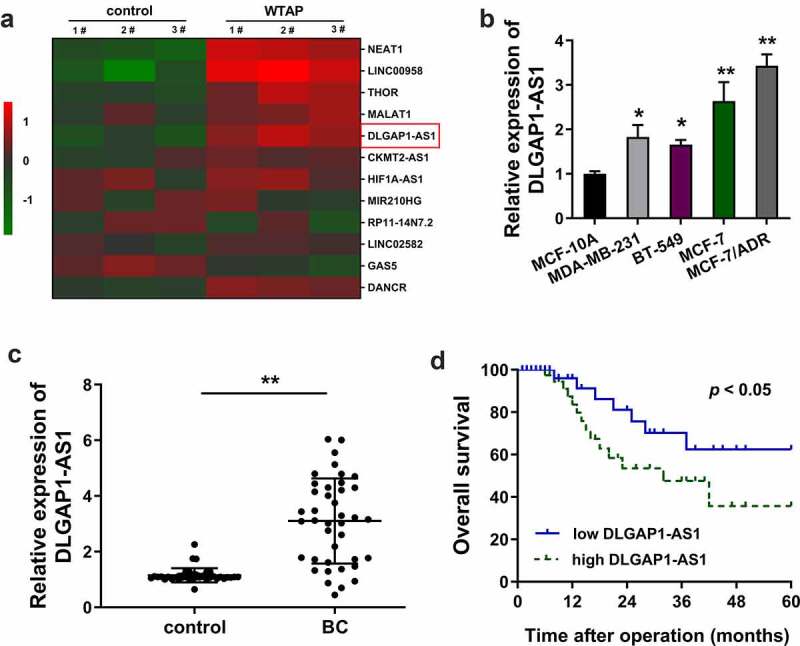
WTAP-induced DLGAP1-AS1 was overexpressed in ADR-resistant BC cells and correlated to unfavorable prognosis for BC patients. (a) Screening analysis using RT-PCR was performed in ADR-resistant BC cells (MCF-7/ADR) transfected with WTAP overexpression plasmids and corresponding controls. Heat map showed the up-regulated or down-regulated lncRNAs. (b) RT-qPCR demonstrated the DLGAP1-AS1 expression in BC cell lines, including MCF-7/ADR cells. (c) RT-qPCR demonstrated the DLGAP1-AS1 expression in BC patients’ population and normal control group. (d) Kaplan-Meier survival curve and log-rank test showed the survival in BC patients with higher DLGAP1-AS1 expression or lower group. **P < 0.01 vs control group
Table 1.
Correlation of DLGAP1-AS1 expression with BC patients’ clinicopathological feature
| DLGAP1-AS1 |
||||
|---|---|---|---|---|
| Total | low (20) | high (20) | p-Value | |
| Age (years) | ||||
| ≥60 | 21 | 9 | 12 | 0.417 |
| <60 | 19 | 11 | 8 | |
| Lymph metastasis | 0.398 | |||
| No | 19 | 8 | 11 | |
| Yes | 21 | 12 | 9 | |
| Tumor size | ||||
| ≥5 cm | 20 | 6 | 14 | 0.104 |
| <5 cm | 20 | 14 | 6 | |
| TNM | ||||
| I–II | 13 | 8 | 5 | 0.016* |
| III–IV | 27 | 12 | 15 | |
| Differentiation | 0.346 | |||
| well, moderate | 21 | 8 | 13 | |
| poor | 19 | 12 | 7 | |
*p < 0.05 represents statistical difference.
3.2. m6A methyltransferase WTAP promoted the stability of DLGAP1-AS1
As a potential origin of DLGAP1-AS1, we found that m6A methyltransferase WTAP might promote the stability of DLGAP1-AS1. MeRIP-Seq found that the m6A peaks were distributed near stop codon, including the CDS and 3ʹ-UTR of RNA (Figure 2(a)). In the 3ʹ-UTR of DLGAP1-AS1, the m6A peak was observable, indicating the remarkable m6A modification in DLGAP1-AS1 (Figure 2(b)). The m6A motif of WTAP was AAGGAC (Figure 2(c)). In the stop codon, the m6A modification site was shown as follows (Figure 2(d)). MeRIP-qPCR showed that WTAP promoted the m6A enrichment of DLGAP1-AS1 (Figure 2(e)). m6A abundance analysis found that WTAP promoted the m6A modification level in ADR-resistant BC cells (MCF-7/ADR) (figure 2(f)). RNA stability analysis found that WTAP accelerated the stability of DLGAP1-AS1 (Figure 2(g)). In summary, these data revealed that m6A methyltransferase WTAP promoted the stability of DLGAP1-AS1 in BC cells.
Figure 2.
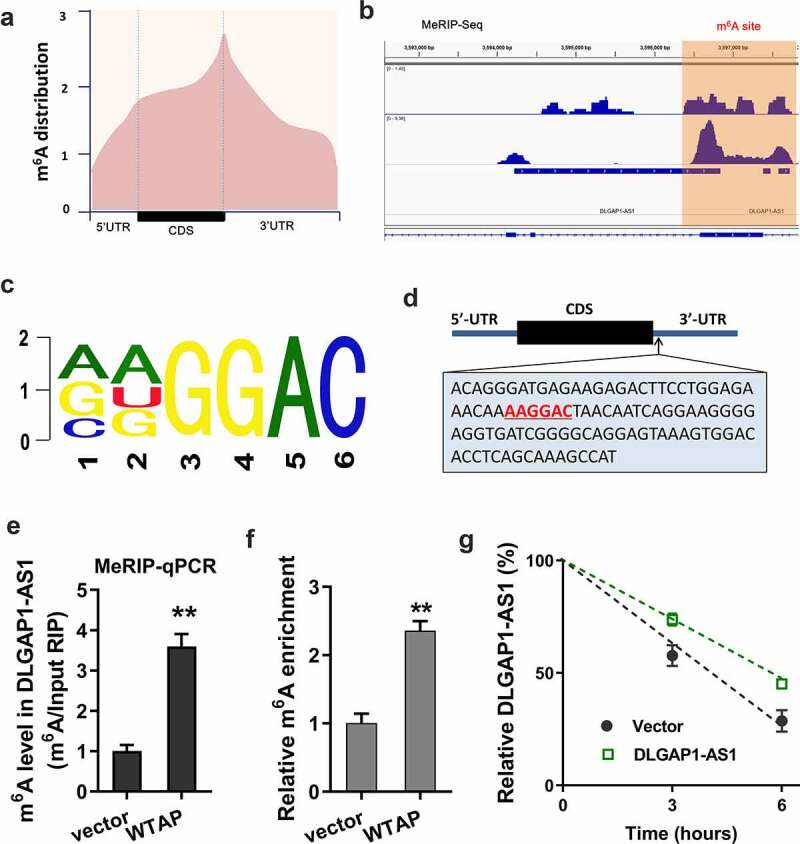
m6A methyltransferase WTAP promoted the stability of DLGAP1-AS1. (a) MeRIP-Seq found that the m6A peaks were distributed near stop codon, including the CDS and 3ʹ-UTR of RNA. (b) IGV view software unveiled the possible m6A sites on the DLGAP1-AS1. (c) The m6A motif of WTAP was AAGGAC. (d) The m6A modification sites were located in the 3ʹ-UTR of DLGAP1-AS1. (e) MeRIP-qPCR was performed to detect the m6A enrichment of DLGAP1-AS1 precipitated by the m6A antibody. (f) m6A abundance analysis detected the m6A modification level in ADR-resistant BC cells (MCF-7/ADR). (g) RNA stability analysis revealed the stability of DLGAP1-AS1. **P < 0.01 vs control group
3.3. DLGAP1-AS1 accelerated the ADR-resistance of BC cells
In order to detect the role of lncRNA DLGAP1-AS1, loss (sh-DLGAP1-AS1) and gain (DLGAP1-AS1 overexpression) of function experiments were performed on ADR-resistance BC cells (MCF-7/ADR) (Figure 3(a)). For the proliferative ability of lncRNA DLGAP1-AS1, CCK8 assay was performed and results showed that knockdown of DLGAP1-AS1 repressed the proliferation of MCF-7/ADR cells; meanwhile, DLGAP1-AS1 overexpression promoted the proliferation (Figure 3(b)). To detect the IC50 of ADR for BC cells, sensitivity analysis was carried out using the CCK8 assays. Results illustrated that knockdown of DLGAP1-AS1 repressed the ADR IC50 of MCF-7/ADR cells; meanwhile, DLGAP1-AS1 overexpression promoted the IC50 (Figure 3(c)). In vivo mice assays illustrated that knockdown of DLGAP1-AS1 repressed the tumor growth ability of MCF-7/ADR cells in vivo (Figure 3(d)). Overall, these data showed that DLGAP1-AS1 accelerated the ADR-resistance of BC cells.
Figure 3.
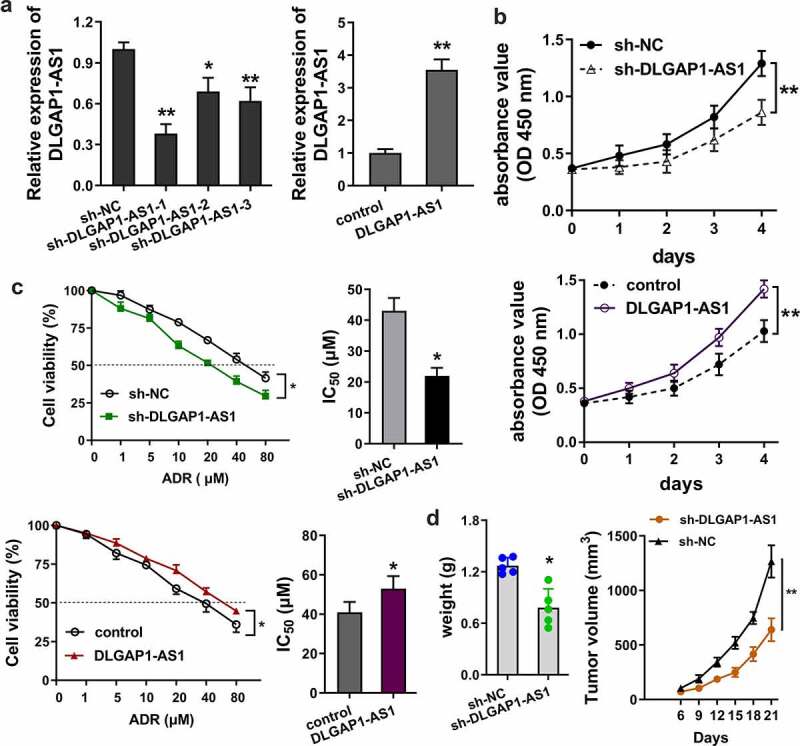
DLGAP1-AS1 accelerated the ADR-resistance of BC cells. (a) Loss (sh-DLGAP1-AS1) and gain (DLGAP1-AS1 overexpression) of function experiments were performed in ADR-resistance BC cells (MCF-7/ADR). (b) CCK8 assay was performed for the proliferative ability of lncRNA DLGAP1-AS1 on MCF-7/ADR cells. (c) ADR sensitivity analysis was e carried out using the CCK8 assays to detect the IC50 of ADR for BC cells in MCF-7/ADR cells transfected with DLGAP1-AS1 knockdown and DLGAP1-AS1 overexpression. (d) In vivo mice assays were performed to illustrate the in vivo tumor growth ability of MCF-7/ADR cells. *P < 0.05, **P < 0.01 vs control group
3.4. DLGAP1-AS1 targeted miR-299-3p/WTAP axis
Firstly, the cellular location of DLGAP1-AS1 was found to be cytoplasmic location (MCF-7/ADR) (Figure 4(a)). Bioinformatic prediction (StarBase, http://starbase.sysu.edu.cn/) inspired that miR-299-3p had the complementary sites with DLGAP1-AS1 wild-type 3ʹ untranslated regions (3ʹ-UTR) (Figure 4(b)). Luciferase reporter assays showed that DLGAP1-AS1 wild type could significantly combine with the miR-299-3p, suggesting the interaction within DLGAP1-AS1 and miR-299-3p (Figure 4(c)). Subsequently, bioinformatic prediction (StarBase) also found that miR-299-3p shared the complementary sites with WTAP mRNA 3ʹ-UTR wild type (Figure 4(d)). Analogously, luciferase reporter assays demonstrated that miR-299-3p significantly interacted with the WTAP mRNA wild type (Figure 4(e)). In MCF-7/ADR cells, RT-PCR demonstrated that WTAP mRNA expression was significantly up-regulated in the DLGAP1-AS1 overexpression transfection, which was decreased in the DLGAP1-AS1 silencing transfection (figure 4(f)). In BC patients’ samples, correlation analysis found that WTAP expression was closely correlated with the DLGAP1-AS1 expression (Figure 4(g)). Collectively, these data demonstrated that DLGAP1-AS1 targeted miR-299-3p/WTAP axis in BC.
Figure 4.
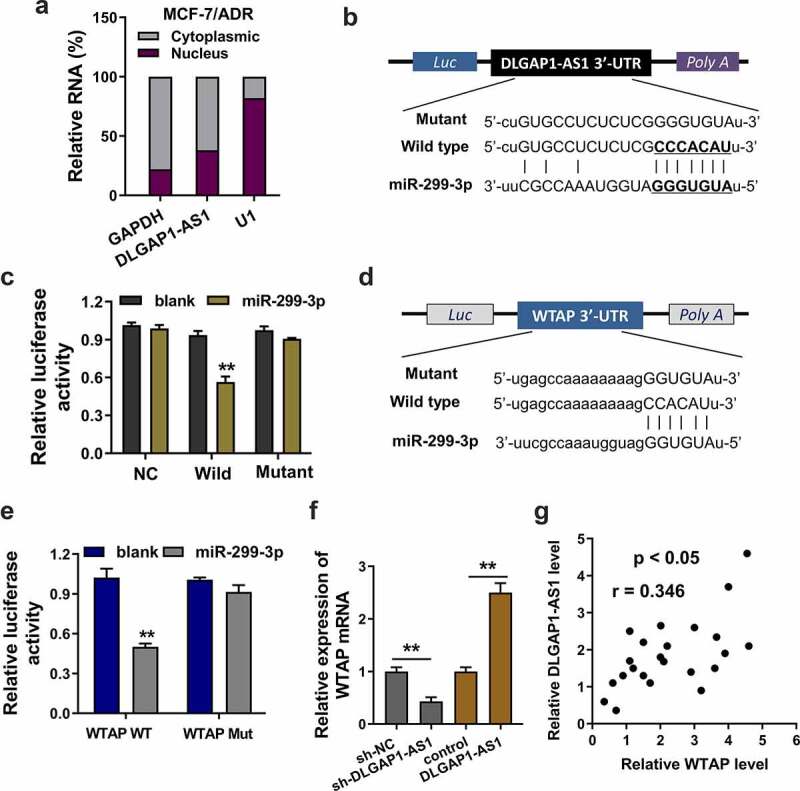
DLGAP1-AS1 targeted miR-299-3p/WTAP axis. (a) Cellular location analysis of DLGAP1-AS1 was detected using subcellular fractionation analysis. (b) Bioinformatic prediction (StarBase, http://starbase.sysu.edu.cn/) inspired the complementary sites of miR-299-3p with DLGAP1-AS1 wild-type 3ʹ-untranslated regions (3ʹ-UTR). (c) Luciferase reporter assays showed the luciferase activity in the co-transfection of DLGAP1-AS1 wild type or mutant with miR-299-3p mimics. (d) Bioinformatic prediction (StarBase) found that miR-299-3p shared the complementary sites with WTAP mRNA 3ʹ-UTR wild type. Mutant sequences were constructed. (e) Luciferase reporter assays demonstrated the luciferase activity in the co-transfection of WTAP mRNA wild type or mutant with miR-299-3p mimics. (f) RT-PCR demonstrated the WTAP mRNA expression in the DLGAP1-AS1 overexpression transfection or DLGAP1-AS1 silencing transfection in MCF-7/ADR cells. (g) Correlation analysis found the WTAP expression correlation with DLGAP1-AS1 expression in BC patients’ samples. *P < 0.05, **P < 0.01 vs control group
4. Discussion
Chemotherapy resistance is a critical obstacle for the human cancer therapy [23]. For BC, adriamycin (ADR) acts as a pivotal first-line drug; however, a remarkable ADR-resistance has been formed in the chemotherapy using ADR [24]. Recent research found that lncRNAs could participate in the regulation of BC ADR resistance.
Initially, screening analysis was carried out to detect the dysregulated lncRNAs in ADR-resistant cells (MCF-7/ADR) transfected with WTAP overexpression or control. Numerous lncRNAs were identified as up or down expressed in ADR treated BC cells. Here, the present research found that lncRNA DLGAP1-AS1 was significantly up-regulated in the BC tissue and cell lines. Given that DLGAP1-AS1 was up-regulated in the MCF-7/ADR cells, we put forward the hypothesis that DLGAP1-AS1 may modulate the ADR-resistance in BC. Functional analysis found that DLGAP1-AS1 overexpression or silencing could enhance or inhibit the IC50 of MCF-7/ADR cells using sensitivity analysis. Besides, results also confirm that DLGAP1-AS1 overexpression or silencing could promote or impair the proliferation and invasion of MCF-7/ADR cells. These data once again support the conclusion that DLGAP1-AS1 acts as an oncogene in the BC tumorigenesis.
As regarding the origin of lncRNA, we found that m6A methyltransferases may significantly accelerate the genesis of lncRNA by activating their transcription. For example, METTL3-mediated m6A modification results in the upregulation of lncRNA LINC00958 by stabilizing the stability of LINC00958 transcript. LINC00958 aggravates the HCC malignant phenotype through sponging miR-3619-5p to upregulate hepatoma-derived growth factor (HDGF) [25]. For another example, m6A reader YTHDF3 facilitates the m6A-modified lncRNA GAS5 degradation, and the GAS5 expressions is negatively correlated with YTHDF3 protein level in colorectal cancer [26]. Therefore, these data supported the phenomenon that m6A methyltransferase activates the lncRNA genesis in BC.
Moreover, lncRNA could regulate the BC tumorigenesis through diverse patterns, including competing endogenous RNA, transcriptional initiation complex and transcription inhibition [27–29]. In this research, we found that DLGAP1-AS1 was mainly located in the cytoplasmic portion of BC cells. Thus, we put forward the hypothesis that DLGAP1-AS1 might function as miRNA sponge to absorb miRNAs, thereby regulating the BC ADR resistance. Bioinformatic prediction inspired that DLGAP1-AS1 might target the miR-299-3p through 3ʹ-UTR binding. Subsequently, miR-299-3p also targeted the WTAP mRNA 3ʹ-UTR. Collectively, DLGAP1-AS1/miR-299-3p/WTAP axis was confirmed in the BC (Figure 5). In human cancers, m6A/lncRNA was found to be an oncogenic factor.
Figure 5.
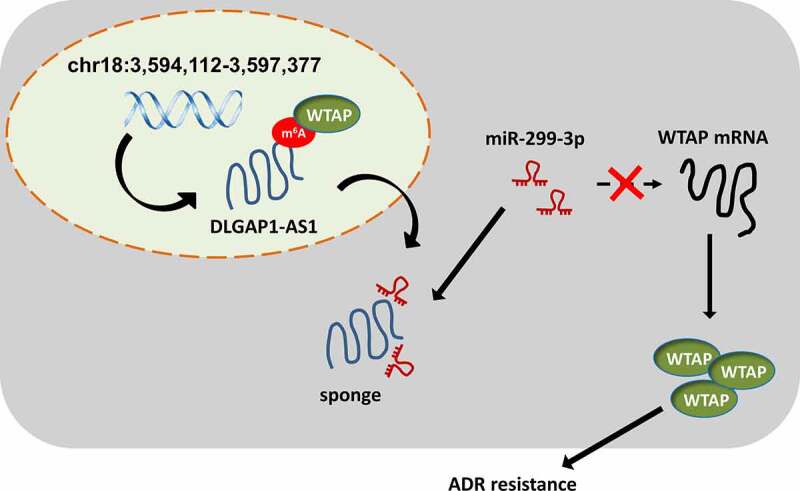
WTAP/DLGAP1-AS1/miR-299-3p positive feedback loop promotes the BC ADR-resistance
5. Conclusion
In conclusion, this research uncovered a novel role of DLGAP1-AS1 in BC ADR-resistance through WTAP/DLGAP1-AS1/miR-299-3p feedback loop. The overexpression of DLGAP1-AS1 was closely correlated with BC development and significantly indicated the prognosis. These findings broaden the knowledge regarding the DLGAP1-AS1 roles in BC and construct potential therapeutic strategies against BC treatment.
Supplementary Material
Funding Statement
The author(s) reported that there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
No research data was shared.
Supplementary materials
Supplemental data for this article can be accessed here
References
- [1].Anestis A, Zoi I, Papavassiliou AG, et al. Androgen receptor in breast cancer-clinical and preclinical research insights. Molecules. 2020;25(2):358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Reddy TP, Rosato RR, Li X, et al. A comprehensive overview of metaplastic breast cancer: clinical features and molecular aberrations. Breast Cancer Res. 2020;BCR 22(1):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].De A, Kuppusamy G.. Metformin in breast cancer: preclinical and clinical evidence. Curr Probl Cancer. 2020;44(1):100488. [DOI] [PubMed] [Google Scholar]
- [4].Ezzati M, Yousefi B, Velaei K, et al. A review on anti-cancer properties of quercetin in breast cancer. Life Sci. 2020;248:117463. [DOI] [PubMed] [Google Scholar]
- [5].Yin L, Duan JJ, Bian XW, et al. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020;BCR 22(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].van der Spek YM, Kroep JR, Tollenaar R, et al. Chemotherapy resistance and stromal targets in breast cancer treatment: a review. Mol Biol Rep. 2020;47(10):8169–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Spring LM, Wander SA, Andre F, et al. Cyclin-dependent kinase 4 and 6 inhibitors for hormone receptor-positive breast cancer: past, present, and future. Lancet. 2020;395(10226):817–827. [DOI] [PubMed] [Google Scholar]
- [8].How CW, Ong YS, Low SS, et al. How far have we explored fungi to fight cancer? Semin Cancer Biol. 2021,S1044-579X(21)00059-6. [DOI] [PubMed] [Google Scholar]
- [9].Wang M, Yu F, Chen X, et al. The underlying mechanisms of noncoding RNAs in the chemoresistance of hepatocellular carcinoma. Mol Ther Nucleic Acids. 2020;21:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang L, Wang H.. Long non-coding RNA in CNS injuries: a new target for therapeutic intervention. Mol Ther Nucleic Acids. 2019;17:754–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Guo T, Gong C, Wu P, et al. LINC00662 promotes hepatocellular carcinoma progression via altering genomic methylation profiles. Cell Death Differ. 2020;27(7):2191–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li Z, Li Y, Wang X, et al. LINC01977 promotes breast cancer progression and chemoresistance to doxorubicin by targeting miR-212-3p/GOLM1 axis. Front Oncol. 2021;11:657094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zheng R, Jia J, Guan L, et al. Long noncoding RNA lnc-LOC645166 promotes Adriamycin resistance via NF-κB/GATA3 axis in breast cancer. Aging (Albany NY). 2020;12(10):8893–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang M, Wang F, Xiang Z, et al. LncRNA XIST promotes chemoresistance of breast cancer cells to doxorubicin by sponging miR-200c-3p to upregulate ANLN. Clin Exp Pharmacol Physiol. 2020;47(8):1464–1472. [DOI] [PubMed] [Google Scholar]
- [15].Hu X, Peng WX, Zhou H, et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27(6):1782–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Xue L, Li J, Lin Y, et al. m6A transferase METTL3-induced lncRNA ABHD11-AS1 promotes the Warburg effect of non-small-cell lung cancer. J Cell Physiol. 2021;236(4):2649–2658. [DOI] [PubMed] [Google Scholar]
- [17].Chen X, Wang YW, Gao P. SPIN1, negatively regulated by miR-148/152, enhances Adriamycin resistance via upregulating drug metabolizing enzymes and transporter in breast cancer. J Exp Clin Cancer Res. 2018;CR 37(1):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Advani P, Hoyne J, Moreno-Aspita A, et al. High-sensitivity troponin T and NT-proBNP kinetics in breast cancer chemotherapy. Chemotherapy. 2017;62(6):334–338. [DOI] [PubMed] [Google Scholar]
- [19].Lin J, Liao S, Li E, et al. circCYFIP2 acts as a sponge of miR-1205 and affects the expression of its target gene E2F1 to regulate gastric cancer metastasis. Mol Ther Nucleic Acids. 2020;21:121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yang J, Lian Y, Yang R, et al. Upregulation of lncRNA LINC00460 facilitates GC progression through epigenetically silencing CCNG2 by EZH2/LSD1 and indicates poor outcomes. Mol Ther Nucleic Acids. 2020;19:1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [21].Li XC, Jin F, Wang BY, et al. The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of CYR61 mRNA. Theranostics. 2019;9(13):3853–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Guo Y, Guo Y, Chen C, et al. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol Cancer. 2021;20(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tan KL, Chia WC, How CW, et al. Benchtop isolation and characterisation of small extracellular vesicles from human mesenchymal stem cells. Mol Biotechnol. 2021;63:780–791. [DOI] [PubMed] [Google Scholar]
- [24].Nedeljković M, Damjanović A. Mechanisms of chemotherapy resistance in triple-negative breast cancer-how we can rise to the challenge. Cells. 2019;8(9):957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zuo X, Chen Z, Gao W, et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020;13(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ni W, Yao S, Zhou Y, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer. 2019;18(1):143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chi C, Wang W, Wang W, et al. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8(9):1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yousefi H, Maheronnaghsh M, Molaei F, et al. Long noncoding RNAs and exosomal lncRNAs: classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene. 2020;39(5):953–974. [DOI] [PubMed] [Google Scholar]
- [29].Li X, Jin F, Li Y. A novel autophagy-related lncRNA prognostic risk model for breast cancer. J Cell Mol Med. 2021;25(1):4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No research data was shared.


