Abstract
Primates are regularly infected by fungal organisms identified as Pneumocystis carinii. They constitute a valuable population for the confirmation of P. carinii host specificity. In this study, the presence of P. carinii was assessed by direct examination and nested PCR at mitochondrial large subunit (mtLSU) rRNA and dihydropteroate synthetase (DHPS) genes in 98 lung tissue samples from captive or wild nonhuman primates. Fifty-nine air samples corresponding to the environment of different primate species in zoological parks were also examined. Cystic forms of P. carinii were detected in smears from 7 lung tissue samples corresponding to 5 New World primate species. Amplifications at the mtLSU rRNA gene were positive for 29 lung tissue samples representing 18 different primate species or subspecies and 2 air samples corresponding to the environment of two simian colonies. Amplifications at the DHPS gene were positive for 8 lung tissue samples representing 6 different primate species. Direct sequencing of nested PCR products demonstrated that a specific mtLSU rRNA and DHPS sequence could be attributed to each primate species or subspecies. No nonhuman primate harbored the human type of P. carinii (P. carinii f. sp. hominis). Genetic divergence in primate-derived P. carinii organisms varied in terms of the phylogenetic divergence existing among the corresponding host species, suggesting coevolution.
Pneumocystis carinii pneumonia (PCP) is still considered one of the most serious fungal respiratory infections that can occur in immunocompromised patients, especially human immunodeficiency virus-infected individuals. Molecular comparisons of various gene sequences (19, 42) clearly demonstrated that the single name P. carinii corresponds in fact to a complex group of eukaryotic organisms which should be assigned to the kingdom Fungi.
Because a continuous in vitro propagation system for P. carinii is still lacking, the basic biology of this fungal group and subsequent epidemiology of PCP remain poorly understood (8, 11, 42). Airborne transmission appears likely. Hospital outbreaks have been documented (27), and well-designed animal studies have provided evidence of direct transmission from host to host via airborne infective particles (9, 26, 41). It has been hypothesized that the main source of P. carinii is patients with PCP. Another source of transmission may be through maternal-infant exposure (9). Domestic or wild animals are not considered sources for humans as P. carinii organisms seem to be characterized by strong host specificity. Several Pneumocystis species may be distinguished, each of them residing in a specific mammalian host (29). Isolates of P. carinii infecting different mammals are divergent at the genetic level (specific karyotypic profiles and gene sequences) (29) and at the phenotypic level (antigenic differences, ultrastructure, and isoenzymatic polymorphism) (36, 46). The concept of close host specificity has been further supported by the failure of most cross-infection experiments (1, 2, 3, 23, 24, 47). However, Sethi (39) described the possibility for human-derived P. carinii to develop in SCID mice, and one recent study suggested transient colonization of owl monkeys (Aotus nancymai) by human-derived P. carinii (6).
In this report, the genetic diversity of P. carinii from primates is examined by analyzing mitochondrial large subunit (mtLSU) rRNA and dihydropteroate synthetase (DHPS) gene sequences. Each of the 18 nonhuman primate species or subspecies which were proved to harbor P. carinii had its own type of organism with specific mtLSU rRNA or DHPS sequences. No P. carinii f. sp. hominis was found in the nonhuman primate lung tissue or air samples examined in the present work.
MATERIALS AND METHODS
Samples for analysis.
Postmortem lung tissues from nonhuman primates were obtained in four French zoological parks (La Palmyre, Jardin des Plantes de Paris, Parc Zoologique de Vincennes, and Parc Zoologique de Mulhouse) and from the Primate Research Center of Strasbourg. Additional lung tissues from wild monkeys were obtained from the ONC (Office National de la Chasse) of French Guyana. The lung tissues were frozen after necropsic examination and stored at −20°C prior to direct examination and DNA extraction. Air samples corresponding to the environment of different primate species in zoological parks were obtained by using the CAP device (Arelco, Fontenay sous Bois, France), as described by Guillot et al. (25). This device sampled airborne particles with a flow rate of 10 liters/min. Particles were impacted on the surface of a rotative cup. Air sampling was performed in the four zoological parks and in the primatology research center of Strasbourg for 24 to 72 h in front of or inside each of the cages where simian colonies were maintained.
Direct examination of lung tissue samples.
Impression smears of cross sections of lungs were stained with toluidine blue O for the detection of cystic forms of P. carinii (10). A small part of the lung tissue (from 300 to 500 mg) was then finely minced, homogenized with crushing, and subjected to sequential membrane filtration (9). The final filtrates were used for direct examination and DNA extraction. Cystic forms of P. carinii were detected and counted in 2.5-μl air-dried smears of the final filtrate, stained with toluidine blue O.
DNA extraction from lung tissue and air samples.
A volume of 100 μl of the final filtrate of lung extract was first frozen at −20°C and digested with proteinase K (Boehringer Mannheim) at a final concentration of 0.34 mg/ml. A phenol-chloroform extraction was then performed with a final precipitation in ethanol. For air samples, the rotative cup (from the CAP apparatus) was washed with 600 μl of extraction buffer (10 mM Tris, 0.5% sodium dodecyl sulfate, 25 mM EDTA, 0.1 M NaCl). DNA was prepared by proteinase K digestion (Boehringer Mannheim) at a final concentration of 0.28 mg/ml, followed by phenol-chloroform extraction.
Primers and PCR amplifications.
The presence of P. carinii DNA in lung tissue and air samples was assessed by nested PCR at the mtLSU rRNA gene and at the DHPS locus. For mtLSU rRNA gene amplification, the primer sets pAZ102-H–pAZ102-E (5′-GAT GGG TGT TTC CAA GCC CA-3′ and 5′-GTG TAC GTT GCA AAG TAC TC-3′) and pAZ102-X/R1–pAZ102-Y/R1 (5′-GGG AAT TCG TGA AAT ACA AAT CGG ACT AGG-3′ and 5′-GGG AAT TCT CAC TTA ATA TTA ATT GGG GAG C-3′) were used (45). The thermocycling conditions for the first PCR round were as follows: each cycle consisted of denaturation for 30 s at 94°C, annealing for 1 min at 50°C, and extension for 2 min at 72°C for 30 cycles. The second round of PCR was performed with 5% (vol/vol) of the first-round mix. The thermocycling conditions for the second PCR round were as follows: each cycle consisted of denaturation for 30 s at 94°C, annealing for 1 min at 55°C, and extension for 2 min at 72°C for 30 cycles.
For the first round of PCR at the DHPS gene, the primer set AHUM-BHUM (5′-GCG CCT ACA CAT ATT ATG GCC ATT TTA AAT C-3′ and 5′-CAT AAA CAT CAT GAA CCC G-3′) was used (31). The thermocycling conditions were as follows: 10 first cycles consisted of denaturation for 30 s at 94°C, annealing for 1 min at 52°C, and extension for 1 min at 72°C; 25 additional cycles consisted of denaturation for 30 s at 94°C, annealing for 1 min at 42°C, and extension for 1 min at 72°C. The second round of PCR was performed with 5% (vol/vol) of the first-round mix and the primer set CPRIM-DPRIM (5′-CCC CCA CTT ATA TCA-3′ and 5′-GGG GGT GTT CAT TCA-3′). The thermocycling conditions for the second PCR round were as follows: each cycle consisted of denaturation for 30 s at 94°C, annealing for 1 min at 50°C, and extension for 1 min at 72°C for 30 cycles. Negative controls were included in each experiment, in both DNA extraction and PCR amplification, to monitor for possible contamination.
Amplification products were purified in a 2% agarose gel (Tris-borate-EDTA buffer) and extracted with the Geneclean II kit (Ozyme, Montigny-le-Bretonneux, France) when nonspecific bands were detected. Amplification products were directly sequenced from both ends by using sets of internal primers on an automated DNA sequencer (GenomeExpress, Montreuil, France). The sequences have been submitted to GenBank.
Molecular phylogenetic analysis.
Nucleotide (mtLSU and DHPS) and derived amino acid (DHPS) sequences were initially aligned with the computer program CLUSTAL X (version 1.63b, December 1997) (44) and then by visual optimization. Only regions without ambiguity were included in the phylogenetic analysis (alignments are available upon request). The aligned sequences were converted to distance matrix (% of differences). For mtLSU rRNA and DHPS sequences, the distance trees (phenetic trees) were generated using the neighbor-joining method (38). Both the branch-and-bound and heuristic search options in the Phylogenetic Analysis Using Parsimony Program (PAUP 4.0; distributed by the Illinois Natural History Survey, Champaign, Ill.) were used for comparison of sequence alignments and generation of parsimonious trees. The strength of the internal branches from the resulting trees was statistically tested by bootstrap analysis (20) from 500 bootstrap replications. For mtLSU rRNA sequence analyses, P. carinii strains from rat (P. carinii f. sp. carinii, GenBank accession number U42914) and from mouse (P. carinii f. sp. muris, GenBank accession number U20169) were chosen as outgroups. For DHPS sequence analyses, P. carinii from mouse (P. carinii f. sp. muris, GenBank accession number U66283) was chosen as the outgroup.
RESULTS
Direct examination and PCR amplification.
A total of 98 lung tissue samples from 33 different primate species were collected (Table 1). Three zoological groups were represented, with 72 lung tissue samples from 15 New World primate species (Platyrrhini), 21 lung tissue samples from 14 Old World primate species (Catarrhini), and 5 lung tissue samples from 4 lemurs (Strepsirhini Lemuridae). Ninety-four lung tissue samples were obtained from French zoological parks and four lung tissue samples were from wild primates in French Guyana (two pale-headed sakis, one red-handed tamarin, and one squirrel monkey). The most frequent causes of death noted at necropsic examination included infectious diseases and trauma. Some monkeys presented pulmonary lesions but no case of pneumocystosis could be diagnosed. Cystic forms of P. carinii were detected in smears from seven lung tissue samples corresponding to five New World primate species (one squirrel monkey, two Geoffroy's marmosets, one red-handed tamarin, one Goeldi's monkey, and two Weddell's tamarins). The organisms (3.5 to 5.9 μm in diameter) were either isolated or disposed in small clusters. Amplification at mtLSU rRNA and DHPS genes was positive with the first round of PCR for all the lung tissue samples in which cystic forms were detected. Additional positive amplification was obtained with the second round of PCR for 22 lung tissue samples (representing 14 different primate species) at the mtLSU rRNA gene and for 1 sample (from a swamp guenon) at the DHPS gene (Table 1).
TABLE 1.
Detection of P. carinii in lung tissue and air samples from 16 primate species and their environment
| Primate species (no. of animals examined)a | Detection of P. carinii in:b
|
||||
|---|---|---|---|---|---|
| Lung tissue samplesc
|
Air samplesd
|
||||
| By direct examination | By mtLSU PCR | By DHPS PCR | By mtLSU PCR | By DHPS PCR | |
| Suborder Platyrrhini (New World monkeys), family Callitrichidae | |||||
| C. goeldii, Goeldi's monkey (4) | 1 | 1 | 1 | − | − |
| C. geoffroyi, Geoffroy's marmoset (10) | 2 | 2 | 2 | − | − |
| C. jacchus, common marmoset (18) | − | 6 | − | + | − |
| S. fuscicolis, Weddell's tamarin (3) | 2 | 3 | 2 | − | − |
| S. imperator, emperor tamarin (2) | − | 1 | − | − | − |
| S. midas midas, red-handed tamarin: captive animals (3); | 1 | 3 | 1 | − | − |
| wild animal (1) | − | 1 | − | ND | ND |
| S. oedipus oedipus, cotton-head tamarin (3) | − | 1 | − | − | − |
| Suborder Platyrrhini (New World monkeys), family Cebidae | |||||
| P. pithecia, pale-headed saki (2 wild animals) | − | 2 | − | ND | ND |
| S. sciureus, squirrel monkey: captive animals (5); | 1 | 1 | 1 | − | − |
| wild animal (1) | − | − | − | ND | ND |
| Suborder Catarrhini (Old World monkeys), family Cercopithecidae | |||||
| A. nigroviridis, swamp guenon (1) | − | 1 | 1 | − | − |
| C. hamlyni, owl-faced guenon (1) | − | 1 | − | − | − |
| C. nictitans, white-nosed guenon (1) | − | 1 | − | − | − |
| M. fascicularis, crab-eating macaque (3) | − | 1 | − | ND | ND |
| M. mulatta, rhesus macaque (2)e | − | 2 | − | ND | ND |
| M. nemestrina, pig-tailed macaque (4) | − | 1 | − | − | − |
| Suborder Strepsirhini (prosimians), family Lemuridae | |||||
| H. griseus, grey gentle lemur (1) | − | 1 | − | − | − |
| M. macaco, black lemur | ND | ND | ND | + | − |
Lung tissue samples and air samples were obtained from 16 additional primate species (Cebus capucinus, Leonpithecus rosalia, Leonpithecus chrysomelas, Callithrix pygmaea, Callithrix kuhli, Cercocebus agilis, Lephocebus aterrimus, Cercopithecus wolfi, Cercopithecus lhoesti, Papio cynocephalus, Colobus abyssinicus, Gorilla gorilla, Mandrillus sphinx, Maki catta, Maki vari, and Phaner furcifer), but direct examination and PCR amplification were systematically negative.
ND, not done.
The results are indicated in numbers of animals in which P. carinii was detected. Minus (−) indicates that none were detected.
Symbols: +, positive amplification; −, no amplification. Air samples were obtained by using the impacter device CAP (Arelco) in front of or inside the cages where simian colonies were maintained.
One rhesus macaque was native to India (Indian subspecies), and the other one was native to China (Chinese subspecies).
A total of 59 air samples were collected from the environment of 31 nonhuman primate species. Amplification at the mtLSU rRNA gene using nested PCR was positive from only two samples: one from the environment of a colony comprising 10 common marmosets (Callithrix jacchus) in La Palmyre zoological park and another one from the environment of a colony comprising 5 black lemurs (Maki macaco) in Vincennes zoological park (Table 1). No amplification at the DHPS gene was obtained from air samples.
PCR products ranged from 313 to 324 bp (first round of PCR at the mtLSU rRNA gene), from 234 to 282 bp (nested PCR at the mtLSU rRNA gene), and from 702 to 720 bp (first round of PCR at the DHPS gene). Amplification was positive with the nested PCR at the DHPS gene for one lung tissue sample (from a swamp guenon), and the PCR product comprised 664 bp.
Mitochondrial LSU rRNA sequence comparison and genetic groupings.
Direct sequencing of PCR products demonstrated that a specific mtLSU rRNA sequence could be attributed to each primate species or subspecies. In the case of the common marmoset (C. jacchus), the PCR amplification for mtLSU rRNA was positive for several lung tissue samples and for one air sample. The corresponding sequences were shown to be identical. In the case of the red-handed tamarin (Saguinus midas midas), positive PCR amplifications were obtained from both captive and wild animals and the corresponding mtLSU rRNA sequences were identical. Multiple sequence types were not detected in a single host species except for the rhesus monkey (Macaca mulatta). On the other hand, the sequence of the human-derived P. carinii was never obtained from lung tissue or air samples examined in the present study.
Eighteen new P. carinii mtLSU rRNA sequences were obtained from nine New World monkey species, six Old World monkey species, and two lemurs. The sequences were identified by the GenBank accession numbers AF362454 (P. carinii from C. jacchus), AF362456 (P. carinii from Callithrix geoffroyi), AF362461 (P. carinii from Callimico goeldii), AF362462 (P. carinii from Saguinus fuscicolis), AF362455 (P. carinii from S. midas midas), AF362453 (P. carinii from Saguinus oedipus oedipus), AF362465 (P. carinii from Saguinus imperator), AF362458 (P. carinii from Saimiri sciureus), AF362470 (P. carinii from Pithecia pithecia), AF362464 (P. carinii from Allenopithecus nigroviridis), AF362457 (P. carinii from Cercopithecus hamlyni), AF362460 (P. carinii from Cercopithecus nictitans), AF362469 (P. carinii from Macaca fascicularis), AF362467 (P. carinii from an M. mulatta Chinese subspecies), AF362468 (P. carinii from an M. mulatta Indian subspecies), AF362466 (P. carinii from Macaca nemestrina), AF362459 (P. carinii from Hapalemur griseus), and AF362463 (P. carinii from M. macaco). In the case of the rhesus monkey, two sequence types were observed according to the subspecies (Chinese or Indian M. mulatta). In the region selected for the phylogenetic analysis, only one base substitution distinguished the two sequence types. A matrix analysis of sequence divergence between P. carinii mtLSU rRNA sequences is depicted in Table 2. Comparison of the mtLSU rRNA aligned sequences was carried out on 287 positions, including gaps, for a total of 21 taxa: 18 original sequence types and 3 sequences already published, P. carinii f. sp. hominis (GenBank S42926), P. carinii f. sp. muris (GenBank U20169), and P. carinii f. sp. carinii (GenBank U42914). P. carinii organisms from Old World primates (including human) were characterized by an insertion at position 36. For members of the genera Allenopithecus and Cercopithecus the insertion was very long (63 bp), whereas for macaques and human-derived P. carinii the insertion was shorter (from 13 to 48 bp). Lemur-derived P. carinii organisms were characterized by a distinct insertion at position 132 (15 bp for the grey gentle lemur and 22 bp for the black lemur).
TABLE 2.
Matrix of mtLSU rRNA sequence divergence for P. carinii from 17 primate species (including P. carinii f. sp. hominis) and rodents
| Source of rRNA | % divergence froma:
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | ||
| 1. P. carinii f. sp. hominis P. carinii from: | |||||||||||||||||||||
| 2. A. nigroviridis | 11.6 | ||||||||||||||||||||
| 3. C. hamlyni | 11.6 | 01.6 | |||||||||||||||||||
| 4. C. nictitans | 13.1 | 02.9 | 02.0 | ||||||||||||||||||
| 5. M. nemestrina | 11.2 | 09.1 | 09.1 | 09.9 | |||||||||||||||||
| 6. M. fascicularis | 11.7 | 09.5 | 09.5 | 10.3 | 00.4 | ||||||||||||||||
| 7. M. mulatta Chinese subsp. | 12.7 | 06.6 | 07.6 | 07.6 | 03.0 | 03.5 | |||||||||||||||
| 8. M. mulatta Indian subsp. | 13.2 | 07.1 | 08.1 | 08.1 | 03.5 | 04.0 | 00.5 | ||||||||||||||
| 9. S. sciureus | 23.8 | 24.9 | 25.4 | 26.4 | 24.9 | 25.4 | 21.4 | 21.9 | |||||||||||||
| 10. P. pithecia | 15.4 | 19.6 | 19.6 | 21.1 | 19.5 | 20.0 | 18.7 | 19.3 | 22.6 | ||||||||||||
| 11. C. jacchus | 18.2 | 19.0 | 20.0 | 20.5 | 18.9 | 19.4 | 18.1 | 18.1 | 25.0 | 13.2 | |||||||||||
| 12. C. geoffroyi | 18.2 | 19.0 | 20.0 | 20.5 | 18.9 | 19.4 | 18.1 | 18.1 | 25.0 | 13.2 | 00.0 | ||||||||||
| 13. C. goeldii | 15.0 | 17.4 | 17.4 | 19.0 | 16.3 | 16.8 | 16.5 | 17.0 | 22.3 | 12.2 | 10.2 | 10.2 | |||||||||
| 14. S. fuscicolis | 15.0 | 17.4 | 17.4 | 19.0 | 16.3 | 16.8 | 16.5 | 17.0 | 23.4 | 13.2 | 09.2 | 09.2 | 01.0 | ||||||||
| 15. S. midas midas | 15.0 | 16.9 | 16.9 | 18.5 | 16.3 | 16.8 | 15.4 | 15.4 | 23.4 | 11.2 | 09.2 | 09.2 | 07.3 | 06.3 | |||||||
| 16. S. imperator | 15.0 | 16.9 | 16.9 | 18.5 | 16.3 | 16.8 | 15.4 | 15.4 | 23.4 | 11.2 | 09.2 | 09.2 | 07.3 | 06.3 | 00.0 | ||||||
| 17. S. oedipus oedipus | 15.5 | 17.4 | 17.4 | 19.0 | 16.8 | 17.3 | 16.0 | 15.4 | 22.9 | 10.2 | 08.7 | 08.7 | 07.3 | 06.3 | 02.4 | 02.4 | |||||
| 18. H. griseus | 22.5 | 22.2 | 22.2 | 23.7 | 19.5 | 20.0 | 21.8 | 22.3 | 28.2 | 19.3 | 22.8 | 22.8 | 19.5 | 19.5 | 21.5 | 21.5 | 20.0 | ||||
| 19. M. macaco | 23.0 | 24.2 | 24.2 | 25.8 | 22.6 | 23.1 | 21.8 | 22.3 | 26.6 | 20.3 | 22.9 | 22.9 | 23.0 | 23.0 | 22.5 | 22.5 | 21.1 | 08.2 | |||
| 20. P. carinii f. sp. muris | 17.4 | 22.9 | 22.9 | 24.5 | 22.7 | 23.1 | 23.1 | 23.7 | 28.0 | 22.8 | 23.6 | 23.6 | 24.3 | 24.3 | 22.3 | 22.3 | 22.3 | 22.7 | 21.3 | ||
| 21. P. carinii f. sp. carinii | 19.2 | 24.6 | 24.6 | 26.2 | 24.9 | 25.3 | 25.0 | 25.5 | 31.9 | 25.4 | 26.3 | 26.3 | 26.9 | 26.9 | 25.4 | 25.4 | 25.9 | 26.4 | 25.7 | 09.4 | |
Numbers 1 to 20 refer to numbered sources of rRNA.
No insertion was detected in mtLSU rRNA sequences of New World primate-derived P. carinii. The two primate species of the genus Callithrix (Geoffroy's marmoset and common marmoset) harbored distinct P. carinii with eight nucleotide differences in the mtLSU rRNA sequences but these differences were located in a region which was not included in the phylogenetic analysis. The same situation was observed for P. carinii from two primate species belonging to the genus Saguinus (Emperor tamarin and red-handed tamarin), with 10 nucleotide differences in the amplified mtLSU rRNA sequences. Within the group composed of primate-derived P. carinii, the maximum divergence (28.2%) was observed between the black lemur-derived and the squirrel monkey-derived P. carinii. The extent of divergence may not be greater between a primate-derived and a nonprimate-derived P. carinii than between primate-derived organisms themselves (Table 2).
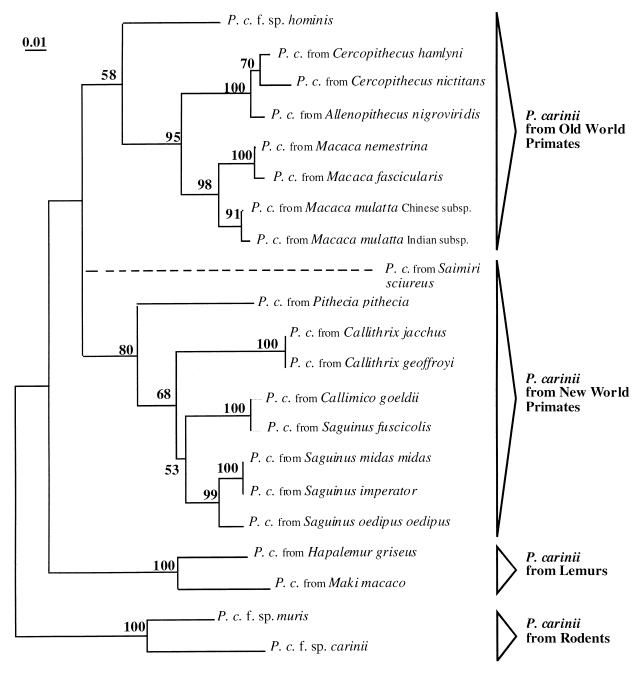
The phylogenetic analysis inferred from mtLSU rRNA sequence comparison demonstrated that genetic groupings within primate-derived P. carinii were in accordance with those of the corresponding hosts. Phylogenetic analyses, neighbor joining, heuristic search, and branch and bound produced topologically similar trees, although statistical support among the branches varied (Fig. 1). All three analyses indicated that primate-derived P. carinii formed a monophyletic group with three distinct clades. The two lemur-derived P. carinii organisms were associated and occupied a basal position. All P. carinii organisms from Old World nonhuman primates were included in the same clade supported by a 95% bootstrap value. Within this group, P. carinii from macaques (Macaca spp.) and from guenons (Allenopithecus and Cercopithecus spp.) were clearly distinguished with branches supported by 98 and 100% bootstrap values, respectively. A branch associating P. carinii f. sp. hominis and Old World primate-derived P. carinii was systematically produced (heuristic search and branch and bound) but with a low bootstrap value (58%). The last major clade included New World monkey-derived P. carinii with a high bootstrap value (80%). Within this group, P. carinii from tamarins (genera Saguinus and Callimico) clustered together. P. carinii organisms from marmosets (genus Callithrix) were also associated. P. carinii from pale-headed sakis (P. pithecia) occupied a paraphyletic position in the New World monkeys group. The exact position of P. carinii from the New World species S. sciureus (squirrel monkey) could not be elucidated.
FIG. 1.
Phylogenetic relationships of P. carinii organisms from primate species inferred from mtLSU rRNA sequences. The phylogram presented resulted from bootstrapped data sets (20) obtained by using parsimony analysis (heuristic search option in PAUP 4.0). This tree was identical to the consensus of 18 most-parsimonious trees generated from the branch and bound algorithm in PAUP 4.0. The percentages above the branches are the frequencies with which a given branch appeared in 500 bootstrap replications. Bootstrap values below 50% are not displayed. The position of the squirrel monkey (S. sciureus)-derived P. carinii was not firmly established (dotted line). Branch lengths correspond to the total nucleotide changes assigned to each branch by PAUP 4.0. P. carinii from rat (P. carinii f. sp. carinii, GenBank no. U42914) and from mouse (P. carinii f. sp. muris, GenBank no. U20169) were chosen as outgroups.
DHPS sequence comparison and genetic groupings.
Direct sequencing of PCR products demonstrated that a specific DHPS sequence could be attributed to each primate species. Six different DHPS sequences were described and corresponded to P. carinii derived from five New World monkey species and one Old World monkey species. The sequences were identified by GenBank accession numbers AF362758 (P. carinii from C. geoffroyi), AF362760 (P. carinii from C. goeldii), AF362761 (P. carinii from S. fuscicolis), AF362762 (P. carinii from S. midas midas), AF362759 (P. carinii from S. sciureus) and AF362757 (P. carinii from A. nigroviridis). The DHPS aligned sequence comprised 217 positions, including gaps for a total of nine taxa: six original sequence types and three sequences already published, P. carinii f. sp. hominis (GenBank U66282), P. carinii f. sp. muris (GenBank U66283), and the sequence from the owl monkey (A. nancymai)-derived P. carinii (6). The last sequence was very short. The lowest divergence was observed between Weddell's tamarin and Goeldi's monkey-derived P. carinii (1.4%). The higher level of divergence was found between the squirrel monkey-derived P. carinii and P. carinii f. sp. muris (16.6%). Point mutations at positions 165 (A/G) and 171 (C/T) related to sulfa drug resistance in P. carinii f. sp. hominis were not detected in any of the DHPS sequences obtained in the present study (31).
The deduced partial amino acid sequences of the DHPS gene from primate-derived P. carinii contain 72 residues. Very low levels of divergence were observed between P. carinii organisms from New World monkeys, Weddell's tamarin, Goeldi's monkey, owl monkey, and Geoffroy's marmoset (1.4%). Higher levels of divergence were found between the squirrel monkey-derived P. carinii and Old World primate-derived P. carinii (9.9%), between the Geoffroy's marmoset-derived P. carinii and P. carinii f. sp. hominis (9.9%), and between P. carinii f. sp. muris and the squirrel monkey and Geoffroy's marmoset-derived P. carinii (16.9%).
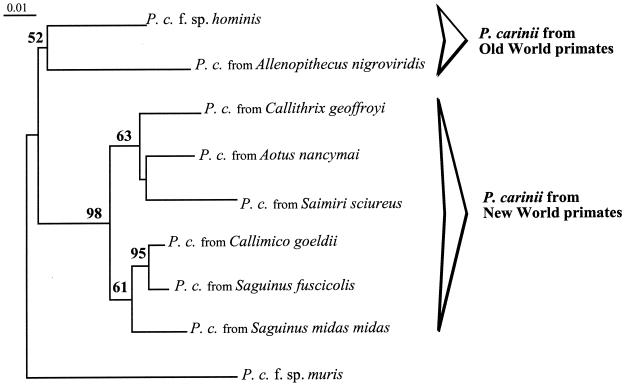
The phylogenetic analyses inferred from DHPS nucleotide sequences yielded identical topologies and confirmed the results obtained with mtLSU rRNA sequence analyses (Fig. 2). The monophyletic group composed of primate-derived P. carinii organisms was clearly divided in two clades. A first clade comprised the two P. carinii organisms from Old World primates (human and swamp guenon-derived P. carinii). A second clade included all P. carinii organisms from New World monkeys. Within this group, P. carinii organisms from tamarins (genus Saguinus) clustered together. As already observed with the analyses of mtLSU rRNA sequences, the branch corresponding to P. carinii organisms from New World monkeys was supported by a higher bootstrap value (98%) than that corresponding to P. carinii organisms from Old World primates (52%). P. carinii from the squirrel monkey had a divergent DHPS sequence. However, it was included in the group of New World monkeys and was systematically associated with the owl monkey-derived P. carinii. When the short sequence of P. carinii from owl monkey (6) was excluded, the phylogenetic analyses could be performed on much longer sequences (629 bp) but the tree topologies remained unchanged, with the squirrel monkey-derived P. carinii included in the New World monkeys P. carinii group.
FIG. 2.
Phylogenetic relationships of P. carinii organisms from primate species inferred from DHPS sequences. The presented phylogram resulted from bootstrapped data sets (20) obtained by using parsimony analysis (heuristic search option in PAUP 4.0). This tree was identical to the consensus of three most-parsimonious trees generated from the branch and bound algorithm in PAUP 4.0. The percentages above the branches are the frequencies with which a given branch appeared in 500 bootstrap replications. Bootstrap values below 50% are not displayed. Branch lengths correspond to the total nucleotide changes assigned to each branch by PAUP 4.0. P. carinii from mouse (P. carinii f. sp. muris, GenBank no. U66283) was chosen as the outgroup.
The phylogenetic analyses inferred from DHPS amino acid sequences were partially in accordance with those inferred from nucleotide sequences. All primate-derived P. carinii organisms were associated and five New World monkey (including the squirrel monkey)-derived P. carinii organisms clustered together. Surprisingly, the red-handed tamarin-derived P. carinii clustered with the swamp guenon-derived P. carinii, and P. carinii f. sp. hominis was paraphyletic to the nonhuman primate-derived P. carinii.
DISCUSSION
P. carinii was originally considered to be a single organism responsible for pulmonary colonization or infection in a very wide range of mammalian hosts. Frenkel was the first author to suspect that the situation might be more complex and to suggest a distinction between human and rodent-derived P. carinii (21). We now know that the single name P. carinii consists in fact of a heterogeneous group of fungal organisms (11, 42). Molecular techniques, including karyotyping, DNA sequence analysis, or single-strand conformation polymorphism, have played a significant role in demonstrating the degree of heterogeneity between P. carinii organisms. To date, the mtLSU rRNA sequence has been reported from P. carinii organisms infecting nine different mammalian species: human, rat (two divergent sequence types), mouse, rabbit, pig, horse, shrew, ferret (five divergent sequence types), and rhesus macaque (17, 23, 37, 46). The genetic diversity of P. carinii was examined at other loci such as the mitochondrial small subunit of rRNA (28), the nuclear ribosomal RNA operon (32, 46), α- and β-tubulin (18, 43), arom (5), thymidilate synthetase (30, 35), DHPS (31), and more recently manganese-dependent superoxide dismutase (16). However, only a small variety of mammalian hosts with P. carinii were examined in former analyses. The present study is the first one to include so many species and to propose a phylogenetic analysis for primate-derived P. carinii. We have used the sequences from two very different types of genes; one is a mitochondrial gene encoding ribosomal RNA and the other is a nuclear gene encoding a protein coding sequence. Both nucleotide sequence analyses provided the same phylogenetic relationships. A few incongruities were observed with the analyses of amino acid DHPS sequences but the number of residues was low.
In 1994, the Pneumocystis workshop (29) proposed a tripartite nomenclature for P. carinii. In accordance with this provisional system we suggest the creation of five new variant names for P. carinii organisms from nonhuman primates (Table 3). The Pneumocystis workshop also proposed a genetic ranking system based on sequence comparison at five loci: mtLSU rRNA, thymidylate synthetase, β-tubulin, arom, and internal transcribed spacers in the nuclear ribosomal RNA genes. Class III corresponded to the highest level of divergence (from 15 to 50%) observed between P. carinii organisms from different mammalian species. Class II divergence ranged from 4 to 7% at the selected loci (except at the much more variable internal transcribed spacer regions) and could be observed between different P. carinii species in the same mammalian host (for example, P. carinii f. sp. carinii and P. carinii f. sp. ratti in rat). The lowest level of divergence (lower than 1%), termed class I, induced a few base substitutions in the sequences of isolates within the same host species.
TABLE 3.
New P. carinii variants from nonhuman primates
| Nonhuman primate varianta | Host species |
|---|---|
| P. carinii f. sp. callimico | C. goeldii, Goeldi's monkey |
| P. carinii f. sp. callithrix | C. geoffroyi, Geoffroy's marmoset |
| P. carinii f. sp. midas | S. midas midas, red-handed tamarin |
| P. carinii f. sp. fuscicolis | S. fuscicolis, Weddell's tamarin |
| P. carinii f. sp. sciureus | S. sciureus, squirrel monkey |
The trinomial names were proposed for Pneumocystis organisms that were confirmed by direct examination as well as genetic amplification (29). Each Pneumocystis organism is specific to a unique primate species.
The results of the present work suggest that in primate-derived P. carinii populations, genetic differences vary in terms of the phylogenetic divergence existing among the host species. In other words, the sequence divergence between two P. carinii subpopulations appears to be correlated with the phylogenetic relationship between the corresponding hosts. When host species belong to different mammalian orders (Insectivora, Lagomorpha, Rodentia, Primates, Carnivora), the class III divergence level is usually observed. Within the order Primates, the divergence level was shown to vary from less than 1% to more than 25%. Thus, a regular, progressive increase in mtLSU rRNA sequence divergence was observed between P. carinii organisms from the crab-eating macaque (M. fascicularis) and P. carinii from progressively distant species: 0.4% (P. carinii from M. nemestrina), 3.5% (P. carinii from M. mulatta), 9.5 to 10.3% (P. carinii from Old World primates belonging to a genus other than Macaca), 16.8 to 25.4% (P. carinii from New World monkeys), and 23.0 to 25.3% (P. carinii from lemurs and rodents).
Class I divergence was revealed in P. carinii organisms from rhesus monkeys. Two closely related mtLSU rRNA sequences were each associated with the Chinese and Indian M. mulatta subspecies. Similar sequences were reported previously by Furuta et al. (23) and Durand-Joly et al. (17), respectively, who noticed an insertion at the beginning of the sequence. In the present work, this insertion appeared as typical to Old World primate-derived P. carinii. Class II divergence was not found among primate-derived P. carinii organisms in this study. This could be due to the fact that in this first approach direct sequencing was used.
Interestingly, all the variants of P. carinii found in lung tissue samples from 16 primate species housed in French zoological parks for many years represented new P. carinii mtLSU rRNA sequences except the two sequences isolated in M. mulatta previously reported. Variants of P. carinii detected in air samples from the environment of two primate species (common marmoset and black lemur) also represented new sequences. These findings suggested a persistent circulation of P. carinii in simian ecosystems, including in captivity. P. carinii f. sp. hominis, the unique species identified in Homo sapiens sapiens (29), the sole primate species dwelling in France at present, was not found in any of the nonhuman primate species examined. The capacity of human-derived P. carinii to infect animals was not the focus of this study. In the past, attempts to infect animals with parasites from humans were often performed using recipient hosts which were latently infected with P. carinii (3, 6, 39), and molecular tools were not available in the oldest studies (48). On the other hand, recent cross-infection experiments, developed by using P. carinii-free recipient hosts and molecular methods to identify the parasite isolates (15), have revealed a strong host specificity in P. carinii strains from rat, mouse, rabbit, ferret, or macaque (1, 2, 3, 23). However, narrow parasite specificity might not be a universal feature of the genus Pneumocystis. For instance, Dermatophyte genera like Trichophyton or Microsporum contain stenoxenic, oligoxenic, or euryxenic species, i.e., with different degrees of parasite host specificity (13). Furthermore, even if human-derived P. carinii natural populations were found to exhibit restricted host specificity, isolates of the parasite might develop in unusual hosts under experimental conditions. For instance, Plasmodium falciparum, a human Apicomplexa protist with strong host specificity, can be propagated in New World primates under experimental conditions (40).
Since culture methods for the routine isolation of P. carinii f. sp. hominis from patients do not yet exist, the alternative of isolation of human-derived parasites using animal models has to be explored. Although in the present work human-derived P. carinii isolates showed relatively high divergence from P. carinii isolates from lemurs or from some neotropical primates (Table 2), on the whole P. carinii f. sp. hominis was found to be phylogenetically closer to most primate-derived P. carinii isolates, especially those from Old World primates, than to rodent-derived P. carinii (Table 2; Fig. 1 and 2). It is noteworthy that current drug testing and therapeutic protocols against PCP are performed using P. carinii strains of rodent origin (4, 7). For these reasons, the host specificity of P. carinii f. sp. hominis should be further explored using nonhuman primates as experimental hosts.
Finally, the results from this study strengthen the view of P. carinii organisms as a new biological group with numerous species (15, 22) widely present in ecosystems (14). Although for almost one century P. carinii was considered to be a unique taxonomic entity, recent research has clearly shown that P. carinii is in fact a group of heterogeneous populations (34, 42, 47) genetically isolated from each other (36) that have undergone a prolonged process of genetic and functional adaptation to each mammal species. This process is probably speciation, and the status of P. carinii natural populations conforms therefore to the biological definition of species (12, 33).
ACKNOWLEDGMENTS
We acknowledge P. Moisson (Parc Zoologique de Mulhouse), J. Rigoulet (Jardin des Plantes de Paris), A. Lécu and F. Ollivet (Parc Zoologique de Vincennes), A. Gessain (Institut Pasteur de Paris), H. Contamin and M. Kazanji (Institut Pasteur de Cayenne), E. André and N. Herrenschmidt (Centre de Primatologie de Strasbourg), and C. Gottini (Muséum National d'Histoire Naturelle) for providing lung tissue samples from captive primates and E. Hansen (Office Nationale de la Chasse de Guyane Française) for providing lung tissue samples from wild primates. We also thank J. P. Hugot (Muséum National d'Histoire Naturelle) for his assistance in sequence analyses and R. Chermette (Ecole Nationale Vétérinaire d'Alfort) for helpful discussions.
This study was developed in the framework of the PRFMMIP project (Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires, French Ministry of Education, Research and Technology) and of the European Key Action “Eurocarinii” (5th PCRD, QLK2-CT2000-01369).
REFERENCES
- 1.Aliouat E M, Mazars E, Dei-Cas E, Cesbron J Y, Camus D. Intranasal inoculation of mouse, rat or rabbit-derived Pneumocystis in SCID mice. J Protozool Res. 1993;3:94–98. [Google Scholar]
- 2.Aliouat E M, Mazars E, Dei-Cas E, Delcourt P, Billault P, Camus D. Pneumocystis cross infection experiments using SCID mice and nude rats as recipient host showed strong host-species specificity. J Eukaryot Microbiol. 1994;41:S71. [PubMed] [Google Scholar]
- 3.Atzori C, Agostoni F, Angeli E, Mainini A, Micheli V, Cargnel A. Pneumocystis carinii host specificity: attempt of cross infection with human derived strains in rats. J Eukaryot Microbiol. 1999;46:S112. [PubMed] [Google Scholar]
- 4.Aviles P, Aliouat E M, Martinez A, Dei-Cas E, Herreros E, Dujardin L, Gargallo-Viola D. In vitro pharmacodynamic parameters of sordarin derivatives in comparison with those of marketed compounds against Pneumocystis carinii isolated from rats. Antimicrob Agents Chemother. 2000;44:1284–1290. doi: 10.1128/aac.44.5.1284-1290.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerji S, Wakefield A E, Alien A G, Maskell D J, Peters S E, Hopkin J M. The cloning and characterization of the arom gene of Pneumocystis carinii. J Gen Microbiol. 1993;139:2901–2914. doi: 10.1099/00221287-139-12-2901. [DOI] [PubMed] [Google Scholar]
- 6.Beard B C, Jennings V M, Teague W G, Carter J L, Mabry J, Moura H, Visvesvara G S, Collins W E, Navin T R. Experimental inoculation of immunosuppressed owl monkeys with Pneumocystis carinii f. sp. hominis. J Eukaryot Microbiol. 1999;46:S113–S115. [PubMed] [Google Scholar]
- 7.Brun Pascaud M, Herreros E, Aliouat E M, Dei-Cas E. Evaluation of drug efficacy by using animal models or in vitro systems. FEMS Immunol Med Microbiol. 1998;22:173–179. doi: 10.1111/j.1574-695X.1998.tb01203.x. [DOI] [PubMed] [Google Scholar]
- 8.Cailliez J C, Séguy N, Denis C M, Aliouat E M, Mazars E, Polonelli L, Camus D, Dei-Cas E. Pneumocystis carinii: an atypical fungal micro-organism. J Med Vet Mycol. 1996;34:227–239. doi: 10.1080/02681219680000401. [DOI] [PubMed] [Google Scholar]
- 9.Ceré N, Polack B, Chanteloup N K, Coudert P. Natural transmission of Pneumocystis carinii in nonimmunosuppressed animals: early contagiousness of experimentally infected rabbits. J Clin Microbiol. 1997;35:2670–2672. doi: 10.1128/jcm.35.10.2670-2672.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalvardjian A M, Grave L A. A new procedure for the identification of Pneumocystis carinii cysts in tissue sections and smears. J Clin Pathol. 1973;16:383–384. doi: 10.1136/jcp.16.4.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cushion M. Chapter 34: Pneumocystis carinii. In: Ajello L, Hay R J, editors. Topley and Wilson's microbiology and microbial infections. 9th. 4. Mycology. London, England: Arnold; 1998. pp. 645–683. [Google Scholar]
- 12.Dei-Cas E. Pneumocystis infections: the iceberg? Med Mycol. 2000;38(Suppl. 1):23–32. [PubMed] [Google Scholar]
- 13.Dei-Cas E, Vernes A. Parasitic adaptation of pathogenic fungi to the mammalian hosts. Crit Rev Microbiol. 1986;13:173–218. doi: 10.3109/10408418609108738. [DOI] [PubMed] [Google Scholar]
- 14.Dei-Cas E, Cailliez J C. Editorial. FEMS Immunol Med Microbiol. 1998;22:1–4. doi: 10.1111/j.1574-695X.1998.tb01181.x. [DOI] [PubMed] [Google Scholar]
- 15.Dei-Cas E, Mazars E, Aliouat E M, Nevez G, Cailliez J C, Camus D. The host-specificity of Pneumocystis carinii. J Mycol Méd. 1998;8:1–6. [Google Scholar]
- 16.Denis C M, Mazars E, Guyot K, Odberg-Ferragut C, Viscogliosi E, Dei-Cas E, Wakefield A E. Genetic divergence at the SODA locus of six different formae speciales of Pneumocystis carinii. Med Mycol. 2000;38:289–300. doi: 10.1080/mmy.38.4.289.300. [DOI] [PubMed] [Google Scholar]
- 17.Durand-Joly I, Wakefield A E, Palmer R J, Denis C M, Creusy C, Fleurisse L, Ricard I, Gut J P, Dei-Cas E. Ultrastructural and molecular characterization of Pneumocystis carinii isolated from a rhesus monkey (Macaca mulatta) Med Mycol. 2000;38:61–72. doi: 10.1080/mmy.38.1.61.72. [DOI] [PubMed] [Google Scholar]
- 18.Edlind T D, Bartlett M S, Weinberg G A, Prah G N, Smith J W. The beta-tubulin gene from rat and human isolates of Pneumocystis carinii. Mol Microbiol. 1992;6:3365–3373. doi: 10.1111/j.1365-2958.1992.tb02204.x. [DOI] [PubMed] [Google Scholar]
- 19.Edman J C, Kovacs J C, Masur H, Santi D V, Helwood H J, Sogin M L. Ribosomal RNA sequence shows Pneumocystis carinii to be a member of the Fungi. Nature. 1988;334:519–522. doi: 10.1038/334519a0. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 21.Frenkel J K. Pneumocystis jiroveci n. sp. from man: morphology, physiology and immunology in relation to pathology. Natl Cancer Inst Monogr. 1976;43:13–30. [PubMed] [Google Scholar]
- 22.Frenkel J K. Pneumocystis pneumonia, an immunodeficiency-dependent disease: a critical historical overview. J Eukaryot Microbiol. 1999;46:S89–S92. [PubMed] [Google Scholar]
- 23.Furuta T, Fujita M, Mukai R, Sakakibara I, Sata T, Miki K, Hayami M, Kojima S, Yoshikawa Y. Severe pulmonary pneumocystosis in simian acquired immunodeficiency syndrome induced by simian immunodeficiency virus: its characterization by the polymerase-chain-reaction method and failure of experimental transmission to immunodeficient animals. Parasitol Res. 1993;79:624–628. doi: 10.1007/BF00932502. [DOI] [PubMed] [Google Scholar]
- 24.Gigliotti F, Harsen A G, Haidaris C G, Haidaris P J. Pneumocystis carinii is not universally transmissible between mammalian species. Infect Immun. 1993;61:2886–2890. doi: 10.1128/iai.61.7.2886-2890.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guillot J, Berthelemy M, Polack B, Laine V, Lacube P, Chermette R, Roux P. Impaction versus filtration for the detection of Pneumocystis carinii DNA in air. J Eukaryot Microbiol. 1999;46:S100–S101. [PubMed] [Google Scholar]
- 26.Hendley J O, Weller T H. Activation and transmission in rats of infection with Pneumocystis. Proc Soc Exp Biol Med. 1971;137:1401–1404. doi: 10.3181/00379727-137-35798. [DOI] [PubMed] [Google Scholar]
- 27.Hennequin C, Page B, Roux P, Legendre C, Kreis H. Outbreak of Pneumocystis carinii pneumonia in a renal transplant unit. Eur J Clin Microbiol Infect Dis. 1995;14:122–126. doi: 10.1007/BF02111870. [DOI] [PubMed] [Google Scholar]
- 28.Hunter J A C, Wakefield A E. Genetic divergence at the mitochondrial small subunit ribosomal RNA gene among isolates of Pneumocystis carinii from five mammalian host species. J Eukaryot Microbiol. 1996;43:S24–S25. doi: 10.1111/j.1550-7408.1996.tb04962.x. [DOI] [PubMed] [Google Scholar]
- 29.Journal of Eukaryotic Microbiology. The Pneumocystis Workshop. J Eukaryot Microbiol. 1994;41:S121–S122. [Google Scholar]
- 30.Keely S, Pai H J, Baughman R, Sidman C, Sunkin S M, Stringer J R, Stringer S L. Pneumocystis species inferred from analysis of multiple genes. J Eukaryot Microbiol. 1994;41:S94. [PubMed] [Google Scholar]
- 31.Lane B R, Ast J C, Flossler P A, Mindell D P, Bartlett M S, Smith J W, Meshnick S R. Dihydropteroate synthetase polymorphism in Pneumocystis carinii. J Infect Dis. 1997;175:482–485. doi: 10.1093/infdis/175.2.482. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Leibowitz M J. Variation and in vitro splicing of group I introns in rRNA genes of Pneumocystis carinii. Nucleic Acids Res. 1993;20:3763–3772. doi: 10.1093/nar/21.10.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayr E. Animal species and evolution. Cambridge, Mass: The Belknap Press of Harvard University Press; 1963. [Google Scholar]
- 34.Mazars E, Dei-Cas E. Epidemiological and taxonomic impact of Pneumocystis biodiversity. FEMS Immunol Med Microbiol. 1998;22:75–80. doi: 10.1111/j.1574-695X.1998.tb01189.x. [DOI] [PubMed] [Google Scholar]
- 35.Mazars E, Odberg Ferragut C, Dei-Cas E, Fourmaux M N, Aliouat E M, Brun Pascaud M, Mougeot G, Camus D. Polymorphism of the thymidylate synthase gene of Pneumocystis carinii from different host species. J Eukaryot Microbiol. 1995;42:26–32. doi: 10.1111/j.1550-7408.1995.tb01536.x. [DOI] [PubMed] [Google Scholar]
- 36.Mazars E, Guyot K, Durand I, Dei-Cas E, Boucher S, Ben Abderrazak S, Banuls A L, Tibayrenc M, Camus D. Isoenzyme diversity in Pneumocystis carinii from rats, mice and rabbits. J Infect Dis. 1997;175:655–660. doi: 10.1093/infdis/175.3.655. [DOI] [PubMed] [Google Scholar]
- 37.Palmer R J, Settnes O P, Lodal J, Wakefield A E. Appl. Environ. Microbiol. 66:4954–4956. 2000. Population structure of rat-derived Pneumocystis carinii in Danish wild rats. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 39.Sethi K K. Multiplication of human-derived P. carinii in severe combined immunodeficient (SCID) mice. Experimentia. 1992;48:63–67. doi: 10.1007/BF01923610. [DOI] [PubMed] [Google Scholar]
- 40.Smyth J D. Introduction to animal parasitology. Cambridge, England: Cambridge University Press; 1994. [Google Scholar]
- 41.Soulez B, Palluault F, Dei-Cas E, Alliouat E M, Camus D. Introduction of Pneumocystis carinii in a colony of SCID mice. J Protozool. 1991;38:S123–S125. [PubMed] [Google Scholar]
- 42.Stringer J R. Pneumocystis carinii: what is it, exactly? Clin Microbiol Rev. 1996;9:489–498. doi: 10.1128/cmr.9.4.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stringer J R, Stringer S L, Zhang J, Baughman R, Smulian A G, Cushion M T. Molecular genetic distinction of Pneumocystis carinii from rats and humans. J Eukaryot Microbiol. 1993;40:733–741. doi: 10.1111/j.1550-7408.1993.tb04468.x. [DOI] [PubMed] [Google Scholar]
- 44.Thompson J D, Gibson T S, Plewniak F, Jeanmougin F, Higgings D G. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wakefield A E. DNA sequences identical to Pneumocystis carinii f. sp. carinii and Pneumocystis carinii f. sp. hominis in samples of air spora. J Clin Microbiol. 1996;34:1754–1759. doi: 10.1128/jcm.34.7.1754-1759.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakefield A E. Genetic heterogeneity in Pneumocystis carinii: an introduction. FEMS Immunol Med Microbiol. 1998;22:5–13. doi: 10.1111/j.1574-695X.1998.tb01182.x. [DOI] [PubMed] [Google Scholar]
- 47.Wakefield A E, Stringer J R, Tamburrini E, Dei Cas E. Genetics, metabolism and host specificity of Pneumocystis carinii. Med Mycol. 1998;36:S183–S193. [PubMed] [Google Scholar]
- 48.Walzer P D. Experimental models of Pneumocystis carinii infection. In: Young L S, editor. Pneumocystis carinii pneumonia. 1st ed. New York, N.Y: M. Dekker, Inc.; 1984. pp. 37–43. [Google Scholar]