ABSTRACT
Diabetic retinopathy (DR) is one of the severe microvascular complications of diabetes. The protective effects of FA on retinal vascular endothelial cells against high glucose levels involve in multiple aspects in DR; however, the underlying mechanism is not fully elucidated. In present study, we investigated the transcriptome as well as genome-wide DNA methylation and hydroxymethylation signature in human retinal microvascular endothelial ACBRI 181 cells cultured within high glucose (HG) medium supplemented with or without FA by RNA-seq, MeDIP-seq, and hMeDIP-seq. Total 3308 differential expressed genes (DEGs) were involved in multiple biological processes and molecular functions containing angiogenesis, inflammation, S-adenosyl methionine metabolism, and hypoxia response. Moreover, the global DNA methylation and hydroxymethylation in ACBRI 181 cells with FA treatment were both compromised compared to HG. Combined with transcriptome data, four subclusters of DEGs with hyper- or hypomethylated promoters were further verified. Unexpectedly, promoters of these 487 genes all displayed a pattern of increased DNA hydroxymethylation. Furthermore, hyperglycemia rat model was established and administered with FA. The DNA methylation and hydroxymethylation changes of selected target genes COL1A1, ITGA7, MMP-14, and VEGFB confirmed by MeDIP-qPCR were consistent with the results in human ACBRI 181 cells. Finally, the presence of activated DNMT1 and TET2 induced by FA was determined in ACBRI 181 cells and hyperglycemia rat. Taken together, this research provided a resource of expression and epigenetic profiles in retinal microvascular endothelial cell, emphasizing a pharmacological mechanism of FA on DNA methylation and hydroxymethylation regulation in retinal microvessel cells of DR.
KEYWORDS: Diabetic retinopathy, folic acid, DNA methylation, DNA hydroxymethylation
Introduction
Diabetes mellitus is defined as the chronic hyperglycemia caused by defects in insulin secretion or insulin action. Type 2 DM has become a severe public health problem accounting for more than 90% worldwide cases. The consequences of uncontrolled glycemic lead to long-term damage, dysfunction, and failure of multiple organs, especially the eyes, kidneys, nerves, heart, and blood vessels [1]. Diabetic retinopathy (DR) is one specific microvasculature complication of DM that affects retinal function and results in severe blindness and relevant retinal diseases [2]. Multifactors including end products of glycosylation, inflammation [3], cytokines and chemokines [4], disruption of proliferator-activated receptor [5], growth factors [6] as well as oxidative stress [7,8] have been implicated in the pathophysiology of diabetic retinopathy.
Folic acid (FA) has been widely used in clinic for DR therapy. Many clinical studies have reported that the patients with proliferative or nonproliferative DR both displayed extremely low FA plasma levels [9]. Correspondingly, FA exerts a protective effect on DR via inhibition of angiogenesis, inflammation and oxidative stress [10]. Biochemically, FA is a form of water-soluble vitamin B, and cannot be only obtained from food. FA participates in the metabolism of purine synthesis and DNA or protein methylation in vivo. One potential role of folate in the DNA methylation process has been acknowledged [11]. Methylation most commonly occurs at cytosines within a 5’-CpG-3’ dinucleotide in human genome. This process is carried on via S-adenosylmethionine (SAM) that is enzymatically transferred a methyl group to the five position of cytosine to generate 5-methylcytosine (5mC) in genomic DNA by DNA methyltransferases (DNMTs). Reversibly, active DNA demethylation in mammalian cells can happen in the absence of DNA replication and cell division. One mechanism of active demethylation involves indirect removal of the methyl group via successive oxidation (5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC)) catalyzed by ten-eleven translocation (TET) family [12]. FA is metabolized to 5-methyltetrahydrofolate first, then converted to tetrahydrofolate through the methionine synthase reaction. This reaction is key to maintaining the flux of methyl groups for the remethylation of homocysteine to methionine via the vitamin B-12–dependent methionine synthase reaction, whereas methionine is the substrate for SAM or AdoMet, a cofactor and methyl group donor for numerous methylation reactions including the methylation of DNA methylation. The 1-carbon pathway, SAM is the major regulator of FA-dependent homocysteine remethylation because it is a potent inhibitor of methylenetetrahydrofolate reductase. Therefore, FA contributes to maintenance of the stability and homeostasis of intracellular DNA methylation and demethylation. However, the association of global DNA methylation with folate status remains controversial in previous disease studies [13–16]. It is noteworthy that low folate status can lead to both hypo- and hypermethylation, resulting in misregulation of this complex system, and the mechanism underlying the therapeutic effect of FA is not fully elucidated.
In current study, we investigated the role of DNA methylation and hydroxymethylation affected by FA in human retinal microvascular endothelial ACBRI 181 cells in vitro and hyperglycemia rat model in vivo, and attempted to unmask the epigenetic regulation on gene expression induced by FA against DR.
Materials and methods
Cell culture
Human primary retinal microvascular endothelial ACBRI 181 cells purchased from Cell Systems Corporation (Kirkland, WA, USA) were cultured in Dulbecco’s modified Eagle medium (DMEM)-F12 supplemented with 20% fetal bovine serum. Confluent cells from the fourth to sixth passage were used for the consequent experiments. For mimicking FA-treated hyperglycemia retinal endothelial cell model, 20 mM D-glucose was added in medium for 96 h first followed by the additional 0.5 μM FA (F8758, Sigma-Aldrich, St Louis, MO, USA) for another 48 h incubation.
Animal study
Adult male Sprague-Dawley rats (200–250 g) fed with food and water ad libitum freely were randomly divided into three groups: hyperglycemia (HG) group (n = 20) only received 50 mg/kg streptozotocin (V900890, Sigma) via intraperitoneal injection per month for total three months; HG-FA group (n = 20) intraperitoneally administered 2 mg/kg FA for 20 days after HG model establishment; negative control (NC) group (n = 10). The rats whose index of blood glucose stably more than 16.7 mmol/L and body weight significantly less than normal control (n = 20) were considered as diabetic model.
Retinal microvascular endothelial cells were isolated as previously described [17]. Rats were euthanized by excessive sodium amobarbital. Eyes were removed and temporarily placed in a dissection pad containing cold PBS. One forceps stabilized the eye by holding the optic nerve, and another forceps pierced through the anterior chamber at the connection between the cornea and the sclera. The sclera was then gently stripped away from the retinal tissue, followed by the removal of vitreous body and the unattached vessels of the hyaloid plexus. Retinal tissue was transferred into new tubes and digested into single cell suspension by 1 mg collagenase type II (#100-0678, Stemcell Technologies, Vancouver, BC, Canada) at 37°C water bath for 30 min, pipetting the digestion mixture up and down every 5 min. After 300 g centrifugation, the supernatant was removed, and the cell pellet was re-suspended by PBS for the subsequent flow cytometry (FACS) assay. Antibodies of APC-CD31 (#551,262, BD Biosciences, Franklin Lakes, NJ, USA) and V450-CD45 (#560,501, BD Biosciences), as well as propidium iodide (Beyotime Biotechnology, Shanghai, China) were used in FACS study. CD31+/CD45− endothelial cell population were considered as retinal endothelial cells. All experimental procedures involving animals were approved by the Institutional Animal Care and Use Committee of Zibo Municipal Hospital.
RNA sequencing (RNA-seq)
1 × 107 ACBRI 181 cells were stored in 1 ml TRIZOL (Thermo Fisher Scientific, Waltham, MA, USA) and grinded in liquid nitrogen, and were added 100 μl chloroform and fully mixed, then centrifuged with highest speed at 4℃ for 15 min. The upper layer was transferred into a new tube, and added the same volume of isopropanol, and centrifuged with highest speed at 4℃ for 10 min. The precipitate was washed by 75% cold ethanol, and dissolved by appropriate volume of DEPC water. The concentration and quality of RNA was measured by Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA) and Agilent bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). About 5 μg of RNA in each group were used for library preparation by NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) following manufacturer’s instructions and were sequenced on an Illumina HiSeq platform 2500 with paired-end 150 bp sequencing (PE10).
The raw data was trimmed adapters and filter out low-quality reads using Trimmomatic, and checked the quality of clean reads using FastQC. Next, clean reads were aligned to the latest mouse genome assembly GRCh38 using Hisat2. The transcripts were assembled and the expression levels were estimated with FPKM values using the StringTie algorithm with default parameters. Differential mRNA and lncRNA expression among the groups were evaluated using an R package Ballgown (log2 fold change (FC) >1 or <-1, p < 0.01), and the significance of differences by the Benjamini & Hochberg (BH) p-value adjustment method were computed. Gene annotation was described by Ensembl genome browser database (http://www.ensembl.org/index.html). The R package ClusterProfiler was used to annotate the differential genes with gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Methylated/hydroxymethylated DNA immunoprecipitation sequencing (MeDIP/hMeDIP-seq)
MeDIP and hMeDIP were performed as described [18]. In brief, genomic DNA of 1 × 107 ACBRI 181 cells was extracted and sheared into 200–500 bp fragments by sonication. The fragmented DNA was immunoprecipitated using antibodies of anti-5-methylcytosine (#91,311, Activemotif, Carlsbad, CA, USA) or anti-5hmC (#39,999, Activemotif). For qPCR assay, primary antibodies of DNMT1 (NB100-56,519, Novus, Centennial, CO, USA), 3A (NB120-13,888, Novus) and 3B (NB300-516, Novus) as well as TET1 (NBP2-15,135, Novus), 2 (NBP2-32,104, Novus), and 3 (NBP2-20,602, Novus) were used. Input or immunoprecipitated DNA was constructed the library using NEBNext Ultra End Repair/dA-Tailing Module (NEB), and carried out sequencing on Illumina HiSeq 2500 with paired-end 50 bp sequencing (PE50).
Trim Galore v0.5.0, Cutadapt v.1.18 (nondefault parameters: – max-n 0 – minimum-length 35) and Trimmomatic v0.38 (nondefault parameters: SLIDINGWINDOW:4:15 LEADING:10 TRAILING:10 MINLEN:35) were used to filter adapters, short reads (length < 35 bp), and low-quality reads. FastQC (with default parameters) was used to ensure high reads quality. Trimmed reads of clean data were aligned to reference genome (assembly GRCm38) using Bowtie2 v2.3.4.1 (with default parameters) and analyzed DNA methylation and hydroxymethylation profiles using methylKit package. Aligned reads with CCGG tag at 5ʹ end were counted. Differentially methylated regions (DMRs) were identified by MethylKit [19], which used a sliding window approach. The window was 200 bp, and the step length was 200 bp. The logistic regression was applied to detect significant DMRs and the screened criteria as methylation difference of ≥10% and p ≤ 0.05. After DMRs were identified, differentially methylated genes (DMGs) located in DMRs were characterized.
Quantitative polymerase chain reaction (qPCR)
Libraries for RNA-seq or (h)MeDIP-seq were treated as template. qPCR was performed using CFX Fast real-time PCR system (Bio-Rad Laboratories, Hercules, CA, USA). The following cycle parameters were used for all experiments: 30 s at 94°C for predenaturation, 20 s at 94°C, 30 s at 60°C, and 30 s at 72°C for total 45 cycles. Table S1 shows the sequences for all primer sets used in these experiments.
Activities of enzymes
The enzyme activities of DNMTs and TETs were quantified in the nuclear fraction (5–15 μg protein) using EpiQuik™ DNA Methyltransferase Activity/Inhi bition (#P-3001, EPIGENTEK, Farmingdale, NY, USA) and Epigenase 5mC-Hydroxylase TET Activit y/Inhibition Assay Kits (#P-3087, EPIGENTEK) in accordance with the manufacturer’s instructions.
Results
To investigate the effects of FA on genomic DNA methylation and hydroxymethylation in retinal microvascular endothelial cells, we prepared human cell model of hyperglycemia and FA treatment, then conducted RNA-seq, MeDIP-seq and hMeDIP-seq to figure out the DNA methylation and hydroxymethylation profiles undergoing FA induction. The further epigenetic mechanism was verified in hyperglycemia rat models treated with FA.
Transcriptional profiling of FA-induced retinal endothelial cells against high glucose
To examine the molecular effect of FA on retinal microvascular endothelium, we conducted RNA-seq to compare the different expressed genes (DEGs) in human primary retinal microvascular endothelial ACBRI 181 cells induced by FA. Appropriate 235.1 M reads of RNA-seq transcriptomic data was generated. In RNA-seq data, 93.1% of the reads were mapped to human genome with an average of 65.2 M aligned reads per sample. Fragments per kilobase of exon model per million mapped fragments (FPKM) values of 13,227 genes more than one were defined as positive expression in all six samples. Furthermore, the strong correlations between two biological duplications in each group indicated a good quality of biological materials and libraries (Figure S1).
Total 1,247 differential expressed genes (DEGs) (FC >1 or < −1, p < 0.01) were observed, in which 867 genes were highly expressed while 380 genes were reduced in high glucose (HG) group compared to negative control (NC), whereas 3,308 differential expressed genes (DEGs) were observed (FC >1 or < −1, p < 0.01), in which 731 genes were highly expressed while 2,577 genes were silenced in FA treatment compared to control (Figure 1a). Here, we majorly paid attention to those intersected 1048 DEGs between HG vs. NC and FA vs. HG, which were involved in multiple biological processes and molecular functions containing angiogenesis, inflammation, S-adenosyl methionine metabolism and hypoxia response (Figure 1b, C). We observed that a large number of genes associated with angiogenesis including ALDH3A1, COL1A1, FGF23, ITGA7, MMP14, THBS1, and VEGFB were all upregulated in HG while decreased in FA treatment (Figure 1d). Taken together, we characterized the overall transcriptional profiling of ACBRI 181 cells under the stimulation of HG and induction by FA.
Figure 1.
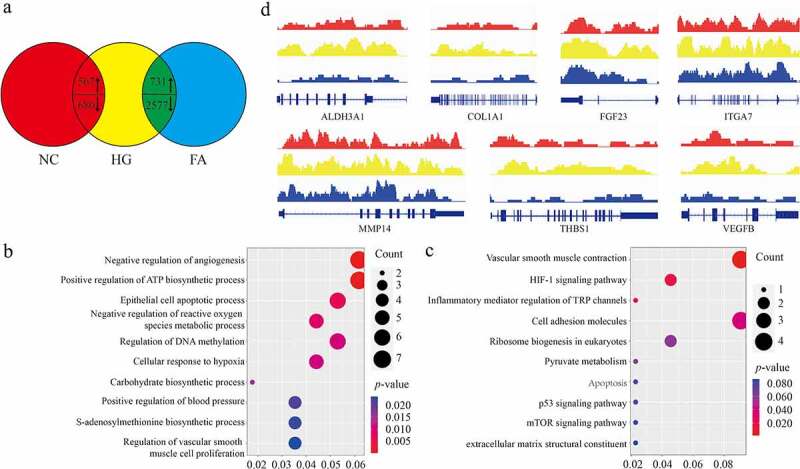
Expression profile of ACBRI 181 cells affected by HG and FA. (a) Venn diagram view of DEGs compared among NC, HG and FA (log2 FC >1 or <-1, p < 0.01). (b) Biological process of GO and (c) KEGG analysis of the involved enriched functions. (d) Gene browser views of transcription of ALDH3A1, COL1A1, FGF23, ITGA7, MMP14, THBS1, and VEGFB. DEG: differential expressed genes; NC: negative control; HG: high glucose/hyperglycemia; FA: folic acid; FC: fold change; GO: gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes
DNA methylation and hydroxymethylation landscape in retinal endothelial cells affected by FA
For the read alignments of every sample, the average of 385,559 annotated CpG sites mapped on human genome occupying 85.2% reads were obtained in MeDIP-seq data, and the average of 410,572 annotated CpG sites mapped on human genome occupying 79.6% reads were obtained in hMeDIP-seq data. For the variance of biological duplication, the Pearson Correlation Coefficients between two biological duplications in each group were all more than 95% (Figure S1). The high unique alignment and stability of experimental repeatability both indicated a good quality of biological materials we harvested.
The global DNA methylation and hydroxymethylation particularly on promoter regions in ACBRI 181 cells were increased in HG compared to NC, while compromised in FA compared to HG (Figure 2a). In current case, we focused on these CpG loci only on gene promoter regions, which largely contributed to transcriptional regulation. Combined with RNA-seq data, four subclusters of DEGs with highly methylated and hydroxymethylated promoters (subcluster A), highly methylated and lowly hydroxymethylated promoters (subcluster B), lowly methylated and highly hydroxymethylated promoters (subcluster C) as well as lowly methylated and hydroxymethylated promoters (subcluster D) were further verified (Figure 2b). A large proportion (2,782/3,308) of DEGs overlapping with subclusters B and C displayed an opposite tendency between transcription and DNA methylation. Unexpectedly, we also observed that 487 genes expression such as COL1A1, MMP14, and ITGA7 were shown an abnormal variance that the trends of transcription, DNA methylation and hydroxymethylation at promoters were positive correlated with each other (Pearson correlation coefficient value = 0.297) by comparison between FA and HG (Figure 2c).
Figure 2.
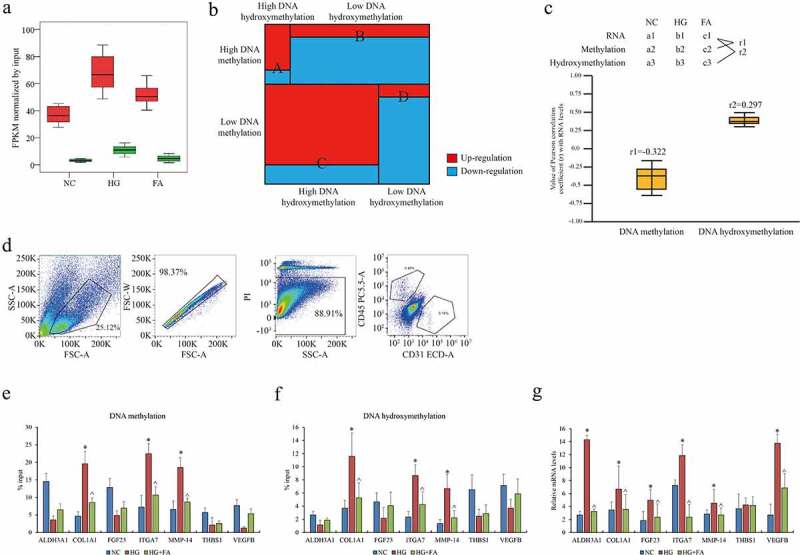
Genome-wide DNA methylation and hydroxymethylation atlas in retinal epithelium. (a) The distribution of DMR (red) and DHMR (green) regions in genomic contexts in ACBRI 181 cells of NC, HG and FA in vitro (log2 FC > 1 or < −1, FDR < 10−4). (b) The subclusters of DEGs with DMR and DHMR. (c) Pearson correlation of gene expression with DNA methylation and DNA hydroxymethylation. The coefficient value of each gene is calculated and averaged. (d) Isolation of primary rat retinal endothelial cells by positive CD31 and negative CD45. (e) DNA methylation and (f) hydroxymethylation at promoters of the target genes and (g) their transcription including ALDH3A1, COL1A1, FGF23, ITGA7, MMP14, THBS1, and VEGFB in rat retinal endothelial cells. qPCR was performed three individual experiments. ‘*’ and ‘^’ represent p value less than 0.05 by comparison between NC and HG, respectively. DMR: differential methylated regions; DHMR: differential hydroxymethylated regions; FPKM: fragments per kilobase of exon model per million mapped fragments; NC: negative control; HG: high glucose/hyperglycemia; FA: folic acid
To validate this epigenetic regulation in vivo, HG rat model was established and administered with FA. CD31+/CD45- retinal endothelial cells were isolated by FACS (Figure 2d) as previously described [17]. The changes of DNA methylation and hydroxymethylation on promoters of the selected target genes ALDH3A1, COL1A1, FGF23, ITGA7, MMP14, THBS1, and VEGFB confirmed by MeDIP- and hMeDIP-qPCR were consistent with the results in human ACBRI 181 cells (Figure 2e, f). We found that DNA methylation and hydroxymethylation were both positively correlated with transcription in COL1A1, ITGA7, and MMP14. Additionally, transcriptional signatures of these genes in HG rat model were verified by qPCR assay. Except THBS1, all genes were significantly upregulated in HG whereas down-regulated in FA addition (Figure 2g). Given genes expression indicated a nonessential epigenetic machinery of DNA methylation and hydroxymethylation on transcriptional regulation. Collectively, the landscapes of genome-wide DNA methylation and hydroxymethylation presented a complicated epigenetic diversity of gene expression regulation in retinal endothelial cells affected by HG and FA in DR in vitro and in vivo.
The effect of DNMT1 and TET2 on DNA methylation and hydroxymethylation in retinal endothelial cells
Finally, the enzymes contributing to this epigenetic event were investigated in this system. The activities of DNMT1, DNMT3A, and DNMT3B for DNA methylation as well as TET1, TET2 and TET3 for DNA hydroxymethylation were examined. Consistent with the alteration of global DNA methylation and hydroxymethylation in vitro, the entire DNMTs and TETs were both activated in HG compared to NC, and suppressed after FA treatment in vitro and in vivo (Figure 3a-b). Furthermore, we examined DNMTs and TETs binding affinity on promoters of MMP14 by ChIP-qPCR assay, and observed that only DNMT1 and TET2 indeed contributed to DNA methylation and hydroxymethylation for the promoters of MMP14 both in vitro and in vivo (Figure 3c-D). Nevertheless, the binding abilities of DNMT1 and TET2 on these gene promoters appeared to be no difference among NC, HG, and FA groups in vitro, or NC, HG, and HG-FA groups in vivo. We summarized the effects of DNMT1 and TET2 on gene expression via DNA methylation and hydroxymethylation in retinal microvascular endothelium.
Figure 3.
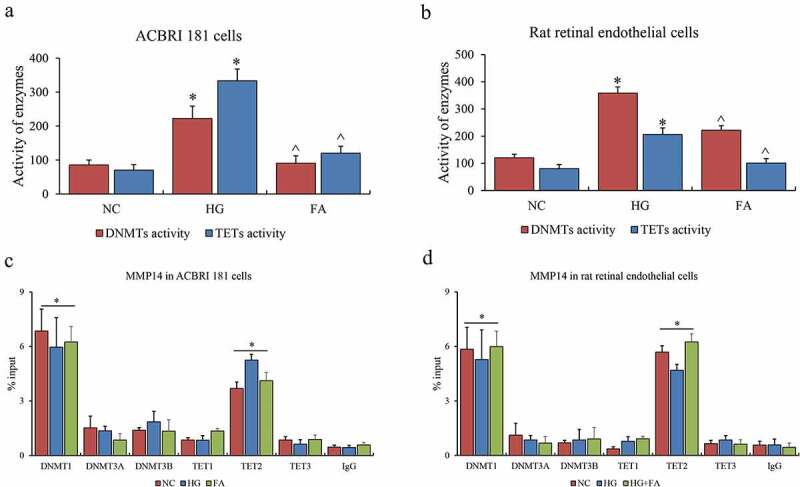
The role of DNMTs and TETs in retinal epithelium. (a) The enzyme activity of DNMTs and TETs in ACBRI 181 cells affected by HG and FA. (b) The enzyme activity of DNMTs and TETs in rat retinal endothelial cells affected by HG and FA. (c) ChIP-qPCR assay of DNMTs on promoter of MMP14. (d) ChIP-qPCR assay of TETs on promoter of MMP14. ChIP-qPCR was performed three individual experiments. ‘*’ and ‘^’ represent p value less than 0.05 by comparison between NC and HG, respectively
Discussion
Epigenetic changes including microRNAs, histone modifications, and methylation of DNA playing a crucial role in gene expression regulation have been demonstrated to involve in DR pathogenesis [20]. DNA methylation as the most widely acknowledged epigenetic modification can cause the heritable and long-term gene silencing. DNA methylation refers to the covalent addition of a methyl (-CH3) group from the SAM to the cytosine of CpG dinucleotides or islands catalyzed by DNMTs. In turn, DNA demethylation as a reversible process appears in specific contexts through active or passive mechanisms [21]. Recent researches have revealed that the methylcytosine can be further successively oxidized toward 5-hydroxymethylcytosine, 5-formylcytosine as well as 5-carboxylcytosine catalyzed by TETs, and the carboxyl group is finally removed by decarboxylase from cytosine, thereby a dynamic cycle of methylation is complete [12]. DNA hydroxymethylation is considered as an essential intermediate of demethylation process step and usually indicates a hallmark of transcriptional activation, which is opposite to the effect of DNA methylation [22]. Therefore, the reverse trend of methylation and hydroxymethylation in the promoter of genes (Sub-cluster B and C) can usually determine their transcriptional activity. The highlight of this study we need to discuss is about the other two groups of gene (subcluster A and D) with the similar trend of methylation and hydroxymethylation at the promoter regions. In our case, very few genes are included in subcluster D, while subcluster A contains COL1A1, ITGA7, MMP-14, and VEGFB that are associated with vascular regeneration and capillary cell apoptosis in DR display a consistent trend with MMP-9 [23]. One previous study has reported a similar epigenotype in mammalian hypothalamus, and suggested that DNA hydroxymethylation is seemingly independent to DNA methylation [24], and we speculate that TET2-mediated DNA hydroxymethylation is supposed to exert a complicated regulatory effect on gene expression beyond the dynamic cycle of methylation.
In our study, the given genome-wide DNA methylation and hydroxymethylation atlas of retinal endothelial cells affected by HG and FA is benefit to advanced understanding the epigenetic alteration in DR pathogenesis. Multiple DEGs showing differential methylation or hydroxymethylation can be studied as the potential therapeutic or diagnostic targets in future research.
Conclusion
In summary, our research provided a resource of expression and epigenetic profiles in retinal micro vascular endothelial cell, emphasizing a pharmacological mechanism of FA on DNA methylation and hydroxymethylation regulation in retinal microvessel cells of DR.
Funding Statement
There is no funding supporting this project.
Research highlights
DNA methylation and hydroxymethylation profiles in retinal vascular endothelial cells.
FA increases DNA methylation and reduces DNA hydroxymethylation globally.
The effects of FA on the activities of DNMTs and TETs.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Availability of Data and Materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
- [1].American Diabetes A. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2011;34(Suppl 1):S62–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Martins T. Diabetic retinopathy: a neuropathy. Einstein. 2020;19:eED6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Meleth AD, Agron E, Chan CC, et al. Serum inflammatory markers in diabetic retinopathy. Invest Ophthalmol Vis Sci. 2005;46(11):4295–4301. [DOI] [PubMed] [Google Scholar]
- [4].Rubsam A, Parikh S, Fort PE. Role of inflammation in diabetic retinopathy. Int J Mol Sci. 2018;19(4):942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Giurdanella G, Lupo G, Gennuso F, et al. Activation of the VEGF-A/ERK/PLA2 axis mediates early retinal endothelial cell damage induced by high glucose: new insight from an in vitro model of diabetic retinopathy. Int J Mol Sci. 2020;21(20):7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dai Y, Wu Z, Wang F, et al. Identification of chemokines and growth factors in proliferative diabetic retinopathy vitreous. Biomed Res Int. 2014;2014:486386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gui F, You Z, Fu S, et al. Endothelial dysfunction in diabetic retinopathy. Front Endocrinol (Lausanne). 2020;11:591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kowluru RA, Mishra M. Oxidative stress, mitochondrial damage and diabetic retinopathy. Biochim Biophys Acta. 2015;1852(11):2474–2483. [DOI] [PubMed] [Google Scholar]
- [9].Malaguarnera G, Gagliano C, Salomone S, et al. Folate status in type 2 diabetic patients with and without retinopathy. Clin Ophthalmol. 2015;9:1437–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lei XW, Li Q, Zhang JZ, et al. The protective roles of folic acid in preventing diabetic retinopathy are potentially associated with suppressions on angiogenesis, inflammation, and oxidative stress. Ophthalmic Res. 2019;62(2):80–92. [DOI] [PubMed] [Google Scholar]
- [11].Crider KS, Yang TP, Berry RJ, et al. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Adv Nutr. 2012;3(1):21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fryer AA, Emes RD, Ismail KM, et al. Quantitative, high-resolution epigenetic profiling of CpG loci identifies associations with cord blood plasma homocysteine and birth weight in humans. Epigenetics. 2011;6(1):86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hoyo C, Murtha AP, Schildkraut JM, et al. Methylation variation at IGF2 differentially methylated regions and maternal folic acid use before and during pregnancy. Epigenetics. 2011;6:928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Heijmans BT, Tobi EW, Stein AD, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences of the United States of America 2008; 105(44):17046–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tobi EW, Lumey LH, Talens RP, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18(21):4046–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chavkin NW, Walsh K, Hirschi KK. Isolation of highly purified and viable retinal endothelial cells. J Vasc Res. 2021;58(1):49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhao X, Tian GG, Fang Q, et al. Comparison of RNA m(6)A and DNA methylation profiles between mouse female germline stem cells and STO cells. Mol Ther Nucleic Acids. 2021;23:431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Akalin A, Kormaksson M, Li S, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13(10):R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shafabakhsh R, Aghadavod E, Ghayour-Mobarhan M, et al. Role of histone modification and DNA methylation in signaling pathways involved in diabetic retinopathy. J Cell Physiol. 2019;234(6):7839–7846. [DOI] [PubMed] [Google Scholar]
- [21].Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11(9):607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Richa R, Sinha RP. Hydroxymethylation of DNA: an epigenetic marker. EXCLI J. 2014;13:592–610. [PMC free article] [PubMed] [Google Scholar]
- [23].Mohammad G, Kowluru RA. Homocysteine disrupts balance between MMP-9 and its tissue inhibitor in diabetic retinopathy: the role of DNA methylation. Int J Mol Sci. 2020;21(5):1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shen Y, Zhou S, Zhao X, et al. Characterization of genome-wide DNA methylation and hydroxymethylation in mouse arcuate nucleus of hypothalamus during puberty process. Front Genet. 2020;11:626536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.


