Abstract
Both Staphylococcus epidermidis and Staphylococcus aureus are important causes of infections associated with catheters and other medical devices. It has recently been shown that not only S. epidermidis but also S. aureus can produce slime and carries the ica operon responsible for slime production. In the operon, coexpression of icaA and icaD is required for full slime synthesis. In this study, the presence of icaA and icaD was determined in a collection of 91 staphylococcal (68 S. epidermidis and 23 S. aureus) strains from intravenous catheter-associated infections, in 10 strains from the skin and mucosa of healthy volunteers, and in two reference strains by a PCR method. Slime-forming ability was tested on Congo red agar plates; 49% of S. epidermidis strains from catheters and, surprisingly, 61% of S. aureus strains were icaA and icaD positive and slime forming. All the saprophytic strains turned out to be negative for both icaA and icaD and also non-slime forming. Two S. aureus and one S. epidermidis strain from catheters, detected as icaA and icaD positive by PCR analysis and as slime forming (black colonies) at 24 h on Congo red agar, at 48 h exhibited tiny red spikes at the center of black colonies. The onset of these variants could not be ascribed to a mutagenic potential of Congo red, which, in the Ames test, was devoid of mutagenicity. PCR analysis showed that these red variants were negative for both icaA and icaD and even lacking the entire icaADBC operon. The data reported indicate an important role of ica genes as a virulence marker in staphylococcal infections from intravenous catheters.
Staphylococcus epidermidis is a saprophyte which is part of the normal mucosa and skin microflora. In recent years, however, S. epidermidis emerged, together with Staphylococcus aureus, as a frequent etiologic agent of infections associated with catheters and other indwelling medical devices. As they possess little intrinsic pathogenic power, staphylococci are usually regarded as opportunistic agents (16, 18). Over the last few years, several studies have been done to elucidate the structures and pathogenetic mechanisms by which staphylococci are able to cause severe and irreducible infections associated with biomaterials (4, 13, 22). As far as S. epidermidis is concerned, polysaccharide slime (also called biofilm) seems to be the most important factor by which it adheres to and colonizes artificial materials (31). As for S. aureus, it was well known, until now, for its ability to express molecules which recognize host matrix proteins (8, 10, 23, 32). It has recently been shown that S. aureus as well as S. epidermidis is capable of forming slime (2, 5, 9, 21, 23).
Recently, the genetic control of slime production has begun to be elucidated (17), first in S. epidermidis and then in S. aureus (9, 21). Synthesis of the capsular polysaccharide is mediated by the ica operon. Upon activation of this operon, a polysaccharide intercellular adhesin is synthesized. This supports cell-to-cell bacterial contacts by means of a multilayered biofilm. The polysaccharide intercellular adhesin is composed of linear β-1,6-linked glucosaminylglycans. It is synthesized in vitro from UDP-N-acetylglucosamine by the enzyme N-acetylglucosaminyltranferase, which is encoded by the intercellular adhesion (ica) locus, in particular by the icaA gene. Sole expression of icaA induces only low enzymatic activity, but coexpression of icaA with icaD leads to a significant increase in activity and is related to phenotypic expression of the capsular polysaccharide (17).
The aim of this study was to determine the occurrence of the icaA and icaD genes for slime production in a collection of staphylococcal clinical isolates by a simple, rapid, and reliable PCR method previously developed in our laboratory. The search for ica genes was carried out in two S. epidermidis reference strains, 68 S. epidermidis isolates from intravenous catheter-associated infections, 23 S. aureus isolates from catheter-associated infections, and 10 S. epidermidis strains from the skin and mucosa of healthy volunteers. Slime-forming ability was evaluated by the Congo red agar (CRA) plate test.
MATERIALS AND METHODS
Bacterial strains.
Two S. epidermidis reference strains were used, the well-known slime-producing strain ATCC 35984 (RP62A) and the non-slime-producing strain ATCC 12228.
The present study focused on 91 staphylococcal isolates from intravenous catheter-associated infections. These included 68 strains of S. epidermidis and 23 strains of S. aureus. A further 10 strains of S. epidermidis isolated from the skin or mucosa of healthy volunteers were also investigated. All isolates were characterized by classic microbiological methods. In particular, the staphylococcal species were identified by the Api-Staph test (Biomérieux, Lyon, France), a biochemical identification kit, and the coagulase test.
Phenotypic characterization of slime-producing ability.
Production of slime from all strains was studied by cultivation of the strains on CRA (15). CRA plates (0.8 g of CRA [Sigma] and 36 g of saccharose [Sigma] to 1 liter of brain heart infusion agar [Oxoid, Basingstoke, Hampshire, England]) were incubated for 24 h at 37°C and subsequently overnight at room temperature. On CRA, slime-producing strains form black colonies, whereas nonproducing strains develop red colonies. In some cases, when pink subcolonies emerged at 48 h in the center of the black colonies, small pink or black samples were picked from both pink and black areas and subcultured again for 24 h on CRA to obtain pure isolates of slime-negative and slime-positive variants.
Ames test.
In order to test if Congo red had mutagenic potential in the CRA plate test, the Ames test was carried out according to the method described by Ames et al. (1) and revised by Maron and Ames (20). Two strains of Salmonella enterica (TA100 and TA1538) were used. Congo red was added to the top agar at a final concentration equal to that used in the CRA test (0.8 g/liter). The experiments were carried out in the absence and in the presence of the rat liver microsomal fraction S9 (Moltox, Annapolis, Md.) at a concentration of 20 μl/plate.
Negative and positive controls for TA100 and TA1538 were run in parallel. The former was intended to evaluate the number of spontaneous revertants, the latter to evaluate the response of the S. enterica strains toward a known mutagen (2-aminofluorene at a concentration of 10 μg/plate). Counting of revertant colonies was carried out after a 48-h incubation at 37°C.
Strain storage.
A single colony of each bacterial strain was seeded in 8 ml of Trypticase soy broth (TSB). After incubation for 24 h at 37°C, the broth culture was fractioned into 1-ml aliquots, which were stored at −80°C.
Solutions for bacterial lysates.
For lysostaphin (Sigma), a stock solution (1 mg/ml in H2O) was stored in small aliquots at −20°C. Before use, the enzyme was diluted 1:10 with H2O to give a 100 μg/ml concentration. For proteinase K (Sigma), a stock solution (1 mg/ml in H2O) was stored in small aliquots at −20°C. Before use, the enzyme was diluted 1:10 with H2O to give a 100 μg/ml concentration. The buffer was 0.1 M Tris-HCl (pH 7.5).
Bacterial DNA extraction.
Bacteria were harvested by centrifuging 100 μl of each broth culture. Cells were resuspended in 45 μl of H2O, 5 μl of lysostaphin solution was added, and samples were incubated at 37°C. After 10 min, 5 μl of proteinase K solution and 150 μl of 0.1 M Tris-HCl (pH 7.5) were added, and incubation proceeded for a further 10 min. Samples were then heated for 5 min at 100°C.
PCR method for amplification of icaA and icaD sequences.
The sequences of icaA and icaD were taken from the GenBank sequence database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) (accession number for the ica operon, U43366).
Primers specific for icaA and icaD were picked on the gene sequences by the Primer3 program (National Institutes of Health National Human Genome Research Institute [http://www.genome.wi.mit.edu/genome_software/other/primer3.html]).
The primers were synthesized by M-Medical Genenco (Florence, Italy). For the detection of icaA, 5′-TCTCTTGCAGGAGCAATCAA was used as the forward primer (primer 1, corresponding to nucleotides 1337 to 1356), and 5′-TCAGGCACTAACATCCAGCA was used as the reverse primer (primer 2, corresponding to nucleotides 1505 to 1524). The two primers include a 188-bp region. For detection of icaD, 5′-ATGGTCAAGCCCAGACAGAG was used as the forward primer (primer 1, corresponding to nucleotides 1963 to 1982), and 5′-CGTGTTTTCAACATTTAATGCAA was used as the reverse primer (primer 2, corresponding to nucleotides 2138 to 2160). The two primers include a 198-bp region. PCR was performed in a DNA thermal cycler (UNO II Thermocycler; Biometra GmbH, Gottingen, Germany). The reaction was in a 25-μl volume containing the above-mentioned primers (1 μM each), together with 150 ng of the extracted DNA, 100 μM each of dATP, dCTP, dGTP, and dTTP, 1 U of Taq DNA polymerase, and buffer (10 mM Tris-HCl [pH 9.0], 50 mM KCl, 0.1% Triton X-100, and 2.5 mM MgCl2). A thermal step program for both icaA and icaD was used, including the following parameters: incubation at 94°C for 5 min, followed by 50 cycles at 94°C for 30 s (denaturation), 55.5°C for 30 s (annealing), 72°C for 30 s (extension), and 72°C for 1 min after conclusion of the 50 cycles. After the first 30 cycles, a further 1 U of Taq DNA polymerase was added. After amplification, 10 μl of the PCR mixture was analyzed by agarose gel electrophoresis (2% agarose in Tris-borate-EDTA). Molecular weight marker kit VI (Boehringer Mannheim) was used.
PCR method for amplification of ica operon.
The presence of the entire ica operon in slime-producing strains and in nonproducing red variants of both S. epidermidis and S. aureus was checked by amplifying a gene product of 2.7 kb, encompassing a region of the icaADBC locus, as described (21).
RESULTS
Detection of slime-producing phenotype of S. epidermidis and S. aureus strains by CRA plate test.
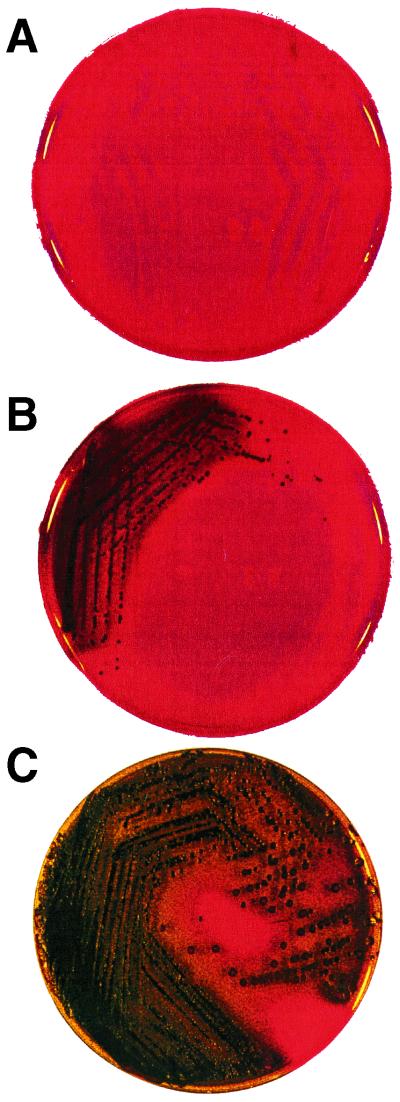
Phenotypic production of slime by all strains under study was assessed by culture on CRA plates. As shown in Fig. 1, slime-producing strains appear as black colonies, and non-slime-producing strains appear as red colonies. S. aureus colonies on CRA were kept under observation for up to 72 h, since the onset of black coloration, indicating slime production, occurs earlier for S. epidermidis than for S. aureus (24 versus 48 to 72 h; unpublished data). Application of Christensen's method to S. aureus requires the addition of glucose for evident slime production to occur (2). The coloration delay found in the CRA method, which uses saccharose as a carbohydrate source, can thus be seen to correspond to the known glucose requirement of S. aureus in Christensen's method. This suggests that S. aureus and S. epidermidis follow different metabolic pathways in the formation of the N-acetylglucosamine slime precursor.
FIG. 1.
CRA plate test. (A) Red colonies of the non-slime-producing S. epidermidis reference strain ATCC 12228. (B) Black colonies of the slime-producing S. epidermidis reference strain ATCC 35984 (RP62A). (C) Black colonies of S. epidermidis strain 13, which exhibited a red central zone at 48 h, indicating the onset of a non-slime-producing phase variant.
Among the clinical isolates, 33 of 68 (48.5%) S. epidermidis strains and 14 of 23 (60.8%) S. aureus strains turned out to be slime producing. All 10 saprophytic S. epidermidis strains from skin and mucosa were non-slime producing.
PCR detection of icaA and icaD.
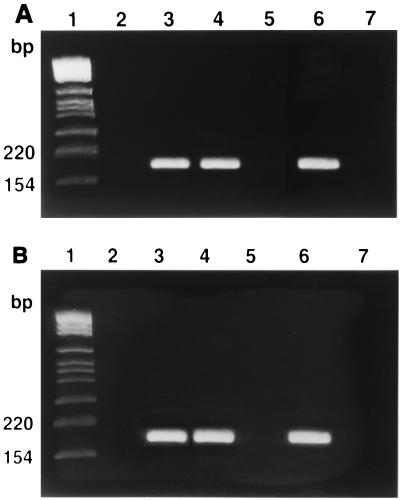
The PCR technique was applied to the 103 staphylococcal strains. As shown in Fig. 2, all strains which were positive for icaA were also positive for icaD. The slime-producing reference strain ATCC 35984 (RP62A) was found to be positive for both genes, giving a 188-bp band for the icaA gene and a 198-bp band for the icaD gene. The non-slime-producing S. epidermidis reference strain ATCC 12228 was negative for both genes. In the saprophytic strains of S. epidermidis from the skin or mucosa of healthy volunteers, neither band was found. Both bands were obtained in slime-producing clinical isolates of both S. epidermidis and S. aureus. Conversely, no band was obtained from non-slime-producing clinical isolates. After determining the sizes of standard bands with the molecular weight marker kit, the image analyzer system assigned the expected lengths to the bands obtained by amplification of the DNA extracted from slime-producing strains. Figure 2 shows the detection of icaA and icaD by PCR.
FIG. 2.
PCR detection of icaA and icaD genes. (A) PCR results with primers for icaA. Lane 1, molecular size markers; lane 2, absence of band with DNA from non-slime-producing S. epidermidis reference strain ATCC 12228; lane 3, 188-bp band obtained with DNA from slime-producing S. epidermidis reference strain ATCC 35984 (RP62A); lane 4, 188-bp band obtained with DNA from slime-producing S. epidermidis strain 13; lane 5, absence of band with DNA from red variant of S. epidermidis strain 13; lane 6, 188-bp band obtained with DNA from slime-producing S. aureus strain 9; lane 7, absence of band with DNA from red variant of S. aureus strain 9; lane 8, negative control (DNA template absent). (B) PCR results with primers for icaD. Lane 1, molecular size markers; lane 2, absence of band with DNA from non-slime-producing S. epidermidis reference strain ATCC 12228; lane 3, 198-bp band obtained with DNA from slime-producing S. epidermidis reference strain ATCC 35984 (RP62A); lane 4, 198-bp band obtained with DNA from slime-producing S. epidermidis strain 13; lane 5, absence of band with DNA from red variant of S. epidermidis strain 13; lane 6, 198-bp band obtained with DNA from slime-producing S. aureus strain 9; lane 7, absence of band with DNA from red variant of S. aureus strain 9; lane 8, negative control (DNA template absent).
Phase variants of S. epidermidis and S. aureus clinical isolates.
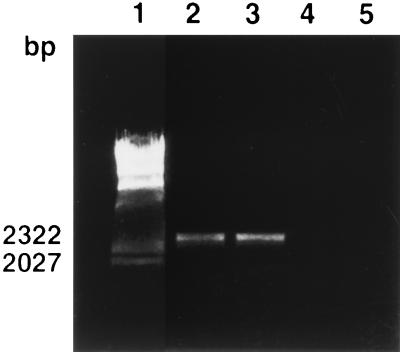
One S. epidermidis and two S. aureus clinical isolates, which at 24 h appeared as black colonies on CRA, at 48 h exhibited tiny pink or red spikes at the center of the black colonies (Table 1 and Fig. 1). Pure black and pure red colonies were obtained on subcultures of samples picked separately from black areas and pink or red spikes, respectively. PCR analysis of these subcolonies showed that the pink and red variants were negative for both icaA and icaD. Moreover, PCR amplification of a 2.7-kb region representing almost the entire ica locus clearly shows that in red variants, not only the icaA and icaD segments were lacking, but even the entire ica locus was absent (Fig. 3).
TABLE 1.
Association between slime production on CRA and presence of icaA and icaD in Staphylococcus strains isolates from catheter infections and from mucosa and skin of healthy volunteers
| Strain | No. of isolates | Origin of infection | Colony color at:
|
Slime production | icaA | icaD | |
|---|---|---|---|---|---|---|---|
| 24 h | 48 h | ||||||
| Reference | |||||||
| ATCC 35984 (RP62A) | Black | Almost black | Strong | + | + | ||
| ATCC 12228 | Red | Red | No | − | − | ||
| S. epidermidis catheter isolates | |||||||
| 1-9-14-20-22-55-60-P13-3561-7P-8P-22P-33P-35P-42AP-47P-49P-2ior-21 | 19 | Catheter | Black | Black | Strong | + | + |
| 13 | 1 | Catheter | Black | Blacka | Strong | + | + |
| 15-B-465-948-38P-52P-54P-58P-7ior-16-28-43-5P | 13 | Catheter | Almost black | Black | Strong | + | + |
| 4-5-7-8-10-22ior-35ior-21-24-61-948-930-0B49-20P-21P-32P | 16 | Catheter | Red | Red | No | − | − |
| 78ior-48P-45P-50P-44P | 5 | Catheter | Red | Deep red | No | − | − |
| 2-11-23-462-470-3P-17P-23P-30P-31P-34P-40P-41P-46P | 14 | Catheter | Deep red | Deep red | No | − | − |
| S. aureus catheter isolates | |||||||
| 3-7-13-14-19 | 4 | Catheter | Black | Black | Strong | + | + |
| 6 | 2 | Catheter | Almost black | Black | Strong | + | + |
| 9-15 | 2 | Catheter | Black | Blacka | Strong | + | + |
| 2-4 | 1 | Catheter | Bordeaux | Black | Strong | + | + |
| 1-17-20-23 | 5 | Catheter | Bordeaux | Almost blackb | Strong | + | + |
| 22 | 1 | Catheter | Red | Deep red | No | − | − |
| 10-12-16-21 | 4 | Catheter | Deep red | Deep red | No | − | − |
| 5-8-11-18 | 4 | Catheter | Red | Red | No | − | − |
| S. epidermidis isolates from healthy volunteers | |||||||
| 798-799-800-801-802-803 | 6 | Mucosa | Deep red | Deep red | No | − | − |
| 901-902-903-904 | 4 | Skin | Deep red | Deep red | No | − | − |
Colonies that were black at 24 h but which exhibited tiny pink or red central spikes at 48 h.
S. aureus colonies kept under further observation for up to 72 h before turning black.
FIG. 3.
PCR detection of ica locus in slime-producing staphylococcal clinical isolates and in their nonproducing red variants. Lane 1, molecular size markers (lambda HindIII digest [Sigma]); lane 2, 2.7-kb band obtained with DNA from slime producer S. epidermidis strain 13; lane 3, 2.7-kb band obtained with DNA from slime producer S. aureus strain 9; lane 4, absence of band with DNA from red variant of S. epidermidis strain 13; lane 5, absence of band with DNA from red variant of S. aureus strain 9.
Lack of mutagenicity of Congo red.
Congo red, when assayed at the concentration used in and under the conditions of the CRA test, was not mutagenic toward the two S. enterica strains tested in either the absence or presence of S9 microsomal extract. The results of the Ames test are shown in Table 2.
TABLE 2.
Ames test for Congo red with two S. enterica test strains
| Addition to agar | Mean no. of revertant colonies/plate ± SD (n = 3)
|
|||
|---|---|---|---|---|
| Strain TA100
|
Strain TA1538
|
|||
| S9 absent | S9 present | S9 absent | S9 present | |
| Congo red | 172 ± 7 | 123 ± 6 | 20 ± 3 | 19 ± 3 |
| Negative control | 165 ± 7 | 119 ± 8 | 21 ± 3 | 20 ± 3 |
| Positive control | 421 ± 18 | 313 ± 11 | 252 ± 25 | 252 ± 25 |
DISCUSSION
Bacterial adhesion has long been considered as a virulence factor contributing to infections associated with catheters and other indwelling medical devices. Interaction of bacteria with biomaterials has been suggested to have a crucial role in conditioning the progress of these severe nosocomial infections (14). The existence of different bacterial molecules of adhesion has been increasingly documented. The possibility was considered that, in the interaction with biomaterials, bacterial cells exhibited various mechanisms, according to environmental conditions and surface characteristics of both material and bacterium. Fletcher and Marshall (12) noted that, with proteolytic enzyme treatment, a Pseudomonas strain could be detached from polystyrene surfaces but not from glass ones, suggesting that different adhesion mechanisms are involved for different materials.
For staphylococcal species, two possible explanations of the ability to colonize artificial materials are the bacterial production of polysaccharide slime and the presence of adhesins for the host matrix proteins that, in vivo, are adsorbed onto the biomaterial surface (8, 10, 23).
Molecular techniques for detection of the gene sequences that encode the adhesion molecules could conveniently be applied in the study of prevalent adhesion mechanisms. From the clinical standpoint, elucidation of the main adhesive mechanisms in periprosthesis infections may help in developing preventive and therapeutic measures, such as antiadhesive coatings or antiadhesin drugs (3, 5, 6, 7, 27, 30).
We have previously described PCR methods for rapid identification of genes encoding the main microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (28) involved in prosthesis adhesion, such as the fibronectin binding protein genes (fnbA and fnbB) (25) and the collagen adhesin gene (cna) (24). Investigation of the presence and expression of cna in a collection of S. aureus strains from orthopedic periprosthesis infections has revealed that the slime-positive strains predominate over the cna-positive ones and that a striking association exists between these two adhesion mechanisms (23).
The data reported here indicate an important role of the ica locus as a virulence marker for clinically significant staphylococcal isolates. Its presence in a high percentage of clinical isolates and its association with the strains' ability to produce slime strongly suggest a role of icaA and icaD in the pathogenetic mechanisms of infection associated with catheters. This should allow routine diagnostic identification of particularly virulent staphylococcus strains. The method recognizes slime-forming strains of both S. epidermidis and S. aureus, requires minimal amounts of DNA, and can thus be readily applied to biopsy material. In the present investigation, staphylococcal strains were retrieved from infected catheters, but in other cases, mainly orthopedic infections, either needle biopsy material or small fragments retrieved from the periprosthesis infected tissues proved to be suitable for PCR amplification. Biopsy material was seeded in TSB, and after incubation for 24 h at 37°C, bacteria were harvested by centrifugation, DNA was extracted, and ica genes were detected by PCR.
For S. epidermidis, the percentage of slime-producing strains (49%) is very close to that found by Muller et al. (46%) for catheter-related infections (26). Ziebuhr et al. (33) have instead reported that 87% of S. epidermidis clinical isolates from catheter infections are slime forming, but the strains were isolated from blood cultures and not directly from catheters, as in our present study. With regard to slime production by S. aureus in catheter-related infections, the data reported provide interesting confirmation of recent observations regarding slime-producing capacities (2, 5) and the presence of ica genes in S. aureus (17). Surprisingly, more slime-producing strains were recorded for S. aureus than for S. epidermidis (61 versus 49%). Thus, far from being peculiar to S. epidermidis, as was once thought, slime production appears to be at least as common in S. aureus. Difficulties in adapting in vitro methods designed for phenotypic study of these abilities in S. epidermidis (11, 15) to S. aureus may have hampered their recognition, as well as the lack, until now, of molecular tests for the detection of genes which control the synthesis of slime. The present results indicate that, among the clinical isolates from catheter infections, 49% of S. epidermidis and 61% of S. aureus are slime producing. These data must be considered reliable and not the outcome of a bias, since the entire catheter specimen is put in culture and the risk of selective isolation of more readily detachable, nonadherent, ica-negative variants can be ruled out. Since, however, adhesion is a prerequisite for the development of an infection, the slime-negative strains must possess other mechanisms which explain their infectious aptitude. The ica-negative, non-slime-producing isolates likely represent strains with alternate means of adhesion, such as MMSCRAMS, relevant in causing catheter-related infections. Actually, in a large collection of clinical isolates from catheter-associated infections, we have found that a large proportion of both S. aureus and S. epidermidis strains, either ica positive or ica negative, harbor the two genes for the fibronectin binding protein (fnbA and fnbB) (unpublished results). This finding reinforces the opinion that mechanisms beside slime production are responsible for bacterial adhesion.
In the present study, PCR analysis also showed that the red variants from three icaA- and icaD-positive, slime-forming isolates from catheters (two S. aureus and one S. epidermidis) were negative for both icaA and icaD and that, moreover, in these variants the entire ica locus seems to be lacking. These data suggest that the phenotypic change may be caused by a deletion of the ica operon rather than an insertion event which inactivates the ica genes. Ziebuhr et al. found that, in S. epidermidis, the insertion of a 1,332-bp sequence element, known as IS256, causes inactivation of icaA gene in non-slime-forming variants of slime-forming S. epidermidis strains (34). A single insertion point has been described in icaA. However, the icaA amplification region selected in this study (nucleotides 1337 to 1524) should be sufficiently distant from the insertion point of IS256 in icaA (at 1708) to ensure that the PCR product is unaffected by this insertion phenomenon. Moreover, no insertion has been described in icaD (34). The present evidence, that in non-slime-producing red variants the ica locus is lacking, could also be explained by the possibility that rare slime-negative mutants, originally present in the slime-positive clinical isolates, emerge as red spikes upon passages and prolonged culture on CRA plates. Moreover, a direct mutagenic effect of Congo red must be ruled out, since, at the concentration employed in the plate test, the dye does not show any effect in the Ames test. This finding is in agreement with a previous observation by Reid et al. (29). These authors showed that Congo red was not mutagenic toward S. enterica TA1538 unless incubated overnight in the presence of rat intestinal bacteria and subsequently assayed in the presence of a postmitochondrial activating system.
In conclusion, we suggest that a study of the presence and expression of genes for adhesion molecules, such as the ica genes, may help in clarifying the relevance of the different adhesion mechanisms in the pathogenesis of infections associated with medical devices. It could also be of value in the development of new preventive and therapeutic measures.
ACKNOWLEDGMENTS
This work was supported by Italian Ministry of Health grant SVE 4-ICS/RF98-610 (Interaction between opportunistic bacteria and biomaterials).
We are grateful to Chiara Vescovini and Roberta Rambaldi for skilled assistance in preparing the manuscript. The technical assistance of Daniela Cavedagna and M. Elena Donati is also gratefully acknowledged. We are also grateful to Marina Cervellati for carrying out the Ames test.
REFERENCES
- 1.Ames B, McCann J, Yamasaki E. Methods for detecting carcinogens and mutagens with the Samonella/mammalian microsome mutagenicity test. Mutat Res. 1975;34:317–364. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- 2.Ammendolia M G, Di Rosa R, Montanaro L, Arciola C R, Baldassarri L. Slime production and expression of the slime-associated antigen by staphylococcal clinical isolates. J Clin Microbiol. 1999;37:3235–3238. doi: 10.1128/jcm.37.10.3235-3238.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An Y H, Blair B K, Martin K L, Friedman R J. Macromolecule surface coating for preventing bacterial adhesion. In: An Y H, Friedmann R J, editors. Handbook of bacterial adhesion: principles, methods and applications. Totowa, NJ: Humana Press Inc.; 2000. pp. 609–625. [Google Scholar]
- 4.An Y H, Friedmann R J. Concise review of mechanisms of bacterial adhesion to biomaterial surfaces. J Biomed Mater Res. 1998;43:338–348. doi: 10.1002/(sici)1097-4636(199823)43:3<338::aid-jbm16>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 5.Arciola C R, Montanaro L, Baldassarri L, Borsetti E, Cavedagna D, Donati M E. Slime production by staphylococci isolated from prosthesis-associated infection. New Microbiol. 1999;22:337–341. [PubMed] [Google Scholar]
- 6.Arciola C R, Montanaro L, Moroni A, Giordano M, Pizzoferrato A, Donati M E. Hydroxyapatite-coated orthopaedic screws as infection resistant material: in vitro study. Biomaterials. 1999;20:323–327. doi: 10.1016/s0142-9612(98)00168-9. [DOI] [PubMed] [Google Scholar]
- 7.Arciola C R, Montanaro L, Caramazza R, Sassoli V, Cavedagna D. Inhibition of bacterial adherence to a high-water-content polymer by a water-soluble, non steroidal, anti-inflammatory drug. J Biomed Mater Res. 1998;42:1–5. doi: 10.1002/(sici)1097-4636(199810)42:1<1::aid-jbm1>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 8.Barth E, Myrvik Q M, Wagner W, Gristina A G. In vitro and in vivo comparative colonization of Staphylococcus aureus and Staphylococcus epidermidis on orthopaedic implant materials. Biomaterials. 1989;10:325–328. doi: 10.1016/0142-9612(89)90073-2. [DOI] [PubMed] [Google Scholar]
- 9.Crampton S E, Gerke C, Schnell N F, Nichols W W, Gotz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cristhensen G D, Simpson W A, Bisno A L, Beachey E H. Adherence of slime-producing Staphyloccus epidermidis to smooth surfaces. Infect Immun. 1982;37:318–326. doi: 10.1128/iai.37.1.318-326.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cristhensen G D, Simpson W A, Younger J J, Baddour L M, Barret F F, Melton D M, Beachey E H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher M, Marshall K C. Bubble contact angle method for evaluating substratum interfacial characteristics and its relevance to bacterial attachment. Appl Environ Microbiol. 1982;44:184–192. doi: 10.1128/aem.44.1.184-192.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster T J, McDevitt D. Molecular basis of adherence of staphylococci to biomaterials. In: Bisno A L, Waldvogel F A, editors. Infection associated with indwelling medical devices. 2nd ed. Washington, D.C.: American Society for Microbiology; 1994. pp. 31–43. [Google Scholar]
- 14.Francois P, Vaudaux P, Foster T J, Lew D P. Host-bacteria interactions in foreign body infections. Infect Control Hosp Epidemiol. 1996;17:514–520. doi: 10.1086/647358. [DOI] [PubMed] [Google Scholar]
- 15.Freeman D J, Falkiner F R, Keane C T. New method for detecting slime production by coagulase-negative staphylococci. J Clin Pathol. 1989;42:872–874. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frère J M, Dubus A, Fonzé E. Pathogens old, new and revived. Nature Biotechnology. 1999;17:7–18. [Google Scholar]
- 17.Gerke C, Kraft A, Sussmuth R, Schweitzer O, Gotz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998;273:18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 18.Huebner J, Goldmann D A. Coagulase-negative staphylococci: role as pathogens. Annu Rev Med. 1999;50:223–236. doi: 10.1146/annurev.med.50.1.223. [DOI] [PubMed] [Google Scholar]
- 19.Kelly C G, Younson J S, Hikmat B Y, Todryk S M, Czisch M, Haris P I, Findall I R, Newby C, Mallet A L, Ma J K, Lehner T A. A syntetic peptide adhesion epitope as a novel antimicrobial agent. Nat Biotechnol. 1999;17:42–47. doi: 10.1038/5213. [DOI] [PubMed] [Google Scholar]
- 20.Maron D, Ames B. Revised method for Salmonella mutagenicity test. Mutat Res. 1983;113:173–215. doi: 10.1016/0165-1161(83)90010-9. [DOI] [PubMed] [Google Scholar]
- 21.McKenney D, Pouliot K L, Wang Y, Murthy V, Ulrich M, Doring G, Lee J C, Goldmann D A, Pier G B. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science. 1999;284:1523–1527. doi: 10.1126/science.284.5419.1523. [DOI] [PubMed] [Google Scholar]
- 22.Montanaro L, Arciola C R. Studying bacterial adhesion to irregular or porous surfaces. In: An Y H, Friedmann R J, editors. Handbook of bacterial adhesion: principles, methods and applications. Totowa, N.J: Humana Press Inc.; 2000. pp. 331–343. [Google Scholar]
- 23.Montanaro L, Arciola C R, Borsetti E, Brigotti M, Baldassarri L. A polymerase chain reaction (PCR) method for the identification of collagen adhesin gene (cna) in Staphylococcus-induced prosthesis infections. New Microbiol. 1998;21:359–363. [PubMed] [Google Scholar]
- 24.Montanaro L, Arciola C R, Borsetti E, Collamati S, Baldassarri L, Montanaro L. Detection of fibronectin-binding protein genes in staphylococcal strains from periprosthesis infections. New Microbiol. 1999;22:331–336. [PubMed] [Google Scholar]
- 25.Montanaro L, Arciola C R, Baldassarri L, Borsetti E. Presence and expression of collagen adhesin gene (cna) and slime production in Staphylococcus aureus strains from orthopaedic prosthesis infections. Biomaterials. 1999;20:1945–1949. doi: 10.1016/s0142-9612(99)00099-x. [DOI] [PubMed] [Google Scholar]
- 26.Muller E, Takeda S, Shiro H, Goldmann D, Pier G B. Occurrence of capsular polysaccharide/adhesin among clinical isolates of coagulase-negative staphylococci. J Infect Dis. 1993;168:1211–1218. doi: 10.1093/infdis/168.5.1211. [DOI] [PubMed] [Google Scholar]
- 27.Nomura S, Lundberg F, Stollenwerk M, Nakamura K, Ljungh A. Adhesion of staphylococci to polymers with and without immobilized heparin in cerebrospinal fluid. J Biomed Mater Res. 1997;38:35–42. doi: 10.1002/(sici)1097-4636(199721)38:1<35::aid-jbm5>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 28.Patti J M, Allen B L, McGavin M J, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 29.Reid T M, Morton K C, Wang C Y, King C M. Conversion of Congo red and 2-azoxyfluorene to mutagens following in vitro reduction by whole-cell rat cecal bacteria. Mutat Res. 1983;117:105–112. doi: 10.1016/0165-1218(83)90157-x. [DOI] [PubMed] [Google Scholar]
- 30.Signas C, Raucci G, Jonsson K, Lindgren P E, Anantharamaiah G M, Hook M, Lindberg M. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc Natl Acad Sci USA. 1989;86:699–703. doi: 10.1073/pnas.86.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tojo M, Yamashita N, Goldmann D A, Pier G B. Isolation and characterization of a capsular polysaccharide adhesin from Staphylococcus epidermidis. J Infect Dis. 1988;157:713–722. doi: 10.1093/infdis/157.4.713. [DOI] [PubMed] [Google Scholar]
- 32.Vaudaux P, Yasuda H, Velazco M I, Huggler E, Ratti I, Waldvogel F A, Lew D P, Proctor A. Role of host and bacterial factors in modulating staphylococcal adhesion to implanted polymer surfaces. J Biomater Appl. 1990;5:134–153. doi: 10.1177/088532829000500204. [DOI] [PubMed] [Google Scholar]
- 33.Ziebuhr W, Heilmann C, Gotz F, Meyer P, Wilms K, Straube E, Hacker J. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 1997;65:890–896. doi: 10.1128/iai.65.3.890-896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ziebuhr W, Krimmer V, Rachid S, Lossner I, Gotz F, Hacker J. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32:345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]