Summary
Invaginations of the mitochondrial inner membrane, termed cristae, are hubs for oxidative phosphorylation. The mitochondrial contact site and cristae organizing system (MICOS) and the dimeric F1Fo-ATP synthase play important roles in controlling cristae architecture. A fraction of the MICOS core subunit Mic10 is found in association with the ATP synthase, yet it is unknown whether this interaction is of relevance for mitochondrial or cellular functions. Here, we established conditions to selectively study the role of Mic10 at the ATP synthase. Mic10 variants impaired in MICOS functions stimulate ATP synthase oligomerization like wild-type Mic10 and promote efficient inner membrane energization, adaptation to non-fermentable carbon sources, and respiratory growth. Mic10's functions in respiratory growth largely depend on Mic10ATPsynthase, not on Mic10MICOS. We conclude that Mic10 plays a dual role as core subunit of MICOS and as partner of the F1Fo-ATP synthase, serving distinct functions in cristae shaping and respiratory adaptation and growth.
Keywords: mitochondria, inner membrane, membrane architecture, MICOS, ATP synthase, cristae organization, respiration, Mic10, metabolic adaptation, membrane potential
Graphical abstract
Highlights
-
•
Dual role of Mic10 of mitochondrial contact site and cristae organizing system (MICOS)
-
•
Mic10 binds to mitochondrial ATP synthase and stabilizes higher order assemblies
-
•
Oligomerization of Mic10 is required for its function in MICOS, not at ATP synthase
-
•
Mic10 binding to ATP synthase supports metabolic adaptation and respiratory growth
Rampelt et al. report that the core component Mic10 of the mitochondrial contact site and cristae organizing system plays a second physiologically important role at the mitochondrial ATP synthase. Mic10 variants that selectively function at the ATP synthase promote efficient metabolic adaptation, respiratory growth, and mitochondrial inner membrane energization.
Introduction
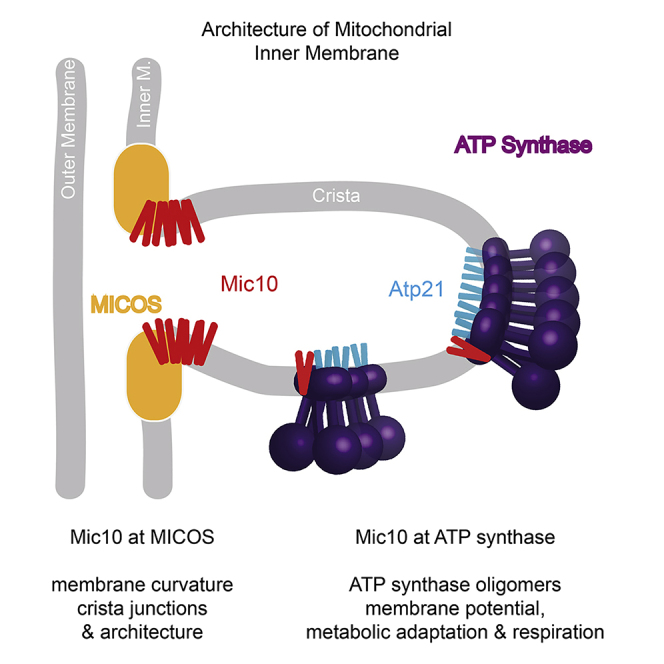
Mitochondria generate the vast majority of the cellular ATP via oxidative phosphorylation that takes place in specialized membrane domains, the cristae. These invaginations of the inner mitochondrial membrane enable optimal efficiency of the respiratory chain both by their topology and by increasing the membrane surface. The organization and shaping of cristae are complex processes involving several machineries, and disruption of the inner membrane architecture is detrimental for mitochondrial physiology and associated with a variety of pathological conditions (Cogliati et al., 2016; Colina-Tenorio et al., 2020; Eramo et al., 2020; Mukherjee et al., 2021). Two major cristae-shaping machineries were reported to function in an antagonistic manner with respect to cristae architecture, the mitochondrial contact site and cristae organizing system (MICOS), and the F1Fo-ATP synthase (Paumard et al., 2002; Rabl et al., 2009; Hoppins et al., 2011; Davies et al., 2012; Harner et al., 2016; Rampelt et al., 2017b; Kojima et al., 2019; Stephan et al., 2020).
The conserved MICOS complex is required for the structural stability of crista junctions, the narrow connections between crista folds and the smooth expanse of the inner membrane localized directly underneath the outer membrane, the inner boundary membrane (von der Malsburg et al., 2011; Harner et al., 2011; Hoppins et al., 2011; Herrmann, 2011; Alkhaja et al., 2012; Guarani et al., 2015; Muñoz-Gómez et al., 2015; Huynen et al., 2016; Kozjak-Pavlovic, 2017; Rampelt et al., 2017b; Wollweber et al., 2017; Kaurov et al., 2018). MICOS consists of six main subunits that are organized into two subcomplexes named after the core subunits Mic60 and Mic10 (Barbot et al., 2015; Bohnert et al., 2015; Friedman et al., 2015). The MIC60 subcomplex contains Mic60 and Mic19 and in Metazoa additionally the Mic19 paralog Mic25. The MIC10 subcomplex consists of Mic10, Mic12, Mic26, and Mic27. Loss of individual MICOS subunits leads to alterations of the inner membrane architecture with a reduced number of crista junctions and the accumulation of internal cristae stacks (John et al., 2005; Rabl et al., 2009; Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011). Both Mic10 and Mic60 are involved in inducing the high membrane curvature at crista junctions, Mic60 via an amphiphilic helix (Hessenberger et al., 2017; Tarasenko et al., 2017) and Mic10 via its wedge-like shape and oligomerization (Barbot et al., 2015; Bohnert et al., 2015). The MIC60 subcomplex additionally interacts with protein complexes of the outer membrane and forms contact sites between the inner and outer membranes that contribute to the efficiency of protein and phospholipid biogenesis pathways and help to position crista junctions in the vicinity of the outer membrane (Xie et al., 2007; Darshi et al., 2011; Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Alkhaja et al., 2012; Bohnert et al., 2012; Körner et al., 2012; Ott et al., 2012; Zerbes et al., 2012; Ding et al., 2015; Friedman et al., 2015; Muñoz-Gómez et al., 2015; Sakowska et al., 2015; Aaltonen et al., 2016; Huynen et al., 2016; Michaud et al., 2016; Stoldt et al., 2019; Tang et al., 2019).
The F1Fo-ATP synthase (complex V) also plays a crucial role in cristae architecture since it forms long rows of dimers that impose a positive membrane curvature (convex when viewed from the matrix) in contrast to the negative membrane curvature induced by MICOS (convex when viewed from the intermembrane space). Dimerization and oligomerization of the ATP synthase are required for the generation of the highly curved rims of cristae as well as for crista tubules (Arnold et al., 1998; Paumard et al., 2002; Arselin et al., 2004; Strauss et al., 2008; Thomas et al., 2008; Davies et al., 2012; Kühlbrandt, 2019). Dimerization relies on the small subunits Atp20 (subunit g) and Atp21 (subunit e/Tim11) that lock the ATP synthase monomers in an angle of up to 90°, resulting in membrane bending by the dimer (Arnold et al., 1998; Bornhövd et al., 2006; Hahn et al., 2016; Guo et al., 2017; Spikes et al., 2020). The higher order assemblies are further stabilized by Atp19 (subunit k; Arnold et al., 1998; Wagner et al., 2010; Guo et al., 2017; Barca et al., 2018; He et al., 2018; Pinke et al., 2020).
Several human pathologies have been described that are the consequence of defects in MICOS or the disturbed formation of higher order assemblies of the ATP synthase (Colina-Tenorio et al., 2020; Eramo et al., 2020; Mukherjee et al., 2021), highlighting the importance of both machineries. Based on their opposing effects on membrane curvature and their differential distribution to crista junctions versus crista rims and tubules, MICOS and the dimeric ATP synthase are thought to perform antagonistic, complementary functions that are crucial in maintaining cristae architecture (Rabl et al., 2009; Hoppins et al., 2011; Harner et al., 2016; Kojima et al., 2019; Stephan et al., 2020). Despite this MICOS-ATP synthase antagonism, a fraction of Mic10 molecules was found to associate with the F1Fo-ATP synthase (Rampelt et al., 2017a; Eydt et al., 2017; Rampelt and van der Laan, 2017; Stephan et al., 2020; Cadena et al., 2021). The Mic10-ATP synthase interaction did not depend on other MICOS subunits yet apparently stabilized higher order assemblies of the ATP synthase (Rampelt et al., 2017a; Eydt et al., 2017). It is unknown whether the interaction of Mic10 with the ATP synthase is of relevance for mitochondrial or cellular functions, particularly as Mic10 and ATP synthase induce opposing membrane curvature. Here, we established experimental conditions to selectively study a MICOS-independent role of Mic10. We report that Mic10ATPsynthase and Mic10MICOS strongly differ in their oligomeric organization and functions. In contrast to the presumed major role of Mic10MICOS, we found that Mic10ATPsynthase is required for efficient metabolic adaptation and respiratory growth.
Results
Mic10 interacts with dimeric ATP synthase in organello
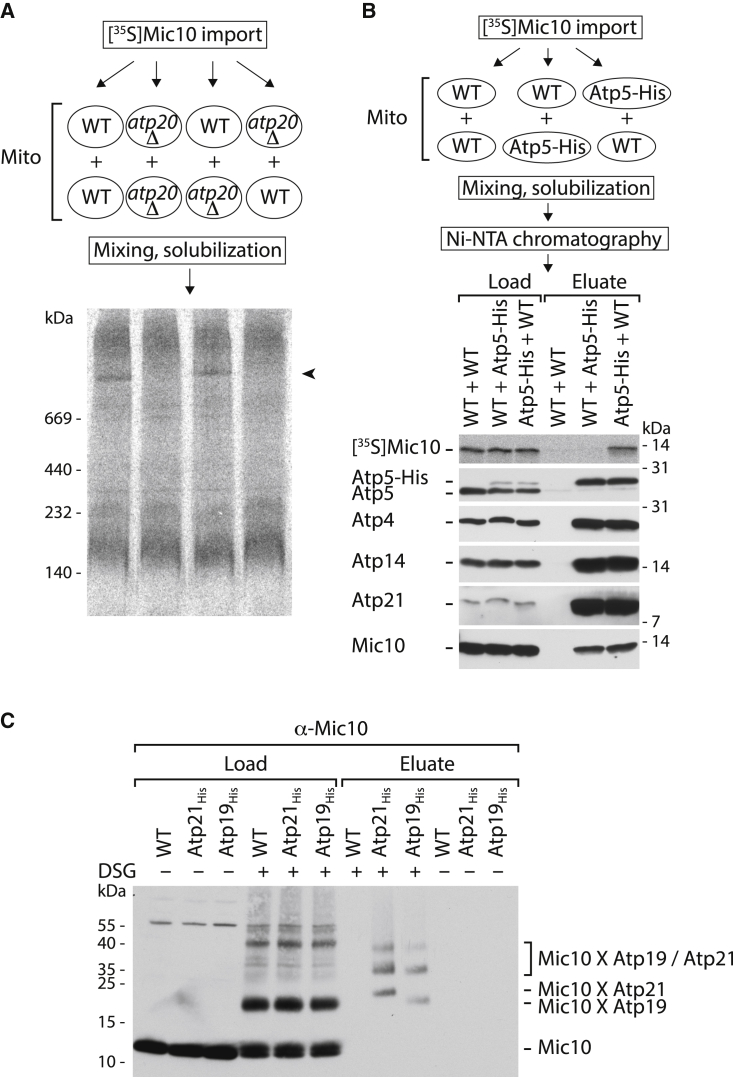
The association of Mic10 with the ATP synthase observed by comigration or copurification upon lysis of mitochondria with non-ionic detergent (Rampelt et al., 2017a; Eydt et al., 2017) may occur in the native mitochondrial system or after lysis of the mitochondrial membranes. We devised assays to distinguish between in organello and post-lysis interactions via blue native electrophoresis (BN-PAGE) and affinity purification. (1) 35S-labeled Mic10 precursor was imported into isolated wild-type (WT) mitochondria or mitochondria lacking the dimerization factor Atp20 (Su g); atp20Δ mitochondria are deficient in stable ATP synthase dimers (Figure S1A; Arnold et al., 1998; Paumard et al., 2002; Arselin et al., 2004; Wagner et al., 2009; Davies et al., 2012). The mitochondria were mixed with untreated atp20Δ or WT mitochondria, solubilized with digitonin, and analyzed by BN-PAGE (Figure 1A). [35S]Mic10 comigrated with the ATP synthase dimer only after import into WT mitochondria, not into atp20Δ mitochondria, excluding a post-lysis interaction of Mic10 with the dimeric ATP synthase in the BN-PAGE analysis. (2) To study copurification of Mic10 with the ATP synthase, we imported [35S]Mic10 into WT mitochondria or mitochondria containing His-tagged Atp5. Upon mixing with mitochondria not containing [35S]Mic10 and solubilization, the ATP synthase was purified by affinity chromatography (Figure 1B). Whereas endogenous Mic10 was copurified in all samples containing Atp5-His mitochondria, [35S]Mic10 was only copurified upon import into Atp5-His mitochondria, demonstrating that Mic10ATPsynthase was formed in organello and not in a post-lysis association. (3) In addition, Mic10 was crosslinked to the dimerization subunit Atp21 (Su e/Tim11) in organello (Rampelt et al., 2017a; Eydt et al., 2017; Figure S1B). This Mic10-Atp21 interaction was not altered by the addition of the divalent cations Mg2+ or Ca2+ (Figure S1C; Mallilankaraman et al., 2012; Petrungaro et al., 2015; Nemani et al., 2018, 2020; Daw et al., 2020). Since Atp21 assembles with monomeric ATP synthase to promote dimerization and oligomerization of the ATP synthase but is also observed as a free subunit (Arnold et al., 1998; Paumard et al., 2002; Arselin et al., 2004; Wagner et al., 2009; Habersetzer et al., 2013), it was unknown to which form the crosslinking occurred. We thus probed for crosslinking of Mic10 to Atp19 (Su k) that preferentially assembles with the dimeric ATP synthase and stabilizes ATP synthase dimers and oligomers (Arnold et al., 1998; Wagner et al., 2010; Guo et al., 2017; He et al., 2018). We treated intact mitochondria with the membrane-permeable crosslinking reagent disuccinimidyl glutarate (DSG) and observed specific crosslinking adducts of Mic10 to Atp19 as well as Atp21 (Figure 1C). Larger crosslinking products containing Mic10, Atp19, and Atp21 indicate the formation of ternary adducts (Figure 1C). Taken together, these results indicate that Mic10 is associated with the fully assembled F1Fo-ATP synthase in intact mitochondria.
Figure 1.
Mic10 associates with F1Fo-ATP synthase in mitochondria
(A) [35S]Mic10 was imported into WT or atp20Δ mitochondria, followed by solubilization together with untreated mitochondria and BN-PAGE. Arrowhead, ATP synthase dimer.
(B) [35S]Mic10 was imported into WT or Atp5-His mitochondria that were solubilized together with untreated mitochondria. [35S]Mic10 was co-isolated with Atp5-His only after import into Atp5-His mitochondria, analyzed by SDS-PAGE. Load, 2.5%; eluate, 100%. F1Fo-ATP synthase subunits: Atp4 (Su b); Atp5 (Su 5/OSCP); Atp14 (Su h); and Atp21 (Su e).
(C) In organello crosslinking followed by affinity purification and SDS-PAGE shows that Mic10 forms direct crosslinks with Atp21 and Atp19. Load, 2.5%; eluate, 100%.
See also Figure S1.
Atp21-Mic10 fusion promotes oligomerization of ATP synthase, but not of Mic10
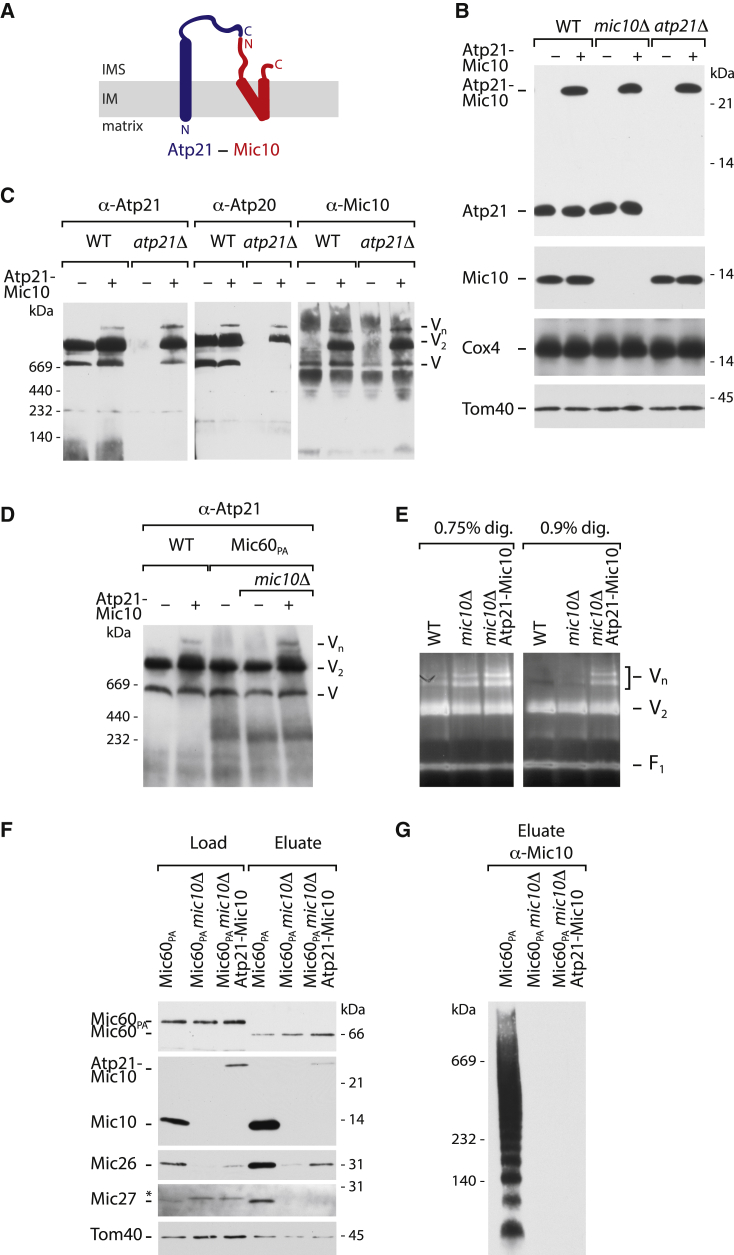
To study possible functions of Mic10ATPsynthase, we searched for conditions to accumulate Mic10 at the ATP synthase and simultaneously disrupt its functions in MICOS. We generated an Atp21-Mic10 fusion protein by connecting the C terminus of Atp21 with the N terminus of Mic10, both located in the intermembrane space (Figure 2A). Plasmid-borne Atp21-Mic10 was stably expressed in WT yeast as well as in strains lacking Atp21 or Mic10 (Figures 2B and S2A). Expression of the fusion protein in atp21Δ yeast restored dimerization of the ATP synthase (Figure 2C; the fusion protein also increased the levels of Atp20 that depend on the presence of Atp21; Arnold et al., 1998; Arselin et al., 2003; Wagner et al., 2009). ATP synthase oligomers in WT mitochondria are destabilized when the inner membrane is disrupted by detergent (Figures 2C and 2D); using very mild solubilization conditions and sensitive ATPase in gel activity staining, oligomers are visible on BN gels (Figure 2E; Arselin et al., 2003; Bornhövd et al., 2006; Strauss et al., 2008). Upon overexpression of Mic10, an increased stability of ATP synthase oligomers was observed (Rampelt et al., 2017a). Importantly, the Atp21-Mic10 fusion protein stabilized the oligomeric forms of the ATP synthase in WT as well as in atp21Δ and mic10Δ mitochondria (Figures 2C–2E). These results indicate that the fusion protein was able to substitute for the functions of Atp21 as well as Mic10 at the ATP synthase.
Figure 2.
Atp21-Mic10 fusion protein functions selectively at the ATP synthase
(A) Topology of Atp21, Mic10, and the fusion protein Atp21-Mic10 in the inner membrane (IM). IMS, intermembrane space.
(B) Expression of Atp21-Mic10 in WT, mic10Δ, and atp21Δ cells, analyzed by SDS-PAGE. Tom40, component of the translocase of the outer membrane (TOM); Cox4, subunit of respiratory chain complex IV.
(C) Native ATP synthase and Mic10-containing complexes in mitochondria isolated from WT or atp21Δ cells with or without Atp21-Mic10 expression were analyzed by BN-PAGE. V, ATP synthase (complex V) monomer; V2, ATP synthase dimer; Vn, oligomeric species of ATP synthase.
(D) The Atp21-Mic10 fusion protein stabilizes ATP synthase oligomers independently of endogenous WT Mic10. Mic60PA, protein-A-tagged Mic60.
(E) Isolated mitochondria were solubilized under very mild conditions in the presence of low digitonin (dig.) concentrations that do not disrupt oligomeric ATP synthase and analyzed by BN-PAGE and in gel ATPase activity staining. F1, F1 particle of ATP synthase.
(F and G) MICOS isolation from mitochondria via protein-A-tagged Mic60 (Mic60PA), analyzed by SDS-PAGE (F) and BN-PAGE (G). Load, 4%; eluate, 100%. Asterisk, non-specific band.
See also Figure S2.
In contrast, the Atp21-Mic10 fusion protein was only poorly copurified with MICOS and did not restore the levels of Mic27 and Mic26 in the absence of Mic10 (Figure 2F; the levels of Mic26 and particularly Mic27 depend on the presence of Mic10; Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Bohnert et al., 2015). Mic10MICOS forms characteristic oligomers, illustrated by formation of a ladder on BN gels upon copurification with Mic60 (Figure 2G; Bohnert et al., 2015; Barbot et al., 2015; Zerbes et al., 2016; Rampelt et al., 2018). Expression of Atp21-Mic10 in mic10Δ yeast, however, did not yield any oligomeric fusion protein copurified with MICOS (Figure 2G). Since oligomerization of Mic10MICOS is required to support crista junction formation (Barbot et al., 2015; Bohnert et al., 2015), these results indicate that Atp21-Mic10 does not support MICOS function.
Taken together, the Atp21-Mic10 fusion protein is functional at the ATP synthase by promoting dimerization and oligomerization of the ATP synthase but neither significantly interacts with nor forms oligomers at MICOS. The biochemical assays thus indicate that the fusion protein recruits Mic10 to the ATP synthase, providing a means to selectively study the role of Mic10 at the ATP synthase.
MICOS-independent role of Mic10 in mitochondrial physiology
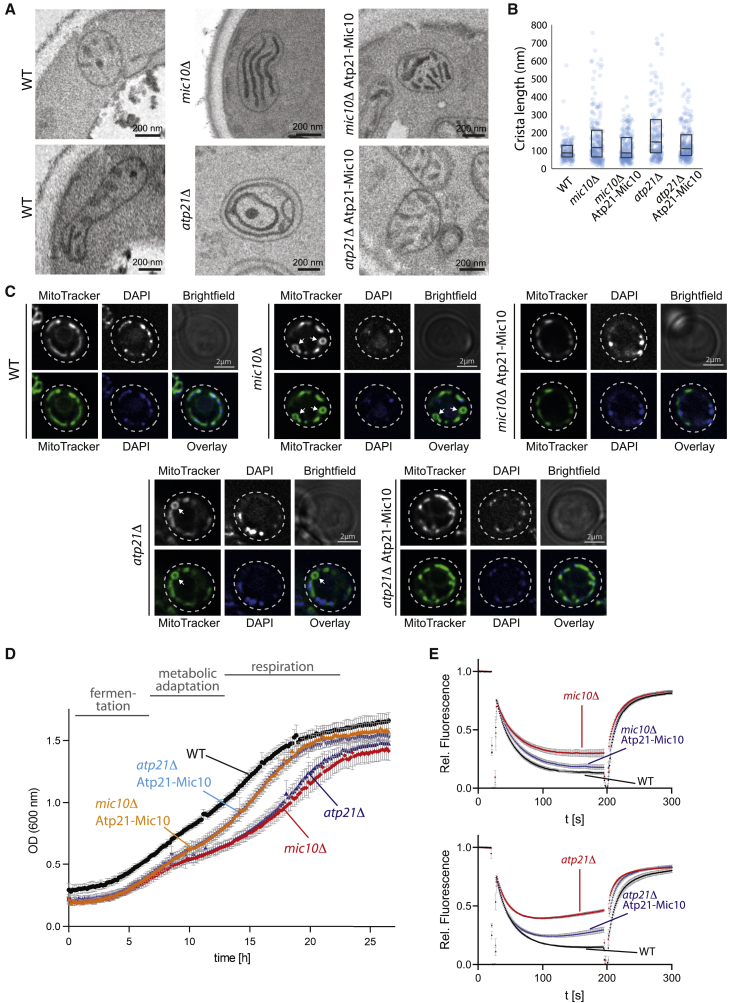
To address possible physiological roles of Mic10ATPsynthase, we analyzed how Atp21-Mic10 affected cristae architecture, mitochondrial morphology, respiratory growth, and energetics. (1) The inner membrane architecture was analyzed by electron microscopy (EM). Expression of Atp21-Mic10 reversed the atp21Δ phenotype of onion-like, septated cristae membranes to a WT-like cristae architecture (Figure 3A, lower panel; Paumard et al., 2002; Arselin et al., 2004; Rabl et al., 2009; Davies et al., 2012), but not the MICOS-deficiency phenotype of mic10Δ yeast. Mitochondria in mic10Δ Atp21-Mic10 cells were characterized by parallel cristae membranes that overwhelmingly lacked crista junctions, similar to those in mic10Δ cells (Figure 3A, upper panel; Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Alkhaja et al., 2012), fully supporting the biochemical findings that Atp21-Mic10 is functional at the ATP synthase, but not at MICOS. (2) The length of inner membranes and cristae is increased in atp21Δ mitochondria as well as mic10Δ mitochondria (Paumard et al., 2002; Arselin et al., 2004; Rabl et al., 2009; von der Malsburg et al., 2011; Harner et al., 2011; Hoppins et al., 2011; Alkhaja et al., 2012; Figure 3B). Upon expression of Atp21-Mic10, the cristae were shifted to a more WT-like average length, also in mic10Δ mitochondria (Figure 3B), suggesting that Mic10ATPsynthase is involved in the regulation of crista length. (3) Deletion mutants of MICOS core components display a fragmented mitochondrial network with enlarged mitochondria; similarly, deletion of Atp21 or Atp20 results in an aberrant mitochondrial network (Everard-Gigot et al., 2005; Rabl et al., 2009; Hoppins et al., 2011; von der Malsburg et al., 2011; Alkhaja et al., 2012; Itoh et al., 2013; Friedman et al., 2015). The aberrant mitochondrial network included spherical structures in mic10Δ yeast and atp21Δ yeast (Figure 3C, arrows). Similar empty regions within mitochondria have been described for MICOS mutants as well as aged mitochondria that display aberrant cristae architecture (Rabl et al., 2009; Itoh et al., 2013; Friedman et al., 2015; Brandt et al., 2017). Expression of Atp21-Mic10 restored the tubular mitochondrial network not only in atp21Δ cells but also in mic10Δ cells (Figure 3C), indicating that the interaction of Mic10 with the ATP synthase is important for formation of the normal mitochondrial network in yeast cells. The EM and live-cell imaging data thus reveal a differentiation of Mic10 functions in mitochondrial membrane architecture and morphology. Formation of the characteristic inner membrane cristae architecture and crista junctions requires the established function of oligomeric Mic10 in MICOS, whereas Mic10 at the ATP synthase is required for maintaining the mitochondrial network organization in cells.
Figure 3.
Mic10ATPsynthase restores physiological functions in mic10Δ cells
(A) EM analysis of yeast cells following diaminobenzidine staining.
(B) Quantitation of crista length in the indicated strains. Interquartile boxplot: first quartile; median; and third quartile are shown. N = 200.
(C) Live-cell fluorescence imaging of the mitochondrial network using MitoTracker dye in WT, mic10Δ, or atp21Δ cells with or without Atp21-Mic10 expression. DAPI, stain for mitochondrial nucleoids.
(D) Growth of WT cells as well as mic10Δ or atp21Δ cells with or without expression of Atp21-Mic10 in selective media with 3% glycerol + 0.1% glucose. Data are represented as mean SD; n = 4. OD, optical density.
(E) The mitochondrial membrane potential was assessed in isolated mitochondria by quenching of a fluorophore whose uptake depends on the membrane potential. Data are represented as mean SEM; n = 8 (upper panel); n = 14 (lower panel).
See also Figure S2.
(4) Mutant strains lacking MICOS core components were reported to display respiratory growth defects in drop tests (Rabl et al., 2009; von der Malsburg et al., 2011; Harner et al., 2011; Hoppins et al., 2011; Alkhaja et al., 2012). Mitochondrial biogenesis and efficient remodeling are particularly important when cells have to switch from fermentation in glucose-rich conditions to respiration upon glucose depletion. Therefore, we compared the growth behavior during fermentation, metabolic adaptation, and respiration by performing growth assays in liquid media that require the cells to adapt from fermentative to respiratory growth. In a non-fermentable carbon source (3% glycerol) supplemented with a small amount of a fermentable carbon source (0.1% glucose), yeast grow fermentatively until the glucose is depleted. In the subsequent growth delay, metabolic remodeling promotes mitochondrial biogenesis and respiration (diauxic shift; Figures 3D and S2B; Di Bartolomeo et al., 2020; Murphy et al., 2015; de Alteriis et al., 2018). When the full respiratory activity is reached, the cells grow faster again. Lack of the MICOS core components Mic10 or Mic60 resulted in defects both in metabolic adaptation and respiratory growth, whereas individual deletions of the other MICOS components did not impair growth (Figure S2B). atp21Δ cells similarly displayed a growth defect during diauxic shift (Figure 3D; Arnold et al., 1998; Paumard et al., 2002; Arselin et al., 2004; Rabl et al., 2009). Unexpectedly, expression of the Atp21-Mic10 fusion protein largely restored the growth of both deletion strains, mic10Δ and atp21Δ (Figure 3D), demonstrating that Mic10ATPsynthase is required for metabolic adaptation and respiratory growth. (5) The inner membrane potential Δψ was assessed by using a Δψ-sensitive fluorophore. Mitochondria lacking Mic10 or Atp21 displayed a decreased Δψ (Figure 3E; Bornhövd et al., 2006; Rabl et al., 2009; von der Malsburg et al., 2011; Wolf et al., 2019). Expression of Atp21-Mic10 largely restored Δψ in mic10Δ and atp21Δ mitochondria (Figure 3E), underscoring the importance of ATP synthase-linked Mic10.
Mic10 thus plays a dual role. Oligomeric Mic10 at MICOS is required for maintaining the native cristae architecture and crista junctions as established previously (Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Alkhaja et al., 2012; Barbot et al., 2015; Bohnert et al., 2015). The interaction of Mic10 with the ATP synthase is important for efficient energization of the inner membrane, metabolic adaptation, respiratory growth, and organization of the mitochondrial network.
Oligomerization-deficient Mic10 supports mitochondrial physiology
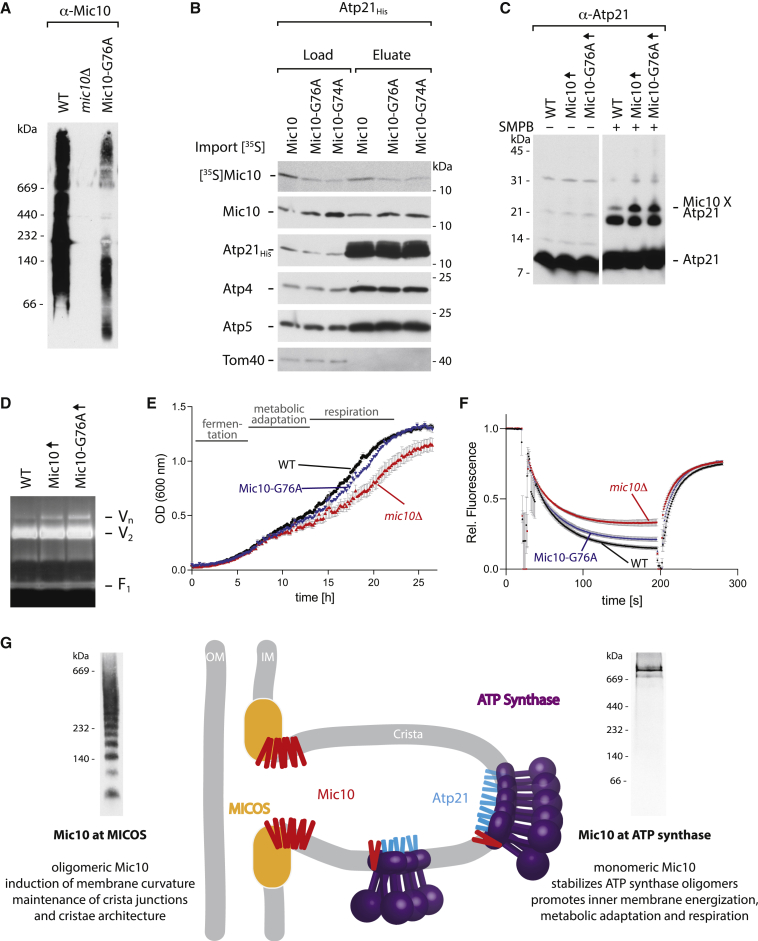
To independently study different functions of Mic10 at the ATP synthase and MICOS, we looked for point mutants that functionally separated Mic10ATPsynthase and Mic10MICOS. Single amino acid substitutions in the conserved glycine motifs of Mic10 do not disturb its association with MICOS but severely impair the oligomerization of Mic10 (Barbot et al., 2015; Bohnert et al., 2015; Figures 4A and S3A). The Mic10 glycine mutants thus cannot support Mic10 function at MICOS, resulting in an altered cristae architecture characteristic for micos deletion mutants (Barbot et al., 2015; Bohnert et al., 2015). To test whether Mic10 glycine mutants interacted with the ATP synthase, we imported 35S-labeled Mic10-G76A and Mic10-G74A into Atp21-His mitochondria, lysed the mitochondria, and performed an affinity purification. The Mic10 point mutants were copurified with Atp21-His with an efficiency comparable to that of WT Mic10 (Figure 4B; taking into account the moderately decreased levels of imported Mic10 mutant forms).
Figure 4.
Oligomerization-deficient Mic10 restores physiological functions in mic10Δ cells
(A) Mic10-containing complexes of mitochondria from WT and mic10Δ cells as well as cells expressing the variant Mic10-G76A, analyzed by BN-PAGE.
(B) Radiolabeled Mic10 or the oligomerization-deficient mutant variants Mic10-G76A or Mic10-G74A were imported into Atp21-His mitochondria, followed by lysis with digitonin and isolation of Atp21-His and interacting proteins. The co-isolation efficiency relative to the levels in the load fraction was not impaired for the Mic10 mutants. Load, 2.5%; eluate, 100%.
(C) In organello crosslinking shows that the band reflecting a direct crosslink between Atp21 and Mic10 is similarly enhanced upon overexpression of Mic10 or Mic10-G76A. SMPB, succinimidyl 4-(p-maleimido-phenyl)butyrate.
(D) Analysis of oligomeric ATP synthase by BN-PAGE and in gel ATPase activity staining using very mild solubilization (0.9% digitonin).
(E) Growth of mic10Δ cells without or with expression of Mic10 (WT) or Mic10-G76A in selective media with 3% glycerol + 0.1% glucose. Data are represented as mean SEM; n = 4.
(F) The mitochondrial membrane potential was assessed in mitochondria isolated from the indicated strains. Data are represented as mean SEM; n = 5.
(G) Dual function of Mic10 at MICOS and the ATP synthase. The majority of Mic10 supports MICOS function and crista junction stability by membrane bending, which relies on Mic10 oligomerization. A distinct Mic10 population interacts with the ATP synthase at substoichiometric amounts and regulates its higher order assemblies. This regulatory role does not require Mic10 oligomerization and is crucial for the maintenance of a healthy membrane potential and mitochondrial network and consequently for efficient metabolic adaptation and respiration. BN-PAGE panels: western blot of Mic10-containing complexes in a Mic60-ProtA eluate (left panel; shown is a shorter exposure of lane 1 of Figure 2G) and autoradiograph of imported radiolabeled Mic10 in an Atp21-His eluate (right panel) are shown.
See also Figures S1 and S3.
We overexpressed Mic10-G76A (Figure S3B) and compared its behavior with that of overexpressed WT Mic10. Whereas the oligomerization of Mic10-G76A was strongly impaired (Figure S3C), the interaction with the ATP synthase was indistinguishable from that of WT Mic10. Upon overexpression, the association of Mic10-G76A with the ATP synthase was enhanced and ATP synthase oligomers were stabilized, as observed for the overexpression of WT Mic10 (Figures 4C, 4D, and S1B; Rampelt et al., 2017a). The biochemical results thus show that Mic10-G76A, which is unable to form Mic10 oligomers, readily interacts with the ATP synthase and promotes its oligomerization.
We compared the growth of mic10Δ yeast upon expression of WT Mic10 or Mic10-G76A. Mic10-G76A efficiently restored metabolic adaptation and respiratory growth to nearly the same efficiency as WT Mic10 (Figure 4E). Similarly, the inner membrane potential was largely restored by the expression of Mic10-G76A (Figure 4F).
Taken together, two independent approaches, expression of an Atp21-Mic10 fusion protein and of an oligomerization-defective Mic10 point mutant, demonstrate that Mic10 not only performs its established role in maintaining crista junctions and inner membrane architecture but plays an important role as direct interaction partner of the ATP synthase. Mic10ATPsynthase stabilizes higher order assemblies of the ATP synthase and is crucial for efficient metabolic adaptation, respiratory growth, and energization of the inner membrane.
Discussion
This study unexpectedly reveals that Mic10's role in metabolic adaptation and respiratory growth mainly depends on its interaction with the presumed antagonistic machinery, the F1Fo-ATP synthase (Rampelt et al., 2017a; Eydt et al., 2017; Stephan et al., 2020; Cadena et al., 2021), and not on its function in the MICOS complex. The role of oligomeric Mic10 in promoting inner membrane bending and stabilizing crista junctions as a core component of MICOS has been well established (Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Alkhaja et al., 2012; Barbot et al., 2015; Bohnert et al., 2015; Muñoz-Gómez et al., 2015; Huynen et al., 2016; Kaurov et al., 2018), and it has been assumed that the resulting aberrant cristae architecture caused the physiological defects of Mic10-deficient cells. We report here that Mic10 fulfills a second role in mitochondria via its interaction with the dimeric ATP synthase, and this interaction is important for mitochondrial energetics and physiology. We were able to selectively study the function of Mic10ATPsynthase by two strategies, by recruiting Mic10 to the dimeric ATP synthase as part of an Atp21-Mic10 fusion protein and by using an oligomerization-deficient Mic10 variant that cannot rescue crista junction stability but interacts with and stabilizes ATP synthase oligomers. Mic10 thus functions in two different populations: as core part of MICOS (Mic10MICOS) and as partner of the ATP synthase (Mic10ATPsynthase; Figure 4G). Mic10MICOS is required for the induction of membrane curvature and the maintenance of crista junctions and consequently for normal cristae architecture. Mic10ATPsynthase favors the formation of higher order assemblies of the ATP synthase and thus plays a crucial role in mitochondrial physiology by promoting an efficient inner membrane energization and adaptation to respiratory growth.
How can Mic10, whose oligomers at crista junctions promote membrane bending in the opposite direction compared with ATP synthase oligomers, enhance the stability of the latter? Mic10 spans the inner membrane in a hairpin topology with an unusually long N-terminal transmembrane segment (∼24 amino acid residues), indicating an asymmetric wedge-like shape (Barbot et al., 2015). This mismatch in the length of the transmembrane helices appears to be a conserved feature of Mic10 (AlphaFold DB; Jumper et al., 2021). Upon oligomerization, Mic10MICOS induces negative membrane curvature in vitro and in organello (Barbot et al., 2015). Monomeric Mic10 is unable to deform membranes by itself, yet its wedge-like shape displays a preference for negatively curved membrane regions. The absolute copy numbers of Mic10 and ATP synthase subunits in yeast (Morgenstern et al., 2017) reveal that Mic10 is present in substoichiometric amounts, approximately 6–8 times less abundant than ATP synthase subunits, such as Atp21 and Atp20 (Figure S3D). Up to 20% of total mitochondrial Mic10 comigrate with the dimeric ATP synthase (Eydt et al., 2017), corresponding to one Mic10ATPsynthase molecule per ∼15–20 ATP synthase dimers. Electron cryotomography studies demonstrate the formation of long rows of 15–20 and more ATP synthase dimers at crista rims and crista tubules (Strauss et al., 2008; Thomas et al., 2008; Davies et al., 2011, 2012; Blum et al., 2019; Kühlbrandt, 2019). Thus, on average, one (or two) Mic10 molecule is associated with these higher order assemblies of the ATP synthase. The transition zone between the pronounced positive membrane curvature of ATP synthase oligomers and flatter membrane areas devoid of ATP synthase includes negative curvature and can lead to membrane tension (Davies et al., 2012; Anselmi et al., 2018). Since Mic10 preferentially localizes to areas of negative membrane curvature, the association of Mic10 with ATP synthase oligomers may alleviate the membrane tension and thus contribute to the stabilization of ATP synthase oligomers in the mitochondrial inner membrane.
In summary, Mic10 functions in two major machineries that control mitochondrial membrane organization, MICOS and the oligomeric ATP synthase, and thus is required for both cristae architecture and an efficient adaptation and growth under respiratory conditions.
Limitations of the study
We cannot exclude that the Atp21-Mic10 fusion protein results in a non-native arrangement of Mic10 at the dimeric ATP synthase, e.g., due to steric effects or an altered stoichiometry. However, since the fusion protein mimics the stabilizing function of Mic10 overexpression on ATP synthase oligomers and promotes metabolic adaptation and respiratory growth, our assays do not indicate disturbing effects on the ATP synthase. An established Mic10 point mutant that selectively functions at the ATP synthase shows a comparable behavior. Technical limitations prevent us from making conclusions on a direct interaction of Mic10 with Atp20. Since Atp21 and Atp20 form close contacts in the dimer (Hahn et al., 2016; Guo et al., 2017; Spikes et al., 2020), the interaction of Mic10 with Atp21 likely brings it in close vicinity of Atp20. However, we did not observe crosslinking of Mic10 to Atp20 with the amino- and sulfhydryl-reactive reagents used, likely because the hydrophilic portion of Atp20 is mainly exposed to the matrix, whereas Mic10 exposes only a short loop of three amino acid residues to the matrix (Bohnert et al., 2015).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal α-Mic10 | Pfanner/Rampelt Labs | GR3343-2, GR3367-7 |
| Rabbit polyclonal α-Mic12 | Pfanner/Rampelt Labs | GR3336-3 |
| Rabbit polyclonal α-Mic26 | Pfanner/Rampelt Labs | GR3335-3 |
| Rabbit polyclonal α-Mic27 (affinity purified) | Pfanner/Rampelt Labs | GR3356-7 |
| Rabbit polyclonal α-Mic60 (affinity purified) | Pfanner/Rampelt Labs | GR857-8 |
| Rabbit polyclonal α-Atp1 | Pfanner/Rampelt Labs | GR5075-4 |
| Rabbit polyclonal α-Atp2 | Pfanner/Rampelt Labs | GR861-3 |
| Rabbit polyclonal α-Atp4 | Pfanner/Rampelt Labs | GR1970-4 |
| Rabbit polyclonal α-Atp5 | Pfanner/Rampelt Labs | GR1546-4 |
| Rabbit polyclonal α-Atp14 | Pfanner/Rampelt Labs | GR1964-5 |
| Rabbit polyclonal α-Atp20 | Pfanner/Rampelt Labs | GR1516-4 |
| Rabbit polyclonal α-Atp21 | Pfanner/Rampelt Labs | 138-9 |
| Rabbit polyclonal α-Cor1 | Pfanner/Rampelt Labs | GR371-6 |
| Rabbit polyclonal α-Cox4 | Pfanner/Rampelt Labs | GR577-4 |
| Rabbit polyclonal α-Tom40 | Pfanner/Rampelt Labs | 168-12 |
| anti-rabbit IgG (whole molecule)-Peroxidase | Sigma-Aldrich | A6154; RRID:AB_258284 |
| Chemicals, peptides, and recombinant proteins | ||
| DSG (disuccinimidyl glutarate) | Thermo Scientific | 20593 |
| SMPB (succinimidyl 4-(p-maleimido-phenyl)butyrate) | Thermo Scientific | 22416 |
| Digitonin | Matrix Biosciences | 60105 |
| [35S]Methionine | PerkinElmer | NEG009T005MC |
| Ni-NTA Agarose | Qiagen | 30230 |
| CNBr-activated Sepharose 4B | GE Healthcare | 17-0430-01 |
| DiSC3(5) (3,3'-Dipropylthiadicarbocyanine Iodide) | Invitrogen | D306 |
| MitoTracker™ Green FM | Invitrogen | M7514 |
| DAPI (4’,6 – Diamidino-2-phenylindole dihydrochloride) | Sigma-Aldrich | D9542 |
| Glutaraldehyde | Sigma | G7651 |
| KMnO4 (potassium permanganate) | Baker Chemicals | 0237 |
| (DAB) diaminobenzidine | Sigma | D-5637 |
| Epon: | ||
| Glycid ether (51.5% w/v) | Serva | 151414 |
| Methylnadic anhydride (47.3% w/v) | Serva | 140573 |
| 2,4,6-Tris (dimethylaminomethyl)phenol (1.2% w/v) | Santa Cruz | F0112 |
| Critical commercial assays | ||
| TNT Quick Coupled Reaction Mix | Promega | L2080 |
| KOD Hot Start Master Mix | Novagen | 71842-3 |
| Experimental models: Organisms/strains | ||
| YPH499 (WT; MATa ura3-52 lys2-801_amber ade2-101_ochre trp1-Δ63 his3-Δ200 leu2-Δ1) | Sikorski and Hieter (1989) | 1501 |
| YPH499 mic10::kanMX4 | Bohnert et al. (2012) | 3047 |
| YPH499 mic60::kanMX4 | Bohnert et al. (2012) | 3092 |
| YPH499 mic12::kanMX4 | Bohnert et al. (2012) | 3035 |
| YPH499 mic19::kanMX4 | Bohnert et al. (2012) | 3037 |
| YPH499 mic26::kanMX4 | Bohnert et al. (2012) | 3045 |
| YPH499 mic27::kanMX4 | Bohnert et al. (2012) | 3039 |
| YPH499 atp19::HIS3MX6 | Wagner et al. (2010) | 2703 |
| YPH499 atp20::kanMX6 | Wagner et al. (2009) | 2702 |
| YPH499 atp21::HIS3MX6 | Wagner et al. (2009) | 2016 |
| YPH499 atp5::ATP5His-HIS3MX6 | this paper | 4941 |
| YPH499 atp19::ATP19His-HIS3MX6 | this paper | 5615 |
| YPH499 atp21::ATP21His-HIS3MX6 | this paper | 5614 |
| YPH499 mic10::kanMX4 pRS425 | this paper | 5616 |
| YPH499 mic10::kanMX4 pRS425-MIC10 | this paper | 3892 |
| YPH499 mic10::kanMX4 pRS425-MIC10-G76A | this paper | 4320 |
| YPH499 pRS426 | this paper | 4557 |
| YPH499 pRS426-MIC10 | this paper | 4558 |
| YPH499 pRS426-MIC10-G76A | this paper | 5617 |
| YPH499 pFL36 | this paper | 5281 |
| YPH499 pFL36-ATP21-MIC10 | this paper | 5618 |
| YPH499 mic10::kanMX4 pFL36 | this paper | 5282 |
| YPH499 mic10::kanMX4 pFL36-ATP21-MIC10 | this paper | 5283 |
| YPH499 atp21::HIS3MX6 pFL36 | this paper | 5284 |
| YPH499 atp21::HIS3MX6 pFL36-ATP21-MIC10 | this paper | 5285 |
| YPH499 mic60::MIC60ProtA-HIS3MX6 pFL36 | this paper | 5619 |
| YPH499 mic60::MIC60ProtA-HIS3MX6 mic10::kanMX4 pFL36 | this paper | 5620 |
| YPH499 mic60::MIC60ProtA-HIS3MX6 mic10::kanMX4 pFL36-ATP21-MIC10 | this paper | 5621 |
| Oligonucleotides | ||
| See Table S1 | ||
| Recombinant DNA | ||
| pGEM4Z-MIC10 | Bohnert et al. (2015) | 2195 |
| pGEM4Z-MIC10-G76A | Pfanner Lab | 2199 |
| pRS425 | Christianson et al. (1992) | X30 |
| pRS425-MIC10 | Bohnert et al. (2015) | 2093 |
| pRS425-MIC10-G74A | Bohnert et al. (2015) | 2095 |
| pRS425-MIC10-G76A | Bohnert et al. (2015) | 2125 |
| pRS426-MIC10 | Bohnert et al. (2015) | 2107 |
| pRS426-MIC10-G76A | this paper | 3183 |
| pFL36 | Bonneaud et al. (1991) | X13 |
| pFL36-ATP21-MIC10 | this paper | 3184 |
| pFL36-MIC10 | Bohnert et al. (2015) | 2237 |
| Software and algorithms | ||
| Adobe Photoshop Elements | Adobe Systems | n/a |
| Adobe Illustrator | Adobe Inc. | n/a |
| Fiji | Schindelin et al. (2012) | n/a |
| Graphpad | Graphpad Software Inc. | n/a |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Heike Rampelt (heike.rampelt@biochemie.uni-freiburg.de).
Materials availability
Plasmids and strains generated in this study may be requested from the lead contact.
Experimental model and subject details
The yeast strains used in this study are derivates of the Saccharomyces cerevisiae wild-type (WT) strain YPH499 (MATa ura3-52 lys2-801_amber ade2-101_ochre trp1-Δ63 his3- Δ200 leu2- Δ1) and are listed in the key resources table. They were cultivated as detailed in the STAR Methods section.
Method details
Yeast growth and mitochondrial isolation
For mitochondrial isolations, growth tests and whole cell extracts, Saccharomyces cerevisiae strains (key resources table) were cultivated at 30°C in YPG media (1% [w/v] yeast extract, 2% [w/v] peptone, 3% [v/v] glycerol) or, in the case of plasmid-containing strains, in selective defined media (0.67% [w/v] yeast nitrogen base, 0.07% [w/v] CSM amino acid mix) with 3% [v/v] glycerol and 0.1% [w/v] glucose. Growth tests were performed at 30°C in a CLARIOstar plate reader (BMG Labtech) using the following parameters: 160 cycles of 10 min each, OD 600 nm measurement by spiral scan averaging, and a combination of linear and double-orbital shaking at 500-600 rpm between measurements. Alternatively, growth tests were performed in a Tecan Spark 10M plate reader with the following parameters: Measurements every 5 min, 10 sec linear shaking at 630 rpm before each measurement, double-orbital shaking at 180 rpm between measurements.
For fluorescence microscopy, strains were grown in selective defined media supplemented with 10 mg/l adenine and with the carbon sources 2% [v/v] lactate and 0.1% [w/v] glucose. For electron microscopy, precultures were grown in synthetic complex medium (van Dijken et al., 1976) with 2% [v/v] lactate and 0.1% [w/v] glucose; no glucose was added to main cultures (Bohnert et al., 2015). To isolate crude mitochondria (Meisinger et al., 2006), cells were pre-treated with DTT buffer (100 mM Tris-H2SO4 pH 9.4, 10 mM DTT) and the cell wall was digested with zymolyase in 20 mM potassium phosphate buffer pH 7.4 with 1.2 M sorbitol. Cells were lysed with a glass Teflon homogenizer in homogenization buffer (10 mM Tris-HCl pH 7.4, 0.6 M sorbitol, 1 mM EDTA, 0.2% bovine serum albumin, 1 mM phenylmethylsulfonyl fluoride (PMSF)), and lysates were cleared 2x by centrifugation at 2000 g. Crude mitochondria were isolated by centrifugation at 17000 g and stored at −80°C in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS-KOH pH 7.2).
Generation of strains and plasmids
Strains with a chromosomal C-terminal His10-tag (Atp5-His, Atp19-His, Atp21-His) were generated by amplification of the tag with a HIS3MX6 cassette from plasmid pFA6a-His10-HIS3MX6 (Meisinger et al., 2001) using the primer sets listed in Table S1. The plasmid pFL36-Atp21-Mic10 (including the MET25 promotor and CYC1 terminator) was generated by AQUA cloning (Beyer et al., 2015) from two PCR products: The plasmid pFL36-Met25P-Mic10-CYC1T (Bohnert et al., 2015) was amplified using primers Mic10-1-F and Met25P-R, and the Atp21 ORF was amplified using primers Met25P-Atp21-F and Fusion-Tim-Mic-R. The plasmid pRS426-Mic10-G76A was generated by Quikchange (Stratagene) PCR from pRS426-Mic10.
Biochemical analysis of mitochondrial proteins
For whole-cell extracts, equal amounts of cells were harvested, washed with water, and treated with 0.1 M NaOH for 5 min at room temperature. Cells were pelleted, resuspended in SDS sample buffer and incubated at 95°C for 5 min prior to SDS-PAGE analysis. For blue native electrophoresis (BN-PAGE) analysis (Stojanovski et al., 2007), mitochondria were solubilized in solubilization buffer (20 mM Tris-HCl pH 7.4, 50 mM NaCl, 10% [v/v] glycerol, 0.1 mM EDTA, 2 mM PMSF, 1% [w/v] digitonin), or in low ionic strength buffer (Wittig et al., 2006) (50 mM imidazole-HCl pH 7.0, 500 mM 6-aminohexanoic acid, 1 mM EDTA, 1 mM PMSF) with 3% [w/v] digitonin at a digitonin to protein ratio of 4:1. For analysis of ATP synthase oligomers, lower digitonin concentrations in low ionic strength buffer were employed as indicated (0.75% digitonin = digitonin/protein ratio 1:1; 0.9% digitonin = digitonin/protein ratio 1.2:1). In gel staining for ATPase activity (Bornhövd et al., 2006) was performed by washing the native gel in water, incubation in ATP buffer (50 mM glycine, 5 mM MgCl2, 20 mM ATP, pH 8.4) and subsequent development with a 10% [w/v] CaCl2 solution. For the pulldown of His-tagged proteins or protein complexes, mitochondria were solubilized for 60–90 min at 4°C in pulldown solubilization buffer (20 mM Tris-HCl pH 7.4, 50 mM NaCl, 10% [v/v] glycerol, 0.1 mM EDTA, 2 mM PMSF, EDTA-free protease inhibitor mix (Roche), 1% [w/v] digitonin) plus 10 mM imidazole pH 7.4. The cleared supernatants were incubated with Ni-NTA agarose (Qiagen) for 90 min at 4°C. After washing the resin 10x with wash buffer (20 mM Tris-HCl pH 7.4, 60 mM NaCl, 10% [v/v] glycerol, 0.1 mM EDTA, 2 mM PMSF, 0.3% [w/v] digitonin) plus 40 mM imidazole pH 7.4, bound proteins were eluted with wash buffer with 250 mM imidazole pH 7.4 and analyzed by SDS-PAGE or BN-PAGE. For the pulldown of Protein A-tagged protein complexes, mitochondria were solubilized in pulldown solubilization buffer for 30–60 min, incubated with IgG sepharose for 1.5-3 h, and the resin was washed 10x with wash buffer. Isolated protein complexes were eluted by treatment with TEV protease (Fisher Scientific) for 2.5 h at 24°C. The His-tagged protease was removed from the eluates by a 30 min incubation with Ni-NTA agarose at 4°C. Samples were further analyzed by SDS-PAGE or BN-PAGE. In organello crosslinking with disuccinimidyl glutarate (DSG) or succinimidyl 4-(p-maleimido-phenyl)butyrate (SMPB) was performed by incubating isolated mitochondria in SEM buffer on ice for 30 min using final crosslinker concentrations of 1.5 mM, while control reactions were incubated with an equal volume of DMSO. To investigate the effects of divalent cations on the interaction, mitochondria were resuspended in SM buffer (250 mM sucrose, 10 mM MOPS-KOH, pH 7.2) including MgCl2 or CaCl2 at the indicated concentrations and then subjected to in organello crosslinking. Excess amino-reactive functional groups of the crosslinkers were quenched by addition of 140 mM Tris-HCl pH 7.4 and mitochondria were re-isolated by centrifugation. SMPB crosslink samples were analyzed by SDS-PAGE. DSG crosslink samples were subjected to a denaturing lysis by solubilization in pulldown solubilization buffer (see above, no digitonin) with 1% [w/v] SDS and 10 mM imidazole and incubation at 95°C for 5 min. After a clearing centrifugation step, mitochondrial lysates were diluted 10-fold into dilution buffer (pulldown solubilization buffer with 0.2% Triton X-100 and 10 mM imidazole) and incubated with Ni-NTA agarose at 4°C for 90 min. Subsequently, the resin was washed 5x with wash buffer with 0.2% Triton X-100 and 40 mM imidazole and eluted using the same buffer but containing 250 mM imidazole pH 7.4. The removal of non-relevant lanes of gel figures by digital processing is indicated by vertical white separating lines.
Protein import into isolated mitochondria
Radiolabeled precursors of Mic10 or Mic10 variants were produced by in vitro transcription/translation with TNT coupled reticulocyte lysate (Promega) using either the plasmids pGEM4Z-Mic10, pGEM4Z-Mic10-G76A or, for radiolabeled Mic10-G74A, a PCR product of the ORF amplified from pRS425-Mic10-G74A using a primer including the SP6 promotor (see Table S1). Import of radiolabeled precursors into isolated mitochondria was performed at 25°C in import buffer (10 mM MOPS-KOH pH 7.2, 3% [w/v] bovine serum albumine, 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 2 mM KH2PO4, 5 mM methionine) with 4 mM NADH, 4 mM ATP, 10 mM creatine phosphate and 0.2 mg/ml creatine kinase. Import reactions were stopped by dissipating the membrane potential and blocking the respiratory chain with AVO mix (8 μM antimycin, 1 μM valinomycin, 20 μM oligomycin). The reaction mixtures were treated with 50 μg/ml proteinase K to remove non-imported precursors and, after inhibition of the protease with 2 mM PMSF, mitochondria were re-isolated, washed with SEM buffer and subjected to pulldown experiments or analyzed by gel electrophoresis and autoradiography.
Live-cell imaging
Exponentially growing cells were attached to 35 mm glass bottom dishes (D35-20 1.5-N [In Vitro Scientific]) pretreated with 1 mg/ml of concanavalin A type IV (Sigma-Aldrich) and live-cell imaging was performed at room temperature. Fluorescence microscopy images were recorded with a DeltaVision Ultra High Resolution microscope (GE Healthcare, Applied Precision) equipped with an UPlanSApo 100x/1.4 oil objective (Olympus), an sCMOS pco.edge camera (PCO), and a seven channel solid state light source (Lumencor). Raw microscopy images were deconvolved using the softWorX deconvolution plugin. Image analysis was performed using Fiji (Schindelin et al., 2012). Images from each figure panel were taken with the same imaging setup and are shown with the same contrast settings. Single focal planes of representative images are shown. Mito Tracker® Green was used to stain mitochondrial networks in live cells. DAPI (4’,6-diamidino-2-phenylindole) was used for mitochondrial DNA staining in live cells.
Electron microscopy
For the ultrastructural analysis of the mitochondria, cells were harvested by centrifugation and fixed for 1 h with 3% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) and incubated for 45 min with 2 mg/ml diaminobenzidine and 0.06% H2O2 in Tris-HCl (pH 7.5). The cells were post-fixed for 20 min with 1.5% KMnO4, post-stained with 0.5% uranyl acetate and dehydrated with graded series of ethanol and embedded in Epon. 70 nm sections were collected on copper grids and inspected with a CM12 transmission electron microscope (Philips) operating at 100 kV. Random images were taken and cristae lengths were measured from 200 mitochondrial cross-sections using Fiji.
Membrane potential assessment
The membrane potential of isolated mitochondria was assessed by potential-dependent quenching of the fluorophore 3,3'-dipropylthiadicarbocyanine iodide (DiSC3(5)) in a Aminco Bowman II fluorescence photometer at 622 nm (excitation) and 670 nm (emission). A saturated solution of DiSC3(5) in ethanol was diluted 1000x into assay buffer (0.6 M sorbitol, 0.1% bovine serum albumine, 10 mM MgCl2, 0.5 mM EDTA, 20 mM potassium phosphate buffer pH 7.2) supplemented with 5 mM malate and 5 mM succinate. After recording the baseline for ca. 20 sec, mitochondria were added and fluorescence quenching was monitored. 1 μM valinomycin was added after ca. 180 sec to uncouple the membrane potential.
Quantification and statistical analysis
In Figure 3B, cristae lengths from 200 mitochondrial cross-sections from images taken at random were measured using Fiji and interquartile box plots were plotted using Excel.
In Figure 3D, mean values of OD measurements from 4 replicates each of the indicated strains were obtained. The error bars represent the standard deviation.
In Figure 3E, upper panel, the measured fluorescence values of 8 replicates from 2 independent experiments were normalized to their start levels and mean values were obtained. The error bars represent the standard error of the mean.
In Figure 3E, lower panel, the measured fluorescence values of 14 replicates from 3 independent experiments were normalized to their start levels and mean values were obtained. The error bars represent the standard error of the mean.
In Figure 4E, mean values of OD measurements from 4 replicates each of the indicated strains were obtained. The error bars represent the standard error of the mean.
In Figure 4F, the measured fluorescence values of 5 replicates were normalized to their start levels and mean values were obtained. The error bars represent the standard error of the mean.
In Figure S2B, mean values of OD measurements from 3 replicates each of the indicated strains were obtained. The error bars represent the standard deviation.
Acknowledgments
We thank Ralf M. Zerbes for discussion and materials. Work included in this study has also been performed in partial fulfillment of the requirements for the doctoral theses of F.W. at Saarland University and M.L. at the University of Freiburg. This work was supported by the Deutsche Forschungsgemeinschaft (PF 202/9-1 – project ID 394024777; project ID 409673687), Sonderforschungsbereiche 746, 894, 1381, and 1177 (project IDs 157660137, 403222702, and 259130777), International Research Training Group 1830 (project ID 202907893), Germany’s Excellence Strategy (CIBSS-EXC2189 – project ID 390939984), the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program under grant agreement no. 769065, the European Union's Horizon 2020 research and innovation programme under grant agreement no. 765912, the Müller-Fahnenberg Stiftung, the Peter und Traudl Engelhorn Stiftung (to H.R.), and a pre-doctoral fellowship of the Boehringer Ingelheim Fonds (to F.W.). This work reflects only the authors' view, and the European Union's Horizon 2020 research and innovation program is not responsible for any use that may be made of the information it contains.
Author contributions
H.R., F.W., M.L., R.d.B., I.P., L.S., and M.B. performed experiments and analyzed data together with T.B., I.v.d.K., C.K., M.v.d.L., and N.P.; H.R., M.v.d.L., and N.P. designed the concepts and supervised the project; H.R., F.W., M.L., and R.d.B. prepared the figures; H.R. and N.P. wrote the manuscript; and all authors discussed results from the experiments and commented on the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 25, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110290.
Contributor Information
Heike Rampelt, Email: heike.rampelt@biochemie.uni-freiburg.de.
Martin van der Laan, Email: martin.van-der-laan@uks.eu.
Nikolaus Pfanner, Email: nikolaus.pfanner@biochemie.uni-freiburg.de.
Supplemental information
Data and code availability
-
•
Required data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aaltonen M.J., Friedman J.R., Osman C., Salin B., di Rago J.-P., Nunnari J., Langer T., Tatsuta T. MICOS and phospholipid transfer by Ups2-Mdm35 organize membrane lipid synthesis in mitochondria. J. Cell Biol. 2016;213:525–534. doi: 10.1083/jcb.201602007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhaja A.K., Jans D.C., Nikolov M., Vukotic M., Lytovchenko O., Ludewig F., Schliebs W., Riedel D., Urlaub H., Jakobs S., et al. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol. Biol. Cell. 2012;23:247–257. doi: 10.1091/mbc.E11-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselmi C., Davies K.M., Faraldo-Gómez J.D. Mitochondrial ATP synthase dimers spontaneously associate due to a long-range membrane-induced force. J. Gen. Physiol. 2018;150:763–770. doi: 10.1085/jgp.201812033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold I., Pfeiffer K., Neupert W., Stuart R.A., Schägger H. Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arselin G., Giraud M.-F., Dautant A., Vaillier J., Brèthes D., Coulary-Salin B., Schaeffer J., Velours J. The GxxxG motif of the transmembrane domain of subunit e is involved in the dimerization/oligomerization of the yeast ATP synthase complex in the mitochondrial membrane. Eur. J. Biochem. 2003;270:1875–1884. doi: 10.1046/j.1432-1033.2003.03557.x. [DOI] [PubMed] [Google Scholar]
- Arselin G., Vaillier J., Salin B., Schaeffer J., Giraud M.-F., Dautant A., Brèthes D., Velours J. The Modulation in Subunits e and g Amounts of Yeast ATP Synthase Modifies Mitochondrial Cristae Morphology. J. Biol. Chem. 2004;279:40392–40399. doi: 10.1074/jbc.M404316200. [DOI] [PubMed] [Google Scholar]
- Barbot M., Jans D.C., Schulz C., Denkert N., Kroppen B., Hoppert M., Jakobs S., Meinecke M. Mic10 oligomerizes to bend mitochondrial inner membranes at cristae junctions. Cell Metab. 2015;21:756–763. doi: 10.1016/j.cmet.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Barca E., Ganetzky R.D., Potluri P., Juanola-Falgarona M., Gai X., Li D., Jalas C., Hirsch Y., Emmanuele V., Tadesse S., et al. USMG5 Ashkenazi Jewish founder mutation impairs mitochondrial complex V dimerization and ATP synthesis. Hum. Mol. Genet. 2018;27:3305–3312. doi: 10.1093/hmg/ddy231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer H.M., Gonschorek P., Samodelov S.L., Meier M., Weber W., Zurbriggen M.D. AQUA cloning: a versatile and simple enzyme-free cloning approach. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum T.B., Hahn A., Meier T., Davies K.M., Kühlbrandt W. Dimers of mitochondrial ATP synthase induce membrane curvature and self-assemble into rows. Proc. Natl. Acad. Sci. U.S.A. 2019;116:4250–4255. doi: 10.1073/pnas.1816556116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert M., Wenz L.-S., Zerbes R.M., Horvath S.E., Stroud D.A., von der Malsburg K., Müller J.M., Oeljeklaus S., Perschil I., Warscheid B., et al. Role of mitochondrial inner membrane organizing system in protein biogenesis of the mitochondrial outer membrane. Mol. Biol. Cell. 2012;23:3948–3956. doi: 10.1091/mbc.E12-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert M., Zerbes R.M., Davies K.M., Mühleip A.W., Rampelt H., Horvath S.E., Boenke T., Kram A., Perschil I., Veenhuis M., et al. Central role of Mic10 in the mitochondrial contact site and cristae organizing system. Cell Metab. 2015;21:747–755. doi: 10.1016/j.cmet.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Bonneaud N., Ozier-Kalogeropoulos O., Li G.Y., Labouesse M., Minvielle-Sebastia L., Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- Bornhövd C., Vogel F., Neupert W., Reichert A.S. Mitochondrial membrane potential is dependent on the oligomeric state of F1F0-ATP synthase supracomplexes. J. Biol. Chem. 2006;281:13990–13998. doi: 10.1074/jbc.M512334200. [DOI] [PubMed] [Google Scholar]
- Brandt T., Mourier A., Tain L.S., Partridge L., Larsson N.-G., Kühlbrandt W. Changes of mitochondrial ultrastructure and function during ageing in mice and Drosophila. eLife. 2017;6:3343. doi: 10.7554/eLife.24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadena L.R., Gahura O., Panicucci B., Zíková A., Hashimi H. Mitochondrial contact site and cristae organization system and F1FO-atp synthase crosstalk is a fundamental property of mitochondrial cristae. mSphere. 2021;6 doi: 10.1128/mSphere.00327-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T.W., Sikorski R.S., Dante M., Shero J.H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cogliati S., Enriquez J.A., Scorrano L. Mitochondrial cristae: where beauty meets functionality. Trends Biochem. Sci. 2016;41:261–273. doi: 10.1016/j.tibs.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Colina-Tenorio L., Horten P., Pfanner N., Rampelt H. Shaping the mitochondrial inner membrane in health and disease. J. Intern. Med. 2020;287:645–664. doi: 10.1111/joim.13031. [DOI] [PubMed] [Google Scholar]
- Darshi M., Mendiola V.L., Mackey M.R., Murphy A.N., Koller A., Perkins G.A., Ellisman M.H., Taylor S.S. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J. Biol. Chem. 2011;286:2918–2932. doi: 10.1074/jbc.M110.171975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies K.M., Anselmi C., Wittig I., Faraldo-Gómez J.D., Kühlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc. Natl. Acad. Sci. U.S.A. 2012;109:13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies K.M., Strauss M., Daum B., Kief J.H., Osiewacz H.D., Rycovska A., Zickermann V., Kühlbrandt W. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw C.C., Ramachandran K., Enslow B.T., Maity S., Bursic B., Novello M.J., Rubannelsonkumar C.S., Mashal A.H., Ravichandran J., Bakewell T.M., et al. Lactate elicits ER-mitochondrial Mg2+ dynamics to integrate cellular metabolism. Cell. 2020;183:474–489.e17. doi: 10.1016/j.cell.2020.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alteriis E., Cartenì F., Parascandola P., Serpa J., Mazzoleni S. Revisiting the Crabtree/Warburg effect in a dynamic perspective: a fitness advantage against sugar-induced cell death. Cell Cycle. 2018;17:688–701. doi: 10.1080/15384101.2018.1442622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bartolomeo F., Malina C., Campbell K., Mormino M., Fuchs J., Vorontsov E., Gustafsson C.M., Nielsen J. Absolute yeast mitochondrial proteome quantification reveals trade-off between biosynthesis and energy generation during diauxic shift. Proc. Natl. Acad. Sci. U.S.A. 2020;117:7524–7535. doi: 10.1073/pnas.1918216117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C., Wu Z., Huang L., Wang Y., Xue J., Chen S., Deng Z., Wang L., Song Z., Chen S. Mitofilin and CHCHD6 physically interact with Sam50 to sustain cristae structure. Sci. Rep. 2015;5:16064. doi: 10.1038/srep16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramo M.J., Lisnyak V., Formosa L.E., Ryan M.T. The “mitochondrial contact site and cristae organising system” (MICOS) in health and human disease. J. Biochem. 2020;167:243–255. doi: 10.1093/jb/mvz111. [DOI] [PubMed] [Google Scholar]
- Everard-Gigot V., Dunn C.D., Dolan B.M., Brunner S., Jensen R.E., Stuart R.A. Functional Analysis of Subunit e of the F1Fo-ATP Synthase of the Yeast Saccharomyces cerevisiae: importance of the N-Terminal Membrane Anchor Region. Eukar. Cell. 2005;4:346–355. doi: 10.1128/EC.4.2.346-355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eydt K., Davies K.M., Behrendt C., Wittig I., Reichert A.S. Cristae architecture is determined by an interplay of the MICOS complex and the F1FO ATP synthase via Mic27 and Mic10. Microb. Cell. 2017;4:259–272. doi: 10.15698/mic2017.08.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Mourier A., Yamada J., McCaffery J.M., Nunnari J. MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. eLife. 2015;4 doi: 10.7554/eLife.07739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarani V., McNeill E.M., Paulo J.A., Huttlin E.L., Fröhlich F., Gygi S.P., Van Vactor D., Harper J.W. QIL1 is a novel mitochondrial protein required for MICOS complex stability and cristae morphology. eLife. 2015;4 doi: 10.7554/eLife.06265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Bueler S.A., Rubinstein J.L. Atomic model for the dimeric FO region of mitochondrial ATP synthase. Science. 2017;358:936–940. doi: 10.1126/science.aao4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habersetzer J., Larrieu I., Priault M., Salin B., Rossignol R., Brèthes D., Paumard P. Human F1F0 atp synthase, mitochondrial ultrastructure and OXPHOS impairment: a (Super-)Complex matter? PLoS One. 2013;8 doi: 10.1371/journal.pone.0075429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A., Parey K., Bublitz M., Mills D.J., Zickermann V., Vonck J., Kühlbrandt W., Meier T. Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol. Cell. 2016;63:445–456. doi: 10.1016/j.molcel.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M.E., Körner C., Walther D., Mokranjac D., Kaesmacher J., Welsch U., Griffith J., Mann M., Reggiori F., Neupert W. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 2011;30:4356–4370. doi: 10.1038/emboj.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M.E., Unger A.-K., Geerts W.J., Mari M., Izawa T., Stenger M., Geimer S., Reggiori F., Westermann B., Neupert W. An evidence based hypothesis on the existence of two pathways of mitochondrial crista formation. eLife. 2016;5 doi: 10.7554/eLife.18853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Ford H.C., Carroll J., Douglas C., Gonzales E., Ding S., Fearnley I.M., Walker J.E. Assembly of the membrane domain of ATP synthase in human mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2018;115:2988–2993. doi: 10.1073/pnas.1722086115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J.M. MINOS is plus: a mitofilin complex for mitochondrial membrane contacts. Dev. Cell. 2011;21:599–600. doi: 10.1016/j.devcel.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Hessenberger M., Zerbes R.M., Rampelt H., Kunz S., Xavier A.H., Purfürst B., Lilie H., Pfanner N., van der Laan M., Daumke O. Regulated membrane remodeling by Mic60 controls formation of mitochondrial crista junctions. Nat. Commun. 2017;8:15258. doi: 10.1038/ncomms15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S.C., Collins S.R., Cassidy-Stone A., Hummel E., DeVay R.M., Lackner L.L., Westermann B., Schuldiner M., Weissman J.S., Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 2011;195:323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen M.A., Mühlmeister M., Gotthardt K., Guerrero-Castillo S., Brandt U. Evolution and structural organization of the mitochondrial contact site (MICOS) complex and the mitochondrial intermembrane space bridging (MIB) complex. Biochim. Biophys. Acta. 2016;1863:91–101. doi: 10.1016/j.bbamcr.2015.10.009. [DOI] [PubMed] [Google Scholar]
- Itoh K., Tamura Y., Iijima M., Sesaki H. Effects of Fcj1-Mos1 and mitochondrial division on aggregation of mitochondrial DNA nucleoids and organelle morphology. Mol. Biol. Cell. 2013;24:1842–1851. doi: 10.1091/mbc.E13-03-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John G.B., Shang Y., Li L., Renken C., Mannella C.A., Selker J.M.L., Rangell L., Bennett M.J., Zha J. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol. Biol. Cell. 2005;16:1543–1554. doi: 10.1091/mbc.E04-08-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. https://alphafold.ebi.ac.uk/ AlphaFold Protein Structure Database. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaurov I., Vancová M., Schimanski B., Cadena L.R., Heller J., Bílý T., Potěšil D., Eichenberger C., Bruce H., Oeljeklaus S., et al. The diverged trypanosome MICOS complex as a hub for mitochondrial cristae shaping and protein import. Curr. Biol. 2018;28:3393–3407.e5. doi: 10.1016/j.cub.2018.09.008. [DOI] [PubMed] [Google Scholar]
- Kojima R., Kakimoto Y., Furuta S., Itoh K., Sesaki H., Endo T., Tamura Y. Maintenance of cardiolipin and crista structure requires cooperative functions of mitochondrial dynamics and phospholipid transport. Cell Rep. 2019;26:518–528.e6. doi: 10.1016/j.celrep.2018.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozjak-Pavlovic V. The MICOS complex of human mitochondria. Cell Tissue Res. 2017;367:83–93. doi: 10.1007/s00441-016-2433-7. [DOI] [PubMed] [Google Scholar]
- Körner C., Barrera M., Dukanovic J., Eydt K., Harner M.E., Rabl R., Vogel F., Rapaport D., Neupert W., Reichert A.S. The C-terminal domain of Fcj1 is required for formation of crista junctions and interacts with the TOB/SAM complex in mitochondria. Mol. Biol. Cell. 2012;23:2143–2155. doi: 10.1091/mbc.E11-10-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühlbrandt W. Structure and mechanisms of F-type ATP synthases. Annu. Rev. Biochem. 2019;88:515–549. doi: 10.1146/annurev-biochem-013118-110903. [DOI] [PubMed] [Google Scholar]
- Mallilankaraman K., Doonan P., Cárdenas C., Chandramoorthy H.C., Müller M., Miller R., Hoffman N.E., Gandhirajan R.K., Molgó J., Birnbaum M.J., et al. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca2+ uptake that regulates cell survival. Cell. 2012;151:630–644. doi: 10.1016/j.cell.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C., Pfanner N., Truscott K.N. Isolation of yeast mitochondria. Methods Mol. Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- Meisinger C., Ryan M.T., Hill K., Model K., Lim J.H., Sickmann A., Müller H., Meyer H.E., Wagner R., Pfanner N. Protein import channel of the outer mitochondrial membrane: a highly stable tom40-tom22 core structure differentially interacts with preproteins, small tom proteins, and import receptors. Mol. Cell. Biol. 2001;21:2337–2348. doi: 10.1128/MCB.21.7.2337-2348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud M., Gros V., Tardif M., Brugière S., Ferro M., Prinz W.A., Toulmay A., Mathur J., Wozny M., Falconet D., et al. AtMic60 is involved in plant mitochondria lipid trafficking and is part of a large complex. Curr. Biol. 2016;26:627–639. doi: 10.1016/j.cub.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern M., Stiller S.B., Lübbert P., Peikert C.D., Dannenmaier S., Drepper F., Weill U., Höß P., Feuerstein R., Gebert M., et al. Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep. 2017;19:2836–2852. doi: 10.1016/j.celrep.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee I., Ghosh M., Meinecke M. MICOS and the mitochondrial inner membrane morphology - when things get out of shape. FEBS Lett. 2021;595:1159–1183. doi: 10.1002/1873-3468.14089. [DOI] [PubMed] [Google Scholar]
- Muñoz-Gómez S.A., Slamovits C.H., Dacks J.B., Baier K.A., Spencer K.D., Wideman J.G. Ancient homology of the mitochondrial contact site and cristae organizing system points to an endosymbiotic origin of mitochondrial cristae. Curr. Biol. 2015;25:1489–1495. doi: 10.1016/j.cub.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Murphy J.P., Stepanova E., Everley R.A., Paulo J.A., Gygi S.P. Comprehensive temporal protein dynamics during the diauxic shift in Saccharomyces cerevisiae. Mol. Cell. Proteomics. 2015;14:2454–2465. doi: 10.1074/mcp.M114.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemani N., Carvalho E., Tomar D., Dong Z., Ketschek A., Breves S.L., Jaña F., Worth A.M., Heffler J., Palaniappan P., et al. MIRO-1 determines mitochondrial shape transition upon GPCR activation and Ca2+ stress. Cell Rep. 2018;23:1005–1019. doi: 10.1016/j.celrep.2018.03.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemani N., Dong Z., Daw C.C., Madaris T.R., Ramachandran K., Enslow B.T., Rubannelsonkumar C.S., Shanmughapriya S., Mallireddigari V., Maity S., et al. Mitochondrial pyruvate and fatty acid flux modulate MICU1-dependent control of MCU activity. Sci. Signal. 2020;13 doi: 10.1126/scisignal.aaz6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott C., Ross K., Straub S.P., Thiede B., Götz M., Goosmann C., Krischke M., Mueller M.J., Krohne G., Rudel T., et al. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol. Cell. Biol. 2012;32:1173–1188. doi: 10.1128/MCB.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumard P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D.M., Brèthes D., di Rago J.-P., Velours J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrungaro C., Zimmermann K.M., Küttner V., Fischer M., Dengjel J., Bogeski I., Riemer J. The Ca2+-dependent release of the Mia40-induced MICU1-MICU2 dimer from MCU regulates mitochondrial Ca2+ uptake. Cell Metab. 2015;22:721–733. doi: 10.1016/j.cmet.2015.08.019. [DOI] [PubMed] [Google Scholar]
- Pinke G., Zhou L., Sazanov L.A. Cryo-EM structure of the entire mammalian F-type ATP synthase. Nat. Struct. Mol. Biol. 2020;27:1077–1085. doi: 10.1038/s41594-020-0503-8. [DOI] [PubMed] [Google Scholar]
- Rabl R., Soubannier V., Scholz R., Vogel F., Mendl N., Vasiljev-Neumeyer A., Körner C., Jagasia R., Keil T., Baumeister W., et al. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J. Cell Biol. 2009;185:1047–1063. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelt H., Bohnert M., Zerbes R.M., Horvath S.E., Warscheid B., Pfanner N., van der Laan M. Mic10, a core subunit of the mitochondrial contact site and cristae organizing system, interacts with the dimeric F1fo-ATP synthase. J. Mol. Biol. 2017;429:1162–1170. doi: 10.1016/j.jmb.2017.03.006. [DOI] [PubMed] [Google Scholar]
- Rampelt H., Zerbes R.M., van der Laan M., Pfanner N. Role of the mitochondrial contact site and cristae organizing system in membrane architecture and dynamics. Biochim. Biophys. Acta. 2017;1864:737–746. doi: 10.1016/j.bbamcr.2016.05.020. [DOI] [PubMed] [Google Scholar]
- Rampelt H., van der Laan M. The Yin & Yang of mitochondrial architecture - interplay of MICOS and F1fo-ATP synthase in cristae formation. Microb. Cell. 2017;4:236–239. doi: 10.15698/mic2017.08.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelt H., Wollweber F., Gerke C., de Boer R., van der Klei I.J., Bohnert M., Pfanner N., van der Laan M. Assembly of the mitochondrial cristae organizer Mic10 is regulated by mic26-mic27 antagonism and cardiolipin. J. Mol. Biol. 2018;430:1883–1890. doi: 10.1016/j.jmb.2018.04.037. [DOI] [PubMed] [Google Scholar]
- Sakowska P., Jans D.C., Mohanraj K., Riedel D., Jakobs S., Chacinska A. The oxidation status of Mic19 regulates MICOS assembly. Mol. Cell. Biol. 2015;35:4222–4337. doi: 10.1128/MCB.00578-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spikes T.E., Montgomery M.G., Walker J.E. Structure of the dimeric ATP synthase from bovine mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2020;117:23519–23526. doi: 10.1073/pnas.2013998117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan T., Brüser C., Deckers M., Steyer A.M., Balzarotti F., Barbot M., Behr T.S., Heim G., Hübner W., Ilgen P., et al. MICOS assembly controls mitochondrial inner membrane remodeling and crista junction redistribution to mediate cristae formation. EMBO J. 2020;39:217–224. doi: 10.15252/embj.2019104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D., Pfanner N., Wiedemann N. Import of proteins into mitochondria. Methods Cell Biol. 2007;80:783–806. doi: 10.1016/S0091-679X(06)80036-1. [DOI] [PubMed] [Google Scholar]
- Stoldt S., Stephan T., Jans D.C., Brüser C., Lange F., Keller-Findeisen J., Riedel D., Hell S.W., Jakobs S. Mic60 exhibits a coordinated clustered distribution along and across yeast and mammalian mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2019;116:9853–9858. doi: 10.1073/pnas.1820364116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss M., Hofhaus G., Schröder R.R., Kühlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27:1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Zhang K., Dong J., Yan C., Hu C., Ji H., Chen L., Chen S., Zhao H., Song Z. Sam50-Mic19-Mic60 axis determines mitochondrial cristae architecture by mediating mitochondrial outer and inner membrane contact. Cell Death Differ. 2019;1763:542. doi: 10.1038/s41418-019-0345-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasenko D., Barbot M., Jans D.C., Kroppen B., Sadowski B., Heim G., Möbius W., Jakobs S., Meinecke M. The MICOS component Mic60 displays a conserved membrane-bending activity that is necessary for normal cristae morphology. J. Cell Biol. 2017;216:889–899. doi: 10.1083/jcb.201609046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Bron P., Weimann T., Dautant A., Giraud M.-F., Paumard P., Salin B., Cavalier A., Velours J., Brèthes D. Supramolecular organization of the yeast F1Fo-ATP synthase. Biol. Cell. 2008;100:591–601. doi: 10.1042/BC20080022. [DOI] [PubMed] [Google Scholar]
- van Dijken J.P., Otto R., Harder W. Growth of Hansenula polymorpha in a methanol-limited chemostat. Physiological responses due to the involvement of methanol oxidase as a key enzyme in methanol metabolism. Arch. Microbiol. 1976;111:137–144. doi: 10.1007/BF00446560. [DOI] [PubMed] [Google Scholar]
- von der Malsburg K., Müller J.M., Bohnert M., Oeljeklaus S., Kwiatkowska P., Becker T., Loniewska-Lwowska A., Wiese S., Rao S., Milenkovic D., et al. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell. 2011;21:694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- Wagner K., Perschil I., Fichter C.D., van der Laan M. Stepwise assembly of dimeric F1Fo-ATP synthase in mitochondria involves the small Fo-subunits k and i. Mol. Biol. Cell. 2010;21:1494–1504. doi: 10.1091/mbc.E09-12-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner K., Rehling P., Sanjuán Szklarz L.K., Taylor R.D., Pfanner N., van der Laan M. Mitochondrial F1Fo-ATP synthase: the small subunits e and g associate with monomeric complexes to trigger dimerization. J. Mol. Biol. 2009;392:855–861. doi: 10.1016/j.jmb.2009.07.059. [DOI] [PubMed] [Google Scholar]
- Wittig I., Braun H.-P., Schägger H. Blue native PAGE. Nat. Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Wolf D.M., Segawa M., Kondadi A.K., Anand R., Bailey S.T., Reichert A.S., Bliek A.M., Shackelford D.B., Liesa M., Shirihai O.S. Individual cristae within the same mitochondrion display different membrane potentials and are functionally independent. EMBO J. 2019;38:1–21. doi: 10.15252/embj.2018101056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollweber F., von der Malsburg K., van der Laan M. Mitochondrial contact site and cristae organizing system: a central player in membrane shaping and crosstalk. Biochim. Biophys. Acta. 2017;1864:1481–1489. doi: 10.1016/j.bbamcr.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Xie J., Marusich M.F., Souda P., Whitelegge J., Capaldi R.A. The mitochondrial inner membrane protein Mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007;581:3545–3549. doi: 10.1016/j.febslet.2007.06.052. [DOI] [PubMed] [Google Scholar]
- Zerbes R.M., Bohnert M., Stroud D.A., von der Malsburg K., Kram A., Oeljeklaus S., Warscheid B., Becker T., Wiedemann N., Veenhuis M., et al. Role of MINOS in mitochondrial membrane architecture: cristae morphology and outer membrane interactions differentially depend on mitofilin domains. J. Mol. Biol. Biol. 2012;422:183–191. doi: 10.1016/j.jmb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Zerbes R.M., Höß P., Pfanner N., van der Laan M., Bohnert M. Distinct roles of Mic12 and Mic27 in the mitochondrial contact site and cristae organizing system. J. Mol. Biol. 2016;428:1485–1492. doi: 10.1016/j.jmb.2016.02.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Required data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.