Abstract
During a 6-year period, 64 of 227 commercially reared cats had microaerobic bacteria isolated from their feces. All the isolates were initially identified as Campylobacter-like organisms based on biochemical and phenotypic characteristics. DNA extractions from 51 of these isolates were subjected to PCR using primers specific for Helicobacter spp. and Campylobacter spp. Of the isolates, 92% (47 of 51 isolates) were positive for Campylobacter spp., 41% (21 of 51 isolates) were positive for Helicobacter spp., 33% (17 of 51 isolates) were positive for both genera, 59% (30 of 51 isolates) were positive only for Campylobacter spp., and 8% (4 of 51) were positive only for Helicobacter spp. Sixteen of the 47 Campylobacter-positive cultures were positive for more than one Campylobacter spp. Based on a species-specific PCR assay, 83% of the isolates were identified as Campylobacter helveticus, 47% of the isolates were identified as Campylobacter upsaliensis, and 6% of the isolates were classified as Campylobacter jejuni. The 1.2-kb PCR products of the 16S rRNA genes of 19 Helicobacter species isolates were subjected to restriction fragment length polymorphism (RFLP) analysis. Of the five different RFLP patterns obtained, two clustered with Helicobacter (“Flexispira”) taxon 8, one clustered with Helicobacter bilis, one clustered with Helicobacter canis, and the remaining pattern was closely related to a novel Helicobacter sp. strain isolated from a woodchuck. The sequence data for the 16S rRNA genes of 10 Helicobacter spp. validated the RFLP-based identification of these isolates. This study demonstrated that biochemical and phenotypic characteristics of microaerobic organisms in cat feces were insufficient to characterize mixed Helicobacter and Campylobacter infections. Molecular structure-based diagnostics using genus- and species-specific PCR, RFLP analysis, and 16S rRNA sequence analysis enabled the identification of multiple microaerobic species in individual animals. The clinical relevance of enteric Helicobacter and Campylobacter coinfection in cats will require further studies.
Cats are recognized reservoirs for enteric Campylobacter spp., including Campylobacter jejuni, Campylobacter coli, Campylobacter upsaliensis, and Campylobacter helveticus (5, 13, 20, 37, 58). C. jejuni and C. coli are among the most frequently encountered human enteric pathogens worldwide (2, 3). C. upsaliensis, a catalase-negative or weakly positive Campylobacter sp. initially isolated from diarrheic or nondiarrheic domestic dogs and cats (20, 48), has also been associated with enteritis (25, 44, 55) and bacteremia (31, 39, 44) in humans. More-serious illnesses, including spontaneous abortion and hemolytic-uremic syndrome, have also been reported for a human infected with C. upsaliensis (27). Campylobacter infections, particularly C. jejuni infections, are zoonotic and are a particular problem among puppies and kittens from shelters (13, 18, 45). C. helveticus, which is closely related to C. upsaliensis, has also been isolated from domestic cats and dogs but has not been linked with human disease (52).
Although Helicobacter spp. are better known as gastric pathogens (7, 10, 18, 19, 28, 41, 43), there has been an increasing interest in enterohepatic Helicobacter spp. isolated from humans and animals. Helicobacter canis has been isolated from normal and diarrheic dogs, cats, and diarrheic humans as well as from the liver of a dog with hepatitis (6, 12, 17, 54); “Flexispira rappini ” strains, which represent at least 10 Helicobacter taxa, have been isolated from feces of mice, sheep, dogs, and humans and have been associated with abortion in sheep (9). Helicobacter pullorum, first isolated from the feces and liver of chickens, also has been cultured from diarrheic humans (53). Helicobacter canadensis, originally misdiagnosed as H. pullorum, has been isolated from Canadian patients with diarrhea (14). Helicobacter cinaedi, isolated from the feces of healthy hamsters, also has been recovered from the inflamed lower bowel and blood of immunocompromised adults and children with diarrhea (11, 24, 57). Recently, mixed infections of Helicobacter spp. and Campylobacter spp. have been noted in diarrheic children residing in developing countries (33). The purpose of this study is to describe for the first time coinfection of enteric Helicobacter spp. and Campylobacter spp. in cats and to provide molecular characterization of novel Helicobacter species in these same animals.
MATERIALS AND METHODS
Animals.
During a 6-year period, 227 purpose-bred cats were obtained from three commercial sources. The cats purchased were from specific-pathogen-free colonies certified to be negative for feline leukemia virus, feline immunodeficiency virus, and feline coronavirus. All cats were evaluated for body condition, appetite, and episodic diarrhea while in quarantine. At the time of fecal culture all cats were clinically healthy.
Bacterial culture.
Rectal swabs from the cats were streaked onto cefoperazone-vancomycin-amphotericin B antibiotic-impregnated media (Remel Laboratories, Lenexa, Kans.) and grown under microaerobic conditions in vented jars containing N2, H2, and CO2 (80:10:10) at 37 and 42°C. The primary isolates were Gram stained and tested for urease, oxidase (Bactidrop; Remel Laboratories), and catalase (3% H2O2). Isolates were also assayed for their ability to hydrolyze hippurate (30). The sensitivity of the isolates to nalidixic acid and cephalothin was tested with antibiotic-impregnated disks. Bacteria identified as Campylobacter-like organisms (CLOs) were frozen at −20°C in 20% glycerol and brucella broth. Bacteria were subsequently reisolated on primary media for molecular characterization.
DNA extraction.
DNA was extracted from individual colonies grown on cefoperazone-vancomycin-amphotericin B plates by using InstaGene matrix (Bio-Rad Laboratories, Hercules, Calif.). Bacteria were resuspended in 1 ml of double-distilled water in a microfuge tube. After centrifugation and removal of the supernatant, 200 μl of InstaGene matrix was added to the pellet and incubated at 56°C for 30 min. The samples were then boiled for 10 min and centrifuged for 5 min at high speed, and then 10 μl of the supernatant was used for the PCR.
PCR amplification.
The nucleotide sequences and the sources of all primers used to amplify the cat fecal isolates are listed in Table 1. PCR amplifications were performed with a Thermal Cycler and an Expand high-fidelity PCR system (Roche Molecular Biochemical, Indianapolis, Ind.). Each reaction mixture (100 μl) contained 1× polymerase buffer, a 0.5 μM concentration of each of two primers, a 200 μM concentration of each deoxyribonucleotide triphosphate, and bovine serum albumin (200 μg/ml). The samples were heated at 94°C for 4 min, briefly centrifuged, and cooled to 58°C. Polymerase (2.5 U) was then added. Amplification was achieved by denaturation at 94°C for 1 min, annealing at 58°C for 2 min, and elongation at 72°C for 2 min. For primers specific for C. helveticus, C. upsaliensis, C. jejuni, and C. coli, the annealing was at 55°C. A 15-μl portion of the sample was then electrophoresed through a 1% agarose gel and followed by ethidium bromide staining and UV illumination.
TABLE 1.
Primers used for this study
| ID | Sequencea | Position | Orientation | Function (reference) |
|---|---|---|---|---|
| C75 | GAGAGTTTGATYCTGGCTCAG | 7–27 | Forward | Sequencing |
| F16 | TAGATACCCYGGTAGTCC | 789–806 | Forward | Sequencing |
| E94 | GAAGGAGGTGWTCCARCCGCA | 1522–1541 | Reverse | Sequencing |
| F20 | CCATTGTARCACGTGTG | 1226–1242 | Reverse | Sequencing |
| C97 | GCTATGACGGGTATCC | 276–291 | Forward | Helicobacter-specific PCR (16) |
| C05 | ACTTCACCCCAGTCGCTG | 1478–1495 | Reverse | |
| C98 | GATTTTACCCCTACACCA | 681–698 | Reverse | Campylobacter-specific PCR (16) |
| C99 | GCGTGGAGGATGACACCT | 402–419 | Forward | |
| Chcu146f | GGGACAACACTTAGAAATGAG | 146–166 | Forward | C. helveticus-specific PCR (35) |
| Ch1371r | CCGTGACATGGCTGATTCAC | 1351–1371 | Reverse | |
| Chcu146f | GGGACAACACTTAGAAATGAG | 146–166 | Forward | C. upsaliensis-specific PCR (35) |
| Cu1024r | CACTTCCGTATCTCTACAGA | 1002–1024 | Reverse | |
| Hip400f | GAAGAGGGTTTGGGTGGTG | 400–418 | Forward | C. jejuni-specific PCR (36) |
| Hip1134r | AGCTAGCTTCGCATAATAACTTG | 1112–1134 | Reverse | |
| Cc18F | GGTATGATTTCTACAAAGCGAG | 18–38 | Forward | C. coli-specific PCR (36) |
| Cc519R | ATAAAAGACTATCGTCGCGTG | 699–519 | Reverse |
Sequences are in the 5′ to 3′ direction.
Restriction fragment length polymorphism of Helicobacter 16S rRNA gene.
Primers C97 and C05 (Table 1) were used to amplify the 1.2-kb PCR fragments from all the Helicobacter species isolates identified in this study. Amplified DNA (20 μl) was digested with 10 U of AluI in the buffer recommended by the enzyme manufacturer at 37°C for 3 h. Restriction patterns were compared after the digested PCR products were separated on a 6% Visigel separation matrix (Stratagene, LaJolla, Calif.).
Cloning and sequencing 16S ribosomal DNA PCR products.
A pGEM-T vector (Promega, Madison, Wis.) was used for cloning the PCR products. The PCR products were purified from a low-melting-point agarose gel with the QIAquick PCR purification kit (Qiagen, Valencia, Calif.). Fifty nanograms of purified PCR product was ligated with 50 ng of pGEM-T vector at 4°C overnight and used to transfer into competent JM109 cells. Ampicillin plates with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and isopropyl-β-d-thiogalactopyranoside (IPTG) were used to select positive clones. Plasmid DNA was isolated from Escherichia coli using a Qiaprep mini spin kit (Qiagen). The 1,600-bp DNA sequences of the 16S rRNA cistrons of two pure isolates and the 1,200-bp sequences of eight PCR products obtained with the use of Helicobacter genus-specific primers were obtained by cycle sequencing. Purified DNA from the PCR and plasmid DNA were sequenced using an ABI prism cycle sequencing kit (BigDye Terminator cycle sequencing kit with AmpliTaq DNA polymerase FS; Perkin-Elmer). The primers listed in Table 1 were used for sequencing. Quarter dye chemistry was used with 80 μM primer and 1.5 μl of DNA in a final volume of 20 μl. Cycle sequencing was performed using an ABI 9700 thermal cycler, with 25 cycles of denaturation at 96°C for 10 s and annealing and extension at 60°C for 4 m. Sequencing reactions were run on an ABI 377 DNA sequencer.
16S rRNA data analysis.
Sequences were first screened by a BLAST analysis comparing them to all entries in GenBank (1). Sequence data were then entered into RNA, a program set for data entry, editing, sequence alignment, secondary structure comparison, similarity matrix generation, and dendrogram construction for 16S rRNA in Microsoft QuickBasic for use with personal computers, and were aligned as previously described (42). Our database contains over 1,000 sequences obtained in our laboratory and over 500 obtained from GenBank. Dendrograms were constructed by the neighbor-joining method (47).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the strains MIT 98-1705-1 and MIT 95-234-6 are AF336947 and AF336948, respectively. Sequences for the 1,200-bp partial sequences are available from the corresponding author.
RESULTS
Campylobacter spp. and Helicobacter spp. commonly colonize the intestines of cats used for biomedical research.
During the 6 years of the survey, 227 fecal samples were collected for culture of CLOs. Sixty-four cats were initially diagnosed as positive for CLOs, based on primary isolation and characterization of bacteria by colony morphology and biochemical tests. The bacteria were slightly curved or spiral-shaped, gram-negative organisms and grew under microaerobic conditions at 37°C. Selected isolates grew at 42°C. Isolates typically were urease negative, oxidase positive, weakly catalase positive or negative, and sensitive to disks containing 30 μg of nalidixic acid and 30 μg of cephalothin.
The overall prevalence rate of CLOs in the feces of cats from three commercial sources was 28%. Forty-nine percent of the samples from one commercial source cultured positive for CLOs, while 18 and 23%, respectively, of samples from the other two sources were culture positive (Table 2). Based on the results of a 3 by 2 χ2 test the prevalence of CLO infection differed significantly among the three sources (χ22 = 12.86, P = 0.002).
TABLE 2.
CLO prevalence in cats from three different commercial sources
| Source of cats | No. of samples | No. positive for CLO by culture | % Positive |
|---|---|---|---|
| A | 22 | 4 | 18 |
| B | 47 | 22 | 47 |
| C | 158 | 38 | 24 |
| Total | 227 | 64 | 28 |
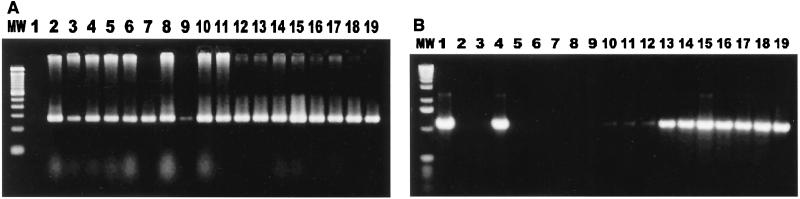
In testing of the 51 isolates which were still retrievable from the original frozen cultures, positive results were obtained for 92% of the isolates (47 of 51 isolates) with the Campylobacter genus-specific primers and for 41% of the isolates (21 of 51 isolates) with the Helicobacter genus-specific primers. Thirty-three percent (17 of 51 isolates) were positive for both Campylobacter and Helicobacter, 59% (30 of 51 isolates) were positive only for Campylobacter, and 8% (4 of 51 isolates) were positive only for Helicobacter (Fig. 1 and Table 3).
FIG. 1.
Cocolonization of cats by Campylobacter spp. and Helicobacter spp. (A) Campylobacter genus-specific primers were used to amplify DNA extracted from cat fecal isolates. Lane MW, 100-bp DNA ladder; lane 1, reagent control; lane 2, Campylobacter-positive control; lanes 3 to 19, isolates from cat feces. (B) Helicobacter genus-specific primers were used to amplify DNA extracted from cat fecal isolates. Lane MW, 1-kb DNA ladder; lane 2, Helicobacter-positive control; lane 3, reagent control; lanes 3 to 19, isolates from cat feces.
TABLE 3.
CLOs isolated from cats
| Year | No. of samples | No. of samples positive by:
|
||
|---|---|---|---|---|
| Culture | Helicobacter-specific PCRa | Campylobacter-specific PCRa | ||
| 1993 | 14 | 8 | 0/2 | 2/2 |
| 1994 | 34 | 7 | 1/2 | 2/2 |
| 1995 | 89 | 41 | 14/39 | 39/39 |
| 1996 | 36 | 1 | 0/1 | 1/1 |
| 1997 | 20 | 0 | 0 | 0 |
| 1998 | 34 | 7 | 6/7 | 3/7 |
| Total | 227 | 64 | 21 | 47 |
Values are number of samples positive per number tested by PCR.
Differentiation of Campylobacter spp. isolated from cat feces.
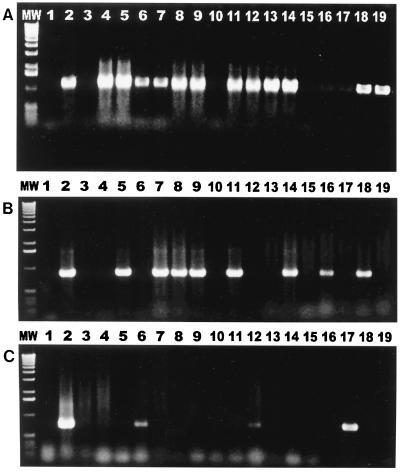
PCR assays specific for C. coli, C. helveticus, C. upsaliensis, and C. jejuni were used to screen the 47 fecal isolates which were positive for Campylobacter spp. by analysis with genus-specific primers. Of the bacterial cultures, 34% (16 of 47 cultures) were found to be mixed cultures of more than one Campylobacter sp. (Fig. 2). Eighty-three percent (39 of 47 cultures) were positive for C. helveticus; 47% (22 of 47 cultures) were positive for C. upsaliensis; and 6% (3 of 47 cultures) were positive for C. jejuni. In tests using C. coli 16S rRNA gene-specific primers, positive results were not obtained for any of the cultures.
FIG. 2.
Results of genus-specific PCR. (A) Primers specific for C. helveticus amplified 16S rRNA PCR products from 12 of 17 cat fecal isolates. Lane MW, 1-kb DNA ladder; lane 1, reagent control; lane 2, positive control; lanes 3 to 19, isolates from cat feces. (B) Primers specific for C. upsaliensis amplified 16S rRNA products from 8 of 17 isolates from cat feces. Lane MW, 1-kb DNA ladder; lane 1, reagent control; lane 2, positive control; lanes 3 to 19, isolates from cat feces. (C) Primers specific for C. jejuni amplified hippuricase gene PCR products from 3 of 17 cat isolates. Lane MW, 1-kb DNA ladder; lane 1, reagent control; lane 2, positive control; lanes 3 to 19, isolates from cat feces.
Restriction fragment length polymorphism (RFLP) analysis of Helicobacter species PCR products from cat isolates.
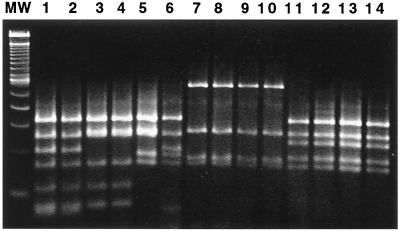
The 1.2-kb products from Helicobacter genus-specific PCR analysis of 19 cat isolates were digested by AluI. Five patterns were observed. Three patterns grouped taxonomically with H. bilis or Helicobacter (“Flexispira”) taxon 8; one was similar to H. canis, and the remaining pattern matched that of a novel Helicobacter sp. isolated from a woodchuck (21) (Fig. 3 and 4).
FIG. 3.
Products (1.2 kb) of PCR using Helicobacter genus-specific primers were digested by AluI and analyzed by electrophoresis on 6% Visigel matrix. Five patterns were observed. Lane MW, 100-bp DNA ladder; lane 1, Helicobacter (“Flexispira”) taxon 8 (ATCC 49317); lane 2, MIT 98-90; lane 3, MIT 95-1850-65; lane 4, MIT 95-513-27; lane 5, H. bilis ATCC 51630; lane 6, MIT 95-234-6; lane 7, H. canis cat isolate (11); lane 8, MIT 95-1114-42; lane 9, MIT 95-1114-46; lane 10, MIT 94-55-4; lane 11, Helicobacter sp. woodchuck isolate (19); lane 12, MIT 95-513-29; lane 13, MIT 94-2635-37; lane 14, MIT 98-1705-4.
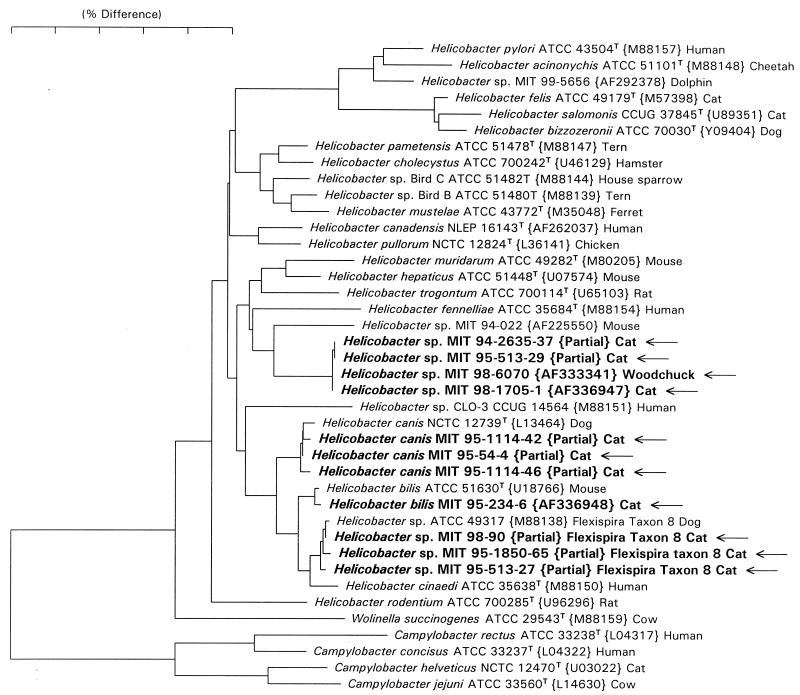
FIG. 4.
Neighbor-joining phylogenetic tree for Helicobacter spp. isolated from feces of cats and reference Campylobacter and Helicobacter species. The scale bar represents phylogenetic distance as estimated using the Jukes Cantor correction. Distances can be determined by adding the lengths of all of the horizontal lines connecting any two species. GenBank accession numbers appear in brackets. The 1,200-bp partial sequences (marked as such) for which accession numbers are not provided were not deposited in GenBank and are available from the corresponding author.
Analysis of 16S rRNA sequences.
Full 16S rRNA sequences were determined for isolates MIT 95-234-6 and MIT 98-1705-1. Isolate MIT 95-234-6 was identical to the sequence of the type strain of H. bilis (GenBank accession no. U18766) except that it did not contain a 187-base intervening sequence (22) in the 198-219 helix, but rather the sequence GGUUUUUC. Isolate MIT 98-1705-1 was identical to a Helicobacter sp. previously isolated from a woodchuck, MIT 98-6070 (GenBank accession no. AF333341). The eight 1,200-base partial sequences clustered into three groups. Clones MIT 95-513-29 and MIT 94-2635-37 differed by 1 base from MIT 98-1705-1. Clones MIT 95-54-4, MIT 95-1114-42, and MIT 95-1114-46 differed from the sequence of the type strain of H. canis (GenBank accession no. L13464) by the following three base changes: G592A, A616G, and A645G (numbering according to the sequence of E. coli). Clones MIT 95-513-27, MIT 95-1850-65 and MIT 98-90 differed from the sequence of Helicobacter (“Flexispira”) taxon 8 (GenBank accession no. M88138) by 0 to 3 bases. A neighbor-joining phylogenetic tree was constructed and is shown in Fig. 4.
DISCUSSION
This study for the first time demonstrated a high prevalence of mixed infections of Campylobacter and Helicobacter species in a large number of clinically healthy cats obtained from three commercial sources located in different geographic regions. CLOs were isolated from 28% of 227 cats, and from 33% of these, mixed cultures of Campylobacter organisms and Helicobacter organisms were obtained. The Campylobacter spp. most frequently isolated from the feces of these cats were C. upsaliensis and C. helveticus; 86% of the cultures contained C. helveticus, while 47% of the cultures contained C. upsaliensis.
The high prevalence of Campylobacter spp. in laboratory-reared cats is consistent with widespread Campylobacter infection observed for domestic animals. C. upsaliensis was first isolated from the feces of healthy and diarrheic dogs in Sweden (63 of 98 [64%] of the Campylobacter strains isolated from the feces of these dogs over 2 years were C. upsaliensis) (48). We have previously documented the presence of C. upsaliensis in cat feces using biochemical characterization and DNA hybridization assays (20). In Switzerland, Campylobacter spp. were isolated from 31% of a group of diarrheic and healthy pet animals; 50% of the isolates from cats were C. upsaliensis. For cats, there was no association between Campylobacter carriage and disease, irrespective of the animals' age. For dogs older than 12 months, there was also no difference in Campylobacter carriage rate between diarrheic and healthy animals. However, 44% of the younger dogs with diarrhea shed Campylobacter species organisms in their feces, more than twice the rate observed for clinically healthy dogs (5). In the United Kingdom, 50% of 156 healthy domestic pets and laboratory animals were positive for Campylobacter spp., with 60% of the cats shedding C. upsaliensis in their feces (38). However, none of the authors of the above-mentioned four studies isolated Helicobacter spp. from the feces of these animals.
C. upsaliensis has also been isolated from the feces of children and adults with diarrhea (25, 44, 55), as well as from the blood of pediatric patients and adults with septicemia (31, 39, 44). Other extraintestinal sites from which this organism has been cultured include a breast abscess (23) and the fetoplacental tissue of an 18-week-pregnant woman who had contact with a household cat. Both isolates had similar sodium dodecyl sulfate gel protein patterns (27). There is other epidemiological evidence that suggests that C. upsaliensis may have zoonotic potential. One study of C. upsaliensis infection reported that four of seven humans infected had animal contact (44). C. upsaliensis was also isolated from a diarrheic patient and his clinically healthy dog (26).
C. helveticus was isolated in a high percentage of cats in this study, which confirmed the results of earlier studies in England, where it was cultured from the feces of healthy cats (52). However, the organism's clinical relevance for pets, if any, has not been reported.
The natural habitat of most Campylobacter spp., including C. jejuni, is the intestinal tract of warm-blooded animals, including birds (40). Campylobacter infection is transmitted to humans from animals either by fecal-oral contact or indirectly by food, milk, or water. Campylobacterosis in humans is largely a result of food-borne infection in which foods of animal origin, particularly poultry, play an important role (8). Domestic animals are common reservoir hosts for C. jejuni, and zoonotic infections have been acquired from pets, including cats with or without diarrhea (4, 8, 13; M. B. Skirrow, G. L. Turnbull, R. E. Walker, and S. E. J. Young, Letter, Lancet i:1188, 1980; A. Svedhem and G. Norkrans, Letter, Lancet i:713–714, 1980). C. jejuni has been isolated from dogs and cats housed in animal shelters in addition to being isolated from dogs and cats used in biomedical research (13, 15, 45). In our study, 4% of cats had C. jejuni in their feces. This prevalence was lower than the 10.7% previously reported for research cats, but it was higher than the 1% C. jejuni isolation rate recorded for pet cats and cats sampled at a humane society shelter (13, 29).
The Helicobacter spp. most frequently isolated from cats in this study were H. canis, Helicobacter (“Flexispira”) taxon 8, and a novel Helicobacter species previously isolated from woodchucks. The novel species shared at least 96% sequence identity with all Helicobacter spp. in the GenBank database but was essentially identical to an isolate from a woodchuck (MIT 98-6070) (21). This novel species shared 97% 16S rRNA sequence homology with a mouse Helicobacter sp. isolate, MIT 94-022. H. canis has been previously reported to have been found in diarrheic cats (12), in a child with gastroenteritis (6), and in dogs with or without diarrhea (54). The organism was also isolated from the liver of a puppy with necrotizing hepatitis (17). Organisms with “Flexispira rappini ” morphology isolated from a number of hosts have been divided into 10 taxa (9). For example, H. bilis was identified, by cloning and sequencing of 16S rRNA, in gall bladders of patients with chronic cholecystitis (16). Although the 16S rRNA sequences of these taxa are very similar, the RFLP patterns may be different (50). To our knowledge, organisms of the Helicobacter (“Flexispira”) taxa (including H. bilis) have not been isolated from cats. However, such organisms have been cultured from the feces of three dogs and their owners, diarrheic children, and rodents (22, 33, 46, 49). They are also increasingly isolated from the blood of immunocompromised patients, including two that had a history of contact with puppies (51, 56).
In our experience, for the best recovery of CLOs, fecal samples should be placed in glycerol medium for transportation. Higher H2 levels (5 to 10%) are required for optimal Helicobacter sp. isolation. Unfortunately, this atmosphere is not available in the commercially available diagnostic kits used for Campylobacter isolation. Identification of multiple species of microaerobic bacteria in the feces of an animal poses a diagnostic challenge, particularly when these microaerobes grow on similar media in comparable atmospheric conditions. Primary isolation of these microaerophilic bacteria may be misleading, because Helicobacter spp. may be present in smaller numbers and grow at a slower rate than Campylobacter spp. The similar phenotypic traits and biochemical profiles of these genera also complicate a diagnosis. Using Campylobacter and Helicobacter genus-specific PCR assays allowed us to distinguish between the two genera. The PCR-RFLP assay was also useful for Helicobacter sp. identification.
Investigators in South Africa have recently published results for a protocol that has been in use in their diagnostic laboratory since 1990 and that allows primary isolation of multiple species of Campylobacter and Helicobacter from the diarrheic specimens of individual children. Filtrates are plated onto antibiotic-free blood agar plates and incubated in an H2-enriched atmosphere (32, 33). The authors not only documented an increase in the number of CLOs isolated but also were able to culture C. upsaliensis for the first time. The authors have reported a 16.2% prevalence of multiple species of CLOs based on primary isolation, biochemical characterization, and serologic confirmation. They frequently recovered between two and five species of CLOs from one stool sample, with C. jejuni (different serotypes), C. coli, C. upsaliensis, Helicobacter fennelliae, and H. cinaedi being commonly isolated (32). Further analysis using the filtration isolation technique with cat and dog feces may yield prevalence rates for mixed Helicobacter and Campylobacter infections even higher than those reported in the present study.
In summary, cats used for biomedical research were commonly colonized with intestinal Helicobacter spp. and Campylobacter spp. Accurate diagnosis of mixed infections with these bacteria may require diagnostic laboratories to incorporate PCR-based assays using Helicobacter and Campylobacter genus- and species-specific primers. This recommendation is supported by a recent study which reported improved sensitivity for PCR compared to conventional culture techniques in identifying mixed infections of Campylobacter spp in cases of human gastroenteritis (34). Although all the CLOs in this study were isolated from clinically healthy cats, some of these species have been linked with diarrheal diseases in humans and animals. The zoonotic importance of intestinal cocolonization with Campylobacter and Helicobacter, as well as their importance in causing disease in cats, other animals, and humans, requires further studies.
ACKNOWLEDGMENTS
This work is supported in part by NIH grants CA-67529 and DK-52413, as well as NIH grants RR-01046 (to J.G.F.) and DE-10374 (to F.E.D.).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaser M J, Reller L B. Campylobacter enteritis. N Engl J Med. 1981;305:1444–1452. doi: 10.1056/NEJM198112103052404. [DOI] [PubMed] [Google Scholar]
- 3.Blaser M J, Taylor D N, Feldman R A. Epidemiology of Campylobacter jejuni infections. Epidemiol Rev. 1983;5:157–176. doi: 10.1093/oxfordjournals.epirev.a036256. [DOI] [PubMed] [Google Scholar]
- 4.Blaser M J, Weiss S H, Barret T J. Campylobacter enteritis associated with a healthy cat. JAMA. 1982;816:247. [PubMed] [Google Scholar]
- 5.Burnens A P, Angeloz-Wick B, Nicolet J. Comparison of Campylobacter carriage rates in diarrheic and healthy pet animals. Zentralbl Vetmed. 1992;39:175–180. doi: 10.1111/j.1439-0450.1992.tb01155.x. [DOI] [PubMed] [Google Scholar]
- 6.Burnens A P, Stanley J, Schaad U B, Nicolet J. Novel Campylobacter-like organism resembling Helicobacter fennelliae isolated from a boy with gastroenteritis and from dogs. J Clin Microbiol. 1993;31:1916–1917. doi: 10.1128/jcm.31.7.1916-1917.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Correa P, Fox J G, Fontham E, Ruiz B, Lin Y, Zavala D, Taylor N S, Mackinley D, de Lima F, Portilla H, Zarama G. Helicobacter pylori and gastric carcinoma: serum antibody prevalence in populations with contrasting cancer risks. Cancer. 1990;66:2569–2574. doi: 10.1002/1097-0142(19901215)66:12<2569::aid-cncr2820661220>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 8.Deming M S, Tauxe R V, Blake B A, Dixon S E, Fowler B S, Jones T S, Lockamy E A, Patton C M, Sikes R O. Campylobacter enteritis at a university: transmission from eating chicken and from cats. Am J Epidemiol. 1987;126:526–534. doi: 10.1093/oxfordjournals.aje.a114685. [DOI] [PubMed] [Google Scholar]
- 9.Dewhirst F E, Fox J G, Mendes E N, Paster B J, Gates C E, Kirkbride C A, Eaton K A. Flexispira rappini strains represent at least ten Helicobacter taxa. Int J Syst Evol Microbiol. 2000;50:1781–1787. doi: 10.1099/00207713-50-5-1781. [DOI] [PubMed] [Google Scholar]
- 10.Eaton K A, Dewhirst F E, Radin M J, Fox J G, Paster B J, Krakowka S, Morgan D R. Helicobacter acinonyx sp. nov., isolated from cheetahs with gastritis. Int J Syst Bacteriol. 1993;43:99–106. doi: 10.1099/00207713-43-1-99. [DOI] [PubMed] [Google Scholar]
- 11.Fennell C L, Totten P A, Quinn T C, Patton D L, Holmes K K, Stamm W E. Characterization of Campylobacter-like organisms isolated from homosexual men. J Infect Dis. 1984;149:58–66. doi: 10.1093/infdis/149.1.58. [DOI] [PubMed] [Google Scholar]
- 12.Foley J E, Marks S, Munson L, Melli A, Dewhirst D E, Yu S, Shen Z, Fox J G. Isolation of Helicobacter canis from a colony of Bengal cats with endemic diarrhea. J Clin Microbiol. 1999;37:3271–3275. doi: 10.1128/jcm.37.10.3271-3275.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox J G, Ackerman J I, Newcomer C E. The prevalence of Campylobacter jejuni in random source cats used in biomedical research. J Infect Dis. 1985;151:743–744. doi: 10.1093/infdis/151.4.743. [DOI] [PubMed] [Google Scholar]
- 14.Fox J G, Chien C C, Dewhirst F E, Paster B J, Shen Z, Melito P L, Woodward D L, Rodgers F G. Helicobacter canadensis sp. nov. isolated from humans with diarrhea: an example of an emerging pathogen. J Clin Microbiol. 2000;38:2546–2549. doi: 10.1128/jcm.38.7.2546-2549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox J G, Claps M C, Taylor N S, Ackerman J I. Campylobacter jejuni/coli in commercially reared beagles: prevalence and serotypes. Lab Anim Sci. 1988;38:262. [PubMed] [Google Scholar]
- 16.Fox J G, Dewhirst F E, Shen Z, Fen Y, Taylor N S, Paster B, Erickson R L, Lau C N, Correa P, Araya J C, Roa I. Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology. 1998;114:755–763. doi: 10.1016/s0016-5085(98)70589-x. [DOI] [PubMed] [Google Scholar]
- 17.Fox J G, Drolet R, Higgins R, Messier S, Yan L, Coleman B E, Paster B J, Dewhirst F E. Helicobacter canis isolated from a dog liver with multifocal necrotizing hepatitis. J Clin Microbiol. 1996;34:2479–2482. doi: 10.1128/jcm.34.10.2479-2482.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox J G, Edrise B M, Cabot E, Beaucage C, Murphy J C, Prostak K S. Campylobacter-like organisms isolated from gastric mucosa of ferrets. Am J Vet Res. 1986;47:236–239. [PubMed] [Google Scholar]
- 19.Fox J G, Lee A. The role of Helicobacter species in newly recognized gastrointestinal tract diseases of animals. Lab Anim Sci. 1997;47:222–255. [PubMed] [Google Scholar]
- 20.Fox J G, Maxwell K O, Taylor N S, Runsick C D, Edmonds P, Brenner D J. “Campylobacter upsaliensis ” isolated from cats as identified by DNA relatedness and biochemical features. J Clin Microbiol. 1989;27:2376–2378. doi: 10.1128/jcm.27.10.2376-2378.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox J G, Xu S, Shen Z, Dangler C A, Cullen J M. A novel Helicobacter sp. isolated from woodchuck livers infected with woodchuck hepatitis virus (WHV) Gastroenterology. 1999;116:A718. [Google Scholar]
- 22.Fox J G, Yan L L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher J C, Mendes E N. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaudreau C, Lamothe F. Campylobacter upsaliensis isolated from a breast abscess. J Clin Microbiol. 1992;30:1354–1356. doi: 10.1128/jcm.30.5.1354-1356.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gebhart C J, Fennell C L, Murtaugh M P, Stamm W E. Campylobacter cinaedi is normal intestinal flora in hamsters. J Clin Microbiol. 1989;27:1692–1694. doi: 10.1128/jcm.27.7.1692-1694.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goossens H, Pot B, Vlaes L, Van den Borre C, Van den Abbeele R, Van Naelten C, Levy J, Cogniau H, Marbehant P, Verhoef J, Kersters K, Butzler J-P, Vandamme P. Characterization and description of “Campylobacter upsaliensis ” isolated from human feces. J Clin Microbiol. 1990;28:1039–1946. doi: 10.1128/jcm.28.5.1039-1046.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goossens H, Vlaes L, Butzler J P, Adnet A, Hanicq P, N′Jufom S, Massart D, de Schrijver G, Blomme W. Campylobacter upsaliensis enteritis associated with canine infections. Lancet. 1991;337:1486–1487. doi: 10.1016/0140-6736(91)93182-9. [DOI] [PubMed] [Google Scholar]
- 27.Gurgan T, Diker K S. Abortion associated with Campylobacter upsaliensis. J Clin Microbiol. 1994;32:3093–3094. doi: 10.1128/jcm.32.12.3093-3094.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanninen M L, Happonen I, Saari S, Jalava K. Culture and characteristics of Helicobacter bizzozeronii, a new canine gastric Helicobacter sp. Int J Syst Bacteriol. 1996;46:160–166. doi: 10.1099/00207713-46-1-160. [DOI] [PubMed] [Google Scholar]
- 29.Hill S, Cheney J M, Taton-Allen G, Reif J, Bruns C, Lappin M. Prevalence of enteric zoonotic organisms in cats. J Am Vet Med Assoc. 2000;216:687–692. doi: 10.2460/javma.2000.216.687. [DOI] [PubMed] [Google Scholar]
- 30.Hwang M-N, Ederer G M. Rapid hippurate hydrolysis method for presumptive identification of group B streptococci. J Clin Microbiol. 1975;1:114–115. doi: 10.1128/jcm.1.1.114-115.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lastovica A J, Le Roux E, Penner J L. Campylobacter upsaliensis isolated from blood cultures of pediatric patients. J Clin Microbiol. 1989;27:657–659. doi: 10.1128/jcm.27.4.657-659.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lastovica A J, le Roux E. Efficient isolation of campylobacteria from stools. J Clin Microbiol. 2000;38:2798–2799. doi: 10.1128/jcm.38.7.2798-2799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lastovica A J, Skirrow M B. Clinical significance of Campylobacter and related species other than Campylobacter jejuni and C. coli. In: Nachamkin I, Blaser M J, editors. Campylobacter. 2nd ed. Washington, D.C.: ASM Press; 2000. pp. 89–120. [Google Scholar]
- 34.Lawson A J, Logan J M J, O'Neill G L, Desai M, Stanley J. Large-scale survey of Campylobacter species in human gastroenteritis by PCR and PCR-enzyme-linked immunosorbent assay. J Clin Microbiol. 1999;37:3860–3864. doi: 10.1128/jcm.37.12.3860-3864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawson A J, Linton D, Stanley J, Owen R J. Polymerase chain reaction detection and speciation of Campylobacter upsaliensis and C. helveticus in human faeces and comparison with culture techniques. J Appl Microbiol. 1997;83:375–380. doi: 10.1046/j.1365-2672.1997.00240.x. [DOI] [PubMed] [Google Scholar]
- 36.Linton D, Lawson A J, Owen R J, Stanley J. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J Clin Microbiol. 1997;35:2568–2572. doi: 10.1128/jcm.35.10.2568-2572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meanger J D, Marshall R B. Campylobacter jejuni infection within a laboratory animal production unit. Lab Anim. 1989;23:126–132. doi: 10.1258/002367789780863637. [DOI] [PubMed] [Google Scholar]
- 38.Moreno G, Griffiths P, Connerton I, Park R. Occurrence of campylobacters in small domestic and laboratory animals. J Appl Bacteriol. 1993;75:49–54. doi: 10.1111/j.1365-2672.1993.tb03406.x. [DOI] [PubMed] [Google Scholar]
- 39.Owen R J, Morgan D D, Costas M, Lastovica A. Identification of Campylobacter upsaliensis and other catalase-negative campylobacters from paediatric blood cultures by numerical analysis of electrophoretic protein patterns. FEMS Microbiol Lett. 1989;49:145–150. doi: 10.1016/0378-1097(89)90029-3. [DOI] [PubMed] [Google Scholar]
- 40.Park R, Griffiths P, Moreno G. Sources and survival of campylobacters: relevance to enteritis and the food industry. Soc Appl Bacteriol Symp Ser. 1991;20:97S–106S. [PubMed] [Google Scholar]
- 41.Parsonnet J, Hanson S, Rodriguez L, Gelb A B, Warnke R A, Jellum E, Orentreich N, Vogelman J H, Friedman G D. Helicobacter pylori infection and gastric MALT lymphoma. N Engl J Med. 1994;330:1267–1271. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- 42.Paster B J, Dewhirst F E. Phylogeny of Campylobacter, wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic acid sequencing. Int J Syst Bacteriol. 1988;38:56–62. [Google Scholar]
- 43.Paster B J, Lee A, Fox J G, Dewhirst F E, Tordoff L A, Fraser G J, O'Rourke J L, Taylor N S, Ferrero R. Phylogeny of Helicobacter felis sp. nov., Helicobacter mustelae, and related bacteria. Int J Syst Bacteriol. 1991;41:31–38. doi: 10.1099/00207713-41-1-31. [DOI] [PubMed] [Google Scholar]
- 44.Patton C M, Shaffer N, Edmonds P, Barrett T J, Lambert M A, Baker C, Perlman D M, Brenner D J. Human disease associated with Campylobacter upsaliensis (catalase negative or weakly positive Campylobacter species) in the United States. J Clin Microbiol. 1989;27:66–73. doi: 10.1128/jcm.27.1.66-73.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prescott J F, Munroe D. Campylobacter jejuni enteritis in man and domestic animals. J Am Vet Med Assoc. 1982;181:1524–1530. [PubMed] [Google Scholar]
- 46.Romero S, Archer J R, Hamacher M E, Bologna S M, Schell R F. Case report of an unclassified microaerophilic bacterium associated with gastroenteritis. J Clin Microbiol. 1988;26:142–143. doi: 10.1128/jcm.26.1.142-143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 48.Sandstedt K, Ursing J, Walder M. Thermotolerant Campylobacter with no or weak catalase activity isolated from dogs. Curr Microbiol. 1983;8:209–213. [Google Scholar]
- 49.Schauer D B, Ghori N, Falkow S. Isolation and characterization of “Flexispira rappini ” from laboratory mice. J Clin Microbiol. 1993;31:2709–2714. doi: 10.1128/jcm.31.10.2709-2714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen Z, Feng Y, Fox J G. Identification of enterohepatic Helicobacter species by restriction fragment length polymorphism analysis of the 16S rRNA gene. Helicobacter. 2000;5:121–128. doi: 10.1046/j.1523-5378.2000.00019.x. [DOI] [PubMed] [Google Scholar]
- 51.Sorlin P, Vandamme P, Nortier J, Hoste B, Rossi C, Pavlof S, Struelens M J. Recurrent “Flexispira rappini” bacteremia in an adult patient undergoing hemodialysis: case report. J Clin Microbiol. 1999;37:1319–1323. doi: 10.1128/jcm.37.5.1319-1323.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanley J, Burnens A, Linton D, On S, Costas M, Owen R. Campylobacter helveticus sp. nov., a new thermophilic species from domestic animals: characterization, and cloning of a species-specific DNA probe. J Gen Microbiol. 1992;138:2293–2303. doi: 10.1099/00221287-138-11-2293. [DOI] [PubMed] [Google Scholar]
- 53.Stanley J, Linton D, Burens A P, Dewhirst F E, On S L W, Porter A, Owen R J, Costas M. Helicobacter pullorum sp. nov.—genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology. 1994;140:3441–3449. doi: 10.1099/13500872-140-12-3441. [DOI] [PubMed] [Google Scholar]
- 54.Stanley J, Linton D, Burens A P, Dewhirst F E, Owen R J, Porter A, On S L W, Costas M. Helicobacter canis sp. nov., a new species from dogs: an integrated study of phenotype and genotype. J Gen Microbiol. 1993;139:2495–2504. doi: 10.1099/00221287-139-10-2495. [DOI] [PubMed] [Google Scholar]
- 55.Taylor D E, Hiratsuka K, Mueller L. Isolation and characterization of catalase-negative and catalase-weak strains of Campylobacter species, including “Campylobacter upsaliensis,” from humans with gastroenteritis. J Clin Microbiol. 1989;27:2042–2045. doi: 10.1128/jcm.27.9.2042-2045.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tee W, Leder K, Karroum E, Dyall-Smith M. “Flexispira rappini” bacteremia in a child with pneumonia. J Clin Microbiol. 1998;36:1679–1682. doi: 10.1128/jcm.36.6.1679-1682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Totten P A, Fennel C L, Tenover F C. Campylobacter cinaedi (sp. nov.) and Campylobacter fennelliae (sp. nov.): two new Campylobacter species associated with enteric disease in homosexual men. J Infect Dis. 1985;151:131–139. doi: 10.1093/infdis/151.1.131. [DOI] [PubMed] [Google Scholar]
- 58.Willard M D, Sugarman B, Walker R D. Gastrointestinal zoonoses. Vet Clin North Am Small Anim Pract. 1987;17:145–178. doi: 10.1016/s0195-5616(87)50610-6. [DOI] [PubMed] [Google Scholar]