ABSTRACT
In recent years, a steady increase has been detected in the incidence of acute cerebral infarction (ACI). ACI is caused by blood flow disruption, leading to high disability and mortality rates. Understanding the underlying molecular mechanisms is critical toward developing effective therapeutic approaches. Circular RNAs (circRNAs) are an important class of non-coding RNAs, which have been implicated in several molecular pathways, and their dysregulation has been described in several disease conditions. Here, we set out to explore the possible regulatory role of circRNAs in ischemic stroke and study their molecular function in disease. First, we applied high-throughput sequencing techniques to identify the differential changes of plasma circRNAs expression in patients with acute cerebral infarction. Next, we used GO and KEGG pathway analysis to predict the function of differentially expressed circRNAs. Moreover, we have assessed the possible interaction between the identified differentially expressed circRNAs and miRNAs. Finally, we have selected and validated five downregulated circRNAs by RT-qPCR. Together, the results of this study provide evidence that circRNAs are potential biomarkers for early diagnosis of cerebral infarction and have to be considered as targets for drug treatment.
KEYWORDS: Acute cerebral infarction, circRNAs, bioinformatics analysis, ischemic stroke
Introduction
Acute cerebral infarction (ACI) is one of the most common and high-incidence cerebrovascular diseases, accounting for approximately 70% of all stroke [1]. ACI is most frequently caused by hypoxia and ischemia of brain tissue, leading to a nerve function damage [2]. Currently, our understanding is rather limited regarding the molecular mechanisms leading to the development of ACI, and we mainly rely on imaging and hemodynamics to evaluate the condition and prognosis [3–5]. Therefore, better understanding of disease-associated changes at a molecular level is necessary in order to find potential intervention or treatment targets.
Circular RNAs (circRNAs) are a highly conserved and stable class of non-coding RNAs, which are found in plant as well as animal cells [6]. CircRNAs have tissue specificity, and they have been described in the regulation of gene expression. Moreover, multiple studies have shown that circRNAs can play a specific role in disease development and progression [7,8]. Mechanistically, circRNAs can enter the blood with exosomes as carriers and stably exist in the peripheral blood. Because it can stably exist in the collected blood samples, the information of tissues and cells can be obtained by detecting the blood samples [9]. In addition, exosomes can cross the blood-brain barrier. Since circRNAs are released from the brain and their presence can be detected in peripheral blood, circRNAs are expected to be suitable biomarkers for auxiliary diagnosis of diseases. In fact, detection of circRNAs would provide an opportunity for a noninvasive examination and early diagnosis of ACI from easily accessible peripheral blood samples.
Here, we hypothesize that some circRNAs are differentially expressed in the body after ACI, and these circRNAs may play an important role in regulating gene expression and influence the disease progression by influencing some specific signal pathways. In order to explore the relationship between circRNAs and ACI, we first quantified the differential changes of plasma circRNA expression in patients with ACI by a high-throughput sequencing technique. The function of differentially expressed circRNAs was predicted by bioinformatics analysis, including GO and KEGG pathway analysis. The possible interaction between circRNAs and miRNAs was explored by predicting their binding. In addition, five downregulated circRNAs were selected for RT-qPCR verification. The purpose of this study is to reveal the regulatory role of circRNAs during ischemic stroke and their interaction with miRNAs by analyzing the expression profile of circRNAs after ischemic stroke. Overall, the results of this study reveal that developing circRNAs are potential novel markers for early diagnosis of cerebral infarction and they should be considered as drug treatment targets.
Materials and methods
Patients
We have collected samples from patients diagnosed with ACI after their admission to the First Affiliated Hospital of Harbin Medical University. Sample collection took place between July 2020 and September 2020. Patients admitted to the hospital due to weakness of one limb were considered for this study. In total, after patients met with all of our inclusion/exclusion criteria (see below), we have been able to collect samples from a total of 3 patients, including two males and one female, with an average age of 61.3 years. Patients had no obvious systemic diseases, except hypertension and diabetes. All patients had clear symptoms of neurological deficits. MRI or CT confirmed the diagnosis of ischemic stroke [10]. In addition, three patients with normal physical examination at the health management center were selected as the normal control group, including one male and two females, with an average age of 61 years.
Inclusion criteria
[1] Meet the general diagnostic criteria of ACI and have exact symptoms of focal neurological deficit, and the diagnosis of ischemic stroke is confirmed by brain MRI (Supplementary Figures 1–3) or CT.
[2] The age is between 55 and 70 years old, and it may be complicated with hypertension and/or diabetes.
[3] The onset time is less than 7 days
Exclusion criteria
Age ≤55 years old or ≥70 years old;
The onset time is more than 7 days; Have a history of other nervous system-related diseases except hypertension and diabetes.
[2] Imaging suggests other intracranial lesions, such as hemorrhage and space occupation.
[3] It is complicated with respiratory, circulatory, blood, and other system diseases or has a history of corresponding diseases in the past.
Sample preparation
After the patient was admitted to hospital, the fasting venous blood of the patient was collected in the morning. Then, plasma was separated by centrifugation, stored in EP tube, labeled, and stored in an ultra-low temperature refrigerator at −80°C until further downstream processing.
Preparation, quality control, and sequencing of RNA library
The qualified samples were enrolled into the group, and the total RNA was extracted after DNA and protein removal. RNA concentrations extracted from samples were measured using a Nano Drop 1000ND analyzer (Thermofisher Scientific, Waltham, MA, USA). The ratio of OD260/OD280 was used as the index of RNA purity. Only samples with OD260/OD280 ratio between 1.8 and 2.1 were considered for downstream processing. A circRNA sequencing library was prepared according to the following steps..
RNA pretreatment. 5 μg of total RNA was used as starting material, and the circRNA enrichment kit (Cloud-SEQ Inc, Shanghai, China) was used for the enrichment of circRNA to obtain the expression profile of circRNAs.
RNA library construction. RNA was pretreated by TruSeq stranded total RNA library prep kit (Illumina, USA). RNA sequencing library was prepared according to the manufacturer's recommendations.
RNA integrity quality control: We have assessed RNA and library quality and quantification using a BioAnalyzer 2100 instrument (Agilent Technologies, USA). In brief, 10 pM library was transformed into single-stranded DNA molecules, which were sequenced using an Illumina 4000 HiSeq sequencer and paired end mode.
Functional go analysis of differential circRNAs
At the time of this study, no special annotation information was available for circRNAs, so we utilized Yunxu organisms to carry out GO functional analysis on the host genes that were found to be linked to differentially expressed circRNAs. GO hits with a p-value <0.05 were considered as statistically significant.
Analysis of KEGG pathway of genes derived from different circRNAs
According to the detected difference in the expression of circRNAs, we carried out a pathway analysis. P-values for each path with different genes (circRNA-derived genes) are indicated. By analyzing the pathways of differentially expressing circRNA-derived genes, we can infer their involved pathways and their biological functions. A P value <0.05 is used as the threshold of significant enrichment.
Prediction of interaction between CircRNAs and microRNA
Previous studies indicated specific interactions between differentially expressed circRNAs and miRNAs, suggesting that circRNAs may function as miRNA sponges. We utilized miRNA target gene prediction software TargetScan (version V7.0) to explore the function of circRNAs. Moreover, miRNA prediction software independently developed by Yunxu Biotechnology Co., Ltd. based on miRanda and TargetScan was also used to predict the binding sites of miRNAs target genes.
Extraction of total RNA from samples
First, whole blood was homogenized after thawing. In brief, 0.2 ml of chloroform (Shanghai Shenggong) was added to every sample homogenized in 750 μl of TRIzol LS Reagent. After shaking the tube vigorously by hand for 15 seconds, samples were incubated at room temperature for 2 to 3 minutes. Next, we centrifuged the samples at 12,000 × g for 15 minutes at 4°C. After centrifugation, the mixed liquid was divided into the lower red phenol-chloroform phase, the middle layer and the upper colorless water phase. The waterish phase, consisting the RNA was transferred to a new centrifuge tube, and 500 μl of isopropanol (Shanghai Shenggong) was added to each sample. After mixing well, samples. Were incubated at 15 to 30°C for 10 minutes, then centrifuged at 12,000 × g at 4°C for 10 minutes, and form on the bottom and side walls of the tube Gelatinous precipitation block. RNA pellets were washed with ethanol and centrifuged again at 7,500 × g at 4°C for 5 minutes. After removing the ethanol, samples were dried on air for 5–10 minutes.Finally, RNA was dissolved by adding RNase-free water to dissolve RNA. The obtained RNA solution was stored at −70°C. The corresponding values at 260 nm and 280 nm were detected by Nano Drop ND 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) to calculate the total amount and quality of RNA.
Different specifications of EP tubes, pipettes, and other experimental consumables used in the process of extracting total RNA all use RNase-free disposable consumables (Shanghai Shenggong), and the operation is carried out on a clean bench icebox. Equipment used in the experiment, such as tweezers, was sterilized at high temperature and high pressure for 2 hours.
Statistical method
Data analysis was carried out using Microsoft Excel (2019). Briefly, the relative expression level of circRNAs was reflected by calculating the relative expression level of target circRNAs in the diseased group compared with the control group. The experimental data were statistically analyzed by SPSS 17.0. Differences with a P < 0.05 was considered statistically significant.
Results
Expression pattern of cyclic ribonucleic acid after acute cerebral infarction
In this study, we set out to investigate the role of circRNAs in ACI. In order to find new biomarkers and therapeutic targets for ACI, we studied the differential changes of plasma circRNAs expression in patients with ACI using high-throughput sequencing techniques.
As shown in Figure 1, a total of 1904 differentially expressed circRNAs were detected in our analysis. Cluster analysis showed that there were significant differences between the experimental group and the control group. We found that 1509 circRNAs were downregulated and 395 were upregulated. Table 1 shows the top 20 differentially expressed circRNAs that were significantly differentially expressed. Among these 20, 10 circRNAs were upregulated and 10 were downregulated. The hits were further thresholded, and those were kept that showed at least a 2-fold change and P-values <0.05. As shown in Figure 2a, the comparison of the two different conditions showed differential expression of circRNAs. Scatter plot results showed robust changes in the expression of circRNAs between the sham control group and the ACI group (Figure 2b). Furthermore, the results of Figure 2c show the distribution of differentially expressed circRNAs in chromosomes. The bar graph based on the gene-derived cyclic ribonucleic acid category shows that 342 exons, 4 antisense, 46 introns, 1 intergene and 13 sense overlapping cyclic ribonucleic acids are upregulated. In contrast, 1301 exons, 2 antisense, 69 introns, 3 intergenes, and 123 sense overlapping cyclic ribonucleic acids were downregulated (Figure 2d).
Figure 1.
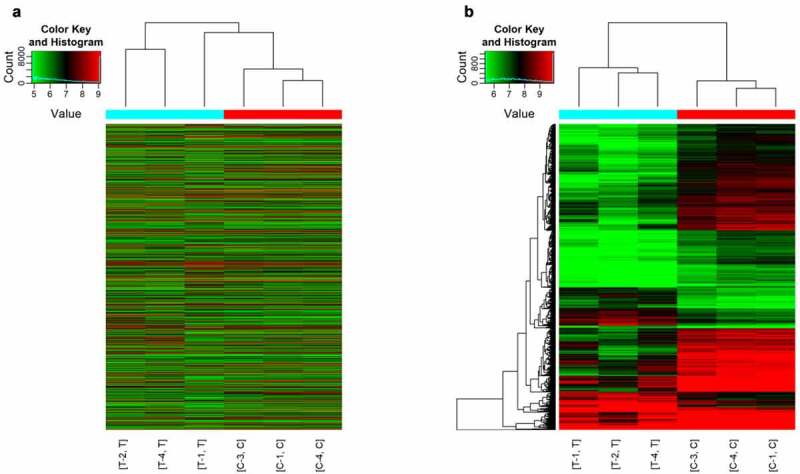
Hierarchical clustering of circRNAs expression. C1/3/4 refers to the peripheral blood samples of sham control group, and T1/2/4 refers to the peripheral blood samples of SCI group. (a) Hierarchical cluster analysis included 13,279 circRNAs in ACI group and sham control group. (b) Hierarchical cluster analysis includes differentially expressed circRNAs (P < 0.05; folding change≥2). There was significant difference between ACI group and sham operation group
Table 1.
Top 10 upregulated and downregulated circRNAs ranked by fold changes after SCI
| circRNA |
FC (abs) |
P-value |
FDR |
chrom |
chrom |
strand |
circRNA_type |
best_transcript |
GeneSymbol |
GeneSymbol |
Regulation |
| hsa_circRNA_103372 | 21.04715 | 0.034768 | 0.195768 | chr3 | chr3 | - | exonic | NM_016291 | IP6K2 | IP6K2 | up |
| hsa_circRNA_405965 | 16.28764 | 0.012321 | 0.129885 | chr2 | chr2 | - | exonic | ENST00000392929 | CCNT2-AS1 | CCNT2-AS1 | up |
| hsa_circRNA_103948 | 13.66356 | 0.037862 | 0.204714 | chr5 | chr5 | + | exonic | NM_021982 | SEC24A | SEC24A | up |
| hsa_circRNA_000102 | 12.03363 | 0.012049 | 0.128922 | chr1 | chr1 | - | intronic | ENST00000357393 | AKNAD1 | AKNAD1 | up |
| hsa_circRNA_001691 | 11.66992 | 0.027711 | 0.176117 | chr7 | chr7 | - | exonic | NM_002137 | HNRNPA2B1 | HNRNPA2B1 | up |
| hsa_circRNA_405641 | 11.58714 | 0.038655 | 0.206399 | chr17 | chr17 | - | intronic | ENST00000269392 | AZI1 | AZI1 | up |
| hsa_circRNA_104939 | 11.54522 | 0.04101 | 0.213631 | chr9 | chr9 | + | exonic | NM_005157 | ABL1 | ABL1 | up |
| hsa_circRNA_074559 | 10.41267 | 0.03915 | 0.207731 | chr5 | chr5 | - | exonic | NM_016221 | DCTN4 | DCTN4 | up |
| hsa_circRNA_008389 | 10.07535 | 0.012801 | 0.131943 | chr1 | chr1 | - | exonic | NM_018198 | DNAJC11 | DNAJC11 | up |
| hsa_circRNA_019744 | 8.532438 | 0.039087 | 0.207485 | chr10 | chr10 | - | exonic | NM_005736 | ACTR1A | ACTR1A | up |
| hsa_circRNA_103887 | 20.1334 | 0.00224 | 0.089408 | chr5 | chr5 | + | exonic | NM_138782 | FCHO2 | FCHO2 | down |
| hsa_circRNA_101836 | 19.86489 | 0.021279 | 0.159215 | chr16 | chr16 | + | exonic | NM_004555 | NFATC3 | NFATC3 | down |
| hsa_circRNA_406791 | 18.30178 | 0.001674 | 0.088954 | chr6 | chr6 | + | exonic | NM_015555 | ZNF451 | ZNF451 | down |
| hsa_circRNA_102995 | 18.21764 | 0.004734 | 0.098011 | chr20 | chr20 | + | exonic | NM_019095 | CRLS1 | CRLS1 | down |
| hsa_circRNA_102183 | 16.39273 | 0.003117 | 0.092018 | chr17 | chr17 | + | exonic | NM_004459 | BPTF | BPTF | down |
| hsa_circRNA_000581 | 16.28442 | 0.001974 | 0.088954 | chr14 | chr14 | + | exonic | NR_038354 | WDR20 | WDR20 | down |
| hsa_circRNA_101837 | 16.22925 | 0.022342 | 0.160614 | chr16 | chr16 | + | exonic | NM_004555 | NFATC3 | NFATC3 | down |
| hsa_circRNA_103572 | 16.15558 | 0.010143 | 0.121564 | chr3 | chr3 | + | exonic | uc003fyk.2 | LRCH3 | LRCH3 | down |
| hsa_circRNA_101771 | 14.87513 | 0.004531 | 0.097171 | chr16 | chr16 | + | exonic | NM_014494 | TNRC6A | TNRC6A | down |
| hsa_circRNA_092476 | 14.86703 | 0.003553 | 0.092018 | chr19 | chr19 | - | exonic | ENST00000596831 | AC004076.9 | AC004076.9 | down |
Figure 2.
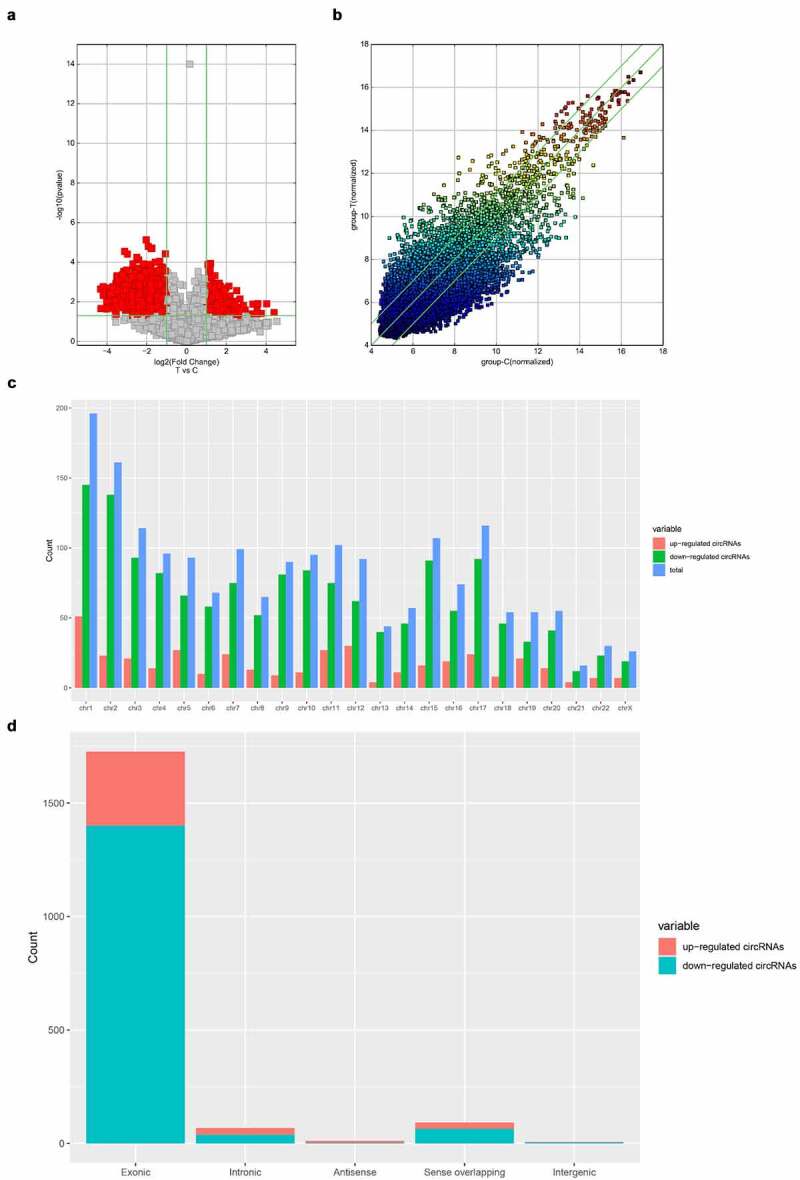
Difference of expression profiles of circRNAs between two groups. (a) Scatter plot show the difference of circRNAs expression between sham operation group and ACI group. The x-axis and y-axis values in the scatter plot are the normalized signal values of samples (log-2 scale) or the average normalized signal values of sample groups (log-2 scale). The green line is the fold change line. The circRNAs above and below the green line represent those circRNAs which show at least 2-fold changes in expression. (b) Volcano map show that cyclic ribonucleic acid was differentially expressed with statistical significance (P < 0.05; folding change ≥ 2). (c) The distribution of circRNAs indicated that the dysregulated circRNAs originated from each chromosome. (d) The classification bar chart of circRNAs based on gene source shows that most cyclic ribonucleic acids changed after ACI are exons
Finally, we predicted the five most likely potential targets. Table 2 lists the five highest-level candidate circRNAs (the first 10 upregulated and downregulated microRNAs) as binding targets for each circRNA.
Table 2.
The five highest-ranking miRNA candidates for top 10 upregulated and downregulated circRNAs
| circRNA |
Predicted miRNA response elements (MREs) | ||||
| MRE1 |
MRE2 |
MRE3 |
MRE4 |
MRE5 |
|
| hsa_circRNA_103372 | hsa-miR-494-5p | hsa-miR-577 | hsa-miR-628-5p | hsa-miR-647 | hsa-miR-526b-5p |
| hsa_circRNA_405965 | hsa-miR-4298 | hsa-miR-3714 | hsa-miR-6072 | hsa-miR-6768-3p | hsa-miR-519d-5p |
| hsa_circRNA_103948 | hsa-miR-421 | hsa-miR-181a-2-3p | hsa-miR-505-3p | hsa-miR-429 | hsa-miR-380-5p |
| hsa_circRNA_000102 | hsa-miR-1296-3p | hsa-miR-194-3p | hsa-miR-3184-3p | hsa-miR-4753-3p | hsa-miR-412-3p |
| hsa_circRNA_001691 | hsa-miR-5193 | hsa-miR-6792-3p | hsa-miR-4691-5p | hsa-miR-6509-3p | hsa-miR-4776-3p |
| hsa_circRNA_405641 | hsa-miR-6808-5p | hsa-miR-6764-5p | hsa-miR-4323 | hsa-miR-6856-5p | hsa-miR-2861 |
| hsa_circRNA_104939 | hsa-miR-193b-3p | hsa-miR-194-3p | hsa-miR-762 | hsa-miR-486-5p | hsa-miR-892b |
| hsa_circRNA_074559 | hsa-miR-3942-5p | hsa-miR-4682 | hsa-miR-7-2-3p | hsa-miR-8076 | hsa-miR-4703-5p |
| hsa_circRNA_008389 | hsa-miR-7111-3p | hsa-miR-6873-3p | hsa-miR-6769b-3p | hsa-miR-760 | hsa-miR-1236-3p |
| hsa_circRNA_019744 | hsa-miR-1301-3p | hsa-miR-6812-5p | hsa-miR-650 | hsa-miR-4739 | hsa-miR-4459 |
| hsa_circRNA_103887 | hsa-miR-890 | hsa-miR-766-5p | hsa-miR-125a-3p | hsa-miR-10b-3p | hsa-miR-571 |
| hsa_circRNA_101836 | hsa-miR-744-5p | hsa-miR-576-3p | hsa-miR-661 | hsa-miR-9-5p | hsa-miR-377-5p |
| hsa_circRNA_406791 | hsa-miR-627-3p | hsa-miR-338-3p | hsa-miR-34 c-5p | hsa-miR-4659a-3p | hsa-miR-3688-5p |
| hsa_circRNA_102995 | hsa-miR-424-5p | hsa-miR-16-5p | hsa-miR-195-5p | hsa-miR-15b-5p | hsa-miR-323a-3p |
| hsa_circRNA_102183 | hsa-miR-218-1-3p | hsa-miR-758-3p | hsa-miR-218-5p | hsa-miR-486-5p | hsa-miR-202-3p |
| hsa_circRNA_000581 | hsa-miR-378a-3p | hsa-miR-185-5p | hsa-miR-378d | hsa-miR-422a | hsa-miR-501-5p |
| hsa_circRNA_101837 | hsa-miR-766-5p | hsa-miR-509-5p | hsa-miR-18b-5p | hsa-miR-18a-5p | hsa-miR-380-3p |
| hsa_circRNA_103572 | hsa-miR-29b-2-5p | hsa-miR-544a | hsa-miR-29a-5p | hsa-miR-518 c-5p | hsa-miR-214-3p |
| hsa_circRNA_101771 | hsa-miR-647 | hsa-miR-370-3p | hsa-miR-520 c-3p | hsa-miR-526b-3p | hsa-miR-520b |
| hsa_circRNA_092476 | hsa-miR-5007-5p | hsa-miR-760 | hsa-miR-4722-5p | hsa-miR-545-3p | hsa-miR-4527 |
Validation by qRT-PCR
After analyzing the data by filtering a high-throughput microarray, we listed 10 differentially expressed circRNAs for further analysis and have been able to validate five by qRT-PCR. As summarized in Figure 3, hsa_circRNA_000581, hsa_circRNA_092476, hsa_circRNA_101836, and hsa_circRNA_102183 in the acute cerebral infarction group were significantly downregulated and hsa_circRNA_103372 was significantly upregulated compared to the control group. These results show that the expression patterns of these candidate circRNAs in microarray and PCR data are similar. Table 3 provides a summary on the variation and calculated p-values. The primers used in this study for quantitative reverse transcription polymerase chain reactions are shown in Table 4.
Figure 3.
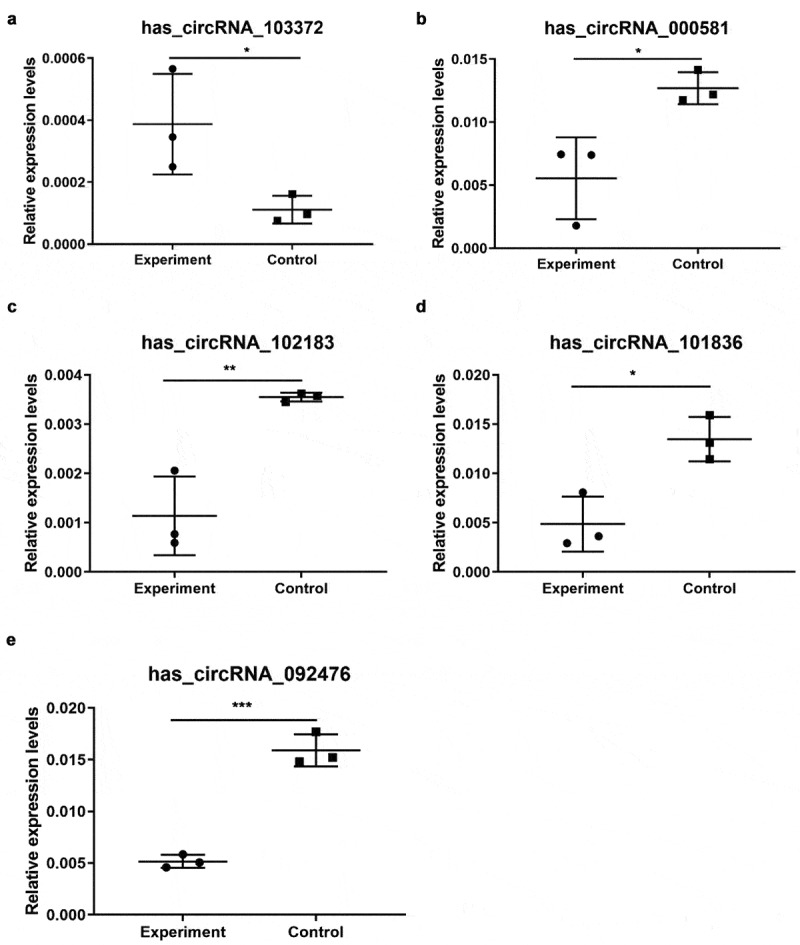
qRT-PCR validation of five selected circRNAs. (a) Relative expression levels of has_circRNA_103372. (b) Relative expression levels of has_circRNA_000581. (c) Relative expression levels of has_circRNA_102183. (d) Relative expression levels of has_circRNA_101836. (e) Relative expression levels of has_circRNA_092476. Compared with the control, rno_circRNA_000581/102,183/101,838/092476 were significantly downregulated. However, rno_circRNA_103372 was significantly upregulated. The data are normalized using the mean ± SEM (*P < 0.05; n = 6 per group)
Table 3.
Comparison of candidate circRNAs expression in microarray and PCR
| CircRNAs |
Microarray | PCR | ||||
| FC (abs) |
P-value |
Regulation |
FC (abs) |
P-value |
Regulation |
|
| hsa_circRNA_000581 | 16.2844189 | 0.001973896 | down | 2.287937 | 0.0238 | down |
| hsa_circRNA_092476 | 14.8670307 | 0.003552554 | down | 3.079344 | 0.0004 | down |
| hsa_circRNA_101836 | 19.8648936 | 0.021279368 | down | 2.766612 | 0.0142 | down |
| hsa_circRNA_102183 | 16.3927337 | 0.003117049 | down | 3.118147 | 0.0066 | down |
| hsa_circRNA_103372 | 21.0471532 | 0.034767906 | up | 3.46493 | 0.0468 | up |
Table 4.
Sequences of primers used for qRT-PCR assay
| Gene name |
Primer sequence |
Ta Opt (◦C) |
Product size (bp) |
| β-actin (H) | F:5ʹ GTGGCCGAGGACTTTGATTG3’ | 60 | 73 |
| R:5ʹCCTGTAACAACGCATCTCATATT3’ | |||
| hsa_circRNA_092476 | F:5ʹ AGCAACAGACTCTCCTGCAGA3’ | 60 | 135 |
| R:5ʹ ACTTGCTGCTTGTGTATCCAG 3’ | |||
| hsa_circRNA_101836 | F:5ʹ CCCATTATGAAACTGAAGGTAGC3’ | 60 | 126 |
| R:5ʹGATGGAGGTGGATCTACATTAAAGA3’ | |||
| hsa_circRNA_102183 | F:5ʹ TCAAATTCAGGCGTTGTTCAA 3’ | 60 | 181 |
| R:5ʹ TCTGCTGCTCCAGTCGTTTT 3’ | |||
| hsa_circRNA_103372 | F:5ʹ GTGTGTGTGGCATGCAGC 3’ | 60 | 104 |
| R:5ʹ TGGGAACTTATATTCCCCTTC 3’ | |||
| hsa_circRNA_000581 | F:5ʹ GCAGGCCAAGTCCAGCTTATA 3’ | 60 | 82 |
| R:5ʹ ACCAACCTCATGGGATTGTTTC 3’ |
CircRNAs/MicroRNA/RNA Interaction
Then, five candidate circRNAs verified by RT-PCR were selected, and the possible interacting miRNA targets were further studied using a computational-based prediction approach. Specifically, we have used starbase to predict possible target miRNAs for each circRNA. The criteria for selecting target genes are clipExpNum >3 and pancancerNum ≥3. Finally, 655 target genes, 13 miRNAs and 5 cirRNAs were obtained. Cytoscape results showed that there were five circRNAs, 13 microribonucleic acids, and 655 microribonucleic acids, showing a large interaction network (Figure 4). CircRNAs can be used as the nucleolar component of microRNA, which usually inhibits the target microRNA. Therefore, circRNAs may upregulate target microRNA indirectly by regulating microRNA.
Figure 4.
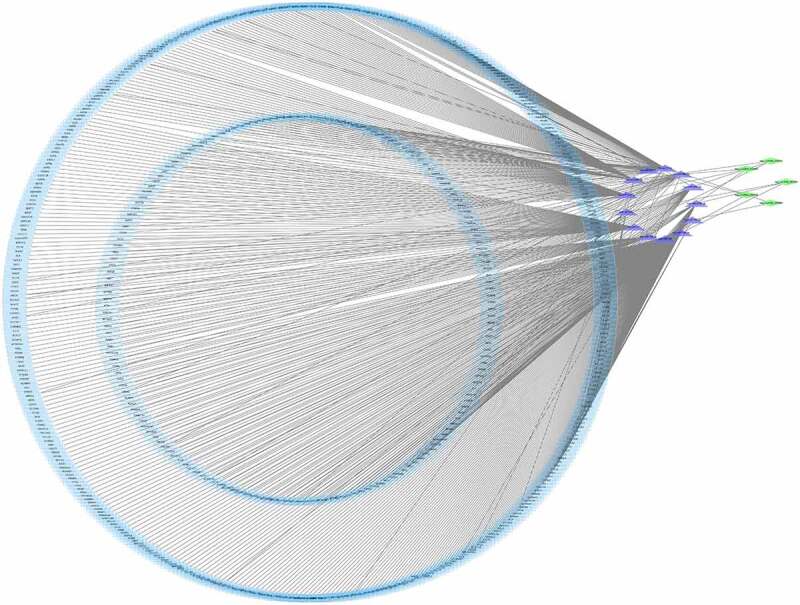
The circRNA/miRNA/mRNA network analysis. The network includes 5 brown nodes circRNAs, 13 red nodes miRNAs and 655 blue nodes mRNAs
Go and pathway analysis of putative target genes
Based on these interactions, five circRNA candidates may play an important role in molecular mechanisms by regulating target molecules. Therefore, we performed GO and KEGG analysis on all target genes, which provided strong evidence for further functional verification of these circRNAs.
Next, we carried out GO bioinformatics analysis, including biological processes, cell compositionxs, and molecular function analysis. For each part, we show the following categories of significant enrichment items: top 10 counts, top 10 times enrichment values, top 10 enrichment scores, and gene ratio values. The results show that [1]: The most significant and meaningful terms in biological process are metabolic process and RNA splicing, which describe a series of biological events (Figure 5a-d) [2]. As far as cell components are concerned, the most important enrichment and meaningful terms are intracellular parts and complexes, describing cell components (Figure 6a-d) [3]. As far as molecular function is concerned, the most important enrichment and meaningful term is the binding event, which indicates the function at the molecular level (Figure 7a-d).
Figure 5.
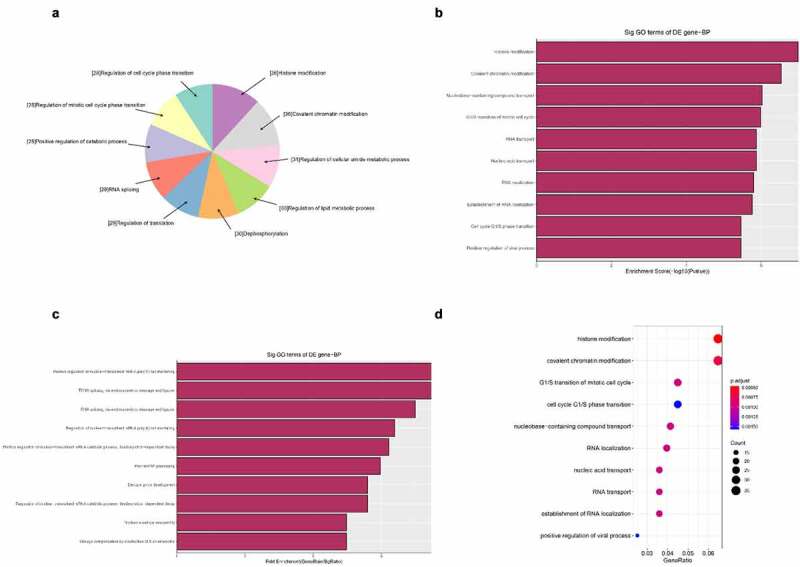
The GO annotations for biological process of target mRNAs regulated by the five candidate circRNAs. Pie (a), bar (b) point charts (c), and dot plot (d) showed the top 10 counts of significant enrichment items
Figure 6.
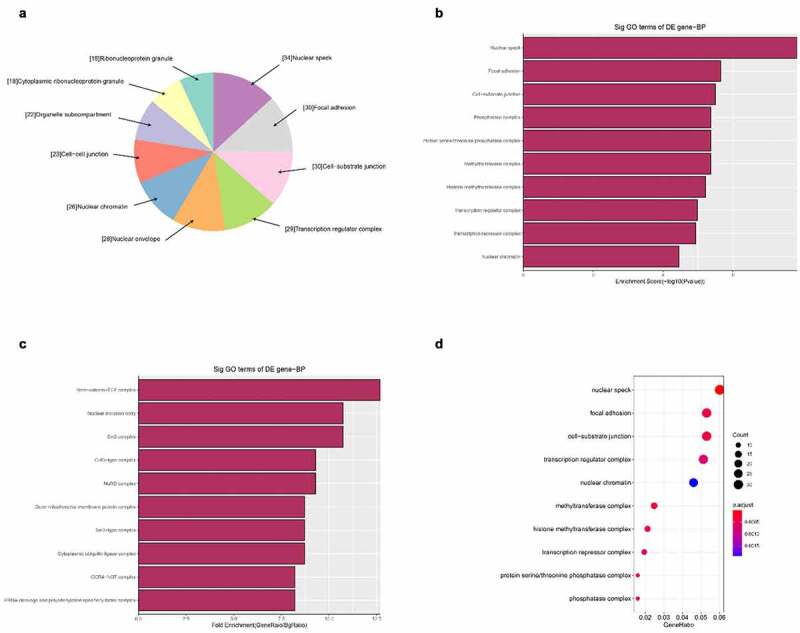
The GO annotations for cellular component of target mRNAs regulated by the five candidate circRNAs. Pie (a), bar (b) point charts (c), and dot plot (d) showed the top 10 counts of significant enrichment items
Figure 7.
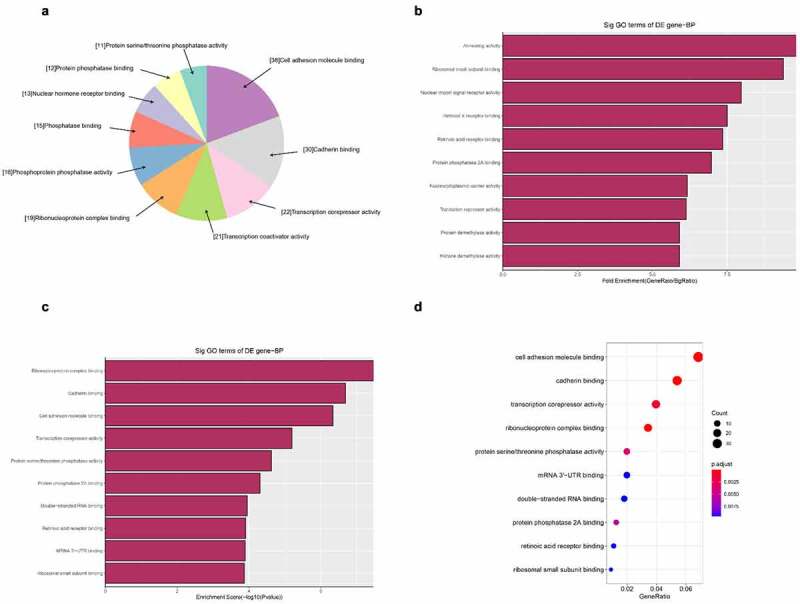
The GO annotations for molecular function of target mRNAs regulated by the five candidate circRNAs. Pie (a), bar (b) point charts (c), and dot plot (d) showing the top 10 counts of significant enrichment items
The top 10 enrichment scores of significant enrichment pathways based on KEGG pathway analysis are shown in Figure 8a. In addition, the dot plot shows the gene ratio values of the top 10 most significant enrichment pathways (Figure 8b). The results showed that these target genes may play an important role in various metabolic activities, and the two most important pathways are the AMPK signaling pathway and the peroxisome-related pathway (Figure 8c).
Figure 8.
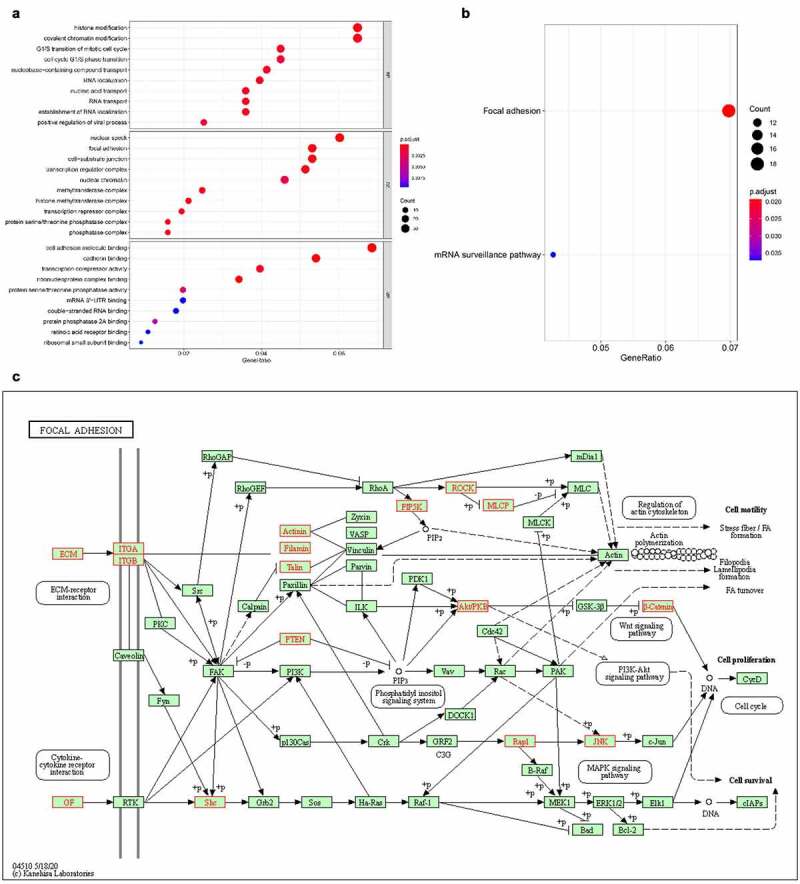
KEGG pathway analysis of five target genes regulated by circRNAs. the bar plot (a), dot plot (b) and related pathway (c) showed the top 10 enrichment score values of the significantly enriched pathway
Discussion
Cells can resist the accumulation of DNA damage by inducing endogenous DNA repair mechanisms, such as base excision repair (BER), nucleotide excision repair (NER), and non-homologous endpoint ligation (NHEJ) [11,12]. Oxidative DNA damage and repair occur within minutes after ischemic stroke and may last up to 6 months after stroke [13]. DNA repair plays an important role in endogenous brain repair during stroke recovery. DNA repair has a profound impact on a wide range of recovery efforts, including neurogenesis [14,15], angiogenesis [16], axon growth [17], and remyelination [18]. However, homocysteine can play a role in local tissues and promote the generation of oxygen-free radicals, while excess oxygen-free radicals will act on nucleic acids, lipids, and other substances in cells to cause oxidation reaction, resulting in cell damage [19,20]. Oxidized low-density lipoprotein is one of the products of the reaction between lipid and oxygen-free radicals, and its content can indirectly reflect the quantity of oxygen-free radicals and the degree of oxidative stress reaction [21,22]. Therefore, circRNAs may play an important role in the process of cerebral infarction through homologous recombination pathways and may be related to dyslipidemia, hyperhomocysteinemia, and other factors related to the onset of cerebral infarction. The specific mechanism of its action needs to be further explored by designing cell and animal experiments.
In this study, we set out to understand the function of circRNAs in ACI. Our results show that circRNAs are differentially expressed in cerebral infarction, and we identify 10 circRNAs, which are significantly upregulated, and 10 circRNAs, which are significantly downregulated. Our analysis proves that circRNAs can regulate many biological processes, cellular components, and molecular functions. Using RT-qPCR verification, we have been able to verify five downregulated circRNAs. Based on our literature search, we found that HSA _ circrna _ 103,372 (p < 0.01), its source gene UIMC1 is significantly enriched in homologous recombination signaling pathway, which indicates that hsa_circRNA_405965 may participate in ACI by affecting oxidative stress through homologous recombination pathway. At the same time, the miRNAs binding sites on differentially expressed circRNAs and the first five putative target miRNAs bound by differentially expressed circRNAs were identified. Taken together, our data provide an important reference for future research on interaction between circRNA-miRNAs in ACI.
In order to further study the potential functions of differentially expressed circRNAs, we performed a preliminarily analysis by annotating the differentially expressed circRNAs. GO analysis of differentially expressed circRNAs showed that UBXN7, the source gene of hsa_circ_103948, is mainly enriched in ubiquitin binding (GO:0043130) and ubiquitin-like protein binding (GO:0032182). The gene SMARCA5 from hsa_circ_000102 and UIMC1 from hsa_circ_405641 are mainly concentrated in double-strand break repair (GO:0006302) and histone binding (GO:0042393). The source group of hsa_circ_104939 is mainly concentrated in the normal junction of microtube polymerization (GO:0031116), microtube nucleation (GO:0007020), and microtube polymerization or depolymerization (GO:0031112). RSRC1, the source gene of hsa_circ_074559, is mainly enriched in replacing mRNA splicing by splices (GO:0000380). MORC3, the source gene of hsa_circ_008389, is mainly concentrated in maintaining the position of protein in nucleus (GO:0051457), negatively regulating the proliferation of fibroblasts (GO:0048147), and maintaining the location of protein in organelles (GO:0072595). UIMC1 is also rich in active regulation of DNA repair (GO:0045739), repair of broken double strands by non-homologous end connection (GO:0006303), etc. The downregulation of differentially expressed circRNAs may participate in the biological process of acute cerebral infarction and play its function through the above aspects, so we speculate that the downregulation of differentially expressed circRNAs may be related to the growth and repair of neurons. KEGG pathway analysis is not limited to gene level, but a higher-level integration analysis of genes, small biological molecules and proteins is presented as a network information map [15]. By analyzing the source genes of differentially expressed circRNAs, we find that we can better understand their biological pathways and evaluate the possible role of these pathways in cerebral infarction. The enrichment of KEGG biological pathway showed that UIMC1, the source gene of hsa_circ_405641, was significantly enriched in homologous recombination signal pathway (http://www.genome.jp/Kegg-bin/show _ pathway? Hsa03440 + 51,720), and homologous recombination plays an important role in genome maintenance and DNA damage in normal growth cells, which can trigger homologous recombination, which is necessary for cell recovery after oxidative stress [23]. After cerebral infarction, oxidative stress may be an important mechanism to cause a series of secondary damages, which is mainly characterized by the increase of oxygen free radicals and oxidative damage to cell structure [24,25], which will lead to the damage of many components including protein, lipid and DNA [26]. Oxidative DNA damage is one of the most harmful consequences of increased oxidative stress in ischemic stroke. When DNA damage is not repaired, it can trigger a variety of death-promoting signaling pathways, which can induce apoptosis and endanger the functional recovery after stroke. However, CircRNAs, a non-coding RNA molecule with multiple biological functions, exists stably in various body fluids and exosomes, and participates in regulating the occurrence and development of various diseases [27]. Studies have shown that the expression profile of circRNAs in brain tissue will change when the brain tissue lacks blood supply, and some circRNAs may participate in the process of cerebral ischemia [28]. In order to explore the potential biological function of differentially expressed circRNAs in the pathogenesis of acute cerebral infarction, this experiment entrusted Shanghai Yunxu Company to support high-throughput sequencing, and provided enrichment analysis of GO function and KEGG signaling pathway by bioinformatics methods. Sequencing results showed that compared with the control group, there were many differentially expressed circRNAs in patients with ACI, which was statistically significant. In this study, we detected 15 downregulated and 3 upregulated circRNAs significantly after cerebral infarction, and these differentially expressed circRNAs may be involved in the occurrence and development of cerebral infarction. Studies have shown that there are many circRNAs expressions in brain tissue, which are widely involved in the regulation of nervous system diseases and the occurrence and development of cardiovascular diseases related to vascular endothelial function [29]. A recent study showed that there were significant differences in the expression of specific circRNAs in brain tissue under ischemia stimulation. In ischemia reperfusion injury model, compared with the control group, the expression of 15 kinds of circRNAs in hippocampus of mice in transient ischemic group changed, among which 3 kinds increased and 12 kinds decreased. Some studies suggest that these circRNAs may play a role as molecular sponges of miRNAs in the process of hypoxic-ischemic brain damage. The upregulated target miRNA of circRNA-015947 were predicted by using prediction software, and the top five predicted values were miR-188-3p, miR-329-5p, miR-3057-3p, miR-5098 and miR-683. These miRNAs have been proved to be involved in the regulation of the pathogenesis of ischemic brain injury [30]. Recent studies have found that after traumatic brain injury (TBI), 8036 circRNAs changed, among which 16 changed significantly (5 were upregulated and 11 were downregulated). Circ RNA chr8 _ 87, 859, 283–87, 904, 548 significantly increased about 4 times in the cerebral cortex around the injured site, and CXCR2 protein was increased by sponge mmu-let-7a-5p to promote neuroinflammation. The results indicated that the increased circrna chr8 _ 87, 859, 283–87, 904, 548 blocked the recovery of nerve function after TBI [31].
At present, most of the research studies on circular RNA related to cerebral infarction are aimed at the cellular level. This study uses the plasma of patients with acute cerebral infarction in clinical practice, which has more practical and specific functions and mechanisms for the subsequent research on circular RNA related to cerebral infarction. Detailed animal experiments and functional verification are carried out, in order to clarify the role of these circRNAs in ACI and study the functional effects of specific circRNA-miRNA interactions. The specific functions of these significantly differentially expressed circRNAs found in this study have not been experimentally explored.
Limitations
There are relatively few samples in this sequencing test, and the sample selection time is slightly single. Although the mixed samples are used in early sequencing and the individual errors can be reduced to a certain extent, the mixed samples will cover up the differential expression among some individuals, resulting in experimental results. There may be bias; Patients with acute cerebral infarction are not grouped according to the location, area, and severity of lesions, which can make the differentially expressed circRNAs more specific; the specific functions of these significantly differentially expressed circRNAs found in this study are still unclear.
Future directions
At present, most of the research studies on circular RNA related to cerebral infarction are aimed at the cellular level. This study uses the plasma from patients with acute cerebral infarction in clinical practice, which has more practical and specific functions and mechanisms for the subsequent research on circular RNA related to cerebral infarction. Detailed animal experiments and functional verification are carried out, in order to clarify the role of these circRNAs in acute cerebral infarction and study the functional effects of specific circRNA-miRNA interactions. In the future, we will further explore the guiding significance of these circRNAs in acute cerebral infarction based on previous work.
Conclusions
In summary, our results suggest that there are 10 differentially expressed circRNAs after acute cerebral infarction, which may upregulate target microRNA indirectly by regulating microRNA.
Supplementary Material
Acknowledgements
None
Funding Statement
The author(s) reported that there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article. https://pan.baidu.com/s/1RbY4bcv8Bu2jiy2YKKp1HA
Highlights
CircRNAs show differential expression after acute cerebral infarction.
circRNAs may upregulate target microRNA indirectly by regulating microRNA.
hsa_circRNA_000581, hsa_circRNA_092476, hsa_circRNA_101836, and hsa_circRNA_102183 are significantly downregulated, and hsa_circRNA_103372 is significantly upregulated in ACI
Target genes of the validated circRNAs play an important role in metabolic processes of the cell.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Naidech AM, Drescher J, Tamul P, et al. Acute physiological derangement is associated with early radiographic cerebral infarction after subarachnoid haemorrhage. J Neurol Neurosurg Psychiatry. 2006;77(12):1340–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Edwards MD, Hughes TAT.. Managing blood pressure in acute cerebral infarction. J Neurol. 2021;268(6):2294–2296. [DOI] [PubMed] [Google Scholar]
- [3].Wu X, Duan Z, Liu Y, et al. Incidental unruptured intracranial aneurysms do not impact outcome in patients with acute cerebral infarction. Front Neurol. 2021;12:613027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tsai CF, Yip PK, Chen CC, et al. Cerebral infarction in acute anemia. J Neurol. 2010;257(12):2044–2051. [DOI] [PubMed] [Google Scholar]
- [5].Nakajo Y, Zhao Q, Enmi JI, et al. Early detection of cerebral infarction after focal ischemia using a new MRI Indicator. Mol Neurobiol. 2019;56(1):658–670. [DOI] [PubMed] [Google Scholar]
- [6].Chen X, Yang T, Wang W, et al. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9(2):588–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wu Z, Sun H, Li J, et al. RNAs in leukemia. Aging (Albany NY). 2019;11(13):4757–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu CX, Li X, Nan F, et al. Structure and degradation of circular RNAs regulate PKR activation in. Immunity I. Cell. 2019;177(4):865–80.e21. [DOI] [PubMed] [Google Scholar]
- [9].Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang W, Cheng J, Zhang Y, et al. Analysis of CT and MRI combined examination for the diagnosis of acute cerebral infarction. J Coll Physicians Surg Pak. 2019;29(9):898–899. [DOI] [PubMed] [Google Scholar]
- [11].Lu Z, Filonov GS, Noto JJ, et al. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA. 2015;21(9):1554–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Franz A, Rabien A, Stephan C, et al. Circular RNAs: a new class of biomarkers as a rising interest in laboratory medicine. Clin Chem Lab Med. 2018;56(12):1992–2003. [DOI] [PubMed] [Google Scholar]
- [13].Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. [DOI] [PubMed] [Google Scholar]
- [14].Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. [DOI] [PubMed] [Google Scholar]
- [15].Li S, Teng S, Xu J, et al. Microarray is an efficient tool for circRNA profiling. Brief Bioinform. 2019;20(4):1420–1433. [DOI] [PubMed] [Google Scholar]
- [16].Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38(16):e100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gupta SK, Garg A, Bär C, Chatterjee S, Foinquinos A, Milting H, et al . Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ Res. 2018;122(2):246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science (New York, NY). 1995;268(5209):415–417. [DOI] [PubMed] [Google Scholar]
- [19].Mohsen MG, Kool ET. The discovery of rolling circle amplification and rolling circle transcription. Acc Chem Res. 2016;49(11):2540–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ning L, Wang X, Xu K, et al. A novel isothermal method using rolling circle reverse transcription for accurate amplification of small RNA sequences. Biochimie. 2019;163:137–141. [DOI] [PubMed] [Google Scholar]
- [21].Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. [DOI] [PubMed] [Google Scholar]
- [22].Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. [DOI] [PubMed] [Google Scholar]
- [24].Fei T, Chen Y, Xiao T, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(26):E5207–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mehta SL, Dempsey RJ, Vemuganti R. Role of circular RNAs in brain development and CNS diseases. Prog Neurobiol. 2020;186:101746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. [DOI] [PubMed] [Google Scholar]
- [27].Zhang P, Chao Z, Zhang R, et al. Circular RNA regulation of myogenesis. Cells. 2019;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang H, Feng C, Jin Y, et al. Identification and characterization of circular RNAs involved in mechanical force-induced periodontal ligament stem cells. J Cell Physiol. 2019;234(7):10166–10177. [DOI] [PubMed] [Google Scholar]
- [29].Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–490. [DOI] [PubMed] [Google Scholar]
- [30].Lei M, Zheng G, Ning Q, et al. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cocquerelle C, Mascrez B, Hétuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article. https://pan.baidu.com/s/1RbY4bcv8Bu2jiy2YKKp1HA


