ABSTRACT
Homoharringtonine (HHT), an Food and Drug Administration (FDA)-approved anti-leukemia drug, exerts anti-tumor activity in several solid tumors, including colorectal cancer (CRC). However, its mechanism of action in CRC progression has not been comprehensively elucidated. The drug-disease targets were obtained using publicly available databases. Protein–protein interaction (PPI) network, Gene ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were performed to reveal the core targets, biological processes and signaling pathways of HHT against CRC. Cell and animal experiments were performed to validate the inhibitory effects of HHT on CRC. A total of 98 overlapping target genes of HHT and CRC were predicted. Through PPI network and topology analysis, we screened out 23 hub genes. Enrichment assays showed 163 biological processes (BP), 18 cell components (CC), 35 molecular functions (MF), and 85 related pathways. Functionally, HHT inhibited CRC cell proliferation, cell cycle progression, colony formation, migration and invasion, and promoted apoptosis. HHT treatment resulted in the inactivation of PI3K/AKT/mTOR signaling in CRC cells. Moreover, activation of PI3K/AKT/mTOR signaling by 740Y-P abated the suppressive effects of HHT on cell malignant phenotypes. Furthermore, HHT repressed CRC tumor growth in nude mice. Our current study demonstrated that HHT repressed CRC progression at least partly by inactivating PI3K/AKT/mTOR signaling pathways, highlighting HHT as a potential therapeutic agent for CRC patients.
KEYWORDS: Network pharmacology, homoharringtonine, colorectal cancer, PI3K/AKT signaling
Introduction
Colorectal cancer (CRC), a kind of gastrointestinal malignancy deriving from either the colon or rectum, ranks third in newly diagnosed cancer cases and is the second most deadly cancer worldwide, with approximately 10.0% of morbidity and 9.4% of mortality, respectively [1]. Despite the stabilizing or decreasing incidence and death of CRC in highly developed countries, there is still a rapidly increasing trend in many low- and middle-income countries in the world [2]. In China, CRC cancer cases and deaths are expected to increase by 63% and 32% from 2016 to 2025 [3]. Familial history, pre-cancerous conditions, obesity, lack of physical exercise, red or processed meat consumption, alcohol drinking and tobacco smoking are major contributing factors to CRC carcinogenesis [4,5]. Surgical excision, chemotherapy, radiotherapy, immunotherapy, and targeted therapy have been applied alone or in combination for CRC management, depending on the clinical scenario [6]. Due to the inconspicuous symptoms of this disease at the early stage, most CRC patients are eventually diagnosed at the distant stages. The 5-year relative survival for CRC patients at distant stage is merely 14% [7]. Therefore, it is of great value to develop novel noninvasive therapies to improve the clinical outcome of advanced CRC patients.
With the characteristics of multi-targeted action, low cytotoxicity and inexpensiveness, nature compounds extracted from traditional Chinese medicine have gained wide attention in anticancer therapy [8,9]. Nature compounds can hinder cell proliferation, promote cell cycle arrest and apoptosis, reduce migration and invasion, and impair metabolism in CRC through regulating numerous molecules and signaling pathways [10]. Homoharringtonine (HHT), an alkaloid extracted from Cephalotaxus, has been used in China to treat patients with chronic myeloid leukemia, acute myeloid leukemia and myelodysplastic syndrome [11]. In addition to hematologic tumors, HHT is recently also found to exhibit anticancer properties in several solid malignancies. For instance, HHT inhibited cell proliferation and migration in hepatocellular carcinoma by regulating EphB4-mediated β-catenin loss [12]. HHT exerted antineoplastic activity in triple negative breast cancer by decreasing anti-apoptotic protein’s abundance [13]. HHT was proved an effective anti-melanoma agent via inducing DNA damage, apoptosis and cell cycle arrest [14]. HHT displayed an anti-tumor activity against lung adenocarcinoma carrying mutant Kras expression by changing the features of immune cells [15]. As for CRC, Park et al. found that HHT repressed viability and induced apoptosis of HCT116 cells via inactivating Wnt/β-Catenin signaling [16]. Shi et al. demonstrated that HHT could decrease LoVo cell growth by targeting EphB4 and its downstream signaling [17]. It is, however, necessary to systematically investigate the detailed pharmacological targets and molecular mechanisms of HHT in treating CRC.
Network pharmacology is a promising approach to establishing ‘drug-target-disease’ network models by integrating system biology, bioinformatics and high-throughput screening technology [18]. Network pharmacology has been widely used for the mechanism research of natural products in different human diseases, including cancer [19,20]. In this study, we aimed to comprehensively elucidate the potential targets, biological processes, and molecular pathways of HHT to treat CRC by using the network pharmacology method. In vitro and in vivo assays further demonstrated that HHT inhibited cell proliferation, migration and invasion, and promoted apoptosis in CRC by inactivating PI3K/AKT/mTOR signaling pathway.
Materials and methods
Collecting candidate targets of HHT
The potential targets of HHT were predicted using available online tools including PharmMpper (http://www.lilab-ecust.cn/pharmmapper/submitfile.html) [21], TargetNet (https://targetnet.scbdd.com) [22] and SwissTargetPrediction (https://www.swisstargetprediction.ch/) [23]. The protein structure of HHT was uploaded to PharmMapper as a mol2 format with limitation to ‘Homo sapiens’, and the targets were screened with ‘normal fit score >0.2’. Canonical smiles were input into TargetNet with AUC ≥ 0.7, and the targets were acquired with ‘probability >0’. Similarly, the targets of HHT were obtained from SwissTargetPrediction with limitation to ‘Homo sapiens’ and ‘probability >0’. After deleting the duplicates, we finally gained the putative targets of HHT.
Identifying targets associated with CRC
We comprehensively retrieved the CRC-related genes from five public databases: GeneCards (https://www.genecards.org/) [24] with ‘relevance score >30’, DisGeNET (http://www.disgenet.org/) [25] with ‘score_gda >0.1’, TTD (http://db.idrblab.net/ttd/) [26], DrugBank (https://www.drugbank.ca/) [27] and GSE110224 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110224) with ‘P < 0.05’ and ‘|log2Fold Change| ≥1’. After removing the duplicates, we finally achieved all putative targets related to CRC. A Venn diagram program was used to identify the target genes shared by HHT and CRC.
Network establishment
‘Compound-Target-Disease (C-T-D)’ network
The CTD interaction was constructed by entering 98 overlapping targets of HHT and CRC into the Cytoscape 3.7.2 software.
‘Protein–Protein Interaction (PPI)’ network
We employed a String database to construct the PPI network of indicated target genes. The TSV document of string-interaction was loaded into Cytoscape software for visualization and analysis. Cytoscape NetworkAnalyzer plug-in was used to analyze the topological parameters of the genes in the PPI network, including the median and maximum degrees of freedom. The nodes with degree between median and maximum degrees were identified as the hub targets of HHT against CRC.
Enrichment analysis of GO and KEGG pathway
GO function annotation and KEGG pathway enrichment of core targets were performed using the bioinformatics tool DAVID (https://david.ncifcrf.gov/). When P < 0.05, GO terms and KEGG pathways are regarded as significant. Bubble charts were produced to visualize the top 20 biological processes (BP), cellular components (CC), molecular functions (MF) and KEGG pathways related to hub genes.
Cell culture
CRC cell lines (LoVo and SW480) were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% FBS and 1% penicillin-streptomycin was used to culture cells at 37°C with 5% CO2. To activate PI3K/AKT pathway, we used a PI3K agonist 740Y-P (MedChem Express, Monmouth Junction, NJ, USA; 15 μM).
Cell Counting Kit-8 (CCK-8) assay
LoVo and SW480 cells were inoculated into 96-well plates with a density of 0.5 × 104 cells/well. After treatment with various concentrations of HHT (Baoji Herbest Biotech Co., Ltd., Shaanxi, China; 0.075, 0.15, 0.3, 0.6, 1.2, 2.4 and 4.8 µM) for 48 h, 10 µL of CCK-8 solution (Beyotime, Shanghai, China) was added to each well. After 3 h of incubation at 37°C, the absorbance at 450 nm was measured. Dose-response curves were plotted to calculate the concentration of HHT inhibiting cell growth by 50% (IC50).
5-Ethynyl-2ʹ-deoxyuridine (EdU) assay
A Cell-LightTM EdU DNA cell proliferation kit (RiboBio, Guangzhou, China) was used to determine cell proliferation and DNA synthesis. Briefly, LoVo and SW480 were treated with HHT (0, 0.15, 0.3 µM) for 48 h. Then, cells were incubated with 25 μM EdU solution, fixed with 4% paraformaldehyde, and permeabilized with 0.5% Triton X-100. After adding Apollo reaction reagent, DAPI dye was further utilized to stain the cell nucleus. EdU-positive cells were finally photographed and counted under a fluorescence microscope.
Colony formation assay
LoVo and SW480 cells (5 × 102 cells/well) were seeded into 6-well plates containing medium supplemented with the designated concentrations of HHT (0, 0.15, 0.3 µM) and cultured for 48 h. After growing in a standard growth medium for 12 days, cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 30 min. The visible colonies were counted in three random fields under a microscope.
Cell cycle assay
LoVo and SW480 cells were treated with HHT (0, 0.15, 0.3 µM) for 48 h. Then, cells were collected and fixed in pre-cold 70% ethanol overnight at 4°C. The following day, cells were resuspended in 500 μL PBS containing 50 μg/ml propidium iodide (PI) and 25 μg/ml RNase A, and incubated for 30 min at room temperature. A FACSCalibur flow cytometry (BD Biosciences, San Jose, CA, USA) was used to measure the cell cycle distribution.
Apoptosis assay
LoVo and SW480 cells were treated with HHT (0, 0.15, 0.3 µM) for 48 h. Subsequently, cells were stained with Annexin V-FITC/PI apoptosis detection kit (Nanjing KeyGen Biotech Co., Ltd., China) following the manufacturer’s protocol. Apoptotic cells were measured using a FACSCalibur flow cytometer (BD Biosciences).
Wound healing assay
LoVo and SW480 cells were maintained in 6-well plates until spreading over the plate. The cell layer was scratched by using a sterile pipette tip to create a wound. After removing the exfoliated cells with PBS, the remaining cells were maintained in a fresh medium containing HHT (0, 0.15, 0.3 µM) for 24 h. At 0 h and 24 h, images were captured with a microscope. The wound width of the cell-free area was determined by ImageJ software.
Transwell invasion assay
LoVo and SW480 cells (3 × 105) in serum-free medium with HHT (0, 0.15, 0.3 µM) were seeded into the top compartment of a Matrigel-coated Transwell chamber (Costar, Corning Inc., NY, USA). The lower compartment was added with medium containing 10% FBS. Cells were then incubated for 24 h at 37°C. After fixed with 4% formaldehyde and stained with 0.1% crystal violet, cells adhering to the lower surface were counted under an optical microscope.
Western blot assay
CRC tissues and cells were lysed by using RIPA buffer. The protein extracts were subjected to 10% SDS-PAGE gels and then transferred onto polyvinylidene fluoride membranes. Thereafter, the membranes were incubated with the primary antibodies against AKT (1:1000; Cat#4691 T), p-AKT (1:1000; Cat#4060S), mTOR (1:1000; Cat#2983S), p-mTOR (1:1000; Cat#5536S), PI3K (1:1000; Cat#4257 T), p-PI3K (1:1000, Cat#4228 T), Bax (1:1000, Cat#ab182734), Bcl-2 (1:1000, Cat#ab196495), MMP-2 (1:1000, Cat#ab97779), MMP-9 (1:1000, Cat#ab228402) and GAPDH (1:1000; Cat#5174) overnight at 4°C. The protein signals were detected after incubation with horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature by SuperSignal West Pico Chemiluminescent ECL system (Thermo Scientific, Waltham, MA, USA). AKT, p-AKT, mTOR, p-mTOR, PI3K, p-PI3K and GAPDH antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). Bax, Bcl-2, MMP-2 and MMP-9 were purchased from Abcam (Cambridge, MA, USA).
In vivo xenograft assays
Animal experiments were approved by the Institutional Animal Care and Use Committee of Guang’anmen Hospital, China Academy of Chinese Medical Sciences. The study procedures strictly complied with the National Institutes of Health guide for the care and use of laboratory animals. Male BALB/c nude mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). LoVo cells (8 × 106/100 μL) were subcutaneously injected into the right flank of mice. When xenograft tumors were visible, mice were randomly assigned into two groups (Control group and HHT group, n = 5/group). Mice in the HHT group were orally administrated with 0.5 mg/kg HHT every day. Mice in the control group were given an equal amount of PBS at the same time points. Tumor dimensions were monitored every 3 days, and tumor volumes were calculated with V = (Length × Width2)/2. On day 23, mice were sacrificed, and tumors were resected for further experiments. Formalin-fixed and paraffin-embedded sections were prepared from the xenograft tumors to perform immunohistochemistry (IHC) assay by using proliferating marker Ki-67 (Abcam; Cat#ab15580) and PCNA (Abcam; Cat#ab92729) antibodies.
Statistical analysis
GraphPad Prism 7 (San Diego, CA, USA) software was employed to plot graphics and analyze data. All experimental data are presented as mean ± SD. One-way analysis of variance (ANOVA) was used for statistical analysis. P < 0.05 indicates a significant difference. All experiments were performed in triplicate.
Results
This study aims to investigate the molecular mechanisms underlying HHT against CRC. First, a network pharmacology approach was applied to predict the candidate targets, biological functions and potential pathways of HHT against CRC. Then, in vitro and in vivo experiments were further performed to validate the functions and mechanisms of HHT in CRC.
Collecting targets associated with CRC and HHT
By searching GeneCards, DisGeNET, TTD, DrugBank and GSE110224, we obtained 1334 CRC-related genes. By retrieving data from PharmMpper, TargetNet and SwissTargetPrediction, a total of 463 targets associated with HHT were acquired. After eliminating the duplicates, 98 overlapping targets were regarded as the potential therapeutic targets of HHT against CRC (Figure 1a).
Figure 1.
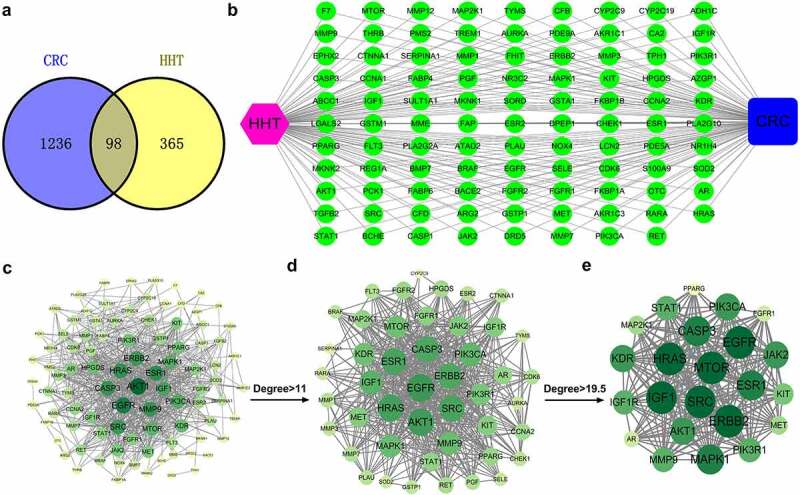
Network pharmacology construction of HHT against CRC. (a) Venn diagram displays the intersection genes of CRC- and HHT-related targets. (b) ‘Compound-Target-Disease’ network consisting of 100 nodes and 196 edges. (c–e) PPI network of common targets and topological analysis
Network construction
By using Cytoscape 3.7.2, a ‘Compound-Target-Disease’ network was established (Figure 1b). To elucidate the interactions among these common genes, we constructed a PPI network consisting of 93 nodes and 687 edges (Figure 1c). Based on the data analysis of network topology, a network of major targets with 46 nodes and 487 edges was generated (Figure 1d). Then, a network of core targets was further generated, with 23 nodes and 225 edges (Figure 1e). The top ten hub targets (ERBB2, EGFR, IGF1, MTOR, SRC, HRAS, MAPK1, CASP3, ESR1 and AKT1) may contribute to the main therapeutic effects of HHT in CRC.
GO function annotation and KEGG pathway enrichment
GO function annotation was conducted to explore the biological functions of 23 core targets involved in HHT against CRC. The results displayed a variety of GO enrichment terms, including 163 biological processes (BP), 18 cell components (CC), and 35 molecular functions (MF) items (Table S1). The top 20 BP, CC and MF items were presented as bubble charts in Figure 2a-C, suggesting that HHT may regulate cell proliferation, migration and apoptosis via protein binding and protein tyrosine kinase activity in plasma membrane, nucleus, cytosol and cytoplasm to exert its anti-cancer properity on CRC. Subsequently, we performed KEGG pathway enrichment of these 23 core targets to elucidate the mechanism of HHT against CRC. A total of 85 related pathways were identified (Table S2). The most significantly enriched pathways included Pathways in cancer, Proteoglycans in cancer, PI3K-AKT signaling pathway, Rap1 signaling pathway and Ras signaling pathway (Figure 2d). In addition, a ‘Target-Pathway’ network was built (Figure S1A). Among the 23 core targets, 15 are associated with PI3K-AKT signaling (Figure S1B). Based on the data mentioned above, we concluded that HHT might exert anticancer effect in CRC by regulating cell proliferation, apoptosis and migration via PI3K-AKT signaling.
Figure 2.

GO and KEGG pathway enrichment analysis of 23 core targets. (a) Enriched GO terms for biological processes (BP). (b) Enriched GO terms for cellular components (CC). (c) Enriched GO terms for molecular functions (MF). (d) Bubble chart of KEGG pathway enrichment results
HHT suppresses cell proliferation and induces apoptosis in CRC
To verify the inhibitory effects of HHT in CRC, we conducted a series of biological function experiments in LoVo and SW480 cells. CCK-8 assay showed that HHT significantly decreased cell viability in a dose-dependent manner (Figure 3a). The IC50 values of HHT were 0.35 μM and 0.4 μM for LoVo and SW480 cells, respectively. Subsequently, cells were treated with HHT (0.15 and 0.3 μM) for 48 h. EdU assays displayed a significant reduction of proliferative cells under the treatment of HHT (Figure 3b). Similarly, HHT treatment resulted in an apparent cell cycle arrest at G0/G1 stage (Figure 3c). Also, cell colony number was significantly declined in the presence of HHT (Figure 3d). As demonstrated by flow cytometry assays, a significant increase of apoptotic rate was observed after treatment with HHT (Figure 3e). Consistently, HHT treatment led to the increase of Bax protein and the decrease of Bcl-2 protein in LoVo and SW480 cells (figure 3f). Taken together, HHT repressed CRC cell proliferation in vitro.
Figure 3.

Effects of HHT on CRC cell proliferation and apoptosis. (a) LoVo and SW480 cells were incubated with different doses of HHT for 48 h, followed by CCK-8 assay of cell viability. (b–e) LoVo and SW480 cells were treated with HHT at 0, 0.15 and 0.3 µM for 48 h, followed by EdU assay of cell proliferation (b), flow cytometry assay of cell cycle (c), colony formation assay (d), flow cytometry assay of apoptosis (e), and Western blot assay of Bax and Bcl-2 proteins (f). **P < 0.01, ***P < 0.001 vs. 0 µM group
HHT inhibits cell migration and invasion in CRC
Next, we performed wound healing and transwell invasion assays to explore the effects of HHT on CRC cell migration and invasion. Compared to the control group, addition of HHT triggered a significant decrease of migration ability in both LoVo and SW480 cells (Figure 4a). Also, HHT treatment effectively suppressed cell invasion capability (Figure 4b). Moreover, the protein levels of MMP-2 and MMP-9 were significantly decreased in LoVo and SW480 cells after treatment with HHT (Figure 4c). These data suggested that HHT lowered CRC cell migration and invasion in vitro.
Figure 4.
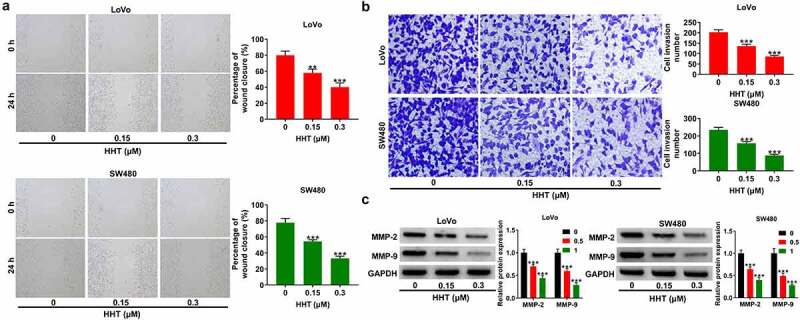
Effects of HHT on CRC cell migration and invasion. (a) Wound-healing assays were performed to determine the migration ability of LoVo and SW480 cells after treatment with HHT (0, 0.15 and 0.3 µM) for 24 h. (b) Transwell assays were used to measure the invasion capability after treatment with HHT (0, 0.15 and 0.3 µM) for 24 h. (c) Western blot assays were used to determine the protein levels of MMP-2 and MMP-9 in LoVo and SW480 cells after treatment with HHT (0, 0.15 and 0.3 µM) for 24 h. **P < 0.01, ***P < 0.001 vs. 0 µM group
HHT inactivates PI3K/AKT/mTOR pathway in CRC cells
To further investigate the influence of HHT on PI3K/AKT signaling, we performed Western blot assays to measure the protein expression of related proteins. As presented in Figure 5a and B, incubation of LoVo and SW480 cells with HHT at 0.15 and 0.3 μM significantly inhibited the phosphorylation level of PI3K, AKT and mTOR. These results indicated the suppressive effect of HHT on PI3K/AKT/mTOR signaling.
Figure 5.

Effects of HHT on PI3K/AKT/mTOR signaling pathway in CRC cells. (a and b) Western blot assays were applied to detect the protein level of p-PI3K, PI3K, p-AKT, AKT, p-mTOR and mTOR in LoVo and SW480 cells after incubation with HHT (0, 0.15 and 0.3 µM) for 48 h. **P < 0.01, ***P < 0.001 vs. 0 µM group
HHT inhibits CRC progression via inactivation of PI3K/AKT pathway
To further clarify whether PI3K/AKT/mTOR pathway was involved in the anticancer effects of HHT on CRC, we used 740Y-P to activate PI3K/AKT signaling in LoVo and SW480 cells in the presence of 0.3 μM HHT. As shown in Figure 6a, HHT-induced inhibition of p-PI3K, p-AKT and p-mTOR was reversed by 740Y-P treatment. Moreover, the proliferation inhibition mediated by HHT was abated due to the intervention of 740Y-P (Figure 6b and C). Consistently, treatment with 740Y-P significantly attenuated HHT-mediated apoptosis of CRC cells (Figure 6d). What is more, the addition of 740Y-P significantly counteracted HHT-induced inhibitory influence on cell migration and invasion (Figure 6e and F). Also, HHT-mediated increase of Bax and decrease of Bcl-2, MMP-2 and MMP-9 were effectively reversed by 740Y-P (Figure 6g). Collectively, activation of PI3K/AKT pathway antagonized the antineoplastic property of HHT in CRC.
Figure 6.
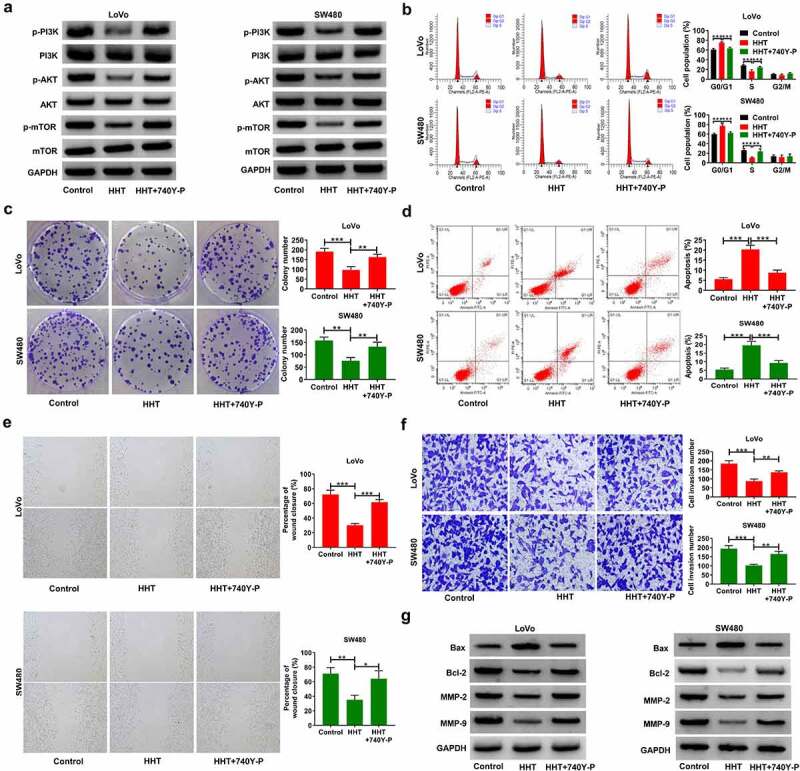
Activation of PI3K/AKT/mTOR signaling attenuates the inhibitory effects of HHT on cell malignant phenotypes in CRC. LoVo and SW480 cells were treated with 0.5 µM HHT alone or combined with 15 μM 740Y-P. (a) Western blot assays of p-PI3K, PI3K, p-AKT, AKT, p-mTOR and mTOR protein levels in treated cells. (b) Cell cycle progression was examined with flow cytomerty analysis. (c) Determination of colony forming potential. (d) Flow cytometry assay of apoptosis in treated cells. (e) Wound healing assays were used to evaluate cell migration. (f) Transwell assays were performed to determine cell invasion. (g) Western blot assays of Bax, Bcl-2, MMP-2 and MMP-9 proteins. *P < 0.05, **P < 0.01, ***P < 0.001 vs. Control or HHT group
HHT suppresses CRC tumor growth and cell proliferation in vivo
A xenograft mouse model was used to detect the effect of HHT on CRC tumor growth. The results showed that compared with the control group, HHT treatment significantly slowed tumor growth (Figure 7a). Moreover, the tumor weight of HTT-treated mice was significantly decreased (Figure 7b and C). As presented by the IHC assay, HHT treatment led to a decrease in Ki-67 and PCNA expression in tumors compared with the control group (Figure 7d). Also, HHT administration significantly suppressed the phosphorylation level of PI3K, AKT and mTOR in xenograft tumors (Figure 7e). Thus, we concluded that HHT repressed CRC tumor growth in vivo.
Figure 7.
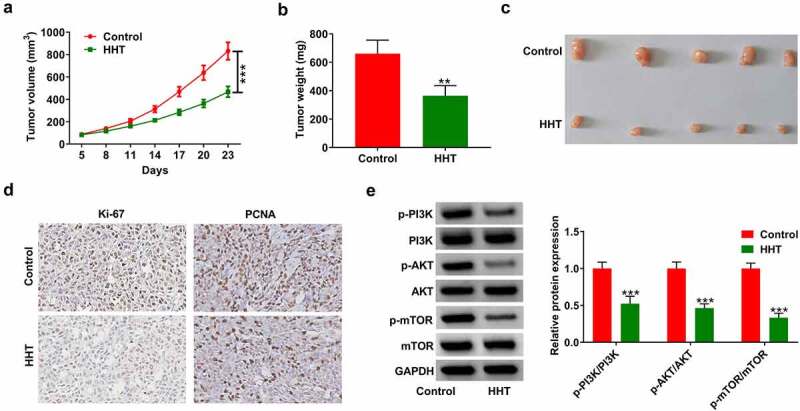
HHT represses CRC tumor growth in vivo. LoVo cells were subcutaneously injected into the right flank of mice. When xenograft tumors were visible, HHT (0.5 mg/kg) or PBS were administrated orally to mice. (a) Tumor size was monitored every 3 days and tumor volume was calculated. (b) Tumor weight was measured at day 23 after cell inoculation. (c) Representative images of removed tumor masses. (d) IHC analysis of Ki-67 and PCNA expression in the excised tumors. (e) Western blot assays of proteins associated with PI3K/AKT/mTOR signaling in tumors
Discussion
In recent years, there have been some reports on the role of HHT in solid tumors. HHT suppressed cell growth and aggravated apoptosis in gefitinib-resistant lung cancer cells by regulating IL-6/JAK/STAT3 signal pathway [28]. HHT abated the malignant phenotypes in hepatocellular carcinoma through activating the Hippo pathway [29]. Although there have been documents reporting the role of HHT in CRC [16,17], the complex biological functions and molecular mechanisms deserve further investigation. In this study, we applied network pharmacology prediction followed by experiment validation to elucidate the mechanism behind the anticancer property of HHT in CRC (Figure 8).
Figure 8.
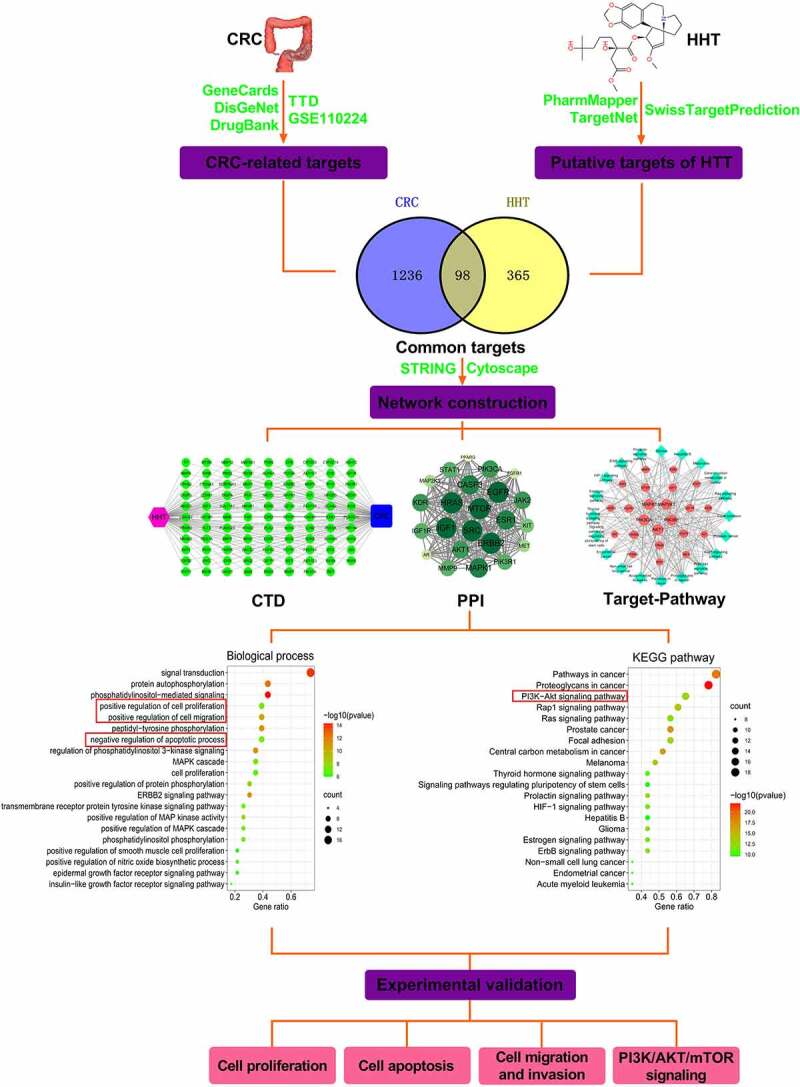
A flow chart of network pharmacology analysis and experiment validation to elucidate the functions and mechanisms of HHT against CRC
By using online public databases, a total of 98 genes were predicted to be shared by both CRC and HHT. According to PPI network and topological screening, we identified 23 core genes potentially involved in the therapeutic effects of HHT in CRC. According to the degree value, we screened out the top 10 key targets (ERBB2, EGFR, IGF1, MTOR, SRC, HRAS, MAPK1, CASP3, ESR1 and AKT1). The mutations and amplifications of ERBB2 (also known as HER2) are present in 7% of colorectal cancers, and ERBB2 is demonstrated as a potential therapeutic target for CRC [30]. EGFR, a transmembrane receptor tyrosine kinase (RTK) belonging to the ERBB family, can activate different downstream pathways, such as RAS-RAF-MAPK, PI3K-PTEN-AKT, and JAK/STAT, thereby promoting cancer cell survival, proliferation, metastasis, and angiogenesis [31]. IGF1 is reported to exert a crucial role in the anti-apoptotic and mitogenic pathway in tumorigenesis by activating PI3K/AKT and MAPK signaling [32]. Epidemiological evidence reveals that IGF1 expression was increased in CRC, and associated with tumor size, distant metastasis and advanced stages [33]. SRC, a non-receptor protein tyrosine kinase, can promote cell proliferation, metastasis, chemoresistance, and formation of cancer stem cells, and targeting Src might be an effective strategy to treat CRC patients [34]. RAS oncogene, including three isoforms (HRAS, KRAS, and NRAS), can regulate cell growth, proliferation, apoptosis, migration, division and differentiation in various human solid cancers [35]. CASP3, a major mediator of apoptosis, not only enhances the sensitivity of CRC cells to chemotherapy and radiotherapy, but also suppresses cell invasion and metastasis [36]. MAPK cascades is involved in gene expression, cell cycle, apoptosis and differentiation in cancer cells, and targeting MAPK is highlighted as a novel therapeutic intervention for cancers [37]. These data indicated that HHT might exert anti-cancer activity in CRC by targeting these hub genes. The key targets predicted by network pharmacology provided a theoretical basis for exploring the multi-targets of HHT against CRC. However, the specific action targets of HHT against CRC need further elucidation by molecular docking and experimental verification.
GO function and KEGG pathway assays indicated that HHT might hinder CRC progression by affecting proliferation, migration and apoptosis via PI3K/AKT signaling. Subsequently, we performed a series of experiments to confirm our hypothesis. Function assays showed that HHT could inhibit cell proliferation, cell cycle, colony formation and induce apoptosis in a dose-dependent manner. Moreover, HHT repressed CRC cell migration and invasion dose-dependently. These data confirmed the inhibitory effect of HHT on cell malignant phenotypes in CRC. HHT administration also slowed tumor growth in nude mice. Compared with the report by Shi et al. [16], we used network pharmacology and experimental validation to comprehensively elucidate the pharmacological targets and pathways of HHT against CRC.
PI3K/AKT signaling is always abnormally activated in human cancers and is closely associated with tumourigenesis, proliferation, apoptosis, metastasis, epithelial-mesenchymal transition (EMT), metabolism and chemoresistance [38,39]. PI3K/AKT/mTOR signaling is demonstrated as an important event in colorectal carcinogenesis, and inhibiting PI3K/AKT/mTOR pathway can treat primary and metastatic CRC [40]. In the current study, we found that HHT treatment suppressed PI3K/AKT/mTOR signaling. After activating PI3K/AKT signaling with 740Y-P, HHT-mediated suppression of cell proliferation, migration and invasion was effectively abated. Furthermore, HHT treatment in nude mice significantly repressed CRC xenograft growth through inhibiting PI3K/AKT/mTOR signaling. These findings elucidated that PI3K/AKT signaling was involved in the anti-tumor role of HHT in CRC. However, the detailed targets by which HHT activated PI3K/AKT/mTOR signaling remain for further investigation. Consistent with our findings, HHT suppressed cell proliferation, migration, invasion, EMT and facilitated apoptosis in hepatocellular carcinoma via inhibiting PI3K/AKT/GSK3β/Slug signaling [41]. PI3K/AKT inhibitor LY294002 potentiated the anti-myeloma activity of HHT both in vitro and in vivo [42]. Natural products have emerged as an exciting adjunctive therapy to treat malignant tumors combined with conventional chemotherapeutic agents [43]. The influence of HHT on multidrug resistance in CRC is required to be further explored.
Conclusion
In conclusion, we applied network pharmacology and experimental validation to explore the pharmacological mechanism of HHT against CRC. HHT suppressed cell proliferation, colony formation, migration and invasion, and induced cell cycle arrest and apoptosis in CRC by inactivating PI3K/AKT/mTOR signaling pathway. This study highlights the therapeutic potential of HHT for CRC patients. More importantly, a combination of network pharmacology and experimental validation offers a powerful approach for investigating natural products’ mechanisms in cancer management.
Supplementary Material
Funding Statement
The authors reported there is no funding associated with the work featured in this article.
Research highlights
The common targets of HHT and CRC are associated with PI3K/AKT signalling.
HHT inhibits cell proliferation, migration and invasion, and induced apoptosis in CRC.
HHT results in the inactivation of PI3K/AKT/mTOR signalling in CRC cells.
HHT suppresses CRC progression in vitro and in vivo by inactivating PI3K/AKT/mTOR pathway.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplemental data for this article can be accessed here
References
- [1].Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. [DOI] [PubMed] [Google Scholar]
- [2].Arnold M, Sierra MS, Laversanne M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. [DOI] [PubMed] [Google Scholar]
- [3].Zhang L, Cao F, Zhang G, et al. Trends in and predictions of colorectal cancer incidence and mortality in China from 1990 to 2025. Front Oncol. 2019;9:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rawla P, Sunkara T, Barsouk A.. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14(2):89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mattiuzzi C, Sanchis-Gomar F, Lippi G.. Concise update on colorectal cancer epidemiology. Ann Transl Med. 2019;7(21):609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet. 2019;394(10207):1467–1480. [DOI] [PubMed] [Google Scholar]
- [7].Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. [DOI] [PubMed] [Google Scholar]
- [8].Dutta S, Mahalanobish S, Saha S, et al. Natural products: an upcoming therapeutic approach to cancer. Food Chem Toxicol. 2019;128:240–255. [DOI] [PubMed] [Google Scholar]
- [9].Goyal S, Gupta N, Chatterjee S, et al. Natural plant extracts as potential therapeutic agents for the treatment of cancer. Curr Top Med Chem. 2017;17(2):96–106. [DOI] [PubMed] [Google Scholar]
- [10].Sun Q, He M, Zhang M, et al. Traditional Chinese medicine and colorectal cancer: implications for drug discovery. Front Pharmacol. 2021;12:685002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lü S, Wang J. Homoharringtonine and omacetaxine for myeloid hematological malignancies. J Hematol Oncol. 2014;7(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhu M, Gong Z, Wu Q, et al. Homoharringtonine suppresses tumor proliferation and migration by regulating EphB4-mediated β-catenin loss in hepatocellular carcinoma. Cell Death Dis. 2020;11(8):632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yakhni M, Briat A, El Guerrab A, et al. Homoharringtonine, an approved anti-leukemia drug, suppresses triple negative breast cancer growth through a rapid reduction of anti-apoptotic protein abundance. Am J Cancer Res. 2019;9(5):1043–1060. [PMC free article] [PubMed] [Google Scholar]
- [14].Tang JF, Li GL, Zhang T, et al. Homoharringtonine inhibits melanoma cells proliferation in vitro and vivo by inducing DNA damage, apoptosis, and G2/M cell cycle arrest. Arch Biochem Biophys. 2021;700:108774. [DOI] [PubMed] [Google Scholar]
- [15].Weng TY, Wu HF, Li CY, et al. Homoharringtonine induced immune alteration for an efficient anti-tumor response in mouse models of non-small cell lung adenocarcinoma expressing Kras mutation. Sci Rep. 2018;8(1):8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Park M, Kwon HJ, Kim SH. Homoharringtonine induces apoptosis in human colorectal carcinoma HCT116 cells via downregulation of Wnt/β-Catenin signaling cascade. Bull Korean Chem Soc. 2019;40(2):196–199. [Google Scholar]
- [17].Shi X, Zhu M, Gong Z, et al. Homoharringtonine suppresses LoVo cell growth by inhibiting EphB4 and the PI3K/AKT and MAPK/EKR1/2 signaling pathways. Food Chem Toxicol. 2020;136:110960. [DOI] [PubMed] [Google Scholar]
- [18].Poornima P, Kumar JD, Zhao Q, et al. Network pharmacology of cancer: from understanding of complex interactomes to the design of multi-target specific therapeutics from nature. Pharmacol Res. 2016;111:290–302. [DOI] [PubMed] [Google Scholar]
- [19].Kibble M, Saarinen N, Tang J, et al. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat Prod Rep. 2015;32(8):1249–1266. [DOI] [PubMed] [Google Scholar]
- [20].Chen L, Ren LQ, Liu Z, et al. Bio-informatics and in vitro experiments reveal the mechanism of schisandrin a against MDA-MB-231 cells. Bioengineered. 2021;12:7678–7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang X, Shen Y, Wang S, et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017;45(W1):W356–W360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yao ZJ, Dong J, Che YJ, et al. TargetNet: a web service for predicting potential drug-target interaction profiling via multi-target SAR models. J Comput Aided Mol Des. 2016;30:413–424. [DOI] [PubMed] [Google Scholar]
- [23].Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47:W357–W364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stelzer G, Rosen N, Plaschkes I, et al. The genecards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinf. 2016;54:1.30.31–31.30.33. [DOI] [PubMed] [Google Scholar]
- [25].Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–D855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang Y, Zhang S, Li F, et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48:D1031–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cao W, Liu Y, Zhang R, et al. Homoharringtonine induces apoptosis and inhibits STAT3 via IL-6/JAK1/STAT3 signal pathway in Gefitinib-resistant lung cancer cells. Sci Rep. 2015;5(1):8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang H, Wang R, Huang D, et al. Homoharringtonine exerts anti-tumor effects in hepatocellular carcinoma through activation of the hippo pathway. Front Pharmacol. 2021;12:592071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pectasides E, Bass AJ. ERBB2 Emerges as a new target for colorectal cancer. Cancer Discov. 2015;5(8):799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhao B, Wang L, Qiu H, et al. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget. 2017;8(3):3980–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].AsghariHanjani N, Vafa M. The role of IGF-1 in obesity, cardiovascular disease, and cancer. Med J Islam Repub Iran. 2019;33:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hu J, Liu X, Chi J, et al. Expressions of IGF-1, ERK, GLUT4, IRS-1 in metabolic syndrome complicated with colorectal cancer and their associations with the clinical characteristics of CRC. Cancer Biomark. 2018;21:883–891. [DOI] [PubMed] [Google Scholar]
- [34].Jin W. Regulation of Src family kinases during colorectal cancer development and its clinical implications. Cancers (Basel). 2020;12:1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kodaz H, Kostek O, Hacioglu MB, et al. Frequency of RAS mutations (KRAS, NRAS, HRAS) in human solid cancer. Breast Cancer. 2017;7:1–7. [Google Scholar]
- [36].Zhou M, Liu X, Li Z, et al. Caspase-3 regulates the migration, invasion and metastasis of colon cancer cells. Int J Cancer. 2018;143(4):921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yuan J, Dong X, Yap J, et al. The MAPK and AMPK signalings: interplay and implication in targeted cancer therapy. J Hematol Oncol. 2020;13:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jiang N, Dai Q, Su X. Role of PI3K/AKT pathway in cancer: the framework of malignant behavior. Mol Biol Rep. 2020;47(6):4587–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Alzahrani AS. PI3K/Akt/mTOR inhibitors in cancer: at the bench and bedside. Semin Cancer Biol. 2019;59:125–132. [DOI] [PubMed] [Google Scholar]
- [40].Narayanankutty A. PI3K/ Akt/ mTOR pathway as a therapeutic target for colorectal cancer: a review of preclinical and clinical evidence. Curr Drug Targets. 2019;20(12):1217–1226. [DOI] [PubMed] [Google Scholar]
- [41].Liu HY, Dong TX, Li ZZ, et al. Homoharringtonine inhibits the progression of hepatocellular carcinoma by suppressing the PI3K/AKT/GSK3β/Slug signaling pathway. Neoplasma. 2021;68(5):924–937. [DOI] [PubMed] [Google Scholar]
- [42].Chen P, Wen X, Wang B, et al. PI3K/Akt inhibitor LY294002 potentiates homoharringtonine antimyeloma activity in myeloma cells adhered to stromal cells and in SCID mouse xenograft. Ann Hematol. 2018;97(5):865–875. [DOI] [PubMed] [Google Scholar]
- [43].Yuan R, Hou Y, Sun W, et al. Natural products to prevent drug resistance in cancer chemotherapy: a review. Ann N Y Acad Sci. 2017;1401(1):19–27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


