Abstract
Thirty-six VanB glycopeptide-resistant Enterococcus faecium isolates were collected from patients in five different hospitals in Taiwan. The vancomycin resistance genes were amplified by the long vanB PCR, which amplifies the 6,373-bp vanB gene cluster including the vanRB2, vanSB2, vanYB2, vanWB2, vanHB2, vanB2, and vanXB2 genes. The deduced amino acid sequences were found to be 95 to 98% homologous to those of the vanB1 gene cluster: VanRB1, 97%; VanSB1, 97%; VanYB1, 96%; VanHB1, 95%; VanB1, 96%; and VanXB1, 98%. Restriction enzyme analysis of the long vanB PCR products revealed that all 36 isolates had the same vanB2-specific pattern. DNA sequence analysis of the vanB2 gene, which is a d-Ala–d-Lac ligase gene, revealed that none of the 36 sequences were identical to the previously published vanB2 sequence. Thirty-one isolates had 1 nucleotide different from the published vanB2 sequence. The sequences of the other five isolates differed from the published vanB2 sequence by 2 or 3 nucleotides. Four isolates with a low or moderate resistance to vancomycin (MIC = 4 to 32 μg/ml) were found to have the same leucine-to-methionine change at amino acid position 308 of the vanB2 gene. The genomic DNAs of all 36 isolates were digested with SmaI and then typed by pulsed-field gel electrophoresis (PFGE). Eight different PFGE types (I to VIII) were observed, and type I was found to be prevalent in all hospitals examined in this study. This result suggests that intra- and interhospital dissemination of this E. faecium strain has occurred in Taiwan.
Glycopeptide resistance in enterococci is genotypically and phenotypically diverse. The VanA and VanB types are the two most common classes of acquired glycopeptide resistance in enterococci. VanA enterococci are resistant to high levels of vancomycin and teicoplanin due to the presence of the vanA gene cluster that includes the vanR, vanS, vanH, vanA, vanX, vanY, and vanZ genes (1). The vanA gene cluster is usually located on transposon Tn1546 or related elements (2). Strains with the VanB phenotype have various levels of resistance to vancomycin but are susceptible to teicoplanin (25). The VanB-type resistance is mediated by the vanB gene cluster that includes the vanRB, vanSB, vanYB, vanW, vanHB, vanB, and vanXB genes (8). The vanB gene cluster may reside on Tn1547, which is flanked by two distantly related insertion sequences, an IS256-like element and IS16, in direct-repeat orientation (24). A novel vanB-containing transposon, Tn5382, was recently described in a vancomycin-resistant Enterococcus faecium (4).
The vanB gene encodes a d-Ala–d-Lac ligase and is classified into three different types, vanB1, vanB2, and vanB3, based on DNA sequence heterogeneity (11, 22). The vanB1 gene was previously referred to as vanB (6, 10). The vanB2 gene was first found in Enterococcus faecalis SF300 (11). The vanB3 gene was found recently in two E. faecalis isolates (22). Its nucleotide sequence differs from that of vanB1 by 5% (22) and from the published vanB2 sequence by 3.6% (22).
We have recently used PCR primers specific for the vanB1 gene (6) to amplify the vancomycin resistance gene from VanB enterococci in Taiwan. The resulting PCR products were found to be 1,100 bp, not the expected 433 bp. The PCR product of one isolate, E. faecium TSGH1, was sequenced and found to contain a portion of the vanHB2 gene, the entire vanB2 gene, and a portion of the vanXB2 gene (GenBank accession no. Z83305). The vanB2 sequence of TSGH1 was found to differ by only 1 nucleotide from that of the published 801-bp vanB2 gene (22). This result suggests that E. faecium TSGH1 harbors a vanB2 gene cluster. We therefore examined all VanB E. faecium isolates that we have collected. The entire vanB gene cluster of each isolate was amplified, and the vanB ligase gene of each isolate was sequenced. Pulsed-field gel electrophoresis (PFGE) was performed to identify the predominant strain of E. faecium in Taiwan.
MATERIALS AND METHODS
Bacterial isolates.
Thirty-six clinical isolates of VanB E. faecium collected from five hospitals (13 isolates from hospital A, 4 from hospital B, 5 from hospital C, 2 from hospital D, and 12 from hospital E) in Taiwan were studied. They were not consecutive vancomycin-resistant enterococcus (VRE) isolates. They were selected because their stock cultures were available and they were previously determined to carry the vanB gene (20). E. faecalis V583 CDC containing vanB1 (26), E. faecium TUH2-18 containing vanB2(7), and E. faecium VRE45 containing vanB3 (7) were used as controls in PCR, PFGE, and DNA sequencing.
Antimicrobial susceptibility assay.
VanB E. faecium isolates were assayed for susceptibility to vancomycin and teicoplanin by the E test (AB Biodisk, Piscataway, N.J.) according to the manufacturer's instructions. E. faecalis ATCC 29212 was used as the control strain for the susceptibility test.
PCR amplification.
Bacterial DNA was isolated as previously described (19). Briefly, the DNA from a loop of cultured bacteria was extracted by boiling the bacteria with lysis buffer (1% Triton X-100, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA) for 30 min. After centrifugation, the DNA in the supernatant was analyzed for the presence of the van gene cluster by PCR. PCRs were performed in the GenAmp PCR system (model 2400; Perkin-Elmer, Norwalk, Conn.). Based on sequence analysis of the vanA and vanB gene clusters, primers VB211F and VB6545R were designed (Table 1) to amplify the 6,373-bp vanB2 gene cluster. This PCR method was named the long vanB PCR. The PCR mixture contained 50 ng of template DNA, PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 2.5 mM MgCl2, 0.001% gelatin), 20 pmol of each PCR primer, 0.4 mM each deoxynucleoside triphosphate, and 2.5 U of TaKaRa Ex Taq DNA polymerase (Takara Shuzo Co., Ltd., Otsu, Shiga, Japan) in a total volume of 100 μl. After a 10-min denaturation at 94°C, the reaction mixture was run through 40 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 55°C, and extension for 6 min at 72°C and then incubated for 10 min at 72°C. After amplification, 5 μl of PCR products was electrophoresed on a 1% agarose gel in TBE buffer (0.0445 M Tris, 0.0445 M boric acid, 0.001 M EDTA) containing ethidium bromide (0.5 μg/ml). The PCR products were purified with the GFX PCR DNA and Gel Band Purification kits (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) according to the manufacturer's instructions.
TABLE 1.
Primers used to characterize the vanB2 gene cluster in this study
| Name | Sequence 5′→3′ | Positiona |
|---|---|---|
| For long vanB PCR | ||
| VB211F | TCTACTTGTCGAGGATGATG | 191–211 |
| VB6545R | AACTTATAGTGCCACCATTC | 6564–6545 |
| For sequencing of vanB2 gene cluster | ||
| VB498F | CAAAGCCATTCAAGATGCGG | 479–498 |
| VB893F | CCTATACGATCATTGTCCTG | 874–893 |
| VB1043R | TATTGTAGTGAAACAGCCCT | 1062–1043 |
| VB1655R | CGGAAATGGTTTTGCCCTGC | 1674–1655 |
| VB2028R | TGCTTCGCGCCTGATCAATG | 2047–2028 |
| VB2047F | CATTGATCAGGCGCGAAGCA | 2028–2047 |
| VB2248R | TCTGTAGTTTTCCTGTAGC | 2256–2248 |
| VB2556F | CTCATACAGGCAGCAGCACT | 2537–2556 |
| VB2596R | TCATCTCCCTGTTCTTTGGC | 2615–2596 |
| VB2944F | CTTGGTCTTGCTGTGGATA | 2926–2944 |
| VB3478F | TTCTTTCTGGTGGCTGGT | 3461–3478 |
| VB3959R | CGGGAGGGGATAACAGATT | 3977–3959 |
| VB4322R | CATAATCCGCAACGCTGTC | 4340–4322 |
| VB4871R | CCGTATGGGGTGTGATGAT | 4889–4871 |
| VB5367R | AAGAATGTAGGCCAGTGA | 5382–5367 |
| VB5382F | CAAATCACTGGCCTACATTC | 5363–5382 |
| VB5454Fb | GTGACAAACCGGAGGCGAGGA | 5434–5454 |
| VB5745F | AGAAAATGCGATGATTACAG | 5726–5745 |
| VB5808R | TCTGCATCCAAGCACCCG | 5825–5808 |
| VB5846Rb | CCGCCATCCTCCTGCAAAAAA | 5866–5846 |
| VB6115F | GGTGGATGGGTATGAGGTG | 6097–6115 |
DNA sequencing.
The long vanB PCR products were sequenced by primer walking using primers designed according to the published sequence of the V583 vanB gene cluster or new sequences obtained in this study (Table 1). The vanB2 ligase genes in various isolates were sequenced using primers VB5382F, VB5367R, VB5745F, and VB5808R (Table 1). The BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems Inc., Foster City, Calif.) was used for sequencing.
Restriction fragment length polymorphism (RFLP) analysis.
Ten microliters of each long vanB PCR product was digested with different restriction enzymes in a total volume of 20 μl. After incubation for 2 h at 37°C, the digested DNA samples were electrophoresed on a 1% agarose gel in TBE buffer containing ethidium bromide (0.5 μg/ml).
Typing of VRE by PFGE.
Preparation of bacterial DNA for PFGE was performed as described by Gouby et al. (12). Bacteria were grown overnight on blood agar plates and then suspended in 10 mM Tris–0.1 mM EDTA solution to a concentration of 10 on the McFarland scale. This bacterial suspension was mixed with an equal volume of molten 1.6% low-melting-point agarose and then cast into plugs. The plugs were treated with lysis solution (6 mM Tris-HCl [pH 7.6], 100 mM EDTA [pH 7.5], 1 M NaCl, 0.2% deoxycholate, 0.5% sodium lauroylsarcosine, 1 mg of lysozyme/ml) at 37°C for 24 h. The solution was subsequently replaced with ESP buffer (0.5 M EDTA, 1% sodium lauroylsarcosine, 0.5 mg of proteinase K/ml) and incubated at 37°C for 24 h. The plugs were then washed once in distilled water and four times in 10 mM Tris–0.1 mM EDTA buffer at 37°C for 30 min each time.
The genomic DNA embedded in the plugs was digested overnight with 50 U of SmaI (New England Biolabs, Beverly, Mass.) in a total reaction volume of 0.3 ml in NEB4 buffer (New England Biolabs) at 25°C. The plugs were then washed in 10 mM Tris–0.1 mM EDTA buffer at 37°C for 1 h, loaded into wells of a 1% agarose gel, and then electrophoresed in 0.5× TBE buffer. A plug containing a 50-kb-to-1-Mb lambda DNA ladder (Bio-Rad Laboratories, Hercules, Calif.) was used as the molecular size marker. PFGE was performed with a CHEF Mapper XA system (Bio-Rad Laboratories) at 14°C under the autoalgorithm mode (gradient, 6 V/cm; run time, 27 h 12 min; included angle, 120; initial switch time, 2.91 s; final switch time, 35.38 s; ramping factor, linear). The gel was stained with ethidium bromide and then photographed. The PFGE results were interpreted according to the criteria of Tenover et al. (27).
Nucleotide sequence accession numbers.
The nucleotide sequences of the whole vanB2 gene cluster of isolate TSGH1 and the vanB2 ligase genes of isolates VRE-1, SLH475, and CG4248 have been deposited in GenBank with accession numbers AF310956, AF310953, AF310954, and AF310957, respectively.
RESULTS
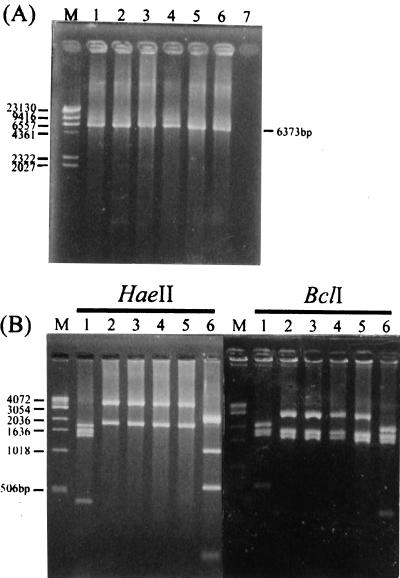
All 36 VanB isolates were amplified by the long vanB PCR (Table 2). The sizes of amplified products were approximately 6,400 bp. The PCR products were digested with HaeII or BclI, and all of the products yielded identical HaeII or BclI digestion patterns that were characteristic of vanB2 but different from those of the vanB1 or the vanB3 gene cluster (Fig. 1).
TABLE 2.
General information for VanB E. faecium strains and isolates
| Origin or hospital | Strain | MIC (μg/ml)
|
Long vanB PCR-RFLP profile | vanB gene type | PFGE type | |
|---|---|---|---|---|---|---|
| Vancomycin | Teicoplanin | |||||
| United States | V583 CDC | 96 | 0.38 | B1 | B1 | IX |
| Norway | TUH2-18 | 12 | 0.38 | B2 | B2 | X |
| United States | VRE45 | ≥256 | 0.5 | B3 | B3 | XI |
| A | TSGH1 | >256 | 1 | B2 | B2a | I |
| A | TSGH2 | >256 | 1 | B2 | B2a | I |
| A | F901 | >256 | 0.25 | B2 | B2a | I |
| A | F909 | >256 | 0.5 | B2 | B2a | I |
| A | F910 | >256 | 0.5 | B2 | B2a | I |
| A | 1716 | >256 | 0.5 | B2 | B2a | II |
| A | 1741 | >256 | 1.5 | B2 | B2a | II |
| A | 1742 | >256 | 2 | B2 | B2a | II |
| A | 1746 | >256 | 1 | B2 | B2a | I |
| A | P6-31 | >256 | 0.75 | B2 | B2a | I |
| A | M908 | >256 | 1 | B2 | B2a | I |
| A | M909 | >256 | 1 | B2 | B2a | I |
| A | M941 | >256 | 1 | B2 | B2a | I |
| B | 16-43 | >256 | 2 | B2 | B2a | I |
| B | 24-24 | >256 | 1.5 | B2 | B2a | I |
| B | Y1 | >256 | 1.5 | B2 | B2a | I |
| B | 2-50 | >256 | 1 | B2 | B2a | I |
| C | VRE-1 | 4 | 1 | B2 | B2b | III |
| C | VRE-3 | 8 | 1 | B2 | B2b | III |
| C | VRE-8 | 32 | 1 | B2 | B2b | IV |
| C | VRE-20 | >256 | 4 | B2 | B2a | I |
| C | CG4248 | 32 | 1 | B2 | B2c | I |
| D | SLH475 | >256 | 1 | B2 | B2d | I |
| D | SLH476 | 3 | 1 | B2 | B2a | V |
| E | CKU-2 | 48 | 0.5 | B2 | B2a | I |
| E | CKU-4 | >256 | 0.5 | B2 | B2a | I |
| E | CKU-6 | >256 | 1 | B2 | B2a | VI |
| E | CKU-8 | 128 | 1 | B2 | B2a | I |
| E | CKU-10 | >256 | 1.5 | B2 | B2a | I |
| E | CKU-11 | >256 | 2 | B2 | B2a | VII |
| E | CKU-12 | >256 | 1 | B2 | B2a | VIII |
| E | CKU-13 | >256 | 1 | B2 | B2a | I |
| E | CKU-14 | >256 | 4 | B2 | B2a | VIII |
| E | CKU-16 | >256 | 4 | B2 | B2a | I |
| E | CKU-20 | >256 | 0.5 | B2 | B2a | I |
| E | CKU-21 | >256 | 4 | B2 | B2a | I |
One base different from the published vanB2 gene sequence of Patel et al. (22).
Two bases different from the published vanB2 gene sequence of Patel et al. (22).
Three bases different from the published vanB2 gene sequence of Patel et al. (22).
One base different from the published vanB2 gene sequence of Patel et al. (22).
FIG. 1.
Analysis of long vanB PCR products. (A) PCR products of VRE harboring the vanB gene cluster amplified with primers VB211 and VB6545R. The sizes of PCR products are indicated on the right-hand side of the gel. Lane M, lambda phage DNA digested with HindIII; lane 1, VanB1 V583; lanes 2 to 5, VanB2 TUH2-18, TSGH1, CG4248, and VRE-1; lane 6, VanB3 VRE45. (B) RFLP analysis of long vanB PCR products by HaeII or BclI digestion. Lane M, 1-kb DNA ladder (Life Technologies, Grand Island, N.Y.); lane 1, VanB1 V583; lanes 2 to 5, VanB2 TUH2-18, TSGH1, CG4248, and VRE-1; lane 6, VanB3 VRE45. The numbers on the left are in base pairs.
The entire 6,400-bp vanB2 gene cluster of isolate TSGH1 was sequenced. This gene cluster was found to contain the 663-bp vanRB2, 1,344-bp vanSB2, 807-bp vanYB2, 828-bp vanWB2, 972-bp vanHB2, 1,029-bp vanB2, and 609-bp vanXB2 genes in addition to the 175-bp vanSB-vanYB intergenic region. The nucleotide sequences of the vanB2 gene cluster were found to be more similar to those of the vanB1 gene cluster (94 to 96% homology) than to those of the vanA gene cluster (48 to 81% homology) and the vanD gene cluster (33 to 71% homology). The deduced amino acid sequences of the vanB2 gene cluster also had a higher degree of homology with those of the vanB1 gene cluster (95 to 97% homology) than to those of the vanA and vanD gene clusters (14 to 76% homology) (Table 3).
TABLE 3.
Homology between the deduced amino acid sequences of proteins encoded by vanB2, vanB, vanA, and vanD gene clusters
| Sequence compared | % Identity
|
|||||
|---|---|---|---|---|---|---|
| VanRB2 | VanSB2 | VanYB2 | VanHB2 | VanB2 | VanXB2 | |
| VanRB1 | 97 | |||||
| VanSB1 | 97 | |||||
| VanYB1 | 96 | |||||
| VanHB1 | 95 | |||||
| VanB1 | 96 | |||||
| VanXB1 | 98 | |||||
| VanR | 44 | |||||
| VanS | 17 | |||||
| VanY | 22 | |||||
| VanH | 67 | |||||
| VanA | 76 | |||||
| VanX | 75 | |||||
| VanRD | 34 | |||||
| VanSD | 15 | |||||
| VanYD | 14 | |||||
| VanHD | 62 | |||||
| VanD | 68 | |||||
| VanXD | 71 | |||||
The vanB2 ligase genes of all 36 isolates were then sequenced. Four types of vanB2 ligase gene sequences, designated TSGH-1, V4248, VRE-1, and SLH475, were observed. Thirty-one isolates belonged to sequence TSGH-1, three belonged to VRE-1, and one each belonged to CG4248 and SLH475 (Table 2). These four sequences differed from each other by only 5 nucleotides at positions 520, 538, 556, 922, and 967. The nucleotides of TSGH-1 at these positions were T, A, C, C, and A, respectively. Those of V4248 were T, G, C, A, and G. Sequence VRE-1 had T, C, A, A, and A at these positions, and SLH475 had G, G, A, C, and A. These sequences differed from that of the vanB1 ligase gene at 44 to 46 (4.3 to 4.5%) nucleotide positions, resulting in 14 or 15 amino acid changes.
The 1,029-bp vanB2 sequences from the 36 isolates were also compared with the previously published 801-bp vanB2 ligase gene sequence (22), which is missing the first 102 and the last 126 bp. Only 1 to 3 nucleotides were found to be different from the published vanB2 ligase gene sequence. The G-to-C change at nucleotide position 538 of the 31 sequences represented by TSGH-1 resulted in a valine-to-leucine change. The A-to-C and C-to-A changes at positions 556 and 922 of the sequence CG4248 resulted in threonine-to-proline and leucine-to-methionine changes at these positions. The VRE-1 sequence had G-to-C and C-to-A changes at positions 538 and 922, causing valine-to-leucine and leucine-to-methionine changes at these positions. The T-to-G change at position 520 of the sequence SLH475 caused a serine-to-alanine change at that position. Four isolates (CG4248, VRE-1, VRE-3, and VRE-8) were found to have a low or moderate resistance to vancomycin (MIC = 4 to 32 μg/ml) (Table 2). Analysis of the sequences revealed that all four of these isolates had a C-to-A change at position 922, resulting in a leucine-to-methionine change at codon 308 of the vanB2 ligase gene.
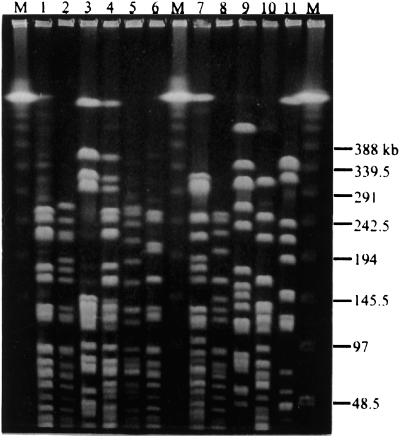
Eight different PFGE types, designated types I to VIII, were observed (Fig. 2). Type I was found to be predominant (Table 2). Twenty-five (69.4%) isolates belonged to type I, three isolates were type II, and two isolates each belonged to types III and VIII. One isolate each belonged to types IV, V, VI, and VII. Type I was also found to be the predominant type in all of the hospitals. Types II and III were found only in hospitals A and C. None of the control strains, one each VanB1 and VanB3 strain from the United States and one VanB2 strain from Norway, belonged to these eight PFGE types (Table 2).
FIG. 2.
PFGE patterns of SmaI digests of chromosomal DNA from VRE isolates. Lanes M, Lambda DNA-PFGE marker ladder; lanes 1 to 6, TSGH1 (type I), 1716 (type II), VRE-1 (type III), VRE-8 (type IV), SLH476 (type V), and CKU-6 (type VI); lanes 7 to 11, CKU-11 (type VII), CKU-12 (type VIII), VanB1 V583 (type IX), VanB2 TUH2-18 (type X), and VanB3 VRE45 (type XI). The numbers on the right are in base pairs.
DISCUSSION
In this study, we found that all 36 VanB E. faecium isolates examined harbored the vanB2 gene cluster. No VanB1 or VanB3 E. faecium were found. These 36 VanB E. faecium isolates were collected from five different hospitals in Taiwan. To our knowledge, this is the first report that all VanB E. faecium isolates from a certain geographical location harbor the vanB2 gene. Previous studies have shown that VanB2 VRE are uncommon in the United States but are quite common in Europe (6, 7, 21). Dahl et al. (7) have recently examined 17 VanB VRE isolates from hospitalized patients in Europe and the United States and showed that only 2 of 9 U.S. isolates but 7 of 8 European isolates harbored the vanB2 gene. McGregor and Young (21) found that 28 of 32 VanB VRE from hospitalized patients in Scotland harbored vanB2. Clark et al. (6) examined 105 clinical isolates of VRE collected from 31 U.S. hospitals in 14 states and found that all 26 VanB VRE isolates harbored the vanB1 gene.
The results of this study also revealed sequence heterogeneity of the vanB ligase genes in different VRE isolates. The vanB2 sequences obtained in this study had a marked difference from those of vanB1 (4.3 to 4.5% difference) and vanB3 (2.8 to 3% difference). In addition, none of the 36 vanB2 ligase gene sequences determined in this study were identical to that of the previously described vanB2 ligase gene (22). There were 1- to 3-nucleotide differences between that sequence and our vanB2 ligase gene sequences. However, the HaeII and BclI digestion patterns of the long vanB PCR products of all 36 isolates are identical. This observation could provide an alternative means for identification of VanB2 VRE for epidemiological or surveillance studies.
Although there are several types of vanB, no correlation has been observed between vanB subtypes and levels of vancomycin resistance (7). A unique finding in this study is that all four VanB2 VRE isolates with a low or moderate resistance to vancomycin (MIC = 4 to 32 μg/ml) had a leucine-to-methionine change at codon 308 of the vanB2 gene. Whether this change actually results in a low or moderate vancomycin resistance in VanB2 VRE remains to be investigated.
The sequence heterogeneity in the vanB ligase genes is in contrast to the reported DNA sequence homogeneity in the vanA ligase gene (13, 14, 28). The sequences of the vanA ligase genes in different VRE isolates appeared to be identical. However, the Tn1546 or Tn1546-like elements that carry the vanA gene clusters are structurally different (13, 14, 28). The structural diversity in Tn1546 elements is caused by deletions or insertions which may have occurred during recombination events (13, 22, 28). VanA VRE have been isolated from a variety of sources, including animals, animal foodstuffs, and humans (14–17, 28). The food chain has been suggested to be a source of VanA VRE (5, 18). To our knowledge, VanB VRE have only been detected in hospitalized patients. The finding that type I E. faecium harboring the vanB2 gene was prevalent (69.4%) suggests that intra- and interhospital dissemination of this VRE has occurred in Taiwan. Because of the frequent occurrence of oxacillin-resistant Staphylococcus aureus, the use of vancomycin has increased tremendously in Taiwan during the last decade. It is possible that the spread of the type I VanB2 E. faecium in Taiwan is the result of unregulated use of vancomycin in humans.
The vanB gene cluster has been found to be associated with various DNA elements ranging from 60 to 250 kb (3, 4, 23, 29). A 64-kb composite transposon (Tn1547), bounded by IS256-like and IS16 elements, has been shown to transpose the vanB gene cluster from the chromosome to a plasmid (23). A 55-MDa transferable plasmid was found to contain both vanB and a gentamicin resistance gene (29). A 27-kb conjugative transposon (Tn5382) which may be part of a 160-kb plasmid containing both the vanB gene and the pbp5 gene, which encodes high-level ampicillin resistance, has also been described (4). The observation that HaeII and BclI restriction maps of the vanB gene cluster in various E. faecium isolates are identical suggests that the entire vanB2 gene cluster is transferred from one isolate to another. In addition to horizontal transfer of the vancomycin resistance gene due to DNA transposition or plasmid transfer, clonal dissemination of a certain strain is another mode of transmission. The finding that the majority (65%) of VanB isolates examined in this study belong to the same PFGE type (type I) is one example of this type of transmission. Based on the results of this study, it is recommended that both the mobile vancomycin resistance determinants and the PFGE types should be determined for epidemiological studies of VRE infections.
ACKNOWLEDGMENTS
We thank Jiunn-Jong Wu, National Cheng-Kung University Medical College; Tsu-Lan Wu, Chang Gung Memorial Hospital; and Po-Ren Hsueh, National Taiwan University Hospital, for supplying VRE isolates. This study was supported by grants from the National Science Council (NSC88-2314-B-016-053 and NSC89-2320-B-016-033), Taiwan, Republic of China.
REFERENCES
- 1.Arthur M, Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 1993;37:1563–1571. doi: 10.1128/aac.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyce J M, Opal S M, Chow J W, Zervos M J, Potter-Bynoe G, Sherman C B, Romulo R L, Fortna S, Medeiros A A. Outbreak of multidrug-resistant Enterococcus faecium with transferable vanB class vancomycin resistance. J Clin Microbiol. 1994;32:1148–1153. doi: 10.1128/jcm.32.5.1148-1153.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carias L L, Rudin S D, Donskey C J, Rice L B. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J Bacteriol. 1998;180:4426–4434. doi: 10.1128/jb.180.17.4426-4434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chadwick P R, Woodford N, Kaczmarski E B, Gray S, Barrell R A, Oppenheim B A. Glycopeptide-resistant enterococci isolated from uncooked meat. Antimicrob Agents Chemother. 1996;38:908–909. doi: 10.1093/jac/38.5.908. [DOI] [PubMed] [Google Scholar]
- 6.Clark N C, Cooksey R C, Hill B C, Swenson J M, Tenover F C. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother. 1993;37:2311–2317. doi: 10.1128/aac.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahl K H, Simonsen G S, Olsvik O, Sundsfjord A. Heterogeneity in the vanB gene cluster of genomically diverse clinical strains of vancomycin-resistant enterococci. Antimicrob Agents Chemother. 1999;43:1105–1110. doi: 10.1128/aac.43.5.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evers S, Courvalin P. Regulation of VanB-type vancomycin resistance gene expression by the VanSB-VanRB two-component regulatory system in Enterococcus faecalis V583. J Bacteriol. 1996;178:1302–1309. doi: 10.1128/jb.178.5.1302-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evers S, Reynolds P E, Courvalin P. Sequence of the vanB and ddl genes encoding D-alanine:D-lactate and D-alanine:D-alanine ligases in vancomycin-resistant Enterococcus faecalis V583. Gene. 1994;140:97–102. doi: 10.1016/0378-1119(94)90737-4. [DOI] [PubMed] [Google Scholar]
- 10.Evers S, Sahm D F, Courvalin P. The vanB gene of vancomycin-resistant Enterococcus faecalis V583 is structurally related to genes encoding D-Ala:D-Ala ligases and glycopeptide-resistance proteins VanA and VanC. Gene. 1993;124:143–144. doi: 10.1016/0378-1119(93)90779-3. [DOI] [PubMed] [Google Scholar]
- 11.Gold H S, Unal S, Cercenado E, Thauvin-Eliopoulos C, Eliopoulos G M, Wennersten C B, Moellering R C., Jr A gene conferring resistance to vancomycin but not teicoplanin in isolates of Enterococcus faecalis and Enterococcus faecium demonstrates homology with vanB, vanA, and vanC genes of enterococci. Antimicrob Agents Chemother. 1993;37:1604–1609. doi: 10.1128/aac.37.8.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gouby A, Neuwirth C, Bourg G, Bouziges N, Carles-Nurit M J, Despaux E, Ramuz M. Epidemiological study by pulsed-field gel electrophoresis of an outbreak of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in a geriatric hospital. J Clin Microbiol. 1994;32:301–305. doi: 10.1128/jcm.32.2.301-305.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handwerger S, Skoble J, Discotto L F, Pucci M J. Heterogeneity of the vanA gene cluster in clinical isolates of enterococci from the northeastern United States. Antimicrob Agents Chemother. 1995;39:362–368. doi: 10.1128/aac.39.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen L B, Ahrens P, Dons L, Jones R N, Hammerum A M, Aarestrup F M. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals and humans. J Clin Microbiol. 1998;36:437–442. doi: 10.1128/jcm.36.2.437-442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jordens J Z, Bates J, Griffiths D T. Faecal carriage and nosocomial spread of vancomycin-resistant Enterococcus faecium. J Antimicrob Chemother. 1994;34:515–528. doi: 10.1093/jac/34.4.515. [DOI] [PubMed] [Google Scholar]
- 16.Klare I, Heier H, Claus H, Bohme G, Marin S, Seltmann G, Hakenbeck R, Antanassova V, Witte W. Enterococcus faecium strains with vanA-mediated high-level glycopeptide resistance isolated from animal foodstuffs and fecal samples of humans in the community. Microb Drug Resist. 1995;1:265–272. doi: 10.1089/mdr.1995.1.265. [DOI] [PubMed] [Google Scholar]
- 17.Klare I, Heier H, Claus H, Witte W. Environmental strains of Enterococcus faecium with inducible high-level resistance to glycopeptides. FEMS Microbiol Lett. 1993;80:23–29. doi: 10.1111/j.1574-6968.1993.tb05930.x. [DOI] [PubMed] [Google Scholar]
- 18.Robredo B, Singh K V, Baquero F, Murray B E, Torres C. Vancomycin-resistant enterococci isolated from animals and food. Int J Food Microbiol. 2000;54:197–204. doi: 10.1016/s0168-1605(99)00195-6. [DOI] [PubMed] [Google Scholar]
- 19.Lu J J, Ben R J, Perng C L, Chi W M, Chu M L, Lee W H. Characterization of the first clinical isolate of vancomycin-resistant Enterococcus faecalis, AH803, in Taiwan. J Formos Med Assoc. 2000;99:178–181. [PubMed] [Google Scholar]
- 20.Lu, J. J., C. L. Perng, T. S. Chiueh, S. Y. Lee, C. H. Chen, F. Y. Chang, C. C. Wang, and W. M. Chi. Detection and typing of vancomycin-resistance genes of enterococci from clinical and nosocomial surveillance specimens by multiplex PCR. Epidemiol. Infect., in press. [DOI] [PMC free article] [PubMed]
- 21.McGregor K F, Young H-K. Identification and characterization of vanB2 glycopeptide resistance elements in enterococci isolated in Scotland. Antimicrob Agents Chemother. 2000;44:2341–2348. doi: 10.1128/aac.44.9.2341-2348.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel R, Uhl J R, Kohner P, Hopkins M K, Cockerill F R., III Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 genes in enterococci. J Clin Microbiol. 1997;35:703–707. doi: 10.1128/jcm.35.3.703-707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quintiliani R J, Courvalin P. Conjugal transfer of the vancomycin resistance determinant vanB between enterococci involves the movement of large genetic elements from chromosome to chromosome. FEMS Microbiol Lett. 1994;119:359–363. doi: 10.1111/j.1574-6968.1994.tb06913.x. [DOI] [PubMed] [Google Scholar]
- 24.Quintiliani R J, Courvalin P. Characterization of Tn1547, a composite transposon flanked by the IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene. 1996;172:1–8. doi: 10.1016/0378-1119(96)00110-2. [DOI] [PubMed] [Google Scholar]
- 25.Quintiliani R J, Evers S, Courvalin P. The vanB gene confers various levels of self-transferable resistance to vancomycin in enterococci. J Infect Dis. 1993;167:1220–1223. doi: 10.1093/infdis/167.5.1220. [DOI] [PubMed] [Google Scholar]
- 26.Sahm D F, Kissinger J, Gilmore M S, Murray P R, Mulder R, Solliday J, Clarke B. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1989;33:1588–1591. doi: 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woodford N, Adebiyi A M, Palepou M F, Cookson B D. Diversity of VanA glycopeptide resistance elements in enterococci from humans and nonhuman sources. Antimicrob Agents Chemother. 1998;42:502–508. doi: 10.1128/aac.42.3.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodford N, Jones B L, Baccus Z, Ludlam H A, Brown D F J. Linkage of vancomycin and high-level gentamicin resistance genes on the same plasmid in a clinical isolate of Enterococcus faecalis. J Antimicrob Chemother. 1995;35:179–184. doi: 10.1093/jac/35.1.179. [DOI] [PubMed] [Google Scholar]