Abstract
The implementation of monoclonal antibody therapeutics during the COVID-19 pandemic altered the selective pressures encountered by SARS-CoV-2, raising the possibility of selection for resistant variants. Within-host viral evolution was reported in treated immunocompromised individuals but whether this signifies a real risk of onward transmission is unclear. We used a regional SARS-CoV-2 sequencing program to monitor lineages with clinically relevant variants in identified patients, which facilitated analysis of parameters potentially relevant to new variant emergence. Here we describe a newly acquired spike E484K mutation detected within the B.1.311 lineage. Multiple individuals in 2 households of the same extended family were infected. The timing and patterns of spread were consistent with de novo emergence of this E484K variant in the bamlanivimab-treated index patient. Our study suggests that the selective pressures introduced by the widespread administration of these antibodies may warrant increased genomic surveillance to identify and mitigate spread of therapy-induced variants.
Keywords: SARS-CoV-2, Drug resistance, Antibody escape mutation, Variant of concern
1. Introduction
Convergent emergence of SARS-CoV-2 mutations in multiple viral lineages with potential for increasing transmissibility, reinfection or reducing vaccine efficacy has caused recent concern [1,2]. In addition, the deployment of several targeted monoclonal antibody therapies [3] and vaccines [4] has introduced numerous selective pressures not previously encountered by SARS-CoV-2. For each of these reasons, improved genomic surveillance will be critical to monitor the spread of these new variants and detect the emergence of new lineages with similarly concerning properties.
The E484K variant in the Spike protein has emerged numerous times in different lineages, including in several emerging variants of concern: B.1.1.7 [5], B.1.351 [6] and P.1 [7]. This suggests that convergent evolution toward some transmission-favoring phenotype may be occurring. The spike protein plays a key role in viral entry [8,9] and is also the target of both naturally developed antibodies [10] as well as synthetic monoclonal antibodies used as treatment or post-exposure prophylaxis [11]. mRNA vaccines elicit a polyclonal antibody response recognizing multiple epitopes on the spike protein but which is dominated by “class 2” antibodies recognizing E484 [12]. Plasma from vaccinated individuals was 1−3-fold less potent in neutralization assays against E484K-pseudotyped virus which correlated with complete elimination of binding against E484K by individual class 2 antibodies [12]. Furthermore, inclusion of class 2 antibodies in cell culture rVSV-SARS-CoV-2 replication assays rapidly drove the emergence of E484K mutations [12].
Therapeutic monoclonal antibodies received Emergency Use Authorization (EUA) in the outpatient setting following trials demonstrating a modest reduction in the risk of hospitalization [13], [14], [15]. Initial authorization of these agents accounted for a drop in clinical utility if the prevailing circulating viral substrains shifted to include variant(s) for which these antibodies had reduced affinity, with continued availability to be based on passive monitoring of sequence data accumulating in repositories. Epidemiologic trends subsequently led to the withdrawal of Bamlanivimab [16] (and later Bamlanivimab/Etesevimab [17]) from widespread clinical use, though the combination was later reinstated and expanded for use as post-exposure prophylaxis as the prevailing circulating variants changed once again [18]. Casirivimab/imdevimab became available under EUA on November 20, 2020 [19] and use for post-exposure prophylaxis was subsequently authorized [20].
The potential for these treatments to select for emergence of antibody resistance mutations has been noted at high frequency (in more than 80% of patients in small case series) in closely-monitored immunocompromised patients [21], [22], [23], but post-administration surveillance in either close contacts or the broader community has not been reported. Between November 2020 and November 2021, our program administered monoclonal antibody therapies of Bamlanivimab, Bamlanivimab/Etesevimab, or Casirivimab/Imdevimab to more than 1,200 COVID-19 positive patients. In parallel, our regional SARS-CoV-2 sequencing program [24], [25], [26] provided the opportunity to detect newly emerging E484K-containing lineages and ascertain the potential epidemiological association, if any, with individuals receiving monoclonal antibody therapy.
2. Materials and methods
2.1. SARS-CoV-2 sequencing
cDNA was generated from residual RNA from diagnostic specimens and sequenced with the Ion AmpliSeq SARS-CoV-2 Panel (Thermo-Fisher; Waltham, MA) as we have previously described [24], [25], [26]. For phylogenetic inference (i.e., to determine the hierarchy of case relationships), sequences were integrated with associated metadata and aligned on a local implementation of NextStrain [27] using augur and displayed via a web browser using auspice.
2.2. E484K mutation rapid tests
A. PCR/Restriction Digest test: cDNA from SARS-CoV-2 specimens was amplified using primers spanning the G23012A polymorphism which encodes E484K: CTTGATTCTAAGGTTGGTGGT and GTAAAGGAAAGTAACAAGTAAAACC. The resultant 157 base pair products were digested with the MseI restriction enzyme which cuts the variant but not the reference allele. Genotypes were determined by band size discrimination using gel electrophoresis.
B. TaqMan Assay: A custom TaqMan SNP genotyping assay (ThermoFisher) was designed to discriminate G v A alleles at 23012. Primers used were GCCGGTAGCACACCTTGT (forward) and GGGTTGGAAACCATATGATTGTAAAGG (reverse). Reporters were AATGGTGTTGAAGGTTT (VIC, reference allele) and AATGGTGTTAAAGGTTT (FAM, variant allele). The assay was run on a Biorad IQ5 real-time PCR machine and NGS-verified reference and variant samples were used to validate allele discrimination.
2.3. Ethical approval
Remnants of diagnostic specimens were sequenced under a protocol approved by the Gundersen Health System IRB (#2-20-03-008) which also allowed access to individual patient data relevant to COVID19 infection.
2.4. Data availability
Sequence data are deposited in GISAID.
3. Results
While performing SARS-CoV-2 surveillance, we sequenced a case from Houston County, Minnesota sampled in late December 2020 which contained a genomic variant, G23012A, encoding an E484K mutation in the Spike protein. Lineage analysis using Pangolin [28] assigned it to B.1.311, a lineage in which Spike:E484K had not been previously detected. This genome was from a child, the third person diagnosed with COVID-19 in a household, whom we designate “P3”. Given concerns surrounding multiple other lineages containing this variant [6,29], we analyzed household contacts of this case.
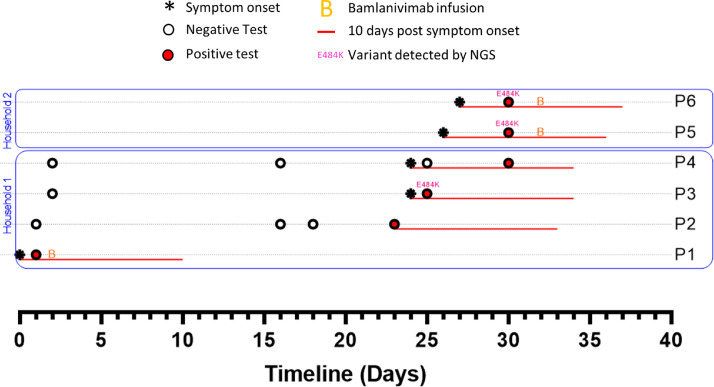
The first case in this family, “P1,” developed symptoms of COVID-19 on a date in the first half of December which we designate as “Day 0” and number other events accordingly (Fig. 1 ). P1 was diagnosed with a rapid COVID-19 test on Day 1. No residual sample was available for sequencing. P1 was treated with bamlanivimab on Day 2. Their disease course was unremarkable and quarantine was discontinued on Day 10. Notably, P1’s medical record provided no indication of any known immunocompromising condition.
Fig. 1.
Timelines of SARS-CoV-2 testing, symptoms and treatments among household and close contacts of a bamlanivimab-treated patient (P1), Genome sequences for P3, P5 and P6 correspond to GISAID accession IDs EPI_ISL_830649, EPI_ISL_861473 and EPI_ISL_861472.
P1’s spouse (P2) tested negative on Days 1, 16 and 18 but developed symptoms and tested positive on Day 23, 13 days after P1 discontinued quarantine. There were 2 children in the household, designated P3 and P4. P3 developed symptoms of COVID-19 (day 24) and tested positive (Day 25). P4 was also symptomatic on Day 24, tested negative on Day 25 but was positive upon repeat testing 5 days later. No residual specimen was available from P2 or P4, but the sample from P3 was sequenced as part of our regional surveillance, and was found to have the E484K-encoding mutation. Two members of the extended family, P5 and P6 (living together nearby), developed symptoms on Days 26 and 27, respectively. Both tested positive on Day 30 and received bamlanivimab on Day 32. Sequencing confirmed that both P5 and P6 had the same B.1.311/E484K substrain.
The observed transmission pattern was consistent with the possibility that the E484K variant emerged in P1 following bamlanivimab treatment, however unambiguously demonstrating this was not possible as we lacked a specimen from this case for sequencing. Nevertheless, several lines of evidence were consistent with this possibility:
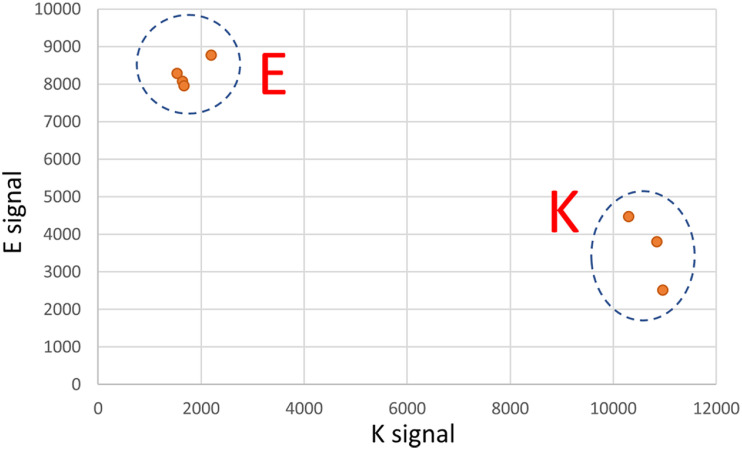
Firstly, despite widespread regional surveillance, we did not detect this B.1.311/E484K lineage in any patient except those associated with this particular household. Mathematical modeling suggests that randomly sampling 5% of positive cases over a period of time should be adequate to detect the presence of a variant of concern if that variant is above 0.1%−1% of the viral population [30]. Our organization was one of several performing COVID-19 testing in the region during this period so genotyping of all positive specimens was not feasible. 287 cases of COVID-19 were diagnosed in that zip code (encompassing the entire city and its rural hinterland) in the last 7 weeks of 2020. We had access to 66 of those specimens which comprised 10%, 15%, 18%, 33%, 16%, 31% and 60% of cases from that zip code in each of those 7 weeks, respectively. 14 of those specimens had been sequenced in our routine surveillance. To more rigorously evaluate whether there was evidence of the E484K substrain outside of this household we used rapid assays (see Methods; discrimination power of Taqman assay shown in Fig. 3) to genotype the remaining 52 specimens. All were wild-type for E484. Accordingly, while we cannot exclude the possibility that this E484K substrain was pre-existing at a very low level, our intense yet incomplete surveillance did not find any evidence of it.
Fig. 3.
Validation of rapid Taqman SNP genotyping assay for the G23012A polymorphism, Clear discrimination between the G allele (encoding E at Spike:484) and the A allele (encoding K at Spike:484) is shown.
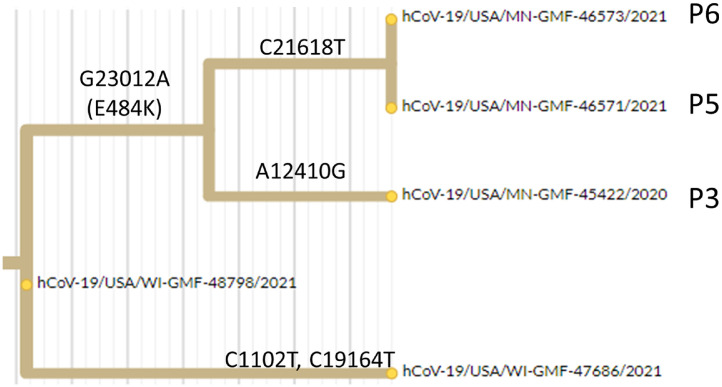
Secondly, we sequenced a case from an adjacent county (WI-GMF-48798) which was the immediate viral ancestor on the B.1.311 lineage to this new E484K containing strain (i.e., it was identical to the 3 sequenced genomes with the exception of the G23012A mutation encoding E484K and the subsequently acquired variants that distinguished the 3 household contacts (Fig. 2 ). Thus, while we cannot formally exclude the possibility that the E484K substrain and its immediate ancestor BOTH emerged elsewhere and arrived in our region in parallel, it seemed plausible that the E484K containing variant of this lineage may have originated locally.
Fig. 2.
Hierarchical relationship between sequenced B.1.311 genomes related to this transmission chain, Excerpt from phylogenetic tree showing acquisition of the E484K encoding mutation (G23012A) on the branch leading to this cluster (P3, P5, P6). A local ancestral sequence to this cluster from an adjacent county is hCoV-19-USA/WI-GMF-48798/2021 (EPI_ISL_942822) and an independent lineage arising from that ancestral strain in the same county is hCoV-19-USA/WI-GMF-47686 (EPI_ISL_942808).
Third, while a 10-day quarantine post-symptom onset may be a reasonable rule-of-thumb for population health guidance, available data demonstrate considerable heterogeneity in the span of time in which culturable virus can be shed in any particular infection, ranging up to 20 days post-symptom onset [31]. Serial viral culture attempts from bamlanivimab-treated patients demonstrated a rise in culturable virus following emergence of mutations at E484 in 3 of 4 evaluated patients [32]. Close contact shedding may also have different dynamics of transmission in a close familial population (e.g., relevance of extrapulmonary shedding from shared bathroom facilities, in line with [33]) with prolonged close contact, as in our study, than perhaps would be expected among the general public.
Fourth, the close timing of symptom onset between P2-P6 makes it less likely that any of these individuals was infected for long enough to transmit the virus to one another, leaving the most likely proposition that P1 acted as the source for each of these transmission events. The number of new sequence variants detected among the 3 sequenced individuals was striking for the degree of genetic diversity demonstrated by sequencing despite such a presumably short transmission chain. This was notably more diversification than we observed in multiple studies of comparable outbreaks of a similar time scale in other households, workplaces, or congregate settings such as skilled nursing facilities (data not shown). The P5 and P6 viral genomes were identical to each other but distinct from P3, ruling out a direct transmission in either direction between P3 and P5/P6. Because P5 and P6 symptom onset dates were only 1 day apart, it seems likely that they were infected by a common source rather than transmitting 1 to the other. The most likely explanation is that each acquired infection from a nearby source with considerable internal viral evolution.
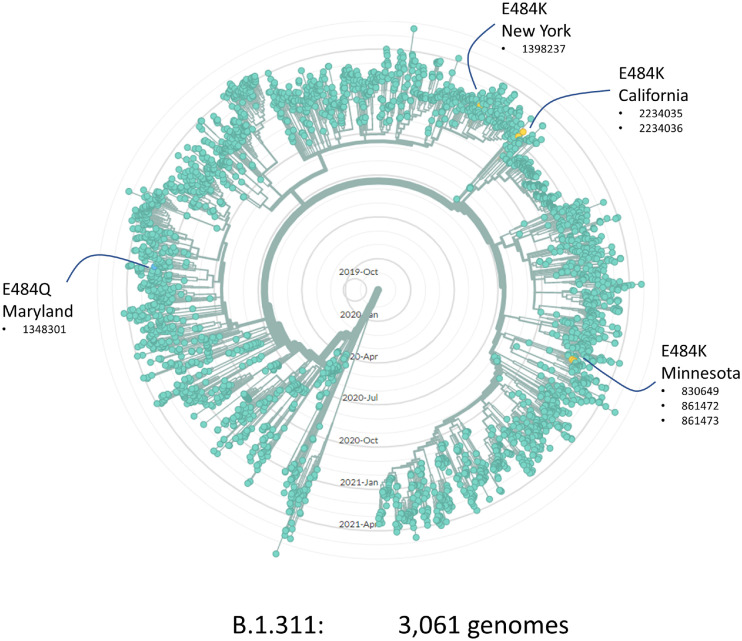
Although broad, our regional surveillance did not have complete coverage of our population leaving open the question of whether additional onward transmission from this cluster might have occurred or whether viral genomes sequenced elsewhere might support or refute our hypothesis. Allowing time for more genomic data to accumulate and be submitted to repositories, we revisited this question in August 2021. We obtained the 3061 genomes from the B.1.311 lineage that had been deposited in GISAID and plotted their relationships on a radial phylogenetic tree. We identified 4 independent emergences of variants at spike E484. In addition to the E484K described in our Minnesota cluster, we found examples in New York (1 sequenced case) and California (2 cases), and an E484Q case in Maryland. No additional genomes on the Minnesota E484K sub-lineage were detected in this analysis or our own longitudinal surveillance. Accordingly, while we cannot exclude any further transmission, it seems unlikely that this particular sub-lineage went on to seed a very large outbreak elsewhere in our region.
4. Discussion
In this study, we describe the probable emergence of a de novo E484K mutation and subsequent transmission to all members of a household and then onward to additional close interpersonal contacts. While the potential for this to occur has been clear for some time in immunocompromised patients, we believe that this is the first genomically-supported demonstration of subsequent transmission of a de novo mutation of concern following the probable selection for this mutation after monoclonal antibody therapy administration to the index case.
Widespread availability under emergency use authorization (EUA) has lowered the barrier to use of monoclonal antibodies, however this has generally not been paralleled by expanded utilization of sequencing to monitor the impact of these treatments on the circulating repertoire of variants. As almost all public health sequencing is performed on deidentified specimens, data on viral strains and epidemiologic connections to specific individuals with prior monoclonal antibody exposure are challenging to link. In contrast, our genomic surveillance was performed with an IRB waiver allowing review of patient records for parameters relevant to COVID-19 disease epidemiology across our service area. This enabled an unusually high-fidelity view of local transmission dynamics and the opportunity to identify clusters of linked cases, such that cases arising subsequent to an intervention (e.g., monoclonal antibody treatment) were straightforward to identify.
It is unclear to what degree the issuance of the original EUA was balanced against the estimation of the hazards of inducing resistant variants in the community. Early trials suggested NGS-detectable resistance mutations in specimens from as many as 10% of treated patients [13], although the potential for onward transmission was not explored. Indeed, detection of such mutations in an individual with a sensitive NGS assay does not necessarily imply that infection-competent virus is being produced at levels sufficient to infect a close contact. In our study, the transmission pattern observed in these households suggests that that is precisely what has occurred.
Widespread use of monoclonal antibodies, especially when regarded as a preference by individuals over vaccination, is suboptimal for several reasons: monoclonal antibodies do not prevent transmission in the very near term, do not cure recipients immediately, provide no clearly durable immunity against reinfection, are linear interventions that scale poorly to populations experiencing exponential disease transmission, and require processes for administration that may divert limited resources from other critical healthcare infrastructure during a crisis. Our study adds the concern that such widespread use may additionally lead to the selection of strains resistant to therapeutic monoclonal antibodies and which might also have some cross-resistance against the dominant antibodies [12] elicited by prior natural or vaccine-acquired immunity.
In assessing the competing risks/benefits we should also consider whether and to what extent this particular risk influences the broader epidemiology of the pandemic. The potential for newly emerging variants such as Alpha, Delta and Omicron to rapidly overtake local strains has been repeatedly demonstrated. Given the widespread use of these agents, if de novo resistance emergence was a very common phenomenon then we might expect to see more frequent examples of independent E484 mutations at clade level than are observed (e.g., Fig. 4 ). Yet even if spontaneously emerging resistance variants do not tend to propagate far from their source, they still have the potential to alter treatment outcomes locally, for example if resistance were to emerge in the early stages of a skilled nursing facility outbreak.
Fig. 4.
Radial phylogenetic tree of 3,061 B.1.311 viral genomes from GISAID (August 2021) showing four independent emergences of sub-lineages with mutations affecting E484, Numbers indicate the GISAID accession number (EPI_ISL_xxxxxx) for each genome.
In summary, our study demonstrates the potential for onward transmission of de novo antibody resistant variants that can emerge in patients treated in the community with single agent monoclonal antibodies. Our work supports the need for additional rapid, scalable surveillance for mutations of concern to be adopted alongside the use of these monoclonal antibodies if their scale of use is to be further expanded.
Author contributions
Study design and conception (PK, AS), Data collection and analysis (PK, CR), Drafting of manuscript and revision (PK, AS, CR), Approval of final manuscript (PK, AS, CR).
Funding
This work was supported by an Emergent Ventures Fast Grant [grant number 2243] to PK and by the Gundersen Medical Foundation. PK holds the Dr. Jon & Betty Kabara Endowed Chair in Precision Oncology.
Declaration of competing interest
The authors declare that they have no conflicts of interest.
References
- 1.Gupta RK. Will SARS-CoV-2 variants of concern affect the promise of vaccines? Nat Rev Immunol. 2021;21:340–341. doi: 10.1038/s41577-021-00556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCormick KD, Jacobs JL, Mellors JW. The emerging plasticity of SARS-CoV-2. Science. 2021;371:1306–1308. doi: 10.1126/science.abg4493. [DOI] [PubMed] [Google Scholar]
- 3.Kreuzberger N, Hirsch C, Chai KL, Tomlinson E, Khosravi Z, Popp M, et al. SARS-CoV-2-neutralising monoclonal antibodies for treatment of COVID-19. Cochrane Database Syst Rev. 2021;9 doi: 10.1002/14651858.CD013825.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alderson J, Batchelor V, O’Hanlon M, Cifuentes L, Richter FC, Kopycinski J, et al. Overview of approved and upcoming vaccines for SARS-CoV-2: a living review. Oxf Open Immunol. 2021;2 doi: 10.1093/oxfimm/iqab010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations [27th January 2021]. Available from: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563.
- 6.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 2020.12.21.20248640. [Google Scholar]
- 7.Faria NR, Morales Claro I, Candido D, Moyeses Franco LA, Andrade PS, Coletti TM, et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings [27th January 2021]. Available from: https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586.
- 8.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okba NMA, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L, Wang P, Nair MS, Yu J, Rapp M, Wang Q, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen P, Nirula A, Heller B, Gottlieb RL, Boscia J, Morris J, et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with covid-19. N Engl J Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottlieb RL, Nirula A, Chen P, Boscia J, Heller B, Morris J, et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, et al. REGEN-COV antibody cocktail clinical outcomes study in covid-19 outpatients. medRxiv. 2021 2021.05.19.21257469. [Google Scholar]
- 16.FDA. Coronavirus (COVID-19) update: FDA revokes emergency use authorization for monoclonal antibody bamlanivimab. Available at: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-monoclonal-antibody-bamlanivimab. Accessed 9/20/2021.
- 17.FDA. Pause in the distribution of bamlanivimab/etesevimab. Available at: https://www.phe.gov/emergency/events/COVID19/investigation-MCM/Bamlanivimab-etesevimab/Pages/bamlanivimab-etesevimab-distribution-pause.aspx. Accessed 9/21/2021.
- 18.FDA. Resumption in use and distribution of bamlanivimab/etesevimab in certain States. Available at: https://www.phe.gov/emergency/events/COVID19/investigation-MCM/Bamlanivimab-etesevimab/Pages/resume-distribution-bamlanivimabetesevimab-all-states-2sept2021.aspx. Accessed 9/22/2021.
- 19.FDA. Coronavirus (COVID-19) update: FDA authorizes monoclonal antibodies for treatment of COVID-19. Available at: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-monoclonal-antibodies-treatment-covid-19.
- 20.FDA. FDA authorizes REGEN-COV monoclonal antibody therapy for post-exposure prophylaxis (prevention) for COVID-19. Available at: https://www.fda.gov/drugs/drug-safety-and-availability/fda-authorizes-regen-cov-monoclonal-antibody-therapy-post-exposure-prophylaxis-prevention-covid-19.
- 21.Jensen B, Luebke N, Feldt T, Keitel V, Brandenburger T, Kindgen-Milles D, et al. Emergence of the E484K mutation in SARS-COV-2-infected immunocompromised patients treated with bamlanivimab in Germany. Lancet Reg Health Eur. 2021;8 doi: 10.1016/j.lanepe.2021.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohr B, Niemann D, Verheyen J. Bamlanivimab treatment leads to rapid selection of immune escape variant carrying E484K mutation in a B.1.1.7 infected and immunosuppressed patient. Clin Infect Dis. 2021;73:2144–2145. doi: 10.1093/cid/ciab392. [DOI] [PubMed] [Google Scholar]
- 23.Peiffer-Smadja N, Bridier-Nahmias A, Ferre VM, Charpentier C, Gare M, Rioux C, et al. Emergence of E484K mutation following bamlanivimab monotherapy among high-risk patients infected with the alpha variant of SARS-CoV-2. Viruses. 2021;13 doi: 10.3390/v13081642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaff BL, Richmond CS, Sabin AP, Athas DM, Adams JC, Meller ME, et al. Outbreak or pseudo-outbreak? integrating SARS-CoV-2 sequencing to validate infection control practices in a dialysis facility. Am J Infect Control. 2021;49:1232–1236. doi: 10.1016/j.ajic.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richmond CS, Sabin AP, Jobe DA, Lovrich SD, Kenny PA. Interregional SARS-CoV-2 spread from a single introduction outbreak in a meat-packing plant in northeast Iowa. medRxiv. 2020 2020.06.08.20125534. [Google Scholar]
- 26.Richmond CS, Sabin AP, Jobe DA, Lovrich SD, Kenny PA. SARS-CoV-2 sequencing reveals rapid transmission from college student clusters resulting in morbidity and deaths in vulnerable populations. medRxiv. 2020 2020.10.12.20210294. [Google Scholar]
- 27.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rambaut A, Holmes EC, O’Toole A, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voloch CM, Silva F Rd, de Almeida LGP, Cardoso CC, Brustolini OJ, Gerber AL, et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. medRxiv. 2020 doi: 10.1128/JVI.00119-21. 2020.12.23.20248598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vavrek D, Speroni L, Curnow KJ, Oberholzer M, Moeder V, Febbo PG. Genomic surveillance at scale is required to detect newly emerging strains at an early timepoint. medRxiv. 2021 2021.01.12.21249613. [Google Scholar]
- 31.Fontana LM, Villamagna AH, Sikka MK, McGregor JC. Understanding viral shedding of severe acute respiratory coronavirus virus 2 (SARS-CoV-2): Review of current literature. Infect Control Hosp Epidemiol. 2021;42:659–668. doi: 10.1017/ice.2020.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boucau J, Chew KW, Choudhary M, Deo R, Regan J, Flynn JP, et al. Monoclonal antibody treatment drives rapid culture conversion in SARS-CoV-2 infection. medRxiv. 2021 doi: 10.1016/j.xcrm.2022.100678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalkeri R, Goebel S, Sharma GD. SARS-CoV-2 shedding from asymptomatic patients: contribution of potential extrapulmonary tissue reservoirs. Am J Trop Med Hyg. 2020;103:18–21. doi: 10.4269/ajtmh.20-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequence data are deposited in GISAID.