Abstract
The discovery of antibiotics more than 80 years ago has led to considerable improvements in human and animal health. Although antibiotic resistance in environmental bacteria is ancient, resistance in human pathogens is thought to be a modern phenomenon that is driven by the clinical use of antibiotics1. Here we show that particular lineages of methicillin-resistant Staphylococcus aureus—a notorious human pathogen—appeared in European hedgehogs in the pre-antibiotic era. Subsequently, these lineages spread within the local hedgehog populations and between hedgehogs and secondary hosts, including livestock and humans. We also demonstrate that the hedgehog dermatophyte Trichophyton erinacei produces two β-lactam antibiotics that provide a natural selective environment in which methicillin-resistant S. aureus isolates have an advantage over susceptible isolates. Together, these results suggest that methicillin resistance emerged in the pre-antibiotic era as a co-evolutionary adaptation of S. aureus to the colonization of dermatophyte-infected hedgehogs. The evolution of clinically relevant antibiotic-resistance genes in wild animals and the connectivity of natural, agricultural and human ecosystems demonstrate that the use of a One Health approach is critical for our understanding and management of antibiotic resistance, which is one of the biggest threats to global health, food security and development.
Subject terms: Natural product synthesis, Population dynamics, Antimicrobial resistance, Bacterial evolution, Infectious-disease epidemiology
Methicillin-resistant strains of Staphylococcus aureus appeared in European hedgehogs in the pre-antibiotic era as a co-evolutionary adaptation to antibiotic-producing dermatophytes and have spread within the local hedgehog populations and between hedgehogs and secondary hosts.
Main
Methicillin-resistant S. aureus (MRSA) is one of the most common antibiotic-resistant bacterial pathogens, causing approximately 171,000 invasive infections each year in Europe alone2. MRSA was first identified in 1960 shortly after the introduction of methicillin (celbenin) as a treatment option against penicillin-resistant S. aureus clones3, but was possibly selected for by the clinical use of penicillin over the previous 20 years4. Methicillin resistance has subsequently emerged in many S. aureus clones around the world, both in hospital and community settings as well as in livestock such as pigs and cattle5,6. This has serious implications for the treatment of severe infections and the World Health Organization now considers MRSA to be an important threat to human health7.
Methicillin resistance in S. aureus is mediated by the mecA and mecC genes, which encode the enzymes penicillin-binding protein 2a (PBP2a) and PBP2c, respectively. mecA and mecC confer resistance to almost all β-lactam antibiotics, including penicillinase-labile penicillins (such as penicillin G), penicillinase-stable penicillins (such as methicillin) and cephalosporins (such as cefoxitin).
Hedgehog surveys from Denmark and Sweden demonstrated a surprisingly high prevalence of MRSA carrying mecC (mecC-MRSA)8,9, raising the possibility that the evolution of these bacteria was driven by natural selection in wildlife, as opposed to clinical use of antibiotics. Historically, mecC-MRSA was first discovered in dairy cows and subsequently in humans10, suggesting that the use of antibiotics in livestock was providing a selective advantage and that human infections were the result of zoonotic transmission. Studies from many different European countries revealed that mecC-MRSA is also present in other domesticated animals such as sheep, goats and horses as well as in a broad range of wild animals, albeit at low frequencies11.
Our hypothesis that the evolution of mecC-MRSA was driven by natural selection is supported by studies from northwestern Europe and New Zealand that showed that hedgehogs are frequently colonized with the dermatophyte T. erinacei, which produces a penicillinase-labile penicillin-like substance that was recently identified as penicillin G12–19. To test our hypothesis, we examined the distribution of mecC-MRSA and other S. aureus isolates in hedgehogs in ten European countries and New Zealand. We sequenced 244 S. aureus isolates from hedgehogs and 913 S. aureus isolates from other sources to infer the evolutionary histories, host dynamics, geographical dispersal patterns and zoonotic potential of the major mecC-MRSA clones in Europe. The potential mechanisms for the natural selection of mecC-MRSA by T. erinacei were assessed by analysing the genome of T. erinacei for β-lactam biosynthetic genes and by screening T. erinacei for the production of β-lactams and antibiotic activity against a panel of S. aureus strains.
The distribution of mecC-MRSA in hedgehogs
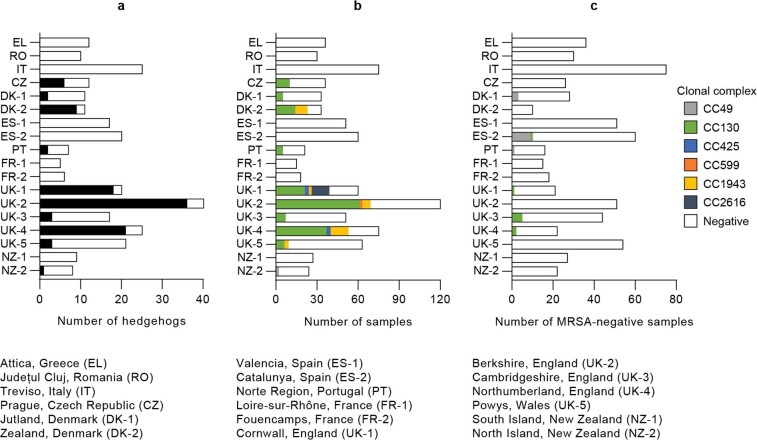
We first examined the geographical distribution and population structure of mecC-MRSA in European hedgehogs, which inhabit large parts of Europe as a result of postglacial expansion from Pleistocene refugia20. European hedgehogs have also become widespread in New Zealand after a series of introductions from the UK between 1869 and 1892 (ref. 21). We analysed 828 samples from the nasal area, skin and feet of 276 hedgehogs originating from 16 wildlife rescue centres in 10 European countries and 2 wildlife rescue centres in New Zealand (Fig. 1 and Extended Data Fig. 1). mecC-MRSA was present in 101 of the 172 hedgehogs (222 out of 516 samples) from England and Wales (66%, 81 out of 123), Czech Republic (50%, 6 out of 12), Denmark (50%, 11 out of 22), Portugal (29%, 2 out of 7) and New Zealand (6%, 1 out of 17), therefore extending the known geographical distribution of mecC-MRSA in hedgehogs (Fig. 1 and Extended Data Fig. 1). By contrast, all 104 hedgehogs (312 samples) from Greece, Romania, Italy, France and Spain tested negative for mecC-MRSA. Whole-genome sequencing showed that the 222 mecC-MRSA isolates belonged to 6 clonal complexes, CC130 (75%), CC1943 (15%), CC2616 (6%), CC425 (3%), CC49 (1%) and CC599 (1%), of which CC130 had the most widespread distribution across western and central Europe (Fig. 1 and Extended Data Fig. 1). We screened all of the MRSA-negative hedgehog samples from our study (n = 606) for the presence of methicillin-susceptible S. aureus (MSSA) isolates belonging to the same clonal complexes as the mecC-MRSA isolates (Extended Data Fig. 1). This led to the identification of 22 MSSA isolates, including 13 CC49 isolates from Spain (n = 9), Denmark (n = 3) and Portugal (n = 1), and 9 CC130 isolates from England (n = 8) and Spain (n = 1).
Fig. 1. Distribution of mecC-MRSA clones in European and New Zealand hedgehog samples.
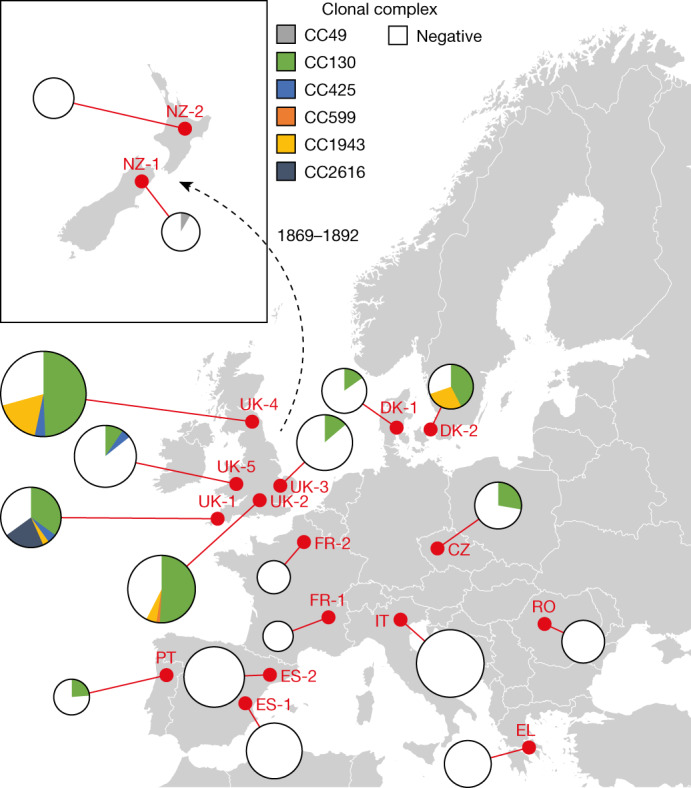
The analysis included 828 samples from the nasal area, skin and feet of 276 hedgehogs originating from 16 wildlife rescue centres in 10 European countries and 2 wildlife rescue centres in New Zealand. The red dots indicate the sampling locations. The pie charts are connected to the sampling locations by a red line. The area of the pie chart is proportional to the number of samples from that location. The introduction of European hedgehogs into New Zealand from the UK between 1869 and 1892 is shown. A detailed description of the results is provided in Extended Data Fig. 1. Maps were provided by Eurostat under a Creative Commons Attribution 4.0 International (CC BY 4.0) licence; the administrative boundaries are copyright of EuroGeographics.
Extended Data Fig. 1. Distribution of mecC-MRSA clones in European and New Zealand hedgehogs.
The analysis included 828 samples from the nasal area, skin and feet of 276 hedgehogs originating from 16 wildlife rescue centres in ten European countries and two wildlife rescue centres in New Zealand. a, Presence of mecC-MRSA in hedgehogs (n = 276). Presence and absence are shown as black and white boxes, respectively. b, Distribution of mecC-MRSA clones in hedgehog samples (n = 828). c, Distribution of MSSA clones in MRSA-negative hedgehog samples (n = 606).
The mecC gene encoding PBP2c is located immediately upstream of a blaZ gene (hereafter, blaZLGA251) on a chromosomally integrated mobile genetic element known as a type XI staphylococcal cassette chromosome mec (SCCmec). PBP2c and the blaZLGA251-encoded penicillinase are orthologues of the PBP2a enzyme and penicillinase produced by other S. aureus clones, although they share only 63% and 65% amino acid identities with each other, respectively10. Penicillinases have a narrower spectrum than PBP2a and PBP2c and provide resistance only to penicillin G and other penicillinase-labile subclasses of penicillin. As expected, blaZLGA251 was present in the 222 mecC-MRSA isolates but absent in the 22 MSSA isolates. However, 14 of the MSSA isolates carried the blaZ gene found in other S. aureus clones (Supplementary Table 1).
Production of β-lactams by T. erinacei
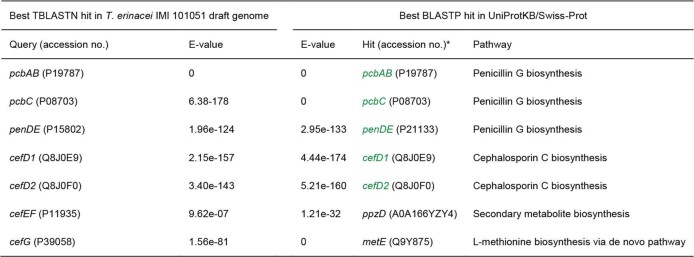
The abundance of mecC-MRSA in hedgehogs led us to speculate that antibiotic production by T. erinacei provides a selective environment in which mecC-MRSA isolates have an advantage over susceptible isolates. Genome sequencing and analysis of the T. erinacei type strain IMI 101051 (ATCC 28443) identified orthologues of pcbAB, pcbC and penDE, which are responsible for key steps in penicillin G production by Penicillium chrysogenum, as well as the Acremonium chrysogenum early cephalosporin C biosynthetic genes cefD1 and cefD2, which are involved in the conversion of isopenicillin N into penicillin N (Fig. 2 and Extended Data Table 1). By contrast, T. erinacei IMI 101051 lacked the A. chrysogenum late cephalosporin C biosynthetic genes cefEF and cefG. P. chrysogenum also carries cefD1 and cefD2 but is nevertheless incapable of producing cephalosporins due to the lack of cefEF and cefG22,23.
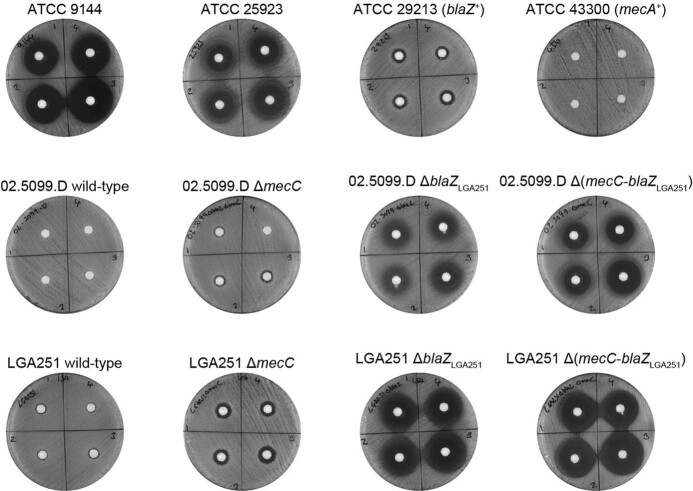
Fig. 2. Penicillin biosynthetic genes and antibiotic activity of T. erinacei IMI 101051.
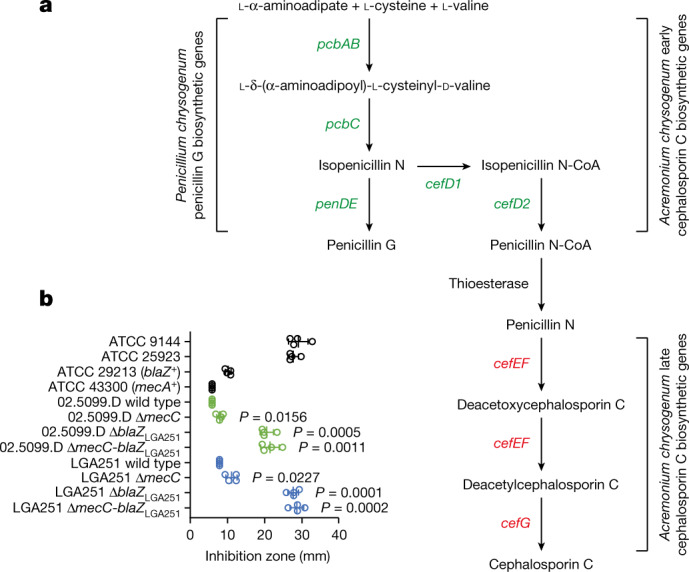
a, Schematic of the key steps in the biosynthesis of penicillin G and cephalosporin C. The presence (green) or absence (red) of T. erinacei penicillin G and cephalosporin C biosynthetic genes is indicated. b, T. erinacei inhibition zones against a collection of S. aureus control strains (black) and two mecC-MRSA wild-type strains belonging to CC130 (green) and CC425 (blue) and their isogenic mutants. Two-tailed paired Student’s t-tests were used to compare inhibition zones of each mutant to the corresponding wild-type strain. Data are mean ± s.d.; n = 4 biologically independent fungal culture extracts. A detailed description of the results is provided in Extended Data Fig. 4.
Extended Data Table 1.
Penicillin biosynthetic genes in T. erinacei IMI 101051
*The in silico identified putative T. erinacei penicillin G and cephalosporin C biosynthetic genes are shown in green
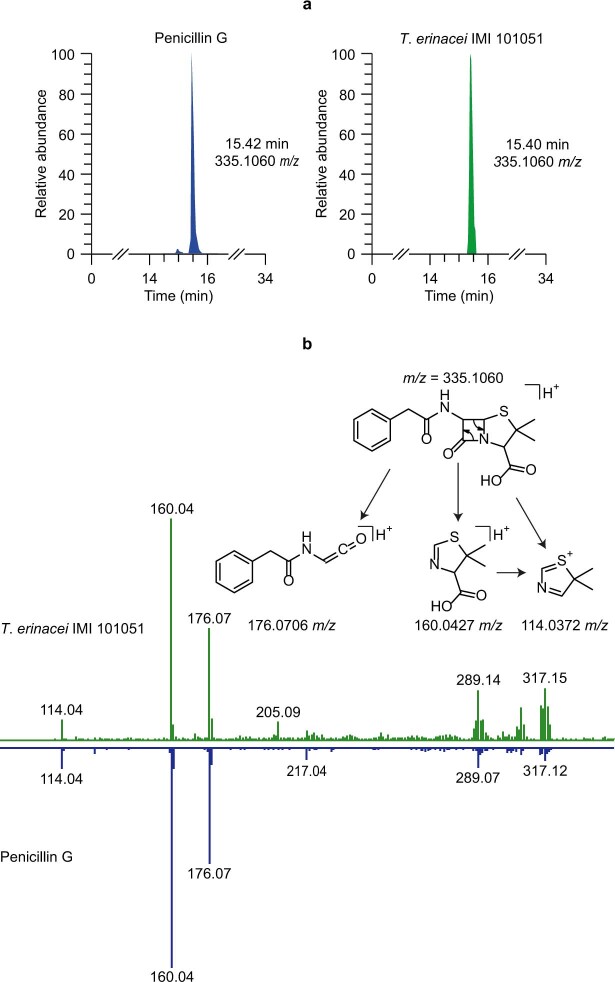
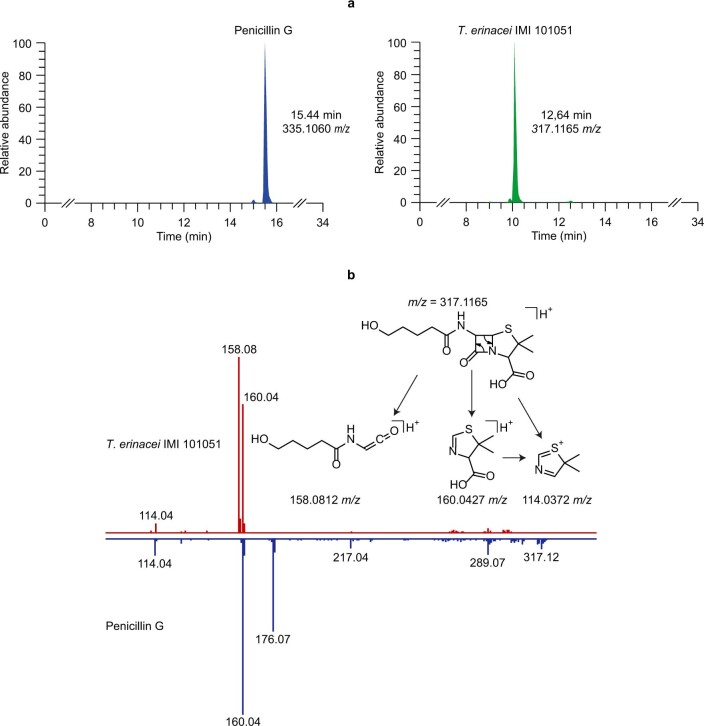
We processed four distinct culture broths of T. erinacei IMI 101051 for metabolic profiling using liquid chromatography–mass spectrometry (LC–MS) and molecular networking analysis. This led to the identification of two β-lactams, penicillin G and 6-(5-hydroxy-n-valeramido)-penicillanic acid (KPN), both of which belong to the penicillin class of antibiotics (Extended Data Figs. 2 and 3 and Supplementary Fig. 1). KPN has to date been found only in culture broths of fungal strains belonging to the genus Paecilomyces24 and differs from penicillin G by having a unique side chain (Extended Data Figs. 2 and 3 and Supplementary Fig. 1). The biosynthetic pathway of KPN is currently unknown.
Extended Data Fig. 2. Detection of penicillin G in T. erinacei IMI 101051 culture broth.
a, Left and right panels show extracted ion chromatogram of a pure standard of penicillin G and in T. erinacei culture broth, respectively. b, Upper and lower panels show MS2 spectra of penicillin G in a T. erinacei culture broth and a pure standard of penicillin G, respectively.
Extended Data Fig. 3. Detection of KPN in T. erinacei IMI 101051 culture broth.
a, Left and right panels show extracted ion chromatogram of a pure standard of penicillin G and KPN in T. erinacei culture broth, respectively. b, Upper and lower panels show MS2 spectra of KPN in a T. erinacei culture broth and a pure standard of penicillin G, respectively.
Selection of mecC-MRSA by T. erinacei
Four culture broths of T. erinacei IMI 101051 were screened for antibiotic activity against a collection of S. aureus control strains. All of the culture broths produced large inhibition zones against two penicillin-susceptible S. aureus strains—ATCC 9144 (Oxford S. aureus) and ATCC 25923—but much smaller zones against the penicillinase-producing S. aureus strain ATCC 29213 and the mecA-positive S. aureus strain ATCC 43300 (Fig. 2 and Extended Data Fig. 4). The role of mecC and blaZLGA251 was assessed by screening the culture broths for antibiotic activity against two mecC-MRSA wild-type strains belonging to CC130 (02.5099.D) and CC425 (LGA251) and their isogenic mutants. The mutants with deleted mecC (ΔmecC), blaZLGA251 (ΔblaZLGA251), and mecC and blaZLGA251 (ΔmecC-blaZLGA251) produced significantly larger inhibition zones compared with the corresponding wild-type strains, although the zones of the ΔblaZLGA251 and ΔmecC-blaZLGA251 mutants were larger compared with the zones of the ΔmecC mutants (Fig. 2 and Extended Data Fig. 4). These results indicate that mecC and blaZLGA251 both contribute to the reduced susceptibility of mecC-MRSA to penicillin G and KPN present in culture broths of T. erinacei IMI 101051.
Extended Data Fig. 4. Antibiotic activity of T. erinacei IMI 101051.
T. erinacei inhibition zones against a collection of S. aureus control strains and two mecC-MRSA wild-type strains belonging to CC130 (02.5099.D) and CC425 (LGA251) and their isogenic mutants. The numbers on the plates refer to each of four biologically independent fungal culture extracts.
Evolutionary history of mecC-MRSA
We sought to infer the evolutionary histories of S. aureus CC130, CC425 and CC1943, which constitute the most successful mecC-MRSA clones in Europe10,11,25. For this purpose, we collected and sequenced 786 mecC-MRSA and 127 MSSA CC130, CC425 and CC1943 isolates selected to represent the known geographical distribution (mainly western and central Europe) and host repertoire (mainly humans, cattle, sheep, goats and wild animals) of each clone (Supplementary Table 1). We used core-genome single-nucleotide polymorphism (SNP) diversity and isolation dates to infer time-scaled phylogenies of these isolates and the 205 mecC-MRSA and 9 MSSA CC130, CC425 and CC1943 isolates collected from hedgehogs (Supplementary Table 1). The sequencing data were processed for pan-genome analysis to identify antibiotic-resistance genes (ARGs) and mobile genetic elements that encode human- and ruminant-specific immune modulators that are involved in host switching events, including a phage-encoded immune evasion cluster-1 (IEC-1) enabling S. aureus to evade the human innate immune response and a staphylococcal pathogenicity island (SaPI)-encoded vwb gene (vwbSaPI), which encodes a von Willebrand factor-binding protein with coagulase activity against ruminant plasma26.
We also sought to infer a time-scaled phylogeny of the 991 type XI SCCmec elements containing the mecC and blaZLGA251 genes but the correlation between root-to-tip distances and isolation dates was too weak with a coefficient of determination R2 = −0.05 (Extended Data Fig. 5). Instead, we used the topology of the type XI SCCmec phylogeny to identify monophyletic mecC-MRSA lineages harbouring orthologous type XI SCCmec elements. The type XI SCCmec elements could be traced back to seven nodes that were connected to each other on a long backbone. Each of the backbone nodes and its orthologous descendants received the same letter designation to reflect their genetic relationship (A to G) (Fig. 3 and Supplementary Fig. 2). Manual mapping of the tips onto the CC130, CC425 and CC1943 phylogenies, and vice versa, enabled us to assign the mecC-MRSA isolates to 16 monophyletic lineages harbouring orthologous type XI SCCmec elements (Fig. 3 and Supplementary Figs. 2–5). The 129 mecC-MRSA CC1943 isolates could be divided into three lineages (C1 to C3), which probably originated in the early-to-late 1800s, long before the first β-lactam—penicillin G—became widely available as a therapeutic option in the 1940s (Fig. 3). The 786 mecC-MRSA CC130 isolates and 76 mecC-MRSA CC425 isolates belonged to 10 and 3 lineages (A1 to A10 and B1 to B3, respectively) (Fig. 3). Several of these lineages also originated in the pre-antibiotic era (Fig. 3). Most mecC-MRSA isolates lacked vwbSaPI (96%, 949 out of 991) and IEC-1 (100%, 990 out of 991) and were genotypically susceptible to non-β-lactam antibiotics (Supplementary Figs. 3–5). The largest mecC-MRSA CC425 lineage (CC425:B3) had a unique evolutionary trajectory with signs of adaptation to ruminants (Supplementary Fig. 4). The basal mecC-MRSA CC425:B3 isolates probably originated in England during the early 1940s and shared epidemiological and genetic characteristics with the other mecC-MRSA lineages: they were associated with multiple hosts, including hedgehogs, cattle and humans, and lacked vwbSaPI and IEC-1. By contrast, their descendants (CC425:B3.1) harboured vwbSaPI, were restricted to cattle and humans in southwest England and probably diverged during the 1960s (date of the most recent common ancestor (MRCA), 1965; 95% confidence interval (CI), 1926–1986). Our analysis revealed that some of the mecC-MRSA lineages (CC130:A2, CC425:B3 and CC1943:C1) carried unique variants of the type XI SCCmec element, supporting that they have evolved through vertical inheritance from the MRCA of each mecC-MRSA lineage, whereas others shared the same type XI SCCmec variant (Fig. 3 and Supplementary Figs. 2–5). The latter findings could be the result of either purifying (negative) selection, convergent evolution, homologous recombination between different mecC-MRSA lineages or horizontal gene transfer.
Extended Data Fig. 5. Root-to-tip linear regression analysis of the type XI SCCmec dataset.
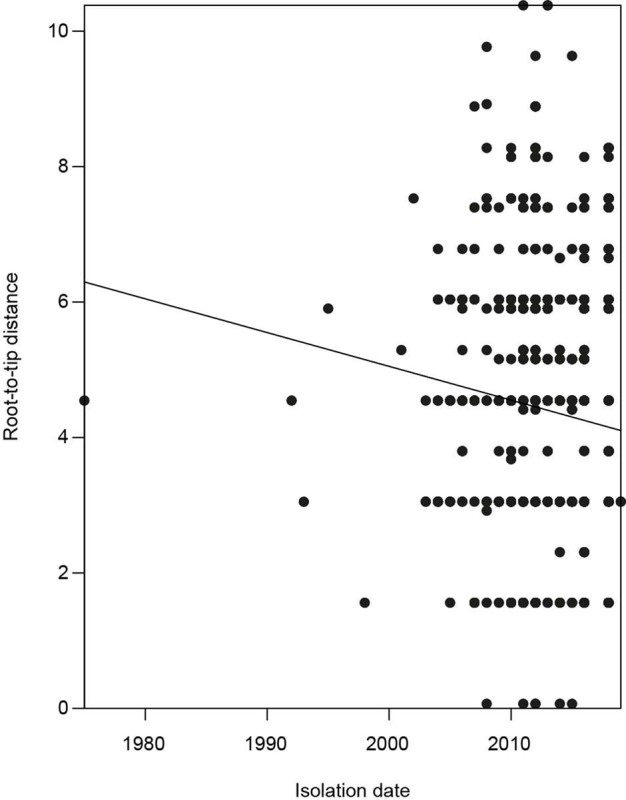
The correlation between root-to-tip distances and isolation dates is very weak with a coefficient of determination R2 = −0.05.
Fig. 3. Timeline of mecC-MRSA CC130, CC425 and CC1943 evolution in Europe.
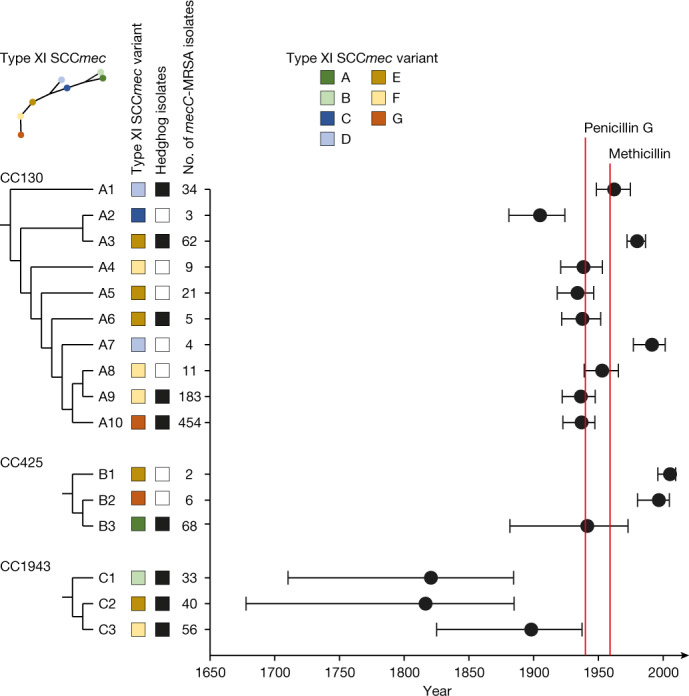
Manual mapping of the tips on the type XI SCCmec phylogeny onto the CC130, CC425 and CC1943 phylogenies, and vice versa, enabled us to assign the mecC-MRSA isolates to 16 monophyletic lineages containing orthologous type XI SCCmec elements (A–G). The trees are redrawn from Supplementary Figs. 2–5 to illustrate the branching order of the different type XI SCCmec variants and mecC-MRSA lineages. Branch lengths are not drawn to scale. The presence and absence of hedgehog isolates in a given lineage are shown as black and white boxes, respectively. A detailed description of the geographical distribution and host range of major mecC-MRSA CC130, CC425 and CC1943 lineages is provided in Extended Data Fig. 7. The estimated date of the most recent common ancestor and 95% confidence interval of each mecC-MRSA lineage are illustrated by filled circles and horizontal lines, respectively. The introduction of penicillin G and methicillin as therapeutic options is indicated by red lines.
To better understand the potential role of horizontal gene transfer in the evolution of the three early mecC-MRSA CC1943 lineages, we determined the smallest number of sublineages that were present at a given time point, which is also the smallest number of acquisition events that could explain the presence of the same type XI SCCmec variant in all sublineages (Extended Data Fig. 6). The analysis showed that mecC-MRSA CC1943 consisted of 15 sublineages in 1940, just before penicillin G became available as a therapeutic option. It has previously been estimated that SCCmec was acquired in a single horizontal gene transfer event in three of the major hospital- and community-associated MRSA clones4,27,28, although multiple introductions have also been reported29. Thus, it is more plausible to assume that the type XI SCCmec element was present in the MRCA of each mecC-MRSA CC1943 lineage rather than assuming horizontal gene transfer into each of the 15 sublineages within a few years at the beginning of the antibiotic era. Vertical inheritance of the type XI SCCmec element is also consistent with the apparent absence of admixture between the different mecC-MRSA CC1943 lineages despite the fact that they are often found in hedgehogs within the same geographical area (Extended Data Fig. 7). Notably, the type XI SCCmec variants found in mecC-MRSA CC1943:C2 (E) and CC1943:C3 (F) were each other’s neighbours on the type XI SCCmec phylogeny, and it is therefore possible that the type XI SCCmec element was acquired even earlier (date of the MRCA, 1737; 95% CI, 1562–1824).
Extended Data Fig. 6. Number of mecC-MRSA CC1943 sublineages.
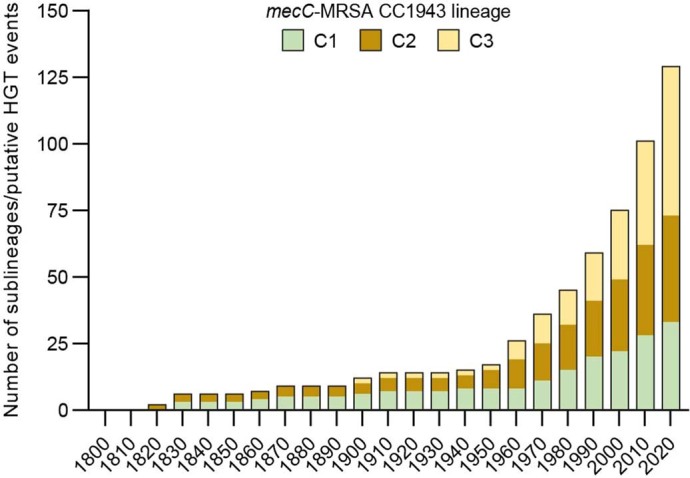
The bars show the number of sublineages of mecC-MRSA CC1943:C1, mecC-MRSA CC1943:C2 and mecC-MRSA CC1943:C3 at different time points.
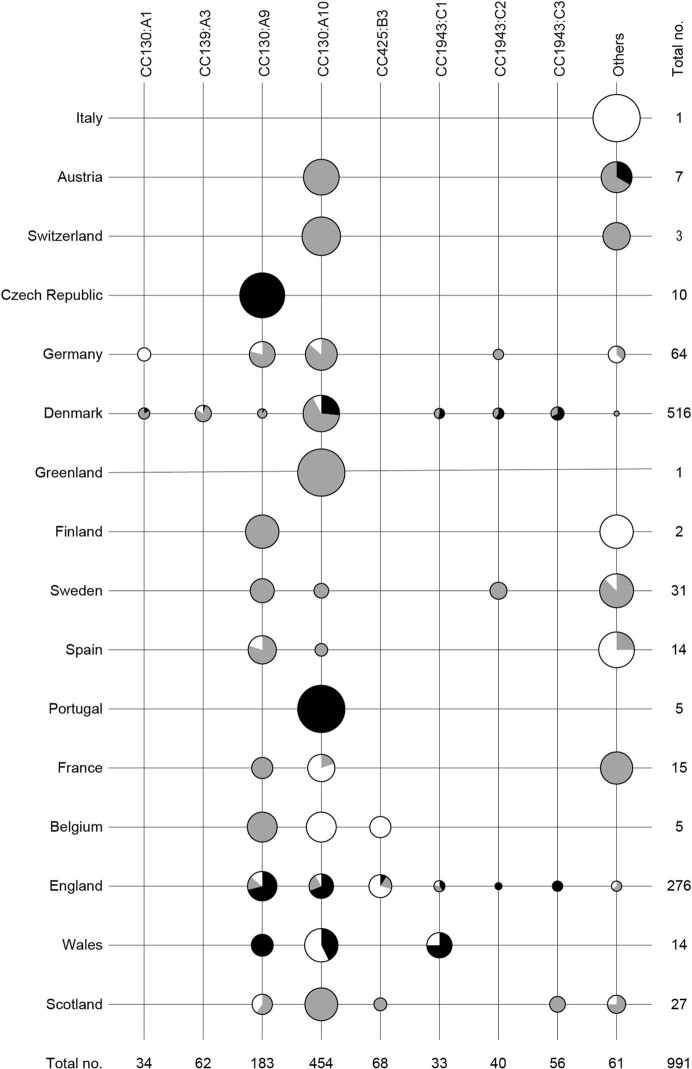
Extended Data Fig. 7. Geographical distribution and host range of major mecC-MRSA CC130, CC425 and CC1943 lineages.
The study collection included 991 mecC-MRSA CC130, CC425 and CC1943 isolates originating from 16 European countries. The countries of sampling and the eight largest (≥25 isolates) mecC-MRSA lineages are shown. Isolates belonging to the eight minor mecC-MRSA lineages are grouped together (others). The pie charts depict the proportion of mecC-MRSA isolates from hedgehogs (black), humans (grey) and other sources (white). The area of the pie chart is proportional to the number of mecC-MRSA isolates from that country.
MSSA isolates comprised 8% (67 out of 851), 47% (68 out of 144) and 0.8% (1 out of 130) of the CC130, CC425 and CC1943 isolates, respectively. The vast majority of the MSSA CC130 and MSSA CC425 isolates, but not the single MSSA CC1943 isolate, were basal to the corresponding mecC-MRSA lineages (Supplementary Figs. 3–5). Most of the basal MSSA CC130 isolates originated from sheep and goats in Italy, France, Spain and Norway and harboured vwbSaPI. Moreover, some of the isolates carried ARGs against antibiotics that are used to treat infections in sheep and goats, including the two tetracycline-resistance genes tet(K) and tet(L), the blaZ gene found in other S. aureus clones, the chloramphenicol resistance gene cat and the macrolide resistance gene erm(C). The earliest branching CC425 lineages were epidemiologically and genetically diverse with respect to host range and the presence/absence of the type XI SCCmec element, vwbSaPI and ARGs, although most originated from wild animals in Spain and lacked the aforementioned genetic determinants. Together, these findings suggest that CC130 and CC425 emerged from distinct ruminant and wildlife reservoirs in Europe and that methicillin resistance is an acquired phenotype within these clones.
Population dynamics of mecC-MRSA
Hedgehogs constitute a large reservoir of mecC-MRSA clones, as demonstrated here and elsewhere8,9, whereas mecC-MRSA isolates are present at much lower frequencies in humans, domesticated animals and other wild animals. Hedgehog isolates were present in 9 out of the 16 mecC-MRSA lineages, including the 8 largest (≥25 isolates) and 3 earliest (200–130 years ago) lineages (Fig. 3). The 2 largest mecC-MRSA CC130 lineages (CC130:A9 and CC130:A10) encompassed 67% (232 out of 344), 65% (339 out of 520) and 43% (66 out of 153) of all mecC-MRSA isolates from hedgehogs, humans and other sources, respectively, and had the broadest geographical ranges in western and central Europe (Extended Data Fig. 7 and Supplementary Figs. 3–5). Several of the major mecC-MRSA CC130 and CC1943 lineages (such as CC130:A9, CC130:A10, CC1943:C1, CC1943:C2 and CC1943:C3) contained isolates that were separated by wide expanses of seawater, reflecting numerous long-distance dispersal events between British and Danish islands and mainland Europe within the past 200 years (Extended Data Fig. 7 and Supplementary Figs. 3–5). By contrast, analysis of the fine-scale population structure of mecC-MRSA isolates from hedgehogs revealed a detailed pattern of diversifications over short distances, resulting in substantial concordance between genetic clusters and geography at the local level (Supplementary Figs. 3–5). The observed clustering is consistent with the limited dispersal capacity of hedgehogs and the effects of habitat fragmentation.
Assuming that hedgehogs act as local reservoirs (sources) of mecC-MRSA in Denmark, theoretical considerations predict that secondary transmission to other hosts (sinks) would lead to similar patterns of genetic variation within these secondary hosts cohabiting the same geographical region. To examine this theory, we determined the local population structures of two broad collections of Danish mecC-MRSA isolates from hedgehogs and humans covering the geographical ranges of two of the three Danish hedgehog subpopulations, namely Jutland (a peninsula connected to continental Europe) and major islands30. Hedgehog and human isolates from the geographical range of the remaining hedgehog subpopulation (the small island of Bornholm) were excluded from the analysis due to their small sample size (nine isolates). Most of the CC130:A10 isolates from Jutland formed a distinct sublineage (CC130:A10.1) together with isolates from other parts of Europe (Supplementary Fig. 3). As a consequence, CC130:A10.1 isolates were treated as a separate group in the analysis. The results revealed distinct patterns of regional dispersal with little overlap between Jutland and the major islands and a notable correlation between the population structures of hedgehog and human isolates at the regional level (P = 0.0149, two-sided Wilcoxon matched-pairs signed-rank test) (Fig. 4 and Extended Data Fig. 8). These findings support the hypothesis that human isolates originate from local hedgehog reservoirs, although it should be noted that the data presented here do not provide evidence for directionality or rule out the involvement of other animal reservoirs (such as livestock) as part of the transmission chains.
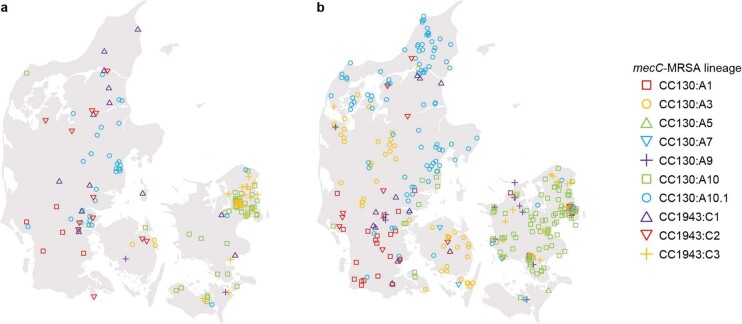
Fig. 4. Population structures of Danish mecC-MRSA isolates from hedgehogs and humans.
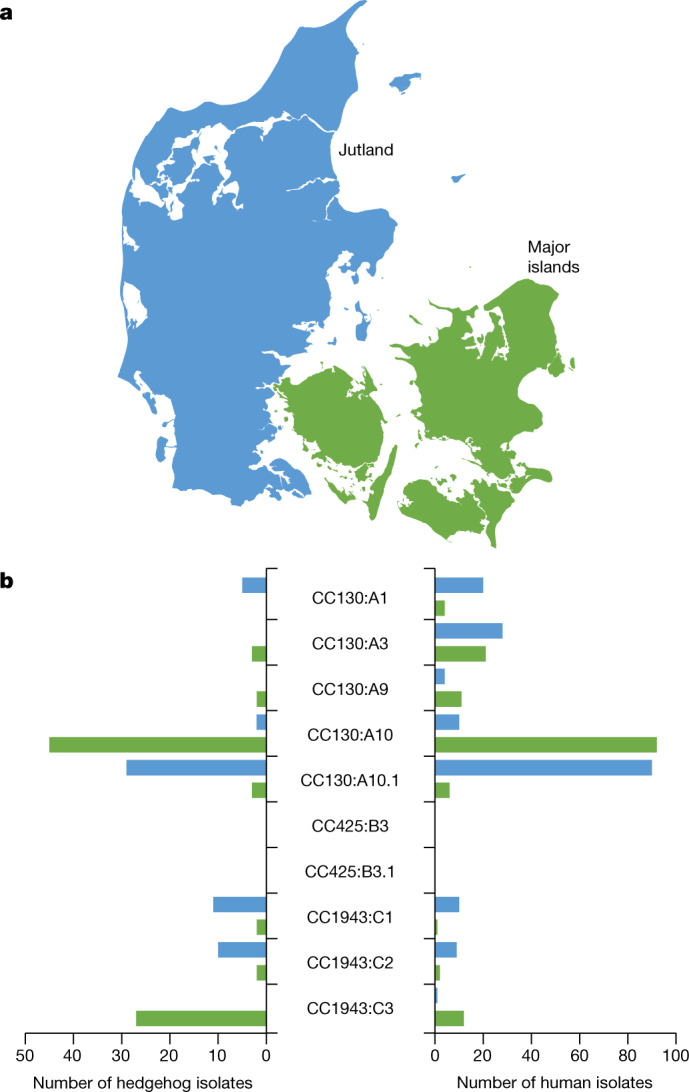
a, The map of Denmark shows the geographical ranges of two of the three hedgehog subpopulations in Jutland and on the major islands. b, The geographical distribution of major mecC-MRSA CC130, CC425 and CC1943 lineages in two broad collections of mecC-MRSA isolates recovered from hedgehogs (n = 141) and humans (n = 327) in Jutland and on the major islands. Hedgehog and human isolates from the remaining hedgehog subpopulation on the small island of Bornholm (not shown) were excluded from the analysis due to their small sample size (n = 9). A detailed map of the sampling locations is provided in Extended Data Fig. 8. Maps were provided by Eurostat under a Creative Commons Attribution 4.0 International (CC BY 4.0) licence; the administrative boundaries are copyright of EuroGeographics.
Extended Data Fig. 8. Sampling locations of Danish mecC-MRSA isolates from hedgehogs and humans.
The maps of Denmark relate to the map shown in Fig. 4. a, mecC-MRSA recovered from hedgehogs (n = 141). b, mecC-MRSA recovered from humans (n = 327). The location of each sample is given at the zip code area level. Maps were provided by Eurostat under a Creative Commons Attribution 4.0 International (CC BY 4.0) licence; the administrative boundaries are copyright of EuroGeographics.
Discussion
This research shows that hedgehogs are a natural reservoir of zoonotic mecC-MRSA lineages that predate the antibiotic era, which is inconsistent with the commonly accepted view that widespread resistance in clinical pathogens is a modern phenomenon that is driven by our use of antibiotics in human and veterinary medicine.
Data on the prevalence of mecC-MRSA in humans and different animal species indicate that hedgehogs are the most likely primary host in some countries. For example, in Denmark, the prevalence in hedgehogs is considerably higher than in cattle (veal calves and bulk tank milk), sheep and goats (61% versus 0.0–1.1%)8,31,32, and the number of human cases is relatively low (3–36 cases per year)25. This is further supported by our finding that mecC-MRSA generally lacks genetic markers of human and ruminant adaptation, with the notable exception of the CC425:B3.1 lineage that has undergone a host jump from hedgehogs to cattle in southwest England. Before this study, dairy cows were considered to be the most likely reservoir of mecC-MRSA and a major source of zoonotic infections in humans. This hypothesis was supported by the fact that β-lactams are commonly used to treat bovine mastitis, as shown by sales data from Denmark and the UK33,34. However, our findings strongly suggest that most mecC-MRSA lineages originate from hedgehogs, although dairy cows and other domesticated animals probably act as intermediate hosts and vectors in zoonotic transmission from hedgehogs to humans, as previously demonstrated35.
Here we show that the T. erinacei type strain IMI 101051 from New Zealand produces two β-lactams—penicillin G and KPN—and that mecC and blaZLGA251 both contribute to reduced susceptibility of mecC-MRSA to these antibiotics. Previous studies have established that T. erinacei is widespread among hedgehogs in New Zealand and northwestern Europe and that isolates from both continents produce a penicillinase-labile penicillin-like substance12–19, and a recent study characterized penicillin G from T. erinacei in Sweden (the presence of other β-lactams was not investigated)19. This suggests that penicillin-producing T. erinacei isolates were circulating in European hedgehogs long before they were introduced into New Zealand in the late 1800s and that methicillin resistance first emerged in Europe as a co-evolutionary adaptation of S. aureus to colonization of hedgehogs. By contrast, it cannot be ruled out that clinical use of antibiotics in humans and livestock has contributed to the evolution of some of the younger mecC-MRSA CC130 and CC425 lineages, although only one of these lineages (CC425:B3) showed signs of adaptation to either of these hosts. Our findings indicate that seven of the younger mecC-MRSA lineages (CC130:A3, CC130:A4, CC130:A5, CC130:A6, CC130:A8, CC130:A9 and CC425:B1) have acquired their type XI SCCmec variants from the early mecC-MRSA CC1943:C2 and CC1943:C3 lineages (Fig. 3 and Supplementary Figs. 2–5). The type XI SCCmec element has also been found at low frequencies in coagulase-negative Staphylococcus species from wild animals and livestock but the evolutionary links between these potential donors and mecC-MRSA remain to be investigated36.
Our analyses suggest that most mecC-MRSA transmission events within hedgehog populations and between hedgehogs and secondary hosts are highly localized. The finding that some human mecC-MRSA isolates probably originate from local hedgehog reservoirs indicates that mecC-MRSA has been a cause of sporadic infections in humans for the past 200 years, more than a century before MRSA was first identified in patients in 1960 (ref. 3). The host interactions that lead to zoonotic transmission probably include direct contact with hedgehogs or contact with secondary animal hosts such as dairy cows, as previously shown for T. erinacei (the cause of ‘hedgehog ringworm’ in humans)13. We also identified several long-distance dispersal events between British and Danish islands and mainland Europe. The connections that bridge geographically isolated hedgehog populations are poorly understood but might involve oversea movements of humans and livestock. Furthermore, a recent report of mecC-MRSA in white storks raises the possibility that migratory birds could be efficient long-distance carriers37.
β-Lactams target PBPs that catalyse carboxypeptidase and transpeptidase reactions during bacterial cell wall synthesis, whereby they inhibit cross-linking of neighbouring peptidoglycan strands. The primary mechanisms of β-lactam resistance in S. aureus are enzymatic cleavage of the amide bond in the β-lactam ring of penicillinase-labile penicillins by blaZ-encoded penicillinases and the production of mecA- or mecC-encoded PBP2a and PBP2c, respectively, with a decreased affinity for a broad spectrum of β-lactams. Our findings support that both mechanisms contribute to protection against penicillin production by T. erinacei in hedgehogs, although it should be noted that the ΔmecC mutants produced smaller inhibition zones than the ΔblaZLGA251 mutants. This might be due to the fact that PBP2c has a relatively high binding affinity for penicillins compared with PBP2a38. It is also possible that PBP2c provides additional ecological benefits, such as protection against cephalosporin-producing fungi and bacteria that occur naturally in all environments39.
mecC and blaZLGA251 have also been found on a pseudo-SCCmec element (ΨSCCmecP5085) in Staphylococcus edaphicus, a soil-dwelling bacterial species isolated from Antarctica40. In contrast to the type XI SCCmec element, ΨSCCmecP5085 lacks the cassette chromosome recombinase (ccr) genes that are responsible for movement (excision and integration) of SCCmec40. Several studies have identified ARGs in ancient and modern samples and bacteria from natural environment such as soil41–43. Yet, the environmental resistome shows limited potential for horizontal gene transfer and, for this reason, the contribution of environmental ARGs to resistance in human pathogens has so far been controversial44. It seems reasonable to assume that the microbiota of wild animals have greater exposure to the environmental resistome than the human microbiota and are therefore more likely to acquire environmental ARGs. Thus, wild animals might represent a hitherto unrecognized conduit through which environmental ARGs can be transferred to clinical pathogens.
We acknowledge some limitations of our study. Although we broadly sampled hedgehogs across Europe and New Zealand, our study represents only a small part of the geographical range and a small number of hedgehog samples in most countries. Thus, the distribution and diversity of mecC-MRSA in Europe and New Zealand might be larger than documented here. We cannot account for the potential effect of transmission within the different wildlife rescue centres. To better understand the transmission dynamics, we examined the frequency of potential transmission events of mecC-MRSA CC130 within the different facilities using a range of maximum pairwise SNP distance thresholds to define a cluster (Extended Data Fig. 9). Using a conservative cut-off of 25 SNPs (transmission age < 6 months)45, 25% (683 out of 2,783) of the mecC-MRSA CC130 isolate pairs collected within the same facility belonged to a potential transmission cluster. It is therefore possible that the prevalence of mecC-MRSA in hedgehogs kept in such facilities is higher than in the wild, although it is also probable that some of these potential transmission events occurred in the wild before admission to the facility. Notably, a previous study found that there is only a slightly lower prevalence of mecC-MRSA in hedgehogs that died in the wild compared with hedgehogs that died while staying in a wildlife rescue centre8, which is consistent with our finding that most hedgehogs acquired mecC-MRSA outside the facility. We were unable to test the hedgehogs for carriage of T. erinacei, because ethical constraints precluded us from collecting appropriate tissues (skin scrapings, hair and spines) for fungal culture, which leaves some important questions unanswered. For example, it remains unclear whether penicillin-producing T. erinacei isolates are present throughout Europe and whether there is a link between their distribution and the geographical range of mecC-MRSA.
Extended Data Fig. 9. Frequency of potential transmission events of mecC-MRSA CC130 isolates within wildlife rescue centres.
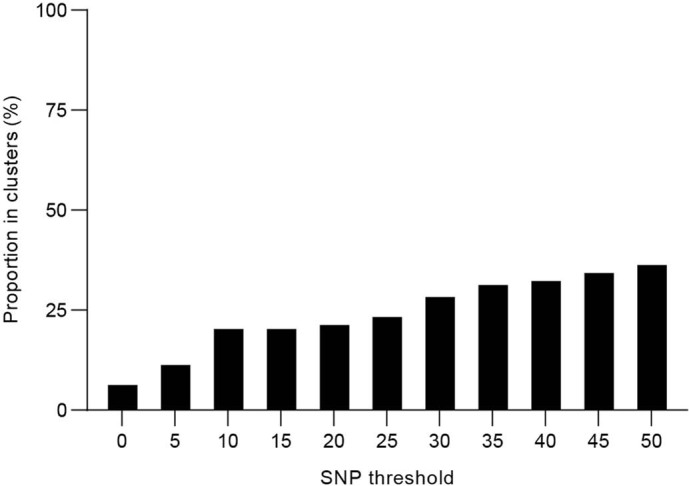
The bars show the proportion of isolates that belong to clusters defined at different pairwise SNP distance thresholds.
In conclusion, we describe the ecological and evolutionary mechanisms that led to the emergence of methicillin resistance in the pre-antibiotic era, possibly as a co-evolutionary adaptation of S. aureus to colonization of dermatophyte-infected hedgehogs. These results underscore the importance of taking a broad One Health perspective on antibiotic resistance that recognises the role of natural selection in wild animals and the connectivity of natural, agricultural and human ecosystems in the evolution and spread of antibiotic-resistant pathogens.
Methods
Hedgehog survey
The aim was to collect hedgehog samples covering the geographical range of hedgehogs in Europe and New Zealand. Personnel at 16 wildlife rescue centres in ten European countries and two wildlife rescue centres in New Zealand were instructed to obtain samples from the nasal area, skin and feet of hedgehogs kept in separate enclosures using FLOQSwabs (Copan). Swabs were stored in liquid Amies medium at ambient temperature and sent to the National Reference Laboratory for Antimicrobial Resistance at Statens Serum Institut in Denmark or the Department of Veterinary Medicine at University of Cambridge in the UK immediately after sample collection. For each swab, a loopful (10 µl) of liquid Amies was inoculated into 5 ml Mueller–Hinton broth (Oxoid) supplemented with 6.5% NaCl and incubated overnight at 35–37 °C. A loopful (10 µl) of enrichment broth was then streaked on a Brilliance MRSA 2 (Oxoid) agar plate followed by incubation at 35–37 °C for 24 h. One presumptive MRSA colony from each plate was subcultured on a blood agar plate at 35–37 °C for 24 h and archived at −80 °C. We also screened all MRSA-negative samples for the presence of MSSA isolates by streaking a loopful (10 µl) of enrichment broth on a SaSelect (Bio-Rad) and Brilliance Staph 24 (Oxoid) agar plate followed by incubation at 35–37 °C for 24 h. One presumptive S. aureus colony from each plate was subcultured on a blood agar plate at 35–37 °C for 24 h and archived at −80 °C.
The hedgehog survey was conducted by staff members of the wildlife rescue centres who collected swab samples in connection with routine checks. In accordance with the Animal Welfare Act 1999 administered by the New Zealand Ministry for Primary Industries and Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes, no ethical approval was required as sample collection did not cause pain, suffering, distress or lasting harm equivalent to, or higher than, that caused by the introduction of a needle in accordance with good veterinary practice or deprived the animal of usual care. Ethical review was undertaken at the Department of Veterinary Medicine, University of Cambridge (CR76).
Bacterial isolates and whole-genome sequencing
A list of the 1,157 isolates used in this study is provided in Supplementary Table 1, including all of the mecC-MRSA (n = 222) and MSSA (n = 22) isolates identified in the hedgehog survey described above and an European collection of mecC-MRSA (n = 786) and MSSA (n = 127) isolates belonging to CC130, CC425 and CC1943, which constitute the most successful mecC-MRSA clones in Europe10,11,25. Most of the Danish isolates originated from two nationwide collections of mecC-MRSA and MSSA isolates recovered from hedgehogs and humans. The Danish collection of hedgehog isolates included 114 mecC-MRSA isolates collected from 188 hedgehogs in a previous study8. We re-examined the 74 MRSA-negative hedgehog samples for the presence of MSSA isolates using the method described above, which led to the inclusion of two MSSA isolates belonging to CC130 (n = 1) and CC425 (n = 1) as well as six mecC-MRSA isolates belonging to CC130 (n = 3) and CC1943 (n = 3) that were missed in the original screening. The Danish collection of human isolates comprised 334 mecC-MRSA and 2 MSSA isolates that were collected from colonized or infected individuals between 1975 and 2016 as part of the national MRSA and S. aureus bacteraemia surveillance programmes. The remaining CC130, CC425 and CC1943 isolates were selected to represent the known geographical distribution (mainly western and central Europe) and host repertoire (mainly humans, cattle, sheep, goats and wild animals) of each clone. Whole-genome sequencing of the 1,157 isolates was performed on different Illumina platforms at the Wellcome Sanger Institute or at Statens Serum Institut. Short-read sequence data are available in the European Nucleotide Archive/NCBI Sequence Read Archive under BioProject IDs PRJEB15105, PRJEB21015, PRJEB2655, PRJEB2755, PRJEB2756, PRJEB28206, PRJEB3174, PRJEB32898, PRJNA596428 and PRJEB43456 and the genome accession numbers are provided in Supplementary Table 1.
Sequence analyses
Draft genomes were de novo assembled using SPAdes (v.3.15)46. Multilocus sequence typing (MLST) was performed by comparing the draft genomes with the S. aureus MLST database47. We used the scn gene (which encodes staphylococcal complement inhibitor-A and an indicator of the IEC-1 element48) in S. aureus strain Newman (GenBank: NC_009641) and the vwbSaPI gene in SaPIbov4 (GenBank: HM211303) as queries in BLASTN searches against the draft genomes, setting length match to 0.9 and similarity match to 0.7. These parameters were chosen to account for allelic diversity of the vwb genes located on SaPIs (sharing 76–100% nucleotide identities with each other)49 while effectively excluding scn homologues located outside the IEC-1 element (sharing 48–61% nucleotide identities with scn)50. Contigs with hits for vwb were analysed using PHASTER51 to ascertain that they were located on SaPIs. ARGs were detected by mapping sequence reads against the ResFinder database52 using the k-mer alignment (KMA) tool (v.1.3)53, setting both length match and similarity match to 0.9.
Phylogenetic analyses of S. aureus CC130, CC425 and CC1943
All 991 mecC-MRSA and 136 MSSA isolates belonging to CC130, CC425 and CC1943 were included. Mapping of sequence reads and SNP calling were performed using NASP (v.1.0)54 as follows: (1) sequence reads were mapped against the reference genome of mecC-MRSA CC425 isolate LGA251 (GenBank: NC_017349) with the Burrows–Wheeler Alignment tool55; (2) SNP calling was achieved using the GATK Unified Genotyper56,57, setting depth of coverage and unambiguously base calls to ≥10× and ≥90%, respectively, and ignoring insertions and deletions; and (3) SNPs contained in repeats were excluded using NUCmer58,59.
Unrooted maximum-likelihood phylogenetic trees were built from core-genome SNP alignments with PhyML (v.3.0)60,61 under the HKY85 substitution model after applying NNI moves to improve the BIONJ starting tree. Putative recombinogenic regions were detected and a recombination-corrected phylogeny was built with ClonalFrameML (v.1.12)62. Time-resolved phylogenies, in which the date of each node is estimated, were constructed with BactDating (v.1.0)63 using an additive uncorrelated relaxed clock model64. Convergence and mixing of the Markov Chain Monte Carlo chains were determined using the R package coda65. The time-resolved phylogenies were rooted as inferred with BactDating (v.1.0)63 to maximize the posterior probability of the tree. Treedater (v.0.5)66 was also applied for the same purpose and found to give similar estimates for the dating of nodes (Supplementary Figs. 6–11 and Source Data of Fig. 3).
The phylogenetic relationships between the type XI SCCmec elements were investigated in a separate analysis. A SNP alignment was generated from precalled SNPs from the 991 mecC-MRSA isolates. SNPs located outside the type XI SCCmec element (corresponding to nucleotide positions 34,403 through 63,839 in mecC-MRSA CC425 isolate LGA251) were manually removed, and the remaining SNPs were used to construct an unrooted maximum-likelihood phylogenetic tree using PhyML (v.3.0)60,61 under the HKY85 substitution model after applying NNI moves to improve the BIONJ starting tree. Putative recombinogenic regions were detected and a recombination-corrected phylogeny was built using ClonalFrameML (v.1.12)62. The tips were manually mapped onto the CC130, CC425 and CC1943 phylogenies, and vice versa, to identify monophyletic mecC-MRSA lineages harbouring orthologous type XI SCCmec elements.
Genome sequencing and analysis of the T. erinacei type strain IMI 101051
DNA extracted from the T. erinacei type strain IMI 101051 was used to prepare a DNA library in accordance with the Nextera XT DNA Library Prep Guide (Illumina) and sequenced on a MiSeq platform (Illumina) with 2 × 251 bp using a MiSeq Reagent Kit v2 (Illumina). Short-read sequencing data were submitted to the European Nucleotide Archive under BioProject PRJEB43453. A draft genome was de novo assembled using SPAdes (v.3.15)46. We used the translated protein sequences of the P. chrysogenum pcbAB, pcbC and penDE genes and the A. chrysogenum cefD1, cefD2, cefEF and cefG genes (Extended Data Table 1) as queries in TBLASTN searches against the draft genome. Hits were used as queries in BLASTP searches against the UniProtKB/Swiss-Prot protein database. Bidirectional best hits with a TBLASTN and BLASTP value of less than 1 × 10−100 in both directions were considered as orthologous gene pairs.
In vitro antibiotic production by the T. erinacei type strain IMI 101051
In vitro antibiotic production by the T. erinacei type strain IMI 101051 was determined using a method modified from Smith and Marples15. In brief, the strain was grown on a Sabouraud dextrose agar plate (Oxoid) for 7 days at 30 °C. The culture was removed using distilled water and a 100-µl suspension was placed in 5 ml Sabouraud dextrose broth (SDB) (Sigma-Aldrich) followed by incubation at 30 °C with shaking at 200 rpm for 3 days. The culture was placed on a Miracloth mesh (Calbiochem) in a 13-cm petri dish containing 35 ml SDB. The plate was incubated at 30 °C for 7 days, after which the mesh containing the fungal mass was placed in a 2-l conical flask containing 75 ml SDB supplemented with 2% glucose. The flask was incubated at 30 °C with shaking at 200 rpm for 7 days. The broth was replaced with fresh 5-strength SDB, and the flask was incubated at 30 °C for another 6 days. The culture medium was transferred into two 50-ml Falcon tubes containing 3 ml Diaion HP-20 resin (Supelco) slurried in 3 ml distilled water. The tubes were placed on a rotator for 1 h, after which the beads were allowed to settle. The supernatant was discarded and antibiotics were eluted from the resin with 9 ml acetone. The eluate was placed on a rotator for 30 min and centrifuged briefly to pellet the resin. The supernatant was transferred to 2-ml Eppendorf tubes, which were placed in a rotary evaporator at 30 °C. This process was repeated until all of the eluate had evaporated to dryness. To prepare bacterial inocula, S. aureus strains were individually grown overnight on blood agar plates (Oxoid) at 37 °C. Colonies were suspended in phosphate-buffered saline (PBS) to a 0.5 McFarland standard, diluted 1:10 in PBS, and streaked evenly on the surface of a Iso-Sensitest agar plate (Oxoid) using a sterile cotton-wool swab. Dried fungal culture extracts were suspended in 200 µl distilled water. Sterile paper discs were impregnated with 10 µl solution and placed on the agar plates. In vitro antibiotic production was assessed by measuring the inhibition zones after overnight incubation at 35 °C.
Metabolic profiling by LC–MS
Dried fungal culture extracts were suspended in 50% methanol to a concentration of 100 mg ml−1, diluted 1:10 in pure methanol and analysed using LC–MS. Metabolic profiles were obtained on a Vanquish UHPLC system (Thermo Fisher Scientific) coupled to a Vanquish diode array detector (Thermo Fisher Scientific) and an Orbitrap Fusion Tribrid high-resolution tandem mass spectrometer (Thermo Fisher Scientific). Chromatographic separation of 5 µl fungal culture extracts was performed on a Luna C18 column (3 μm, 3 × 150 mm) (Phenomenex) using a linear mobile phase gradient from 0% methanol, 90% water and 10% acetonitrile containing 1% (v/v) formic acid to 90% methanol, 0% water and 10% acetonitrile containing 1% (v/v) formic acid over 20 min at a flow rate of 400 μl min−1. Ultraviolet data were recorded between 210 nm and 550 nm. For comparative purposes, a pure standard of penicillin G (Sigma-Aldrich) was prepared at a final concentration of 9.4 μg ml−1 and analysed using LC–MS.
Mass spectra were obtained in the positive and negative ionization modes using the full scan and data-dependent MS2 and MS3 acquisition modes. Full scan total ion current chromatograms were obtained over the range of 125–1,800 m/z using a spray voltage of +3.5 kV and −2.5 kV for the positive and negative ionization modes, respectively. Three different scan events were recorded for the data-dependent acquisition modes as follows: (1) MS2 of the most intense ion in the full scan acquisition mode; (2) MS3 of the most intense ion in the MS2 spectra; and (3) MS3 of the second-most intense ion in the MS2 spectra. Additional parameters for the MS included the following: full scan resolution set to 60,000 (full-width at half-maximum, FWHM), capillary temperature set to 350 °C, ion transfer tube temperature set to 325 °C, RF lens set to 50%, automatic gain control target set to 4.0 × 105 (full scan) or 1.0 × 104 (MS2 and MS3), intensity threshold set to 1.0 × 104, collision-induced dissociation energy set to 35 eV, activation Q set to 0.25 and isolation window set to 4 m/z. Nitrogen was used as the drying, nebulizer and fragmentation gas.
Molecular networking analysis of LC–MS data
A molecular network was created using the Global Natural Products Social Molecular Networking (GNPS) workflow67. Chromatographic raw data in the positive ionization mode were transformed to the mzXML format using the ProteoWizard command-line utility msConvert68. The data were then filtered by removing all MS2 fragment ions within ±17 Da of the precursor m/z value. The precursor ion mass and MS2 fragment ion tolerances were set to 2.0 Da and 0.5 Da, respectively, to enable more comprehensive comparisons with the GNPS database. MS2 spectra were filtered by choosing the top six fragment ions in the ±50-Da window throughout the spectrum. A network was then created considering a cosine score above 0.7 and more than six matched peaks to link different nodes through edges. Edges between two nodes were kept in the network if each of the nodes appeared in each other’s respective top ten most similar nodes. Finally, the maximum size of a molecular family was set to 100. The spectra in the network were then searched against spectral libraries in GNPS, filtering the library spectra in the same manner as the input data. All matches between network spectra and library spectra were required to have a score above 0.7 and at least six matched peaks.
Statistics
Statistical analyses were performed using GraphPad Prism (v.8.3) (GraphPad Software). We used two-tailed paired Student’s t-tests to compare inhibition zones of each mutant to the corresponding mecC-MRSA wild-type strain and a two-tailed Wilcoxon matched-pairs signed rank test to compare population structures of mecC-MRSA at the lineage level.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-021-04265-w.
Supplementary information
Supplementary Figs. 1–11 and their accompanying legends.
Information about the 1,157 S. aureus isolates used in this study.
Supplementary Data 1–10 and their accompanying legends.
Source data
Acknowledgements
We thank M. Ganoti, F. Attila-Zoltán, M. Dugar, Z. Pokorná, D. Madsen, S. M. Gamborg, R. Molina-López, C. Rodrigues, T. Vieira, J. Viricel, A. Fingar, K. South, G. Prince, H. Gasser, S. Sebright, N. Ennew, C. Catchpole, E. Acton, N. Acton, K. Horrey and P. Loague for providing hedgehog samples; A. Medina, L. R. H. Kildevang, P. T. Hansen, and S. M. Johansson for technical assistance during the analysis of hedgehog samples; A. E. Henius for help with Figs. 1, 4 and Extended Data Fig. 8; and V. Bortolaia for reading the manuscript. This work is dedicated to V. R. Simpson, who passed away during the study. B.Č.B. was supported by a grant from the Czech University of Life Sciences Prague (no. IGA 20213106). X.D. was funded by a grant from the National Institute for Health Research (NIHR) Health Protection Research Unit in Genomics and Enabling Data (no. NIHR200892). M.A.H. was supported by grants from the Medical Research Council (nos. G1001787/1, MR/N002660/1 and MR/P007201/1) and the Economic and Social Research Council (no. ES/S000186/1). E.M.H. was supported by a UK Research and Innovation (UKRI) Fellowship (no. MR/S00291X/1). We acknowledge Eurostat for the provision of maps, which are licensed under the Creative Commons Attribution 4.0 International (CC BY 4.0) licence, and administrative boundaries, which are copyright of EuroGeographics.
Extended data figures and tables
Author contributions
J.L., M.A.H., E.M.H. and A.R.L. initiated and designed the study. J.L., C.L.R., B.Č.B., P.H. and S.L.R. directed the hedgehog survey. J.L., C.L.R. X.B. and Ø.A. analysed hedgehog samples. C.L.R., I.L., H.K., P.A., R.H., A.D., S.V., R.L.S., A.P., C.E.-G., F.M.A., L.J.L., S.M.N., F. Laurent, K.B., B.W., C.K., C.C., F. Layer, G.W., W.W., I.S., P.M., H.J.J., H.d.L., E.C., F.G.-G., S.B., S.H., V.P., C.J.T., A.S.W., B.P., M.D.C., M.J.E., S.J.P., D.J.S., N.F.H., J.A.L., G.F.E., G.F., G.K.P., M.A.H. and A.R.L. provided bacterial isolates from other sources. X.B., C.B., M.S., F.J.E.M., G.K.P. and M.A.H. conducted whole-genome sequencing of bacterial isolates. J.L., C.L.R., R.N.S., J.J.W., J.P., M.T.G.H., X.D., M.A.H. and E.M.H. performed sequence and phylogenetic analyses of bacterial isolates. J.L., M.C.A. and R.K.H. performed genome sequencing and analysis of T. erinacei IMI 101051. C.L.R., X.B. and D.J.S. investigated in vitro antibiotic production by T. erinacei IMI 101051. N.J.S. prepared samples, performed LC–MS analysis and guided the identification of the penicillin-like substances. G.F.P.-G. analysed LC–MS data, performed molecular networking analysis and identified the two penicillin-like substances. M.S.J.S. coordinated and supervised the LC–MS analysis. J.L. wrote the manuscript with considerable input from C.L.R., X.B., N.J.S., G.F.P.-G., X.D., M.A.H., E.M.H. and A.R.L. All of the authors reviewed the manuscript.
Peer review information
Nature thanks Ross Fitzgerald and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
LC–MS data in mzXML format and molecular networking results are available at the MassIVE repository under identifier MSV000087335. S. aureus short-read sequence data have been deposited in the European Nucleotide Archive/NCBI Sequence Read Archive under BioProject IDs PRJEB15105, PRJEB21015, PRJEB2655, PRJEB2755, PRJEB2756, PRJEB28206, PRJEB3174, PRJEB32898, PRJNA596428 and PRJEB43456 and the genome accession numbers are provided in Supplementary Table 1. T. erinacei type strain IMI 101051 short-read sequence data have been deposited in the European Nucleotide Archive under BioProject PRJEB43453. Tree and genealogy files in Newick format are provided in Supplementary Data 1–10. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jesper Larsen, Claire L. Raisen
These authors jointly supervised this work: Mark A. Holmes, Ewan M. Harrison, Anders R. Larsen
Change history
2/24/2022
Extended Data Figure 1 was updated to correct an error in panel c.
Extended data
is available for this paper at 10.1038/s41586-021-04265-w.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-021-04265-w.
References
- 1.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Centre for Disease Prevention and Control, European Medicines Agencies. The Bacterial Challenge: Time to React. A Call to Narrow the Gap Between Multidrug-Resistant Bacteria in the EU and the Development of New Antibacterial Agentshttps://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/0909_TER_The_Bacterial_Challenge_Time_to_React.pdf (2009).
- 3.Jevons MP. “Celbenin”—resistant Staphylococci. Br. Med. J. 1961;1:124–125. [Google Scholar]
- 4.Harkins CP, et al. Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol. 2017;18:130. doi: 10.1186/s13059-017-1252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers HF, DeLeo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price LB, et al. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio. 2012;3:e00305-11. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibioticshttp://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua=1 (WHO, 2017).
- 8.Rasmussen SL, et al. European hedgehogs (Erinaceus europaeus) as a natural reservoir of methicillin-resistant Staphylococcus aureus carrying mecC in Denmark. PLoS ONE. 2019;14:e0222031. doi: 10.1371/journal.pone.0222031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bengtsson B, et al. High occurrence of mecC-MRSA in wild hedgehogs (Erinaceus europaeus) in Sweden. Vet. Microbiol. 2017;207:103–107. doi: 10.1016/j.vetmic.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 10.García-Álvarez L, et al. Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect. Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paterson GK, Harrison EM, Holmes MA. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2014;22:42–47. doi: 10.1016/j.tim.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marples MJ, Smith JMB. The hedgehog as a source of human ringworm. Nature. 1960;188:867–868. doi: 10.1038/188867b0. [DOI] [PubMed] [Google Scholar]
- 13.English MP, Evans CD, Hewitt M, Warin RP. “Hedgehog ringworm”. Br. Med. J. 1962;1:149–151. doi: 10.1136/bmj.1.5272.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith JMB, Marples MJ. A natural reservoir of penicillin-resistant strains of Staphylococcus aureus. Nature. 1964;201:844. doi: 10.1038/201844a0. [DOI] [PubMed] [Google Scholar]
- 15.Smith JMB, Marples MJ. Dermatophyte lesions in the hedgehog as a reservoir of penicillin-resistant staphylococci. J. Hyg. 1965;63:293–303. doi: 10.1017/s0022172400045174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith JMB. Staphylococcus aureus strains associated with the hedgehog Erinaceus europaeus. J. Hyg. Camb. 1965;63:293–303. doi: 10.1017/s0022172400045162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris P, English MP. Trichophyton mentagrophytes var. erinacei in British hedgehogs. Sabouraudia. 1969;7:122–128. [PubMed] [Google Scholar]
- 18.Le Barzic C, et al. Detection and control of dermatophytosis in wild European hedgehogs (Erinaceus europaeus) admitted to a French wildlife rehabilitation centre. J. Fungi. 2021;7:74. doi: 10.3390/jof7020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dube F, Söderlund R, Salomonsson ML, Troell K, Börjesson S. Benzylpenicillin-producing Trichophyton erinacei and methicillin resistant Staphylococcus aureus carrying the mecC gene on European hedgehogs: a pilot-study. BMC Microbiol. 2021;21:212. doi: 10.1186/s12866-021-02260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 21.Brockie RE. Distribution and abundance of the hedgehog (Erinaceus europaeus) L. in New Zealand, 1869–1973. N. Z. J. Zool. 1975;2:445–462. [Google Scholar]
- 22.van den Berg MA, et al. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat. Biotechnol. 2008;26:1161–1168. doi: 10.1038/nbt.1498. [DOI] [PubMed] [Google Scholar]
- 23.Ullán RV, Campoy S, Casqueiro J, Fernández FJ, Martín JF. Deacetylcephalosporin C production in Penicillium chrysogenum by expression of the isopenicillin N epimerization, ring expansion, and acetylation genes. Chem. Biol. 2007;14:329–339. doi: 10.1016/j.chembiol.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Kitano K, et al. A novel penicillin produced by strains of the genus. Paecilomyces. J. Ferment. Technol. 1976;54:705–711. [Google Scholar]
- 25.Petersen A, et al. Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin. Microbiol. Infect. 2013;19:E16–E22. doi: 10.1111/1469-0691.12036. [DOI] [PubMed] [Google Scholar]
- 26.Richardson EJ, et al. Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat. Ecol. Evol. 2018;2:1468–1478. doi: 10.1038/s41559-018-0617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holden MTG, et al. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 2013;23:653–664. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strauß L, et al. Origin, evolution, and global transmission of community-acquired Staphylococcus aureus ST8. Proc. Natl Acad. Sci. USA. 2017;114:E10596–E10604. doi: 10.1073/pnas.1702472114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nübel U, et al. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc. Natl Acad. Sci. USA. 2008;105:14130–14135. doi: 10.1073/pnas.0804178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasmussen SL, Nielsen JL, Jones OR, Berg TB, Pertoldi C. Genetic structure of the European hedgehog (Erinaceus europaeus) in Denmark. PLoS ONE. 2020;15:e0227205. doi: 10.1371/journal.pone.0227205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen JE, et al. LA-MRSA CC398 in dairy cattle and veal calf farms indicates spillover from pig production. Front. Microbiol. 2019;10:2733. doi: 10.3389/fmicb.2019.02733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eriksson J, Espinosa-Gongora C, Stamphøj I, Larsen AR, Guardabassi L. Carriage frequency, diversity and methicillin resistance of in Danish small ruminants. Vet. Microbiol. 2013;163:110–115. doi: 10.1016/j.vetmic.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 33.Danish Integrated Antimicrobial Resistance Monitoring and Research Programme. DANMAP 2019: Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria From Food Animals, Food, and Humans in DENMARKhttps://www.danmap.org/-/media/Sites/danmap/Downloads/Reports/2019/DANMAP_2019.ashx?la=da&hash=AA1939EB449203EF0684440AC1477FFCE2156BA5 (2020).
- 34.Veterinary Medicines Directorate. UK Veterinary Antibiotic Resistance and Sales Surveillance Reporthttps://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/950126/UK-VARSS_2019_Report__2020-TPaccessible.pdf (2020).
- 35.Harrison EM, et al. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol. Med. 2013;5:509–515. doi: 10.1002/emmm.201202413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loncaric I, et al. Characterization of mecC gene-carrying coagulase-negative Staphylococcus spp. isolated from various animals. Vet. Microbiol. 2019;230:138–144. doi: 10.1016/j.vetmic.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 37.Gómez P, et al. Detection of MRSA ST3061-t843-mecC and ST398-t011-mecA in white stork nestlings exposed to human residues. J. Antimicrob. Chemother. 2016;71:53–57. doi: 10.1093/jac/dkv314. [DOI] [PubMed] [Google Scholar]
- 38.Kim C, et al. Properties of a novel PBP2A protein homolog from Staphylococcus aureus strain LGA251 and its contribution to the β-lactam-resistant phenotype. J. Biol. Chem. 2012;287:36854–36863. doi: 10.1074/jbc.M112.395962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tahlan K, Jensen SE. Origins of the β-lactam rings in natural products. J. Antibiot. 2013;66:401–419. doi: 10.1038/ja.2013.24. [DOI] [PubMed] [Google Scholar]
- 40.Pantůček R, et al. Staphylococcus edaphicus sp. nov. isolated in Antarctica harbors the mecC gene and genomic islands with a suspected role in adaptation to extreme environment. Appl. Environ. Microbiol. 2018;84:e01746–17. doi: 10.1128/AEM.01746-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D’Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 42.Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J. 2009;3:243–251. doi: 10.1038/ismej.2008.86. [DOI] [PubMed] [Google Scholar]
- 43.Forsberg KJ, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forsberg KJ, et al. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014;509:612–616. doi: 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coll F, et al. Definition of a genetic relatedness cutoff to exclude recent transmission of meticillin-resistant Staphylococcus aureus: a genomic epidemiology analysis. Lancet Microbe. 2020;1:e328–e335. doi: 10.1016/S2666-5247(20)30149-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its application to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Wamel WJ, Rooijakkers SH, Ruyken M, van Kessel KP, Strijp JA. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 2006;188:1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viana, D. et al. Adaptation of Staphylococcus aureus to ruminant and equine hosts involved SaPI-carried variants of von Willebrand factor-binding protein. Mol. Microbiol. 77, 1583–1594 (2010). [DOI] [PubMed]
- 50.Rooijakkers SHM, et al. Staphylococcal complement inhibitor: structure and active sites. J. Immunol. 2007;179:2989–2998. doi: 10.4049/jimmunol.179.5.2989. [DOI] [PubMed] [Google Scholar]
- 51.Arndt D, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bortolaia V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clausen PTLC, Aarestrup FM, Lund O. Rapid and precise alignment of raw reads against redundant database with KMA. BMC Bioinform. 2018;19:397. doi: 10.1186/s12859-018-2336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sahl JW, et al. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb. Genom. 2016;2:e000074. doi: 10.1099/mgen.0.000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Durbin R. Fast and accurate short read alignment with Burrow-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guindon S, Gasquel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 61.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 62.Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genome. PLoS Comput. Biol. 2015;11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Didelot X, et al. Bayesian inference of ancestral dates on bacterial phylogenetic trees. Nucleic Acids Res. 2018;46:e134. doi: 10.1093/nar/gky783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Didelot X, Siveroni I, Volz EM. Additive uncorrelated relaxed clock models for the dating of genomic epidemiology phylogenies. Mol. Biol. Evol. 2021;38:307–317. doi: 10.1093/molbev/msaa193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Plummer M, Best N, Cowles K, Vines K. CODA: convergence diagnosis and output analysis for MCMC. R News. 2006;6:7–11. [Google Scholar]
- 66.Volz EM, Frost SD. Scalable relaxed clock phylogenetic dating. Virus Evol. 2017;3:vex025. [Google Scholar]
- 67.Wang M, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adusumilli R, Mallick P. Data conversion with ProteoWizard msConvert. Methods Mol. Biol. 2017;1550:339–368. doi: 10.1007/978-1-4939-6747-6_23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1–11 and their accompanying legends.
Information about the 1,157 S. aureus isolates used in this study.
Supplementary Data 1–10 and their accompanying legends.
Data Availability Statement
LC–MS data in mzXML format and molecular networking results are available at the MassIVE repository under identifier MSV000087335. S. aureus short-read sequence data have been deposited in the European Nucleotide Archive/NCBI Sequence Read Archive under BioProject IDs PRJEB15105, PRJEB21015, PRJEB2655, PRJEB2755, PRJEB2756, PRJEB28206, PRJEB3174, PRJEB32898, PRJNA596428 and PRJEB43456 and the genome accession numbers are provided in Supplementary Table 1. T. erinacei type strain IMI 101051 short-read sequence data have been deposited in the European Nucleotide Archive under BioProject PRJEB43453. Tree and genealogy files in Newick format are provided in Supplementary Data 1–10. Source data are provided with this paper.