Abstract
Background & Aims
Sphingosine 1-phosphate receptors (S1PRs) are a group of G-protein–coupled receptors that confer a broad range of functional effects in chronic inflammatory and metabolic diseases. S1PRs also may mediate the development of nonalcoholic steatohepatitis (NASH), but the specific subtypes involved and the mechanism of action are unclear.
Methods
We investigated which type of S1PR isoforms is activated in various murine models of NASH. The mechanism of action of S1PR4 was examined in hepatic macrophages isolated from high-fat, high-cholesterol diet (HFHCD)-fed mice. We developed a selective S1PR4 functional antagonist by screening the fingolimod (2-amino-2-[2-(4- n -octylphenyl)ethyl]-1,3- propanediol hydrochloride)-like sphingolipid-focused library.
Results
The livers of various mouse models of NASH as well as hepatic macrophages showed high expression of S1pr4. Moreover, in a cohort of NASH patients, expression of S1PR4 was 6-fold higher than those of healthy controls. S1pr4+/- mice were protected from HFHCD-induced NASH and hepatic fibrosis without changes in steatosis. S1pr4 depletion in hepatic macrophages inhibited lipopolysaccharide-mediated Ca++ release and deactivated the Nod-like receptor pyrin domain-containning protein 3 (NLRP3) inflammasome. S1P increased the expression of S1pr4 in hepatic macrophages and activated NLRP3 inflammasome through inositol trisphosphate/inositol trisphosphate–receptor–dependent [Ca++] signaling. To further clarify the biological function of S1PR4, we developed SLB736, a novel selective functional antagonist of SIPR4. Similar to S1pr4+/- mice, administration of SLB736 to HFHCD-fed mice prevented the development of NASH and hepatic fibrosis, but not steatosis, by deactivating the NLRP3 inflammasome.
Conclusions
S1PR4 may be a new therapeutic target for NASH that mediates the activation of NLRP3 inflammasome in hepatic macrophages.
Keywords: Hepatic Macrophages, Ca++, Functional Antagonist, S1P
Abbreviations used in this paper: ATP, adenosine triphosphate; BM, bone marrow; CDA+HFD, choline-deficient; L-amino acid-defined, high-fat diet; EGFP, enhanced green fluorescent protein; ELISA, enzyme-linked immunosorbent assay; ER, endoplasmic reticulum; FBS, fetal bovine serum; FTY720, fingolimod; FTY720-P, fingolimod-phosphate; HFHCD, high-fat; high-cholesterol diet, HSC; hepatic stellate cell, IL-1β; interleukin-1β, IP3, inositol trisphosphate; IP3R, inositol trisphosphate receptor; LPS, lipopolysaccharide; MCDD, methionine- and choline-deficient diet; mRNA, messenger RNA; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; NF-κB, nuclear factor-κB; NLRP3, NLR family pyrin domain containing 3; PBS, phosphate-buffered saline; PLC, phospholipase C; S1P, sphingosine 1-phosphate; S1PR, S1P receptor; shRNA, short hairpin RNA; SK, sphingosine kinase; TG, triglyceride; WD, Western diet; WT, wild-type
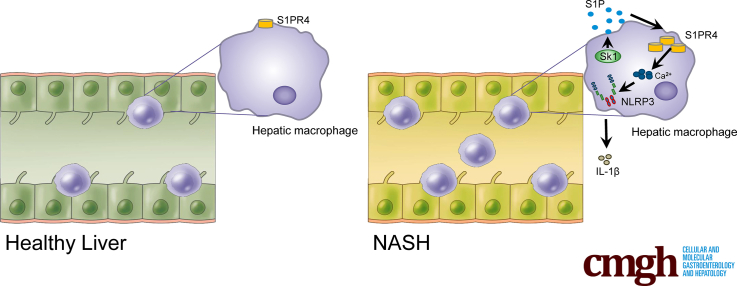
Graphical abstract
Summary.
Type 4 sphingosine-1-phosphate receptor, which mediates the activation of the Nod-like receptor (NLR) family pyrin domain containing 3 inflammasome, emerges as a new therapeutic target for nonalcoholic steatohepatitis. We developed a selective functional antagonist for type 4 sphingosine-1-phosphate receptor (SLB736), which protected mice against nonalcoholic steatohepatitis and fibrosis.
Nonalcoholic fatty liver disease (NAFLD) has become a major health issue worldwide.1 Approximately 10%–20% of patients with NAFLD develop nonalcoholic steatohepatitis (NASH), an advanced stage of NAFLD that subsequently may progress to liver cirrhosis and hepatocellular carcinoma.
The mechanism by which simple steatosis progresses to NASH and liver fibrosis is not completely understood, and an effective treatment for halting the progression of NASH has yet to be discovered.2,3 Lipotoxic hepatocyte death may be the primary lesion that causes liver inflammation and fibrosis.4, 5, 6 Damage-associated molecular patterns released from dying hepatocytes may activate hepatic macrophages, and secretion of proinflammatory and fibrogenic cytokines from macrophages promotes hepatic stellate cell (HSC) activation.7
Sphingosine 1-phosphate (S1P) is a bioactive sphingolipid that influences a wide range of important cellular processes by activating 5 G-protein–coupled receptors (S1PR1–5).8 Receptor-mediated S1P signaling has become an attractive therapeutic target in several diseases such as chronic inflammatory disease, autoimmunity, cancer, and metabolic disease.9, 10, 11, 12 In the liver, S1PR2 participates in cholestasis-induced liver injury13,14 and in chronic liver damage of different etiologies, including bile duct ligation, as well as in methionine–choline–deficient diet (MCDD) and high-fat diet (HFD) feeding, or carbon tetrachloride–mediated liver injury and fibrosis.15 S1PR1 and S1PR3 are involved in HSC motility and activation16 and play a crucial role in the angiogenic process required for fibrosis development.17 Targeting S1PRs was shown to be a promising strategy for treating NASH after the recent preclinical success of 2-amino-2-[2-(4- n -octylphenyl)ethyl]-1,3- propanediol hydrochloride (fingolimod; FTY720),18 a drug developed for multiple sclerosis.19 FTY720, a nonselective modulator of S1PRs (S1PR1, 3, 4, and 5), has been shown to prevent the development of alcoholic liver disease,20 NAFLD,21 and NASH18 in murine models. However, the widespread use of FTY720 in NASH has been hampered by its lymphopenic effects.
In the present study, we identified S1PR4 as a novel player in the pathogenesis of NASH by examining various murine models of NASH, as well as a cohort of NASH patients. We found that S1pr4 expression was significantly higher in the liver of various diet-induced murine models of NASH. S1pr4 heterozygous knockout (S1pr4+/-) mice were protected from high-fat, high-cholesterol diet (HFHCD)-induced NASH and hepatic fibrosis by showing minimal activation of the NLR family pyrin domain containing 3 (NLRP3) inflammasome in hepatic macrophages. To provide further insights into the biological role of S1PR4, we developed and characterized SLB736 as a S1PR4-selective modulator, which acted as a functional antagonist of S1PR4. SLB736 was effective in preventing the development of NASH and fibrosis via inhibiting the activation of the NLRP3 inflammasome in hepatic macrophages. Collectively, our results suggest that S1PR4 is a potential target for the treatment of NASH and hepatic fibrosis.
Results
S1PR4 Expression Is Increased in the Liver of Diet-Induced Murine Models of NASH and in NASH Patients
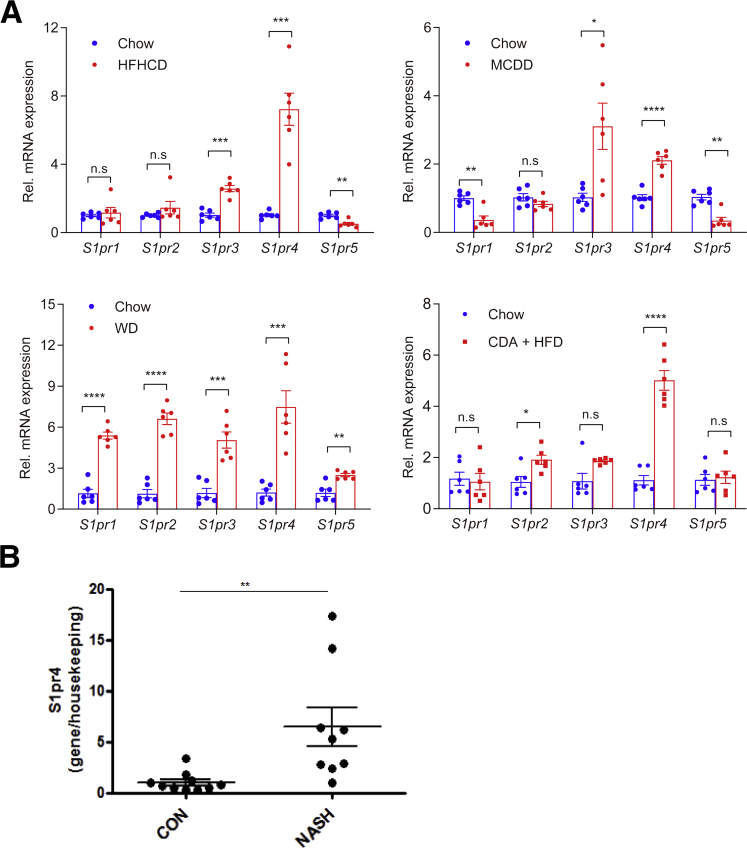
We first investigated which type of S1PR isoform is activated in murine models of NASH. HFHCD feeding is one of the animal models that closely resembles the clinical characteristics of NASH.5,22,23 We also used the MCDD, Western diet (WD), and choline-deficient, L-amino acid-defined (CDA)+HFD.24 Interestingly, S1pr4 was the only isoform that consistently showed increased messenger RNA (mRNA) expression in the livers of mice fed HFHCD, MCDD, WD, or CDA+HFD; the expression of S1pr1 and S1pr2 were increased only in mice fed a WD, and S1pr3 expression was increased only in mice fed HFHCD, MCDD, or WD (Figure 1A). To validate the clinical relevance of the earlier-described findings, we examined the S1PR4 expression in the livers of patients with NASH. Similar to the animal models of NASH, NASH patients showed higher hepatic levels of S1PR4 expression compared with healthy controls (Figure 1B and Table 1).
Figure 1.
Hepatic S1pr4 expression is uniquely increased in the liver of murine NASH models. (A) Diet was administered to mice for 12 weeks for HFHCD, 8 weeks for MCDD, 16 weeks for WD, and 6 weeks for CDA+HFD. (A) Hepatic S1pr mRNA expression in HFHCD-, MCDD-, WD-, and CDA+HFD–induced dietary models of NASH (n = 6). (B) Hepatic mRNA expression levels of S1PR4 in the liver samples of patients with NASH/cirrhosis undergoing liver transplantation (n = 9). Surgical specimens of the donor livers were used as controls (n = 10). All data are shown as means ± SEM. Data were analyzed by Student two-tailed unpaired t test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. CON, control; Rel., relative.
Table 1.
Clinical and Analytical Characteristics of NASH Patients Undergoing Liver Transplantation and Controls (Donors)
| Human samples | Age, y | Sex, M/F | Weight, kg | BMI, kg/m2 | AST, IU/L | ALT, IU/L | GGT, IU/L | ALP, IU/L | Bilirubin, mg/dL | INR | Total cholesterol, mg/dL | TG, mg/dL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor 1 | 42 | M | 90 | 27.8 | 30 | 30 | 30 | NA | NA | 1.0 | NA | NA |
| Donor 2 | 70 | F | NA | NA | 58 | 25 | 18 | NA | NA | 1.1 | NA | NA |
| Donor 3 | 67 | M | 85 | 29.4 | NA | NA | NA | NA | NA | NA | NA | NA |
| Donor 4 | 76 | M | 75 | 27.5 | 20 | 35 | 21 | NA | NA | 1 | NA | NA |
| Donor 5 | 62 | M | 80 | 26.4 | 117 | 66 | NA | NA | NA | NA | NA | NA |
| Donor 6 | 75 | F | 78 | 28.7 | 19 | 16 | 11 | NA | NA | NA | NA | NA |
| Donor 7 | 77 | M | 70 | 27.3 | 14 | 15 | 27 | NA | NA | NA | NA | NA |
| Donor 8 | 28 | M | 58 | 17.9 | 50 | 30 | 23 | NA | NA | 1.0 | NA | NA |
| Donor 9 | 58 | M | 90 | 31.1 | 5 | 23 | 36 | NA | NA | NA | NA | NA |
| Donor 10 | 74 | M | 82 | 26.8 | 9 | 12 | 40 | NA | NA | 1.0 | NA | NA |
| Patient 1 | 46 | M | 78 | 26.1 | NA | 76 | 735 | 298 | 1.2 | 4.68 | 192 | 212 |
| Patient 2 | 57 | M | 93 | 28.7 | 20 | 13 | 23 | 203 | 1.3 | 2 | 58 | 45 |
| Patient 3 | 56 | F | 69 | 27.3 | 18 | 6 | 42 | 116 | 0.6 | 1.36 | 183 | 172 |
| Patient 4 | 66 | M | 77 | 24.9 | 14 | 1 | 22 | 84 | 0.6 | 1.2 | NA | NA |
| Patient 5 | 62 | F | 80 | 27.1 | 56 | 26 | 154 | 158 | 1.2 | 1.2 | 183 | 180 |
| Patient 6 | 60 | F | 100 | 35.9 | 45 | 25 | 183 | 152 | 1.4 | 1.4 | 79 | 70 |
| Patient 7 | 67 | M | 95 | 33.7 | 37 | 20 | 76 | 199 | 3.2 | 1.4 | 59 | 47 |
| Patient 8 | 58 | F | 95 | 33.3 | 27 | 11 | 27 | 92 | 3.7 | 1.9 | 39 | 29 |
| Patient 9 | 64 | F | 79 | 32.9 | 105 | 68 | 20 | 270 | 5.3 | 2.1 | 140 | 53 |
ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; F, female; GGT, γ-glutamyltransferase; INR, international normalized ratio; M, male; NA, not applicable; TG, triglyceride.
S1PR4 Is the Key Mediator of NASH Development
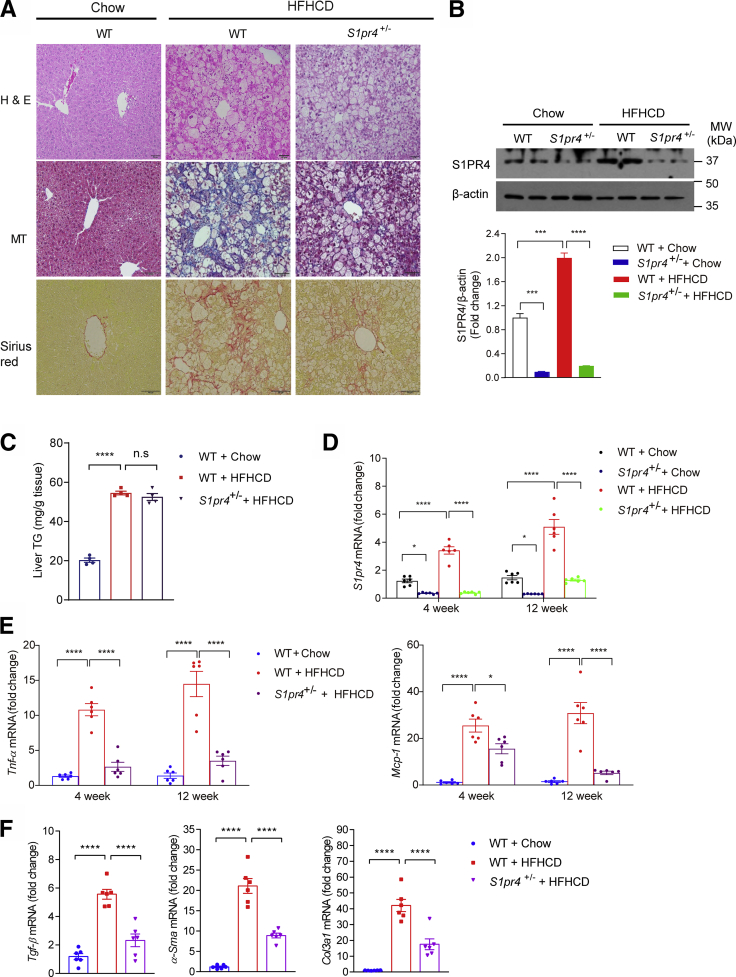
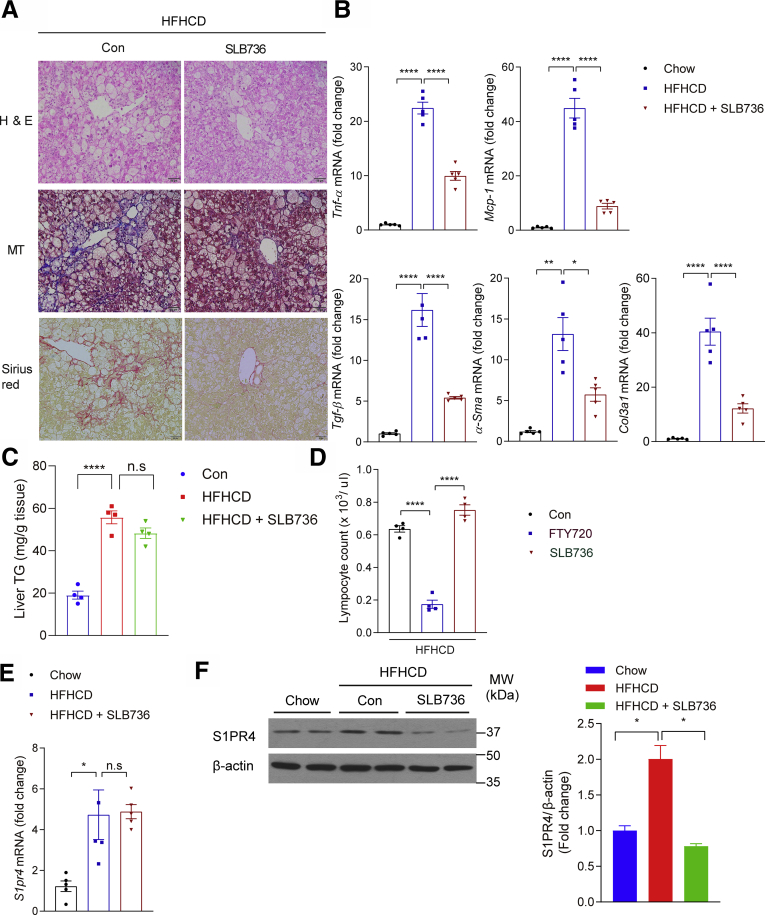
We thus tested the possible involvement of S1PR4 in the development of NASH by using genetic modulation. HFHCD-fed heterozygous S1pr4 knockout mice (S1pr4+/- mice) showed significantly lower degrees of hepatic inflammation and fibrosis compared with HFHCD-fed wild-type (WT) mice (Figure 2A and B). However, the degree of hepatic steatosis was similar regardless of the S1pr4 genotype (Figure 2C and D). In addition, we observed that the expression levels of both S1pr4 and genes involved in inflammation (ie, Tnf-α and Mcp-1) were higher in mice fed HFHCD for 4 and 12 weeks than in control mice. On the other hand, these changes were decreased significantly in the livers of S1pr4+/- mice (Figure 2D and E). In addition, S1pr4 knockdown ameliorated HFHCD-induced liver fibrosis and reduced the expression of Tgf-β, α-Sma, and Col3a1 (Figure 2F). Collectively, these data indicate that S1PR4 is a critical mediator of the development of NASH.
Figure 2.
S1PR4 is a critical mediator of hepatic inflammation and fibrosis.S1pr4+/- mice were fed a chow diet or HFHCD for 12 weeks. (A) Representative H&E, Masson's trichrome, and Sirius Red staining in liver tissues. Scale bar: 50 μm. (B) Hepatic S1PR4 protein expression in the S1pr4+/- mice fed with HFHCD (n = 6). The S1PR4 protein was normalized to β-actin. (C) Liver TG content (n = 4). (D) Hepatic S1pr4 mRNA expression and (E) relative mRNA expression levels of the genes associated with inflammation (Tnf-α, Mcp-1) (n = 6). S1pr4+/- mice were fed chow or HFHCD for 4 or 12 weeks. (F) Relative mRNA expression of fibrosis (Tgf-β, α-Sma, Col3al) (n = 6). S1pr4+/- mice were fed a chow diet or HFHCD for 12 weeks. All data are shown as means ± SEM. (B–F) Data were analyzed by one-way analysis of variance with Bonferroni correction. ∗P < .05, ∗∗∗P < .0005, and ∗∗∗∗P < .0001. MW, molecular weight.
NLRP3 Deficiency Prevents Diet-Induced NASH and Fibrosis, but Not Hepatic Steatosis
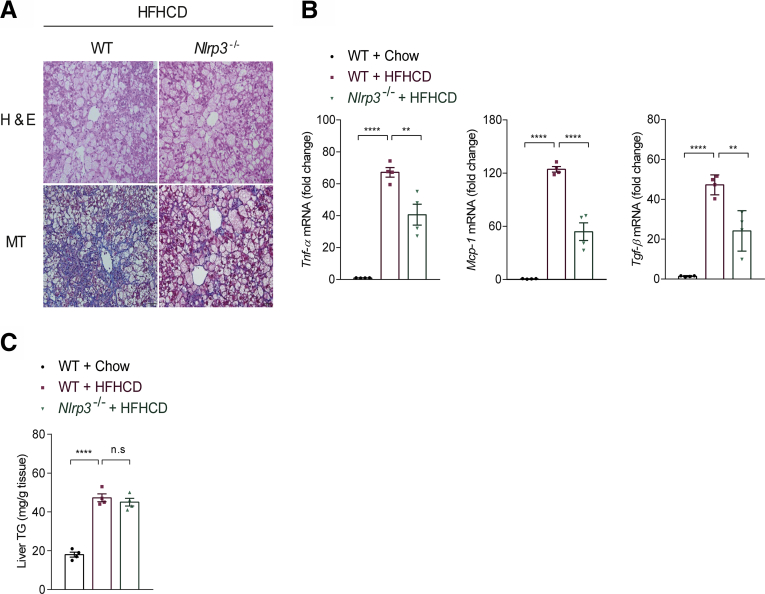
NLRP3 inflammasome is involved in the pathogenesis of various inflammatory and metabolic diseases including arthritis, diabetes, and atherosclerosis.25,26 Recent evidence also has suggested that NLRP3 inflammasome activation in hepatic macrophages (Kupffer cells and monocyte-derived macrophages) is an important contributor to NASH and liver fibrosis,27 and blockade by a small molecule that antagonizes NLRP3 inflammasome or interleukin (IL-1β) was shown to reduce liver inflammation and fibrosis in alcoholic steatohepatitis and NASH in mice.28,29 Similar to previous results,28,30 genetic knockout of Nlrp3 reduced the expression of Tnf-a and Mcp-1 as well as Tgf-β, but did not affect the content of hepatic triglyceride (TG) levels in HFHCD-fed mice (Figure 3). Thus, similar to S1PR4, NLRP3 mediates the inflammation and fibrosis response in NASH without modulating steatosis.
Figure 3.
Nlrp3-/-mice are protected from the development of NASH. (A) Representative H&E and Masson's trichrome staining of the livers of WT and Nlrp3-/- mice fed HFHCD for 12 weeks. Scale bar: 50 μm. (B) Relative mRNA expression levels of the genes associated with inflammation and fibrosis in the liver (n = 4). (C) TG levels in the livers of WT mice fed chow or HFHCD, and Nlrp3-/- mice fed HFHCD (n = 4). All data are shown as means ± SEM. (B and C) Data were analyzed by one-way analysis of variance with Bonferroni correction. ∗∗P < .01, ∗∗∗∗P < .0001.
S1PR4 Is Necessary for Activation of the NLRP3 Inflammasome in Hepatic Macrophages
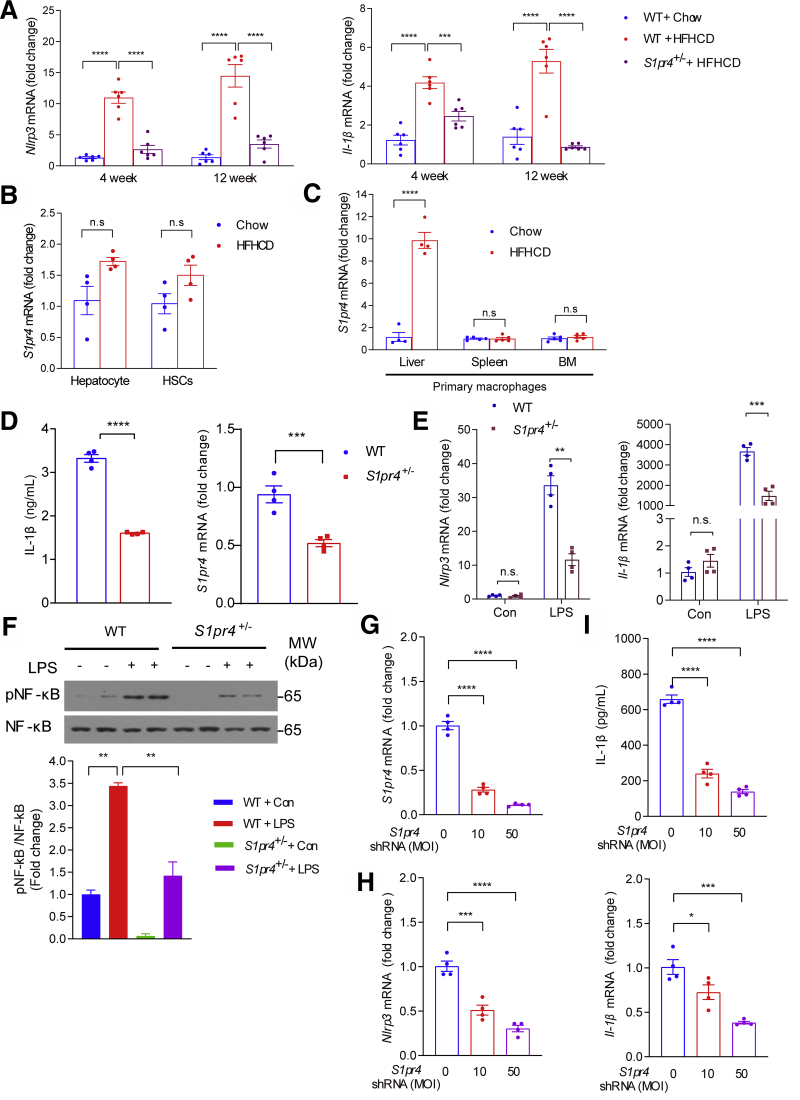
We next evaluated the role of the NLRP3 inflammasome in the S1pr4+/- mice. Compared with HFHCD-fed WT mice, HFHCD-fed S1pr4+/- mice showed significantly decreased NLRP3 inflammasome markers, such as Nlrp3 and Il-1β in the liver (Figure 4A). S1PR4 was reported to be expressed specifically in myeloid cells such as dendritic cells and macrophages31,32; however, its role in the pathogenesis of NASH is largely unknown. To address which cell types are responsible for the up-regulation of S1PR4, we examined the expression levels of S1pr4 in the liver, primary hepatocytes, hepatic macrophages, and HSCs. Significant increases in the expression of S1pr4 by HFHCD were not observed in hepatocytes and HSCs (Figure 4B); in contrast, S1pr4 expression was significantly higher in hepatic macrophages isolated from HFHCD-fed mice for 4 weeks than those isolated from control mice (Figure 4C). We next examined whether S1pr4 expression is increased in resident macrophages in other tissues. Spleen and bone marrow (BM) macrophages isolated from HFHCD-fed mice showed similar S1pr4 mRNA levels compared with those of control-fed mice (Figure 4C). These data suggest that the increase of S1PR4 from hepatic macrophages is an important event in the development of hepatic inflammation and fibrosis.
Figure 4.
S1PR4 depletion decreases NLRP3 inflammasome activation in hepatic macrophages. (A) Relative hepatic mRNA expression levels of the genes associated with the components of NLRP3 inflammasome (n = 6). S1pr4+/- mice were fed chow or HFHCD for 4 or 12 weeks. (B) mRNA expression of S1pr4 in primary hepatocytes and HSCs from mice fed chow diet or HFHCD for 4 weeks (n = 4). (C) mRNA expression of S1pr4 in macrophages isolated from the liver, spleen, and BM. S1pr4+/- mice were fed chow or HFHCD for 4 weeks (n = 4). (D–F) S1pr4 depletion decreases NLRP3 inflammasome activation in hepatic macrophages. (D) Hepatic macrophages isolated from S1pr4+/- mice were stimulated with LPS (100 ng/mL) for 3 hours followed by ATP (1 mmol/L) for 30 minutes. Cell culture media were collected and IL-1β levels were measured by ELISA (n = 4). (E) Relative mRNA expression levels of Nlrp3 and Il-1β (n = 4) and (F) representative Western blots of NF-κB phosphorylation (pNF-κB) and corresponding quantification (n = 3). S1pr4+/- hepatic macrophages were stimulated with LPS (100 ng/mL) for (E) 3 hours or (F) 30 minutes. (G–I) Dose-dependent effect of S1pr4 shRNA transfection on the NLRP3 inflammasome in hepatic macrophages. Relative mRNA expression levels of (G) S1pr4 and (H) Nlrp3 and Il-1β are shown. (I) IL-1β in the cell culture media after transfection of normal hepatic macrophages with the indicated multiplicity of infection (MOI) of S1pr4 shRNA. Hepatic macrophages were infected with lentiviral vectors coding S1pr4 shRNA at an MOI of 0, 10, and 50. After 48 hours, hepatic macrophages were stimulated with (G and H) LPS (100 ng/mL) for 3 hours, (I) followed by stimulation with ATP (1 mmol/L) for 30 minutes. All data are shown as means ± SEM. (A and F–I) Data were analyzed by one-way analysis of variance with Bonferroni correction. (B–E) Data were analyzed by Student two-tailed unpaired t test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. Con, control; MW, molecular weight.
To explore the relationship between high S1pr4 expression in hepatic macrophages and NLRP3 inflammasome, we isolated hepatic macrophages from S1pr4+/- mice. Hepatic macrophages from S1pr4+/- mice had a significantly lower degree of lipopolysaccharide (LPS)- and adenosine triphosphate (ATP)-induced increases in IL-1β production (Figure 4D). These results suggest that S1PR4 is necessary for NLRP3 inflammasome activation in hepatic macrophages. The activation of NLRP3 inflammasome is achieved through 2 sequential steps: signal 1 (priming) and signal 2 (activation)33: signal 1 is provided by microbial molecules or endogenous cytokines and leads to the up-regulation of NLRP3 and Il-1β through the activation of the transcription factor nuclear factor-κB (NF-κB), and signal 2 is triggered by ATP, pore-forming toxins, viral RNA, and particulate matters.
Interestingly, LPS-induced increases in Nlrp3 and Il-1β were significantly nullified in S1pr4+/- hepatic macrophages (Figure 4E). In addition, the phosphorylation of NF-κB in LPS-primed S1pr4+/- hepatic macrophages was decreased (Figure 4F), suggesting that S1PR4 activates the NLRP3 inflammasome from signal 1. Considering that S1PR4 proteins were reduced markedly in the livers of S1pr4+/- mice compared with wild-type mice (Figure 2B), we further investigated the inhibitory effects of an S1pr4-targeting lentivirus-mediated short hairpin RNA (shRNA) on the inflammasome activation in the hepatic macrophages. When compared with the nontargeting control shRNA, S1pr4 shRNA significantly reduced S1pr4 mRNA in a dose-dependent manner (Figure 4G). S1pr4 silencing significantly diminished the LPS-induced increases in Nlrp3 and Il-1β in a dose-dependent manner (Figure 4H). In addition, increases in IL-1β concentration induced by LPS and ATP were abrogated by S1pr4 silencing (Figure 4I).
S1PR4-Dependent Calcium Release From Endoplasmic Reticulum Plays a Pivotal Role in the NLRP3 Inflammasome Activation in Hepatic Macrophages
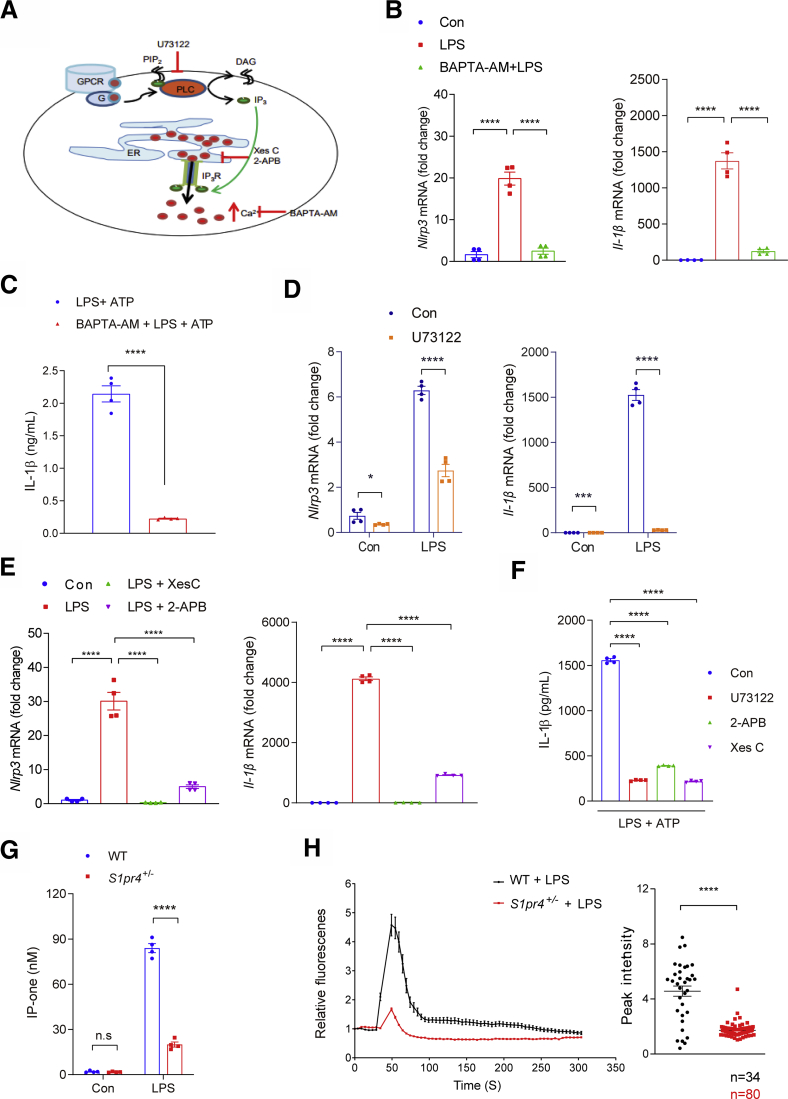
Intracellular ions such as K+, Ca++, and Cl- have significant roles in the activation of the NLRP3 inflammasome.34 Among them, intracellular Ca++ signaling plays one of the major roles in the activation of NLRP3 inflammasomes.35 Accordingly, treatment of hepatic macrophages with the [Ca++] chelator 1,2-Bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetrakis(acetoxymethyl ester) (BAPTA-AM; A1076, sigma-aldrich, St. Louis, MO), significantly decreased the IL-1β production in response to LPS and ATP stimulation, as well as the LPS-induced increases in the expression of Nlrp3 and Il-1β (Figure 5A–C).
Figure 5.
Intracellular calcium signaling is necessary for S1PR4-dependent activation of NLRP3 inflammasome in hepatic macrophages. (A) Schematic illustration of inhibitor of PLC and IP3. (B and C) Effect of calcium depletion on LPS-induced activation of NLRP3 inflammasome. Relative mRNA expression levels of (B) Nlrp3 and Il-1β in the cells and (C) IL-1β secretion into the culture medium (n = 4). (B) Hepatic macrophages were pretreated with BAPTA-AM (10 μmol/L) for 30 minutes and then treated with LPS (100 ng/mL) for 3 hours. (C) In another set of experiments, LPS-primed hepatic macrophages were treated with ATP (1 mmol/L) for 30 minutes. (D–F) Inhibition of LPS-induced NLRP3 inflammasome pathway by suppression of the PLC/IP3/IP3R axis. (D) Relative mRNA expression levels of Nlrp3 and Il-1β. Hepatic macrophages were treated with (D) 10 μmol/L U73122, (E) 5 μmol/L Xes C or 100 μmol/L 2-APB (n = 4). (F) IL-1β in the culture medium. Hepatic macrophages pretreated with U73122 (10 μmol/L), Xes C (5 μmol/L), or 2-APB (100 μmol/L) for 30 minutes and dimethyl sulfoxide as vehicle control. Hepatic macrophages were treated with 1 mmol/L ATP for 30 minutes after 3 hours of LPS priming (100 ng/mL). IL-1β secreted in the culture supernatants was quantified by ELISA (n = 4). (G and H) S1PR4-dependent calcium release from ER plays a pivotal role in the priming of NLRP3 inflammasome in hepatic macrophages. (G) Levels of IP-one in S1pr4+/- hepatic macrophages (n = 4). (H) Effect of S1PR4 on LPS-mediated [Ca++] release. WT or S1pr4+/- hepatic macrophages were incubated with Fluo-4/AM followed by stimulation with LPS for 2 hours. [Ca++] was analyzed by time-lapse confocal microscopy (left panel). Quantification of LPS-induced peak fluorescent intensities (right panel). All data are shown as means ± SEM. (C, D, G, and H) Data were analyzed by Student two-tailed unpaired t test. (B, E, and F) Data were analyzed by one-way analysis of variance with Bonferroni correction. ∗P < .05, ∗∗∗P < .001, and ∗∗∗∗P < .0001. Con, control; DAG, diacylglycerol; G, G-protein; GPCR, G protein-coupled receptor.
A previous study indicated that phospholipase C (PLC)-dependent changes in [Ca++] are the downstream signaling of S1PR4.36 Activation of PLC triggers the release of inositol trisphosphate (IP3) from phosphatidylinositol 4, 5-bisphosphate, and [Ca++] is released to the cytosol when IP3 interacts with IP3 receptor (IP3R) located at the endoplasmic reticulum (ER) membrane.37 In our experimental setting, treatment with the PLC inhibitor U73122 (1-[6-[((17β)-3-Methoxyestra-1,3,5[10]-trien-17-yl)amino]hexyl]-1H-pyrrole-2,5-dione) or IP3R inhibitors Xestospongin C (Xes-c) and 2-Aminoethyl diphenylborinate (2-APB) significantly decreased the LPS-mediated increases in the expression levels of Nlrp3 and Il-1β (Figure 5A, D, and E), as well as the production of IL-1β in response to LPS and ATP stimulation (Figure 5F). Taken together, these results indicate that increases in [Ca++] release from the ER through the PLC/IP3R axis play an important role in the activation of the NLRP3 inflammasome in hepatic macrophages.35
The LPS-induced increase in the level of IP3 , the product of PLC,38 was decreased significantly in S1pr4+/- cells (Figure 5G). Consistently, measurement of [Ca++] showed that LPS treatment in hepatic macrophages induced a robust increase in [Ca++] (Figure 5H), which is in line with previously reported data.39 On the other hand, LPS-induced [Ca++] release was decreased significantly in S1pr4+/- hepatic macrophages (Figure 5H). These results collectively indicate that S1PR4 is required for the calcium signaling associated with the NLRP3 inflammasome activation.
S1P Activates the NLRP3 Inflammasome by the S1PR4/PLC/IP3 Axis
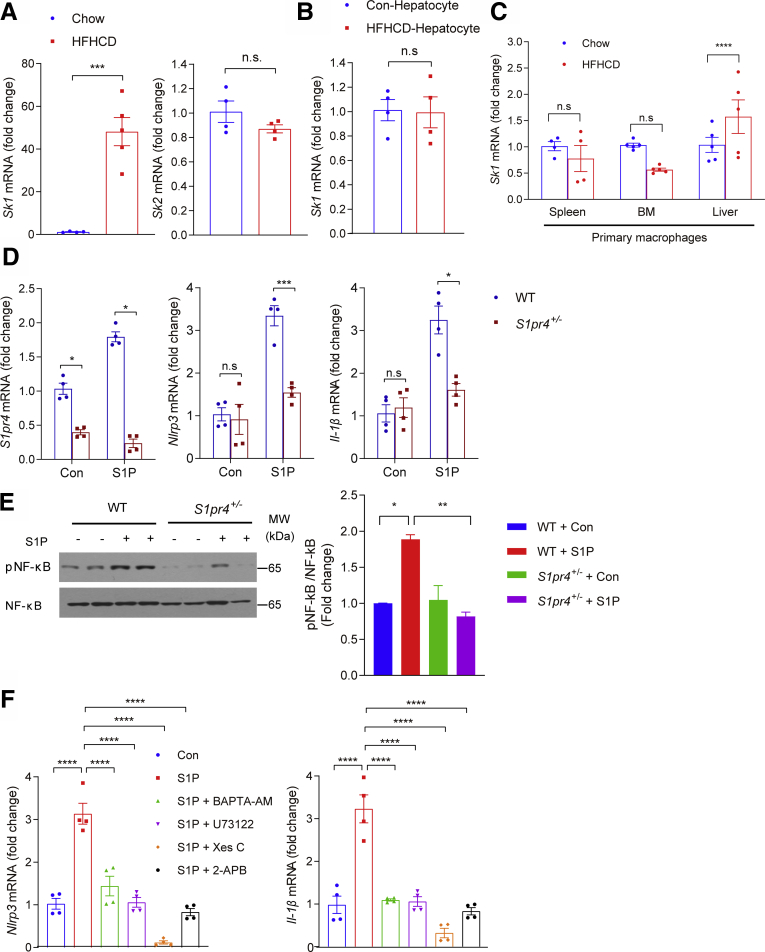
We examined the possible role of the S1P/S1PR4 axis in the activation of the NLRP3 inflammasome. S1P has been shown to contribute to nonalcoholic fatty liver disease and liver fibrosis.18,40 Sphingosine kinases (SKs; eg, SK1 and SK2) catalyze the formation of S1P from the precursor sphingosine.8 Interestingly, expression of Sk1 was increased markedly in the livers but not in hepatocytes of HFHCD-fed mice (Figure 6A and B). On the other hand, Sk1 mRNA expression was increased significantly in hepatic macrophages, whereas the macrophages isolated from the spleen and BM were not affected significantly by HFHCD feeding (Figure 6C). In contrast to Sk1, Sk2 expression was not increased in the livers of HFHCD-fed mice (Figure 6A). These data suggest that increased S1P levels in the hepatic macrophages may induce hepatic inflammation. To further explore the role of S1P on the activation of the NLRP3 inflammasome, hepatic macrophages were treated with S1P. S1P significantly increased the expression level of S1pr4 in hepatic macrophages (Figure 6D). S1P also increased the expression levels of Nlrp3 and Il-1β significantly, an effect that was dampened in S1pr4+/- hepatic macrophages (Figure 6D). S1P also stimulated the phosphorylation of NF-κB in hepatic macrophages, and this was reduced in S1pr4+/- hepatic macrophages (Figure 6E). Pretreatment with BAPTA-AM, U73122, XesC, or 2-APB significantly reduced the S1P-mediated induction of Nlrp3 and Il-1β expression (Figure 6F). These results suggest that extracellular S1P may act as a modulator of the NLRP3 inflammasome in hepatic macrophages through the PLC/IP3/IP3R signaling axis.
Figure 6.
S1P activates NLRP3 inflammasome through S1PR4. (A) Sk1 and Sk2 mRNA expression in the livers of mice fed HFHCD for 12 weeks (n = 4). (B and C) Sk1 mRNA expression in (B) primary hepatocytes and (C) in macrophages from mice fed HFHCD for 4 weeks (n = 4–5). Macrophages were isolated from the liver, spleen, and BM. (D and E) S1pr4 knockdown decreases the S1P-induced NLRP3 inflammasome activation. (D) The mRNA expression of S1pr4, Nlrp3, and Il-1β. Serum-starved S1pr4+/- hepatic macrophages were treated with S1P (1 μmol/L) for 2 hours (n = 4). (E) Representative Western blots of NF-κB phosphorylation (pNF-κB) and corresponding quantification (n = 3). S1pr4+/- hepatic macrophages were treated with S1P (1 μmol/L) for 30 minutes. (F) mRNA expression of Nlrp3 and Il-1β. Serum-starved hepatic macrophages were treated with BAPTA-AM (10 μmol/L), U73122 (10 μmol/L), Xes C (5 μmol/L), or 2-APB (100 μmol/L), and treated with S1P (1 μmol/L) (n = 4). All data are shown as means ± SEM. (A–D) Data were analyzed by Student two-tailed unpaired t test. (E and F) Data were analyzed by one-way analysis of variance followed by Bonferroni correction. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. Con, control; MW, molecular weight.
Development of a Novel Functional Antagonist for S1PR4
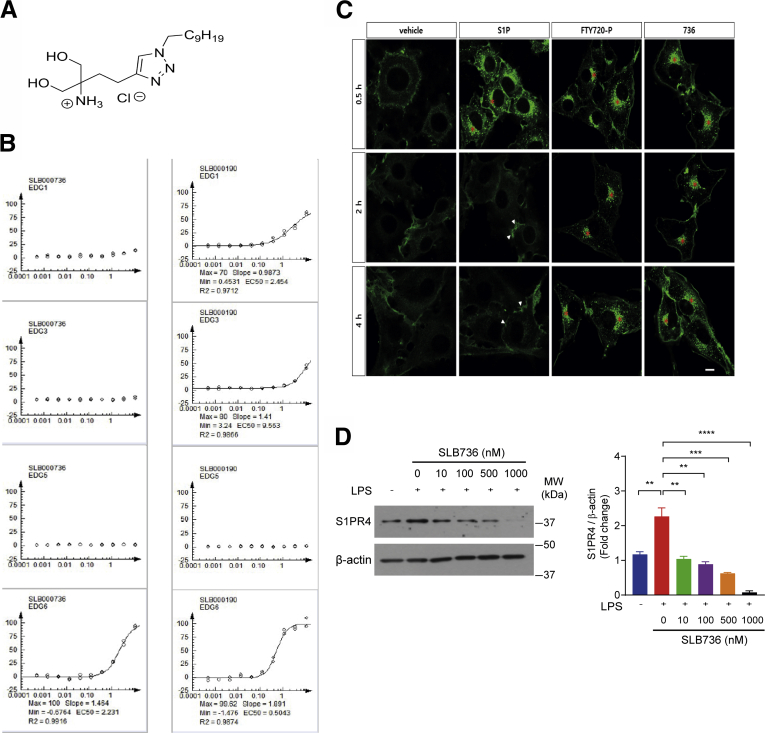
To further clarify the biological function of S1PR4, we developed a chemical probe that selectively modulates S1PR4. We envisioned that introducing additional heteroatoms into the structure of FTY720 could change the selectivity for the S1P subtype by referring to the X-ray crystal structure of S1PR141 and previous structural studies of S1PR4.42 Accordingly, we designed and synthesized several types of heteroatom-containing analogues of FTY720. Among the designed analogues, a triazole-containing compound SLB736 (Figure 7A) showed a selective agonistic activity against S1PR4 (EDG6; endothelial differentiation G-protein coupled receptor 6) based on β-arrestin recruitment assay (Figure 7B). The unique action of the currently used drugs targeting S1PRs, including FTY720, is the functional antagonism for S1PR1,43 in which S1PR1 irreversibly is internalized and degraded upon binding with the phosphorylated active form of FTY720 (FTY720-P).43,44 Down-regulation of S1PR1 on T lymphocytes mediates the immunomodulatory effect of FTY720.45 However, the possible functional antagonistic effects of FTY720-P or SLB736 on S1PR4 have not been investigated. We reasoned that the agonistic activity of SLB736 for S1PR4 as observed in β-arrestin recruitment assay may reflect functional antagonism. Therefore, we examined the localization of S1PR4 at different time points after the activation of S1PR4 in C6 glioma cells bearing enhanced green fluorescent protein (EGFP)-conjugated S1PR4. S1PR4 was internalized by brief exposure (0.5 h) to S1P, FTY720-P, or SLB736. Although S1PR4 quickly was recycled to the cell surface after the removal of S1P, S1PR4 remained in the cytoplasm for 2 and 4 hours after the removal of FTY720-P or SLB736 (Figure 7C). These data indicate that SLB736 and FTY720-P act as functional antagonists for S1PR4. Consistently, even at fairly low concentrations, SLB736 decreased the protein levels of S1PR4 in LPS-primed hepatic macrophages in a dose-dependent manner (Figure 7D).
Figure 7.
SLB736 acts as a functional antagonist of S1PR4. (A) Structure of SLB736. (B) Isoform-specific S1PR activity of SLB736. β-arrestin PathHunter assay was performed in the clonal S1PR/HEK293 cell line in the presence of SLB736 (left) and FTY720-P (right). EDG 1 = S1PR1, EDG5 = S1PR2, EDG3 = S1PR3, EDG6 = S1PR4. (C) S1PR4 internalization and recycling were assessed in C6 glioma cells overexpressing EGFP-fused S1PR4. Cells were exposed to vehicle (0.1% BSA), S1P (100 nmol/L), FTY720-P (1 μmol/L), or SLB736 (1 μmol/L) for 0.5 hours and fixed. Cells exposed to reagents for 0.5 hours were washed and further incubated with vehicle in the presence of cyclohexamide for up to 4 hours. Asterisks or arrowheads indicate cytosolic locations or the cell surface. Scale bar: 10 μm. (D) Protein level of S1PR4 in hepatic macrophages. The S1PR4 protein was normalized to β-actin. Hepatic macrophages were pretreated with SLB736 at the indicated dose and then treated with 100 ng/mL of LPS for 24 hours (n = 4). All data are shown as means ± SEM. (D) Data were analyzed by one-way analysis of variance followed by Bonferroni correction. ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. EC50, 50% Effective Concentration; Max, maximum; Min, minimum; MW, molecular weight.
SLB736 Prevents the Development of NASH and Fibrosis
Administration of SLB736 to HFHCD-fed mice prevented the development of NASH and hepatic fibrosis (Figure 8A and B). Conversely, liver TG content was not altered significantly after SLB736 administration (Figure 8C), suggesting that SLB736 treatment prevents diet-induced liver injury, inflammation, and fibrosis, but not steatosis.
Figure 8.
SLB736 treatment prevents HFHCD-induced NASH. (A) Representative H&E, Masson's trichrome, and Sirius Red staining of the livers. Scale bars: 50 μm. SLB736 (1 mg/kg/d) was administered for 12 weeks in HFHCD-fed mice. (B) Relative mRNA expression levels of the genes associated with inflammation (Tnf-α, Mcp-1) and fibrosis (Tgf-β, α-Sma, Col3al) (n = 5). (C) Liver TG contents (n = 4). (D) Lymphocyte counts in the blood. FTY720 or SLB736 (1 mg/kg/d) were administered for 12 weeks in HFHCD-fed mice (n = 4). (E and F) mRNA expression of (E) S1pr4 and (F) representative Western blots of S1PR4 and corresponding quantification in the liver of HFHCD-fed mice treated with SLB736 (n = 4). All data are shown as means ± SEM. (B–F) Data were analyzed by one-way analysis of variance followed by Bonferroni correction. ∗P < .05, ∗∗P < .01, and ∗∗∗∗P < .0001. Con, control; MW, molecular weight.
Interestingly, the administration of SLB736 did not reduce the number of lymphocytes, which is a well-known adverse effect of FTY720 through its effect on S1PR1 (Figure 8D).46 Although treatment with SLB736 did not significantly reduce the diet-induced increases in the mRNA level of S1pr4 (Figure 8E), the protein level of S1PR4 was decreased significantly upon treatment with SLB736 (Figure 8F), thus signifying that SLB736 carries functional antagonistic roles on S1PR4 in vivo. Whether SLB736 promotes S1PR4 protein instability or degradation remains to be established.
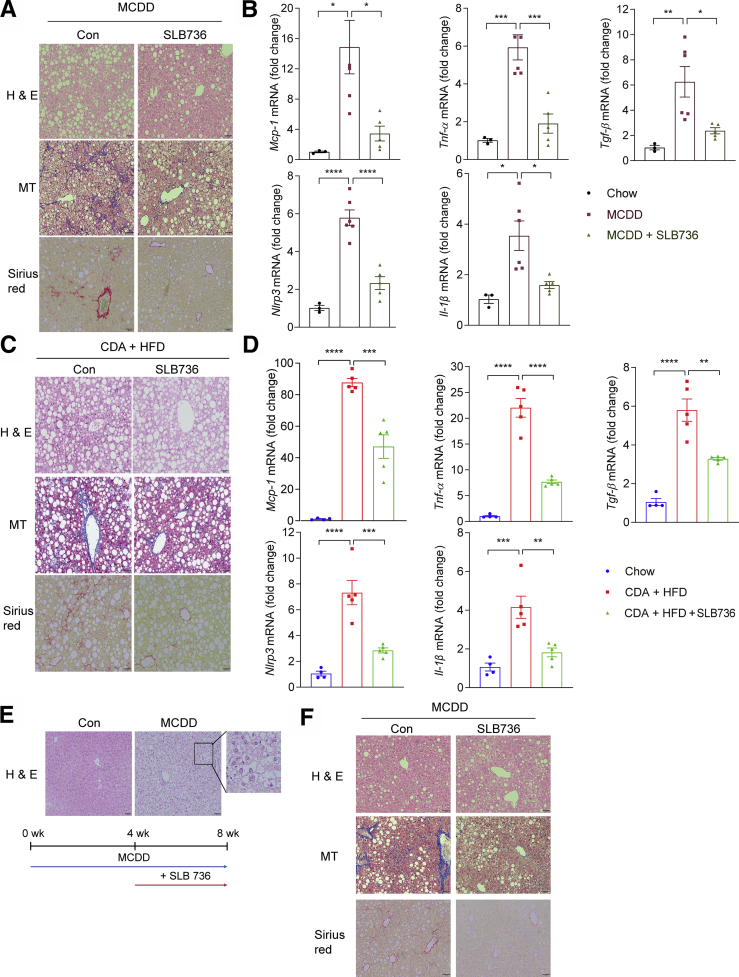
We further investigated whether SLB736 shows a similar preventive effect in other diet-induced murine models of NASH. We found that similar to HFHCD-fed mice, mice fed MCDD or CDA+HFD developed NASH along with increases in the expression of inflammation and inflammasome markers, which effectively were nullified by the administration of SLB736 (Figure 9A–D). To further show the therapeutic effect of SLB736, we administered SLB736 to mice fed MCDD for 4 weeks, a time point at which hepatic steatosis was evident (Figure 9E). Administration of SLB736 for 4 weeks ameliorated NASH and fibrosis in these mice (Figure 9F). Thus, SLB736 was effective in preventing the development of NASH in different nutritional models.
Figure 9.
SLB736 prevents NASH in other animal models and slowsthe progression to NASH and fibrosis. (A) Representative H&E, Masson's trichrome, and Sirius Red staining of the livers of MCDD-fed mice with or without SLB736 treatment for 8 weeks. Scale bars: 50 μm. (B) Relative mRNA expression levels of inflammation, fibrosis, and inflammasome markers in the livers of chow-fed mice and MCDD-fed mice with or without SLB736 (n = 3–6). (C) Representative H&E, MT, and Sirius Red staining of the livers of CDA+HFD–fed mice with or without SLB736 treatment for 6 weeks. Scale bars: 50 μm. (D) Relative mRNA expression levels of markers for inflammation, fibrosis, and inflammasome in the livers of chow-fed mice and CDA+HFD–fed mice with or without SLB736 treatment (n = 4–5). (E) Representative H&E of livers of MCDD-fed mice for 4 weeks. Scale bars: 50 μm. Inlet shows the lipid accumulation in hepatocytes. Lower panel: Schematic schedule for observing the therapeutic effect of SLB736 in MCDD-fed mice. (F) Representative H&E, MT, and Sirius Red staining of the livers. Scale bars: 50 μm. After feeding MCDD for 4 weeks, SLB736 (1 mg/kg/d) was administrated for 4 weeks with MCDD. All data are shown as means ± SEM. (B and D) Data were analyzed by one-way analysis of variance followed by Bonferroni correction. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. Con, control.
SLB736 Suppresses the NLRP3 Inflammasome in Hepatic Macrophages
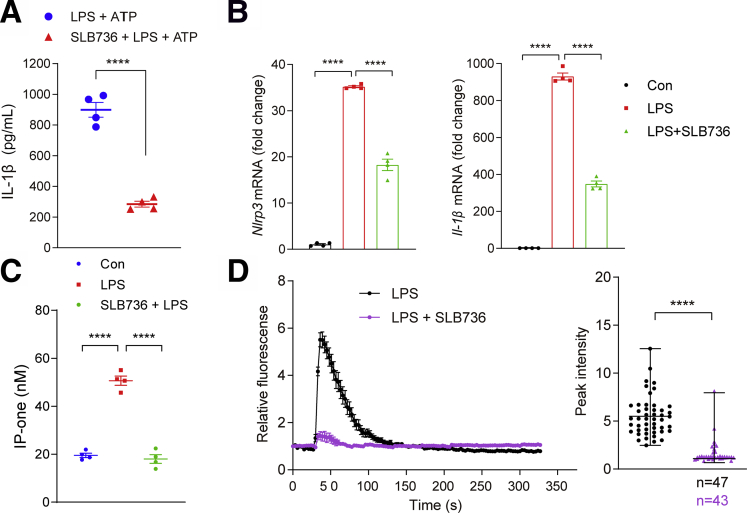
We next checked the effect of SLB736 on the NLRP3 inflammasome in hepatic macrophages. As expected, SLB736 decreased the production of IL-1β in response to LPS and ATP, and the expression of Nlrp3 and Il-1β in response to LPS (Figure 10A and B). Consistently, LPS-induced increase in the level of IP3 (IP-one), the product of PLC, was decreased significantly in cells treated with SLB736 (Figure 10C). Pretreatment with SLB736 inhibited the LPS-induced [Ca++] release in hepatic macrophages (Figure 10D). These data suggest that SLB736 acts as a functional antagonist of S1PR4, thereby leading to the inactivation of NLRP3 inflammasome and prevention of NASH development.
Figure 10.
SLB736 decreases NLRP3 inflammasome activation in hepatic macrophages. (A) IL-1β levels in the culture media of hepatic macrophages. Hepatic macrophages were pretreated with 1 μmol/L SLB736 for 2 hours, and treated with LPS 100 ng/mL for 3 hours followed by ATP (1 mmol/L) for 30 minutes. (B) Relative mRNA expression of Nlrp3 and Il-1β. Hepatic macrophages were pretreated with 1 μmol/L SLB736 for 2 hours, and treated with LPS 100 ng/mL for 3 hours (n = 4). (C) IP-one levels in hepatic macrophages. Cells were pretreated with 1 μmol/L SLB736 or vehicle for 2 hours (n = 4). The levels of IP-one in cell lysates was measured by an ELISA kit. (D) Effect of SLB736 on LPS-mediated [Ca++] release. Hepatic macrophages were pretreated with 1 μmol/L SLB736 or vehicle for 2 hours. Cells then were incubated with Fluo-4/AM followed by stimulation with LPS. [Ca++] was analyzed by time-lapse confocal microscopy (left panel). Quantification of LPS-induced peak fluorescent intensities (right panel). All data are shown as means ± SEM. (A and D) Data were analyzed by Student two-tailed unpaired t test. (B and C) Data were analyzed by one-way analysis of variance with Bonferroni correction. ∗∗∗∗P < .0001. Con, control.
Discussion
S1PR4 is expressed specifically in myeloid cells such as dendritic cells and macrophages.31,32 S1PR4 also is required for the differentiation of plasmacytoid dendritic cells31 and regulates the production of interferon-α thereof.47 However, compared with other S1PRs, our knowledge of the physiological relevance of S1PR4 has been modest.45
We found that the S1PR4 expression was significantly higher in the livers of both diet-induced NASH animal models and patients with NASH undergoing liver transplantation. This is in line with a previous study that reported the up-regulation of S1PR4 in human samples of liver cirrhosis.48 In addition, our data showed that genetic depletion of S1pr4 protected the mice against hepatic inflammation and fibrosis. Hepatocyte death and inflammation are the critical trigger of NASH, which sequentially activates HSCs.7,49 Interestingly, S1pr4 expression was induced significantly by HFHCD feeding in hepatic macrophages, whereas it was not detected in hepatocytes and HSCs. A recent study reported that S1PR2 is involved in the NLRP3 inflammasome activation in hepatic macrophages during chronic liver injury.15 In the present study, we found that S1PR4 in hepatic macrophages also plays an important role in the pathogenesis of NASH by activating the NLRP3 inflammasome.
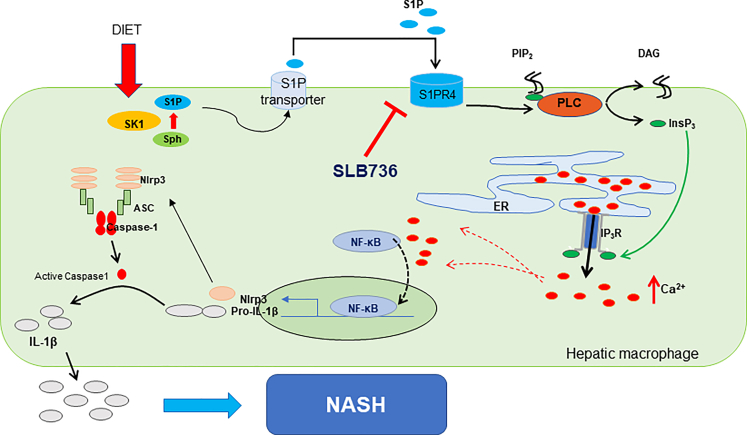
As a major reservoir of intracellular [Ca++], the ER plays a critical role in the regulation of intracellular [Ca++] regulation.37 Activation of the IP3R, a Ca++-release channel on the ER surface, is triggered by IP3, a product of PLC-mediated phosphatidylinositol 4, 5-bisphosphate cleavage. We found that LPS sequentially activated PLC and IP3R in hepatic macrophages to increase [Ca++] and to activate the NLRP3 inflammasome, and that this reaction was abrogated by genetic depletion of S1pr4 or treatment with SLB736. In addition to LPS, we found that S1P also can activate the NLRP3 inflammasome. Interestingly, expression of Sk1 but not Sk2 was induced selectively in hepatic macrophages by HFHCD feeding, and S1P increased the expression of S1pr4 in hepatic macrophages. Accordingly, a previous study showed that the overloading of saturated fatty acids induces Sk1 in hepatocytes to initiate proinflammatory signaling.50 On the other hand, in HFHCD-fed mice, Sk1 expression was induced in hepatic macrophages, but not in hepatocytes. We thus suggest that S1P produced by SK1 from hepatic macrophages induces S1PR4 in a paracrine manner to activate the NLRP3 inflammasome (Figure 11).
Figure 11.
Conceptual model showing the role of the SK1/S1PR4 axis in the pathogenesis of NASH. S1P produced by SK1 from hepatic macrophages induces S1PR4 in a paracrine manner, which is necessary for the [Ca++]-dependent priming of the NLRP3 inflammasome. ASC, Apoptosis-associated speck-like protein containing a C-terminal caspase-recruitment domain; DAG, diacylglycerol; Sph, sphingosine.
In our study, we used mice with heterozygous mutation of S1PR4 considering that homozygous mutation of S1PR4 leads to embryonic lethality. Interestingly, some studies reported the use of adult S1pr4 homozygote knockout mice.31,51,52 Among them, 1 study used S1pr4 knockout mice that were bred in a BALB/C background,52 whereas our mice were bred in a C57BL/6J background. Therefore, it may be possible to study the effects of S1pr4 homozygous knockout in NASH by using BALB/C mice, albeit the results may have limited implications owing to the distinct genetic and immunologic characteristics of BALB/C mice. Nevertheless, our study showed the importance of S1PR4 in NASH because the deletion of 1 copy of the S1PR4 gene was effective in preventing hepatic inflammation and fibrosis in nutritional NASH. We also studied the effect of S1PR4 modulation by shRNA-induced knockdown, which resulted in dose-dependent protection against LPS- or ATP-induced inflammasome activation in primary hepatic macrophages. Our findings should be confirmed in future studies using WT, S1pr4+/-, and S1pr4-/- mice to determine whether S1PR4 confers a protective effect on hepatic inflammation and fibrosis in a dose-dependent manner in vivo.
An important highlight of the present study was the design and synthesis of heteroatom-containing FTY720 analogues with the aim of obtaining high S1PR4 activity and subtype selectivity. Among the synthesized analogues, SLB736 showed an excellent subtype selectivity and functional antagonism for S1PR4. The administration of SLB736 prevented the development of NASH and fibrosis. Interestingly, SLB736 did not affect the expression of S1PR4 at the mRNA level, although the protein levels of S1PR4 in the liver were decreased significantly by SLB736. Notably, SLB736 did not induce lymphopenia, a potentially serious side effect of FTY720 arising from its action on S1PR1,46,53 highlighting the relevance of SLB736 as a novel therapy for NASH.
An interesting finding of our study was that both S1pr4 heterozygous knockout and SLB736 treatment did not affect the degree of hepatic steatosis in HFHCD-fed mice, while effectively preventing diet-induced liver injury, inflammation, and fibrosis. This may be explained by the fact that the primary effect of SLB736 is on NLRP3, which is dominant in hepatic macrophages. Accordingly, similar findings also were observed in Nlrp3 knockout mice.
NAFLD occurs mostly in obese individuals, and insulin resistance and deregulation of lipid metabolism increase the risk of NAFLD and NASH.1 Although lifestyle modification is the first-line treatment for patients with NASH, it usually is unsuccessful. Therefore, many agents for the treatment of NASH by targeting different pathways are under development,2 and several compounds have shown promising histologic results in phase IIa studies.3,54 However, it was pointed out that histologic NASH is not an independent predictor of long-term mortality and that the stage of fibrosis is the only robust and independent predictor of liver-related mortality.3,55 In this regard, targeting NLRP3 inflammasome activation, which plays a central role in hepatic inflammation and fibrosis, increasingly is recognized as a promising strategy for developing an efficient therapy against NASH.28,29 In accordance, our study showed that SLB736 was effective in preventing the development of NASH and fibrosis by deactivating the NLRP3 inflammasome.
Materials and Methods
All authors had access to the study data and reviewed and approved the final manuscript.
Mice and Diet
Mice were housed at ambient temperature (22°C ± 1°C) with a 12:12-hour light-dark cycle and free access to water and food. All animal use and experiment protocols were approved by the Institutional Animal Care and Use Committee of the Asan Institute for Life Sciences (Seoul, Korea).
Eight-week-old male C57BL/6J mice were fed either normal chow diet (12% energy from fat), CDA+HFD containing 60 kcal fat and 0.1% methionine (A06071302; Research Diets, New Brunswick, NJ) for 6 weeks, MCDD (Dyets, Inc, Bethlehem, PA) for 8 weeks, or HFHCD (60% energy from fat and 2.5% cholesterol; Dyets, Inc) for 12 weeks. In another group, mice were fed a WD (TD.120330, 0.2% cholesterol + 22% hydrogenated vegetable oil, Envigo RMS, Inc, Indianapolis, IN) supplemented with high-fructose syrup in the drinking water for 16 weeks.56 After the indicated time of diet feeding, mice were fasted for 5 hours in the morning before they were killed.
S1pr4+/- mice were purchased from Jackson Laboratories (mouse strain 005799; Bar Harbor, ME). Eight-week-old male S1pr4+/- mice and their littermate controls (S1pr4+/+) were fed either a normal chow diet or HFHCD for 4 or 12 weeks. Considering the description on the MGI (mouse genome informatics) website (1333809) on the embryonic lethality of the homozygous mutation of the S1pr4 gene, we used heterozygote mice for this study and observed the deletion of 1 copy of the S1pr4 gene. Nlrp3 knockout mice were generated using the transcription activator-like effector nucleases method.57
Human Liver Samples
Human liver samples were obtained from the liver explants of donors and recipients diagnosed with NASH/liver cirrhosis undergoing liver transplantation at the Liver Transplantation Unit of the Hospital Clinic (Barcelona, Spain). Healthy liver tissues were obtained from the surgical specimens of donor livers used for transplantation. Biopsy of the resected livers from the recipients was performed immediately after the hepatectomy and the samples were fixed in formalin for histologic examination. The clinical data of the patients are presented in Table 1. The protocol was approved by the Hospital Clinic/University of Barcelona Ethics Committee (HCB/2012/8011) of the Hospital Clinic (Barcelona, Spain).
Isolation of Hepatocytes and Hepatic Macrophages
Hepatocytes and hepatic macrophages were isolated from mice by collagenase digestion, gradient centrifugation, and selective adherence,58 with modifications. Briefly, the mice were anesthetized and the peritoneal cavity was opened; the livers were perfused with Ca++ and Mg++-free Hank's balanced salt solution (LB 003-04; Welgene, Daegu, Korea) containing collagenase (17101-015; Gibco, Carlsbad, CA) and trypsin inhibitor (T2011; Sigma-Aldrich, St. Louis, MO). The digested livers were removed and placed in 60-mm Petri dishes. The livers were frittered with forceps in RPMI1640 (LM 011-01; Welgene) supplemented with 10% (vol/vol) fetal bovine serum (FBS) (16000-044; Gibco). The cell suspensions were filtered through a sterile Falcon 100-μm nylon cell strainer (352360; Corning Inc, NY) to remove undigested tissues and connective tissues. The cells were centrifuged at 50 × g for 3 minutes at room temperature. For isolation of hepatocytes, the pellet was resuspended in Dulbecco’s modified Eagle medium with 36% Percoll (Sigma-Aldrich), and then centrifuged at 50 × g for 10 minutes at room temperature. The pellet then was washed 2 times with phosphate-buffered saline (PBS). Finally, the hepatocytes were plated on collagen-coated tissue culture dishes in Dulbecco’s modified Eagle medium cell culture medium supplemented with 10% FBS and 1% penicillin/streptomycin solution, and kept in a humidified cell culture incubator with 5% CO2 at 37ºC.
For isolation of hepatic macrophages, the supernatants were transferred to clean 50-mL tubes. The supernatants were centrifuged at 1600 rpm (4°C) for 10 minutes, and the cell pellets were resuspended in 20% OptiPrep and gently layered on OptiPrep gradient (20%, 11.5%, and Hank's balanced salt solution) and centrifuged at 3000 rpm at 4°C for 17 minutes with the brake option off. Subsequently, the upper layers were removed and the cell fraction between 20% OptiPrep and 11.5% OptiPrep gradient were collected without contamination from the pellets. The collected layers were washed twice with RPMI1640 supplemented with 10% (vol/vol) FBS, and plated into 12-well or 24-well tissue culture plates. At 10 minutes after seeding, nonadherent cells (cell debris or blood cells) were removed by aspiration and fresh media were added. The next day, the cells were washed twice with 1× PBS, and the attached hepatic macrophages were cultured for another 48 hours, at which point they were ready for experimental use.
Hepatic macrophages were identified by flow cytometry using a monoclonal anti-F4/80 antibody. Briefly, after 48 hours of culture, macrophages were detached by incubation with 0.25% trypsin for 5 minutes, and pelleted by centrifugation for 5 minutes at 100 rpm/min. The cells then were incubated with anti-F4/80 antibody (clone BM8)-conjugated phycoerythrin (12-4801-82; Invitrogen, Carlsbad, CA) for 30 minutes at 4°C (1:200 dilution). The data were collected using FACSCanto2 (BD Bioscience, San Jose, CA) and analyzed with FlowJo software (BD Bioscience, San Jose, CA).
Isolation of Macrophages From the Spleen and BM
Spleens were excised and digested for 30 minutes with collagenase (Sigma-Aldrich) at 37°C while shaking. Cell suspensions were filtered through a 70-μm sieve and centrifuged at 450 × g for 5 minutes. Femurs were collected in RPMI, and bone marrow cells were flushed from the femurs and then depleted of red blood cells using red blood cell lysis buffer (R7757; Sigma-Aldrich). For macrophage staining, cells were incubated with Fc (fragment crystallizable) Block (101302; BioLegend, San Diego, CA) for 10 minutes in ice, and then washed and stained with anti-F4/80 antibody (12-4801-82; Invitrogen) and anti-CD45.2 antibody (45-0454-82; Invitrogen) for 30 minutes on ice in the dark. Macrophages were sorted as live CD45.2 and F4/80 double-positive cells into RPMI supplemented with 20% FBS using FACS Aria2 (BD Bioscience).
Primary HSC Isolation
Selective macrophage depletion was achieved with a single intraperitoneal injection of clodronate (20 mg/mL) according to the manufacturer’s instructions (FormuMax Scientific, Inc, Sunnyvale, CA). After 24 hours, primary HSCs were isolated using the same protocol used in the isolation of hepatic macrophages, and the cell fraction between the upper layer and the 20% OptiPrep gradient were collected without contamination from the pellets. After centrifugation, the cells were seeded into culture plates in Dulbecco's modified Eagle medium containing fetal serum at 37°C. The culture medium was changed and the RNA was isolated from primary HSCs.59
Histologic Analysis
Liver tissue samples were fixed in 10% neutral buffered formalin and embedded in paraffin. Serial sections (5-μm thick) were stained with H&E, Masson’s Trichrome, or Sirius Red, as appropriate.
Liver TG Contents
TG content in the livers was determined in duplicate using the triglyceride kit (GPO-Trinder; Sigma-Aldrich).
Real-Time Polymerase Chain Reaction Analysis
Total RNA isolated from each sample was reverse-transcribed and the target complementary DNA levels were quantified by real-time polymerase chain reaction analysis using gene-specific primers (Table 2). Total RNA was isolated using TRIzol (Invitrogen), and 1 μg of each sample was reverse-transcribed with random primers using the Reverse Aid M-MuLV Reverse-Transcription Kit (Fermentas, Amherst, NY). The relative expression levels of each gene were normalized to that of 18S rRNA, Tbp, or ACTB.
Table 2.
Reverse-Transcription PCR Primers
| Primer | Forward | Reverse |
|---|---|---|
| Reverse-transcription PCR primers (mice) | ||
| Tbp | CCTTCACCAATGACTCCTATGAC | CAAGTTTACAGCCAAGATTCAC |
| 18S rRNA | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGC |
| Tnf-α | GAGAAAGTCAACCTCCTCTCTG | GAAGACTCCTCCCAGGTATATG |
| Mcp-1 | ACGGCATGGATCTCAAAGAC | AGATAGCAAATCGGCTGACG |
| Tgf-β | TATAGCAACAATTCCTGGCG | CCTGTATTCCGTCTCCTTG |
| α-Sma | ACTGGGACGACATGGAAAAG | GTTCAGTGGTGCCTCTGTCA |
| Col3a1 | GGGTTTCCCTGGTCCTAAAG | CCTGGTTTCCCATTTTCTCC |
| Nlrp3 | ATTACCCGCCCGAGAAAGG | TCGCAGCAAAGATCCACACAG |
| Il-1β | TCTTTGAAGTTGACGGACCC | TGAGTGATACTGCCTGCCTG |
| S1pr1 | ATGGTGTCCACTAGCATCCC | CGATGTTCAACTTGCCTGTGTAG |
| S1pr2 | ATGGGCGGCTTATACTCAGAG | GCGCAGCACAAGATGATGAT |
| S1pr3 | ACTCTCCGGGAACATTACGAT | CAAGACGATGAAGCTACAGGTG |
| S1pr4 | GGGTGTACTACTGCCTGCTG | AGCAGACTGAAGGTGGATGC |
| S1pr5 | GCTTTGGTTTGCGCGTGAG | GGCGTCCTAAGCAGTTCCAG |
| Sk1 | CCATCCAGAAACCCCTGTGT | ACCTGCTCGTACCCAGCATAGT |
| Sk2 | AGACGGGCTGCTTTACGAG | CAGGGGAGGACACCAATG |
| Reverse-transcription PCR primers (human) | ||
| S1PR4 | GACGCTGGGTCTACTATTGCC | CCTCCCGTAGGAACCACTG |
| ACTB | TGGTGATGGAGGAGGTTTAGTAAGT | AACCAATAAAACCTACTCCTCCCTTAA |
Western Blot Analysis
Cell and liver samples were homogenized in lysis buffer (50 mmol/L Tris, pH 7.4, 150 mmol/L KCl, 4 mmol/L EDTA, 4 mmol/L ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid and 1% NP-40 containing protease [04693132001; Roche, Carlsbad, CA] and phosphatase [04906837001; Roche] inhibitor mixture tablets) at 4°C for 30 minutes. The resulting protein (40–50 μg) was subjected to immunoblotting with primary antibodies: antibodies against phosphorylated NF-κB and NF-κB were purchased from Cell Signaling (Danvers, MA). Anti-S1PR4 antibody was purchased from Novus (Centennial, CO). β-actin (A5441; Sigma-Aldrich) was used as housekeeping control (Table 3). The signal intensities of protein bands were quantified with ImageJ software (National Institutes of Health, Bethesda, MD) and normalized using the intensity of the loading control.
Table 3.
Antibodies
| NF-κB | Cell Signaling Technology | Cat# 6956, RRID:AB_10828935 |
| Phospho–NF-κB | Cell Signaling Technology | Cat# 3033, RRID:AB_331284 |
| S1PR4 | Novus | Cat# NBP1-00795, RRID:AB_1503063 |
| β-actin | Sigma-Aldrich | Cat# A5441, RRID:AB_476744 |
| HRP goat anti-mouse IgG | BioLegend | Cat# 405306, RRID:AB_315009 |
| HRP donkey anti-rabbit IgG | BioLegend | Cat# 406401, RRID:AB_2099368 |
| F4/80 | Invitrogen | Cat# 12-4801-82, RRID:AB_465923 |
| CD45.2 | Invitrogen | Cat# 45-0454-82 RRID: AB_953590 |
HRP, horseradish peroxidase.
IL-1β Measurement
Mouse IL-1β in cell culture supernatants was measured using the mouse IL-1β/IL1F2 Quantikine enzyme-linked immunosorbent assay (ELISA) kit (DY401; R&D Systems, Minneapolis, MN).
Lentiviral-Silencing S1pr4
The shRNA sequences for S1pr4 were as follows: Forward, 5′- GCC TGC TGA ACA TCA CAC TGA TCA AGA GTC AGT GTG ATG TTC AGC AGG CTT TTT TG-3′; Reverse: 5′- CAA AAA AGC CTG CTG AAC ATC ACA CTG ACT CTT GAT CAG TGT GAT GTT CAG CAG GC-3′. S1pr4 shRNA was subcloned into the pCDH-MCS lentiviral vector (CD513B-1; System Biosciences, Mountain View, CA) and the plasmids were transfected in Lenti-X 293T cells (632180; Clontech, Mountain View, CA), along with the packaging plasmids pMDLg/pRRE (12251; Addgene, Cambridge, MA) and pRSV-Rev (12253; Addgene) and the envelope plasmid pCMV-VSV-G (8454, Addgene) using Lipofectamine 3000 (L30000015; Invitrogen). Hepatic macrophages were infected with lentiviral vectors coding S1pr4 shRNA for 12 hours at a range of multiplicity of infection (ie, 0, 10, and 50). The medium was changed after 12 hours.
Intracellular IP-One Measurement
PLC activity was tested with the IP-one ELISA (72IP1PEA; Cisbio, Bedford, MA), in which hepatic macrophages were stimulated with LPS or S1P and then the cell culture medium was replaced with fresh medium. Intracellular IP-one, a surrogate measure for the level of inositol triphosphate, was measured after treatment with LiCl (50 mmol/L) to prevent the degradation of IP-one into myo-inositol. The level of inositol triphosphate in cell lysates was measured using an ELISA.
Calcium Analysis by Confocal Microscopy
Hepatic macrophages were plated on a 35-mm imaging dish (81156; Ibidi, Gräfelfing, Germany) at a density of 0.1 × 106 cells and incubated with Fluo-4/AM (F36206; Invitrogen). Images of untreated cells were acquired at t = 0, and the cells were treated with 1 μg/mL LPS or 1 mmol/L ATP in RPMI1640. The cells were imaged for 5 minutes at 5-second intervals on a Zeiss LSM780 Confocal Imaging System (Carl Zeiss, Oberkochen, Germany) using the 488-nm laser and emission in the range of 500–600 nm. The images were analyzed using Zen 2012 SP5 software (Carl Zeiss, Oberkochen, Germany) by creating surfaces to encompass the volume of each cell. The absolute intensity for all cells in a field at different time points was obtained, and normalized to t = 0 to calculate the fold increases in intensity. Data are shown as the relative intensity of cells in a field.
Reagents for Calcium Signaling
BAPTA-AM (A1076;Sigma-Aldrich), U73122 (U6756; Sigma-Aldrich), 2-APB (D9754; Sigma-Aldrich), and Xes-c (X2628; Sigma-Aldrich) were used for detecting calcium signaling.
S1P Treatment in Hepatic Macrophages
Hepatic macrophages were serum-starved for 6 hours and then stimulated with 1 μmol/L S1P (S9666; Sigma-Aldrich) for 2 hours.
Synthesis of SLB736
The chemical and spectroscopic data are as follows: melting point 130 °C; 1H NMR (400 MHz, CD3OD) δ 8.35 (s, 1H), 4.53 (t, J = 7.2 Hz, 2H), 3.69 (s, 4H), 2.97 (td, J = 4.3, 8.0 Hz, 2H), 2.11 (td, J = 4.3, 8.0 Hz, 2H), 2.00–1.95 (m, 2H), 1.35–1.28 (m, 14H), 0.89 (t, J = 6.8 Hz, 3H); 13C NMR (100 MHz, CD3OD) δ 146.5, 127.3, 63.0 (2C), 62.6, 54.3, 33.8, 32.0, 31.5, 31.4, 31.3, 31.2, 30.8, 28.1, 24.5, 19.9, 15.2; IR (neat) υmax = 3180, 2918, 2851, 2421, 1599, 1454, 1080, 1063, 958, and 715 (cm–1); HRMS (FAB) calculated for C17H35N4O2 [M−Cl−]+ 327.2760, found 327.2762. (NMR: nuclear magnetic resonance; s: singlet; d: doublet; t: triplet; td: triplet of doublets; m: multiplet; H: proton; J: coupling constant; C: carbon; IR: infrared spectroscopy; υmax: lambda max; HRMS: high-resolution mass spectrometry; FAB: fast atom bombardment).
Treatment With SLB736 or FTY720 In Vivo
Mice were administered SLB736, FTY720 (1 mg/kg body weight each), or vehicle (0.9% NaCl) via oral gavage every day for 5 d/wk for the indicated periods. After the indicated period of treatment, the mice were fasted overnight and killed. The liver tissues were quickly removed and kept frozen at -70°C for subsequent analysis.
SLB736 Treatment In Vitro
Hepatic macrophages were treated with chemicals at the indicated doses or sterile water (control) for 2 hours. After washing twice with PBS, the cells were stimulated with LPS (L2880; Sigma-Aldrich) at a concentration of 100 ng/mL for 3 hours, and then 1 mmol/L ATP (A6419; Sigma-Aldrich) was added for 30 minutes.
Determination of S1PR4 Localization in a C6 Glioma Cell Line
Stable C6 glioma cells expressing EGFP-conjugated S1PR4 were prepared by infection with retrovirus bearing S1PR4–EGFP fusion construct (kindly provided by Dr Jerold Chun at Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA). S1PR4 internalization and recycling were assessed as previously described.44 In brief, cells were plated on poly-L-lysine (100 μg/mL)-coated coverslips, cultivated, serum-deprived, and then used for experiments. The cells were treated with vehicle (0.1% fatty acid-free bovine serum albumin), S1P, FTY720-P, or SLB736 for 0.5 hours; in some cases, the cells were washed and further incubated in the presence of cycloheximide (5 μg/mL) for 2 or 4 hours. At the end of each experiment, the cells were fixed in 4% paraformaldehyde and mounted with Vectashield (H-1000; Vector Laboratories, Burlingame, CA). S1PR4 localization in cells was assessed by detecting the EGFP signal using laser scanning confocal microscopy (Eclipse A1+; Nikon, Tokyo, Japan).
S1PR β-Arrestin Assay
β-arrestin recruitment assays for S1PR activity were performed by DiscoveRx (Fremont, CA).
Blood Lymphocyte Measurement
Blood lymphocytes were counted using an automated hematology analyzer (ADVIA 2120i 53; Siemens Healthcare Diagnostics, Tarrytown, NY).
Statistical Analysis
Data are expressed as means ± SEM. Unpaired two-tailed Student t tests were used to compare variables between groups, and one-way analysis of variance was used to compare variables among multiple groups. Bonferroni correction was applied for post hoc analysis of the multiple comparisons. All statistical tests were conducted according to two-sided sample sizes and were determined on the basis of previous experiments that used similar methodologies. For all experiments, the stated replicates are biological replicates. Statistical analysis and graphing were performed using IBM SPSS Statistics for Windows version 22.0 (IBM Corp, Armonk, NY) or GraphPad Prism 7 (GraphPad Software, La Jolla, CA).
Acknowledgments
The authors thank Joon Seo Lim, PhD, ELS, from the Scientific Publications Team at Asan Medical Center for his editorial assistance in preparing this manuscript.
CRediT Authorship Contributions
Chung Hwan Hong (Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Writing – original draft: Equal)
Myoung Seok Ko (Data curation: Lead; Formal analysis: Lead; Supervision: Equal)
Jae Hyun Kim (Data curation: Lead; Formal analysis: Lead; Methodology: Lead; Resources: Lead)
Hyunkyung Cho (Data curation: Equal; Methodology: Equal; Resources: Equal)
Chi-Ho Lee (Data curation: Equal; Formal analysis: Equal; Methodology: Equal)
Ji Eun Yoon (Data curation: Equal; Investigation: Equal; Methodology: Equal)
Ji-Young Yun (Data curation: Supporting; Methodology: Supporting; Validation: Supporting)
In-Jeoung Baek (Methodology: Equal; Resources: Equal; Validation: Equal)
Jung Eun Jang (Investigation: Equal; Supervision: Equal; Validation: Equal; Writing – original draft: Equal)
Seung Eun Lee (Investigation: Equal; Supervision: Equal)
Yun Kyung Cho (Investigation: Supporting; Supervision: Equal; Validation: Equal)
Ji Yeon Baek (Investigation: Equal; Validation: Equal)
Soo Jin Oh (Data curation: Equal; Resources: Equal; Visualization: Equal)
Bong Yong Lee (Supervision: Equal; Writing – review & editing: Equal)
Joon Seo Lim (Validation: Equal; Visualization: Equal; Writing – review & editing: Equal)
Jongkook Lee (Conceptualization: Equal; Resources: Equal; Writing – review & editing: Equal)
Sean M. Hartig (Writing – review & editing: Equal)
Laura Conde de la Rosa (Data curation: Equal; Formal analysis: Equal; Resources: Lead)
Carmen Garcia-Ruiz (Data curation: Equal; Methodology: Equal; Resources: Lead)
Ki-Up Lee (Conceptualization: Lead; Funding acquisition: Lead; Writing – original draft: Lead)
Jose C. Fernández-Checa (Data curation: Lead; Formal analysis: Lead; Funding acquisition: Supporting; Resources: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Ji Woong Choi (Methodology: Lead; Resources: Lead; Software: Lead; Writing – original draft: Lead)
Sanghee Kim (Conceptualization: Lead; Funding acquisition: Lead; Methodology: Lead; Resources: Lead; Writing – original draft: Lead)
Eun Hee Koh (Conceptualization: Lead; Formal analysis: Lead; Funding acquisition: Lead; Supervision: Lead; Writing – original draft: Lead)
Footnotes
Conflicts of interest These authors disclose the following: Eun Hee Koh, Sanghee Kim, and Ki-Up Lee have filed a provisional patent application in the Korea patent office (10-2017-00040139: composition for preventing and treating nonalcoholic steatohepatitis by targeting S1PR4). The remaining authors disclose no conflict.
FundingSupported by the National Research Foundation of Korea, funded by the Ministry of Education, Science, and Technology, Korea, grants 2017R1E1A1A01073206 (K.-U.L.), 2020R1A2B5B02098524 (E.H.K.), 2019R1C1C1003567 (J.-Y.Y.), and 2021R1A2C1005520 (J.W.C.), the Korea Drug Development Fund funded by the Ministry of Science and ICT, Ministry of Trade, Industry, and Energy, and Ministry of Health and Welfare grant KDDF-201406-03 (K.-U.L.), and Asan Institute for Life Sciences, Korea, grant 2020IP0016-1 (E.H.K.). This work also was supported by the Mid-Career Researcher Programs grant NRF-2019R1A2C2009905 (S.K.) of the National Research Foundation of Korea, funded by the Government of Korea (MSIP); and by grants PID2019-111669RB-I00 and PID2020-115055RB-I00 from Plan Nacional de Investigación y Desarrollo, Spain, by the Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas, Instituto de Salud Carlos III, Spain; by grant P50AA011999 from the Southern California Research Center for Alcoholic Liver and Pancreatic Diseases and Cirrhosis funded by National Institute of Alcohol Abuse and Alcoholism; by Agency for Administration of University and Research Grants of the Generalitat de Cataluña SGR-2017-1112 and the “ER stress-mitochondrial cholesterol axis in obesity-associated insulin resistance and comorbidities” Ayudas Fundación Banco Bilbao Vizcaya Argentaria a Equipos de Investigación Científica 2017, the Red Nacional 2018-102799-T de Enfermedades Metabólicas y Cáncer, and by project 201916/31 Contribution of mitochondrial oxysterol and bile acid metabolism to liver carcinogenesis 2019 by Fundació Marato Televisión 3. None of the funders had a role in the study design, collection, analysis, or interpretation of data.
Contributor Information
Jose C. Fernández-Checa, Email: checa229@yahoo.com.
Ji Woong Choi, Email: pharmchoi@gachon.ac.kr.
Sanghee Kim, Email: pennkim@snu.ac.kr.
Eun Hee Koh, Email: ehk@amc.seoul.kr.
References
- 1.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lassailly G., Caiazzo R., Pattou F., Mathurin P. Perspectives on treatment for nonalcoholic steatohepatitis. Gastroenterology. 2016;150:1835–1848. doi: 10.1053/j.gastro.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Younossi Z.M., Loomba R., Rinella M.E., Bugianesi E., Marchesini G., Neuschwander-Tetri B.A., Serfaty L., Negro F., Caldwell S.H., Ratziu V., Corey K.E., Friedman S.L., Abdelmalek M.F., Harrison S.A., Sanyal A.J., Lavine J.E., Mathurin P., Charlton M.R., Chalasani N.P., Anstee Q.M., Kowdley K.V., George J., Goodman Z.D., Lindor K. Current and future therapeutic regimens for nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology. 2018;68:361–371. doi: 10.1002/hep.29724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machado M.V., Diehl A.M. Pathogenesis of nonalcoholic steatohepatitis. Gastroenterology. 2016;150:1769–1777. doi: 10.1053/j.gastro.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koh E.H., Yoon J.E., Ko M.S., Leem J., Yun J.Y., Hong C.H., Cho Y.K., Lee S.E., Jang J.E., Baek J.Y., Yoo H.J., Kim S.J., Sung C.O., Lim J.S., Jeong W.I., Back S.H., Baek I.J., Torres S., Solsona-Vilarrasa E., Conde de la Rosa L., Garcia-Ruiz C., Feldstein A.E., Fernandez-Checa J.C., Lee K.U. Sphingomyelin synthase 1 mediates hepatocyte pyroptosis to trigger non-alcoholic steatohepatitis. Gut. 2021;70:1954–1964. doi: 10.1136/gutjnl-2020-322509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grabherr F., Grander C., Effenberger M., Adolph T.E., Tilg H. Gut dysfunction and non-alcoholic fatty liver disease. Front Endocrinol (Lausanne) 2019;10:611. doi: 10.3389/fendo.2019.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seki E., Schwabe R.F. Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology. 2015;61:1066–1079. doi: 10.1002/hep.27332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cartier A., Hla T. Sphingosine 1-phosphate: lipid signaling in pathology and therapy. Science. 2019;366 doi: 10.1126/science.aar5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F., Okamoto Y., Inoki I., Yoshioka K., Du W., Qi X., Takuwa N., Gonda K., Yamamoto Y., Ohkawa R., Nishiuchi T., Sugimoto N., Yatomi Y., Mitsumori K., Asano M., Kinoshita M., Takuwa Y. Sphingosine-1-phosphate receptor-2 deficiency leads to inhibition of macrophage proinflammatory activities and atherosclerosis in apoE-deficient mice. J Clin Invest. 2010;120:3979–3995. doi: 10.1172/JCI42315. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Lee S.Y., Hong I.K., Kim B.R., Shim S.M., Sung Lee J., Lee H.Y., Soo Choi C., Kim B.K., Park T.S. Activation of sphingosine kinase 2 by endoplasmic reticulum stress ameliorates hepatic steatosis and insulin resistance in mice. Hepatology. 2015;62:135–146. doi: 10.1002/hep.27804. [DOI] [PubMed] [Google Scholar]
- 11.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18:33–50. doi: 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai H.C., Han M.H. Sphingosine-1-phosphate (S1P) and S1P signaling pathway: therapeutic targets in autoimmunity and inflammation. Drugs. 2016;76:1067–1079. doi: 10.1007/s40265-016-0603-2. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y., Aoki H., Yang J., Peng K., Liu R., Li X., Qiang X., Sun L., Gurley E.C., Lai G., Zhang L., Liang G., Nagahashi M., Takabe K., Pandak W.M., Hylemon P.B., Zhou H. The role of sphingosine 1-phosphate receptor 2 in bile-acid-induced cholangiocyte proliferation and cholestasis-induced liver injury in mice. Hepatology. 2017;65:2005–2018. doi: 10.1002/hep.29076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao Y., Liu R., Li X., Gurley E.C., Hylemon P.B., Lu Y., Zhou H., Cai W. Long noncoding RNA H19 Contributes to cholangiocyte proliferation and cholestatic liver fibrosis in biliary atresia. Hepatology. 2019;70:1658–1673. doi: 10.1002/hep.30698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou L., Yang L., Chang N., Zhao X., Zhou X., Dong C., Liu F., Yang L., Li L. Macrophage sphingosine 1-phosphate receptor 2 blockade attenuates liver inflammation and fibrogenesis triggered by NLRP3 inflammasome. Front Immunol. 2020;11:1149. doi: 10.3389/fimmu.2020.01149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X., Yue S., Li C., Yang L., You H., Li L. Essential roles of sphingosine 1-phosphate receptor types 1 and 3 in human hepatic stellate cells motility and activation. J Cell Physiol. 2011;226:2370–2377. doi: 10.1002/jcp.22572. [DOI] [PubMed] [Google Scholar]
- 17.Yang L., Yue S., Yang L., Liu X., Han Z., Zhang Y., Li L. Sphingosine kinase/sphingosine 1-phosphate (S1P)/S1P receptor axis is involved in liver fibrosis-associated angiogenesis. J Hepatol. 2013;59:114–123. doi: 10.1016/j.jhep.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 18.Mauer A.S., Hirsova P., Maiers J.L., Shah V.H., Malhi H. Inhibition of sphingosine 1-phosphate signaling ameliorates murine nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2017;312:G300–G313. doi: 10.1152/ajpgi.00222.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calabresi P.A., Radue E.W., Goodin D., Jeffery D., Rammohan K.W., Reder A.T., Vollmer T., Agius M.A., Kappos L., Stites T., Li B., Cappiello L., von Rosenstiel P., Lublin F.D. Safety and efficacy of fingolimod in patients with relapsing-remitting multiple sclerosis (FREEDOMS II): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Neurol. 2014;13:545–556. doi: 10.1016/S1474-4422(14)70049-3. [DOI] [PubMed] [Google Scholar]
- 20.Moro-Sibilot L., Blanc P., Taillardet M., Bardel E., Couillault C., Boschetti G., Traverse-Glehen A., Defrance T., Kaiserlian D., Dubois B. Mouse and human liver contain immunoglobulin A-secreting cells originating from Peyer's patches and directed against intestinal antigens. Gastroenterology. 2016;151:311–323. doi: 10.1053/j.gastro.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 21.Rohrbach T.D., Asgharpour A., Maczis M.A., Montefusco D., Cowart L.A., Bedossa P., Sanyal A.J., Spiegel S. FTY720/fingolimod decreases hepatic steatosis and expression of fatty acid synthase in diet-induced nonalcoholic fatty liver disease in mice. J Lipid Res. 2019;60:1311–1322. doi: 10.1194/jlr.M093799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jang J.E., Park H.S., Yoo H.J., Baek I.J., Yoon J.E., Ko M.S., Kim A.R., Kim H.S., Park H.S., Lee S.E., Kim S.W., Kim S.J., Leem J., Kang Y.M., Jung M.K., Pack C.G., Kim C.J., Sung C.O., Lee I.K., Park J.Y., Fernandez-Checa J.C., Koh E.H., Lee K.U. Protective role of endogenous plasmalogens against hepatic steatosis and steatohepatitis in mice. Hepatology. 2017;66:416–431. doi: 10.1002/hep.29039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farrell G., Schattenberg J.M., Leclercq I., Yeh M.M., Goldin R., Teoh N., Schuppan D. Mouse models of nonalcoholic steatohepatitis: toward optimization of their relevance to human nonalcoholic steatohepatitis. Hepatology. 2019;69:2241–2257. doi: 10.1002/hep.30333. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto M., Hada N., Sakamaki Y., Uno A., Shiga T., Tanaka C., Ito T., Katsume A., Sudoh M. An improved mouse model that rapidly develops fibrosis in non-alcoholic steatohepatitis. Int J Exp Pathol. 2013;94:93–103. doi: 10.1111/iep.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strowig T., Henao-Mejia J., Elinav E., Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 26.Davis B.K., Wen H., Ting J.P. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol. 2011;29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwabe R.F., Tabas I., Pajvani U.B. Mechanisms of fibrosis development in nonalcoholic steatohepatitis. Gastroenterology. 2020;158:1913–1928. doi: 10.1053/j.gastro.2019.11.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mridha A.R., Wree A., Robertson A.A.B., Yeh M.M., Johnson C.D., Van Rooyen D.M., Haczeyni F., Teoh N.C., Savard C., Ioannou G.N., Masters S.L., Schroder K., Cooper M.A., Feldstein A.E., Farrell G.C. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J Hepatol. 2017;66:1037–1046. doi: 10.1016/j.jhep.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tilg H., Moschen A.R., Szabo G. Interleukin-1 and inflammasomes in alcoholic liver disease/acute alcoholic hepatitis and nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. Hepatology. 2016;64:955–965. doi: 10.1002/hep.28456. [DOI] [PubMed] [Google Scholar]
- 30.Wree A., McGeough M.D., Inzaugarat M.E., Eguchi A., Schuster S., Johnson C.D., Pena C.A., Geisler L.J., Papouchado B.G., Hoffman H.M., Feldstein A.E. NLRP3 inflammasome driven liver injury and fibrosis: roles of IL-17 and TNF in mice. Hepatology. 2018;67:736–749. doi: 10.1002/hep.29523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dillmann C., Mora J., Olesch C., Brune B., Weigert A. S1PR4 is required for plasmacytoid dendritic cell differentiation. Biol Chem. 2015;396:775–782. doi: 10.1515/hsz-2014-0271. [DOI] [PubMed] [Google Scholar]
- 32.Schuster C., Huard A., Sirait-Fischer E., Dillmann C., Brune B., Weigert A. S1PR4-dependent CCL2 production promotes macrophage recruitment in a murine psoriasis model. Eur J Immunol. 2020;50:839–845. doi: 10.1002/eji.201948349. [DOI] [PubMed] [Google Scholar]
- 33.He Y., Hara H., Nunez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci. 2016;41:1012–1021. doi: 10.1016/j.tibs.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong T., Yang Y., Jin T., Jiang W., Zhou R. Orchestration of NLRP3 inflammasome activation by ion fluxes. Trends Immunol. 2018;39:393–406. doi: 10.1016/j.it.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Lee G.S., Subramanian N., Kim A.I., Aksentijevich I., Goldbach-Mansky R., Sacks D.B., Germain R.N., Kastner D.L., Chae J.J. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature. 2012;492:123–127. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamazaki Y., Kon J., Sato K., Tomura H., Sato M., Yoneya T., Okazaki H., Okajima F., Ohta H. Edg-6 as a putative sphingosine 1-phosphate receptor coupling to Ca(2+) signaling pathway. Biochem Biophys Res Commun. 2000;268:583–589. doi: 10.1006/bbrc.2000.2162. [DOI] [PubMed] [Google Scholar]
- 37.Raffaello A., Mammucari C., Gherardi G., Rizzuto R. Calcium at the center of cell signaling: interplay between endoplasmic reticulum, mitochondria, and lysosomes. Trends Biochem Sci. 2016;41:1035–1049. doi: 10.1016/j.tibs.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiang C.Y., Veckman V., Limmer K., David M. Phospholipase Cgamma-2 and intracellular calcium are required for lipopolysaccharide-induced Toll-like receptor 4 (TLR4) endocytosis and interferon regulatory factor 3 (IRF3) activation. J Biol Chem. 2012;287:3704–3709. doi: 10.1074/jbc.C111.328559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schappe M.S., Szteyn K., Stremska M.E., Mendu S.K., Downs T.K., Seegren P.V., Mahoney M.A., Dixit S., Krupa J.K., Stipes E.J., Rogers J.S., Adamson S.E., Leitinger N., Desai B.N. Chanzyme TRPM7 mediates the Ca(2+) influx essential for lipopolysaccharide-induced Toll-like receptor 4 endocytosis and macrophage activation. Immunity. 2018;48:59–74 e5. doi: 10.1016/j.immuni.2017.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohrbach T., Maceyka M., Spiegel S. Sphingosine kinase and sphingosine-1-phosphate in liver pathobiology. Crit Rev Biochem Mol Biol. 2017;52:543–553. doi: 10.1080/10409238.2017.1337706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanson M.A., Roth C.B., Jo E., Griffith M.T., Scott F.L., Reinhart G., Desale H., Clemons B., Cahalan S.M., Schuerer S.C., Sanna M.G., Han G.W., Kuhn P., Rosen H., Stevens R.C. Crystal structure of a lipid G protein-coupled receptor. Science. 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pham T.C., Fells J.I., Sr, Osborne D.A., North E.J., Naor M.M., Parrill A.L. Molecular recognition in the sphingosine 1-phosphate receptor family. J Mol Graph Model. 2008;26:1189–1201. doi: 10.1016/j.jmgm.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oo M.L., Thangada S., Wu M.T., Liu C.H., Macdonald T.L., Lynch K.R., Lin C.Y., Hla T. Immunosuppressive and anti-angiogenic sphingosine 1-phosphate receptor-1 agonists induce ubiquitinylation and proteasomal degradation of the receptor. J Biol Chem. 2007;282:9082–9089. doi: 10.1074/jbc.M610318200. [DOI] [PubMed] [Google Scholar]
- 44.Choi J.W., Gardell S.E., Herr D.R., Rivera R., Lee C.W., Noguchi K., Teo S.T., Yung Y.C., Lu M., Kennedy G., Chun J. FTY720 (fingolimod) efficacy in an animal model of multiple sclerosis requires astrocyte sphingosine 1-phosphate receptor 1 (S1P1) modulation. Proc Natl Acad Sci U S A. 2011;108:751–756. doi: 10.1073/pnas.1014154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stepanovska B., Huwiler A. Targeting the S1P receptor signaling pathways as a promising approach for treatment of autoimmune and inflammatory diseases. Pharmacol Res. 2020;154 doi: 10.1016/j.phrs.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 46.Enriquez-Marulanda A., Valderrama-Chaparro J., Parrado L., Diego Velez J., Maria Granados A., Luis Orozco J., Quinones J. Cerebral toxoplasmosis in an MS patient receiving Fingolimod. Mult Scler Relat Disord. 2017;18:106–108. doi: 10.1016/j.msard.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Dillmann C., Ringel C., Ringleb J., Mora J., Olesch C., Fink A.F., Roberts E., Brune B., Weigert A. S1PR4 signaling attenuates ILT 7 internalization to limit IFN-alpha production by human plasmacytoid dendritic cells. J Immunol. 2016;196:1579–1590. doi: 10.4049/jimmunol.1403168. [DOI] [PubMed] [Google Scholar]
- 48.Kim K.M., Han C.Y., Kim J.Y., Cho S.S., Kim Y.S., Koo J.H., Lee J.M., Lim S.C., Kang K.W., Kim J.S., Hwang S.J., Ki S.H., Kim S.G. Galpha12 overexpression induced by miR-16 dysregulation contributes to liver fibrosis by promoting autophagy in hepatic stellate cells. J Hepatol. 2018;68:493–504. doi: 10.1016/j.jhep.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parthasarathy G., Revelo X., Malhi H. Pathogenesis of nonalcoholic steatohepatitis: an overview. Hepatol Commun. 2020;4:478–492. doi: 10.1002/hep4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geng T., Sutter A., Harland M.D., Law B.A., Ross J.S., Lewin D., Palanisamy A., Russo S.B., Chavin K.D., Cowart L.A. SphK1 mediates hepatic inflammation in a mouse model of NASH induced by high saturated fat feeding and initiates proinflammatory signaling in hepatocytes. J Lipid Res. 2015;56:2359–2371. doi: 10.1194/jlr.M063511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olesch C., Sirait-Fischer E., Berkefeld M., Fink A.F., Susen R.M., Ritter B., Michels B.E., Steinhilber D., Greten F.R., Savai R., Takeda K., Brune B., Weigert A. S1PR4 ablation reduces tumor growth and improves chemotherapy via CD8+ T cell expansion. J Clin Invest. 2020;130:5461–5476. doi: 10.1172/JCI136928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schulze T., Golfier S., Tabeling C., Rabel K., Graler M.H., Witzenrath M., Lipp M. Sphingosine-1-phosphate receptor 4 (S1P(4)) deficiency profoundly affects dendritic cell function and TH17-cell differentiation in a murine model. FASEB J. 2011;25:4024–4036. doi: 10.1096/fj.10-179028. [DOI] [PubMed] [Google Scholar]
- 53.Matloubian M., Lo C.G., Cinamon G., Lesneski M.J., Xu Y., Brinkmann V., Allende M.L., Proia R.L., Cyster J.G. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 54.Konerman M.A., Jones J.C., Harrison S.A. Pharmacotherapy for NASH: current and emerging. J Hepatol. 2018;68:362–375. doi: 10.1016/j.jhep.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 55.Taylor R.S., Taylor R.J., Bayliss S., Hagstrom H., Nasr P., Schattenberg J.M., Ishigami M., Toyoda H., Wai-Sun Wong V., Peleg N., Shlomai A., Sebastiani G., Seko Y., Bhala N., Younossi Z.M., Anstee Q.M., McPherson S., Newsome P.N. Association between fibrosis stage and outcomes of patients with nonalcoholic fatty liver disease: a systematic review and meta-analysis. Gastroenterology. 2020;158:1611–1625.e12. doi: 10.1053/j.gastro.2020.01.043. [DOI] [PubMed] [Google Scholar]
- 56.Machado M.V., Michelotti G.A., Xie G., Almeida Pereira T., Boursier J., Bohnic B., Guy C.D., Diehl A.M. Mouse models of diet-induced nonalcoholic steatohepatitis reproduce the heterogeneity of the human disease. PLoS One. 2015;10 doi: 10.1371/journal.pone.0127991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sung Y.H., Baek I.J., Kim D.H., Jeon J., Lee J., Lee K., Jeong D., Kim J.S., Lee H.W. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol. 2013;31:23–24. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- 58.Li P.Z., Li J.Z., Li M., Gong J.P., He K. An efficient method to isolate and culture mouse Kupffer cells. Immunol Lett. 2014;158:52–56. doi: 10.1016/j.imlet.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 59.Choi W.M., Eun H.S., Lee Y.S., Kim S.J., Kim M.H., Lee J.H., Shim Y.R., Kim H.H., Kim Y.E., Yi H.S., Jeong W.I. Experimental applications of in situ liver perfusion machinery for the study of liver disease. Mol Cells. 2019;42:45–55. doi: 10.14348/molcells.2018.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]