Abstract
Advances in quantitative biomarker development have accelerated new forms of data-driven insights for patients with cancer. However, most approaches are limited to a single mode of data, leaving integrated approaches across modalities relatively underdeveloped. Multimodal integration of advanced molecular diagnostics, radiological and histological imaging, and codified clinical data presents opportunities to advance precision oncology beyond genomics and standard molecular techniques. Yet most medical datasets are still too sparse to be useful for the training of modern machine learning techniques, and significant challenges remain before this is remedied. Combined efforts of data engineering, computational methods for analysis of heterogeneous data, and instantiation of synergistic data models in biomedical research are required for success. In this Perspective, we offer our opinions on synthesizing complementary modalities of data with emerging multimodal artificial intelligence methods. Advancing along this direction will result in a re-imagined class of multimodal biomarkers to propel the field of precision oncology in the coming decade.
Introduction
As patients with cancer traverse diagnostic, treatment, and monitoring processes, physicians order a suite of diagnostics across distinct modalities to guide management. A significant opportunity thus emerges to aggregate, integrate, and analyse these complementary digital assets across large patient populations to discover multimodal prognostic features, learning from the collective history of large cohorts of patients to inform better management of future patients. For example, genomic profiling of tumor tissue has significantly enhanced clinical decision-making, and the genomic data produced in turn yield a rich molecular repository for further study 1. This leads to further understanding of the cancer genome, drug sensitivity 2 and resistance mechanisms, 3 and prognostic associations 4,5. During and after treatment, serial radiological imaging, such as positron emission tomography (PET) and computerized tomography (CT), quantifies tumor burden in response to intervention, yielding digital archives for large-scale machine learning [G] (ML). Pathology specimens depicting cell morphology, tissue architecture, and tumor–immune interfaces also are increasingly digitized 6. Other modalities in development, such as cell-free DNA analysis and serial laboratory medical tests of biochemical and metabolic analytes, provide longitudinal read-outs of tumor progression and recurrence 7-11.
We contend that integrated anatomical, histological, and molecular measurements approach a comprehensive description of the state of a cancer, resulting in an effective ‘digital biobank’ 12 for each patient. However, at present, even when these data are available, they are rarely integrated, and few advances have been reported that computationally exploit the research discovery potential of large-scale, multi-modal integration. Artificial intelligence [G] (AI) and ML techniques have enormous potential to convert data into a new generation of diagnostic and prognostic models and to drive clinical and biological discovery, but the potential of these techniques often goes unrealized in biomedical contexts, where research-ready datasets are sparse. Cultural and infrastructural changes toward scaled research-ready data archives and development of multimodal ML methods will advance our understanding of the statistical relationships among diagnostic modalities and the contextual relevance of each. Repurposing aggregated, multimodal data—the digital biobanks—therefore presents opportunities to develop next-generation, data-driven biomarkers [G] to advance patient stratification and personalised cancer care.
The central premise of multimodal data integration is that orthogonally derived data complement one another, thereby augmenting information content beyond that of any individual modality. Concretely, modalities with fully mutual information would not yield improved multimodal performance compared to each modality alone. Modalities with fully orthogonal information, conversely, would dramatically improve inference. For example, radiological scans and pathological specimens describe tumors spatially at different scales and thus are expected to describe disparate elements of tumor biology. Each modality is incomplete and often noisy, but integrating weak signals across modalities can overcome noise in any one modality and more accurately infer response variables of interest, such as risk of relapse or treatment failure.
To exemplify this premise, we will focus on four major modalities in cancer data: histopathology, radiology, genomics, and clinical information (Figure 1). While rapid progress using deep learning [G] (DL) and other ML methods has been made in each of these individual modalities, major unresolved questions about multimodal data integration remain. What are the latent relationships and underlying causal mechanisms at the molecular, cellular, and anatomical scales? Can rational multimodal predictive models enhance clinical outcomes for patients with cancer? Can cancer research exploit advances in computational methods and AI models to realise new insights from multimodal data integration? How much data is enough to realise such generalisable predictive models? How can annotations produced during routine clinical care and focused research studies be repurposed to train robust models? How can we fully engage and academically credit both clinicians and data scientists in collaborative studies? How do we establish data infrastructures to enable meaningful and rapid scientific advances while preserving the integrity of patient consent? Herein, we explore these questions through literature review and by developing a blueprint for navigating the infrastructural, methodological, and cultural challenges along the path to achieving robust multimodal data integration in cancer research.
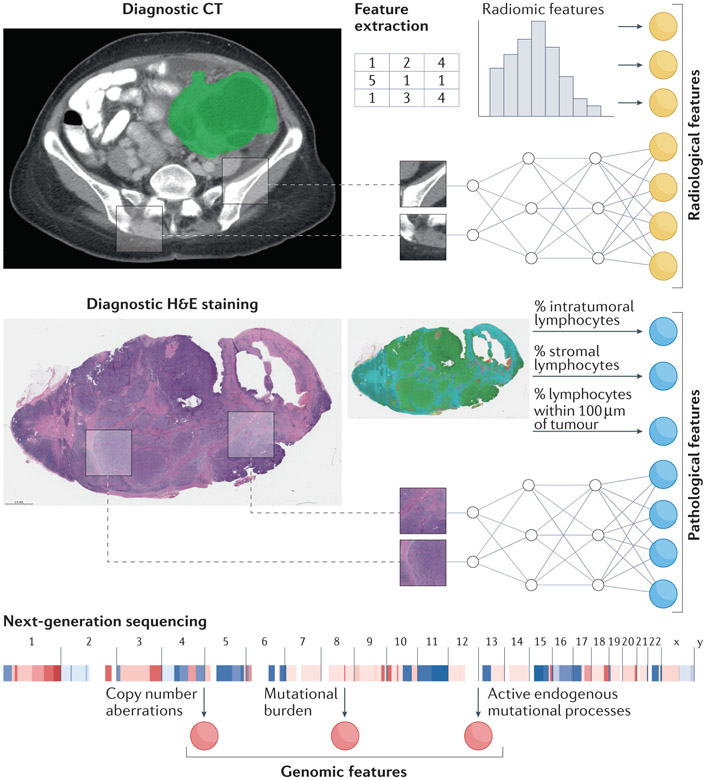
Figure 1. Example data modalities for integration include radiology, histopathology, and genomic information.
Image feature extraction involves choosing deep learning or engineered features. CT, computed tomography; H&E, hematoxylin & eosin.
Unimodal machine learning methods
Cancer imaging data have been exploited to predict molecular features of tumors and to discover new prognostic associations with clinical outcomes, and we refer readers to a number of excellent reviews in these areas 13-15. In radiology specifically, previous work analyzed features manually extracted by radiologists, such as the VASARI (Visually Accessible Rembrandt Images) set of imaging features for glioma, and their association with clinical outcomes and molecular biomarkers 16. However, such features are highly prone to inter-reader variability, and the laborious nature of extraction limits cohort size. As radiology data are digital by construction, automatically extracting deterministic, quantitative features is tractable 17. These features have been associated with clinical outcomes, such as response to immune checkpoint blockade (ICB) in pan-cancer analyses 18, residual tumor volume after resection in ovarian cancer 19, and progression of disease in pediatric optic pathway glioma 20. Furthermore, when cohorts are sufficiently large, convolutional neural networks [G] (CNNs), a type of deep neural network [G] (DNN) [Box 1] have been shown to predict isocitrate dehydrogenase 1 (IDH1) mutational status of glioma from magnetic resonance imaging (MRI), pathological grade of prostate cancer from MRI, epidermal growth factor receptor (EGFR) mutational status of lung adenocarcinoma from CT, and BRCA1 or BRCA2 mutational status of breast cancer from full-field digital mammography 21-25. Three-dimensional CNNs have shown success in stratifying patients with non-small-cell lung cancer (NSCLC) by overall survival (OS) 26 and empirically outperformed two-dimensional CNNs in other radiology tasks, such as diagnosing appendicitis 27. The relative performance of DL versus conventional ML-based methods on human-defined (‘engineered’) features is largely determined by cohort size.
Box 1. Deep learning architectures.
Machine learning (ML) can be divided broadly into unsupervised learning and supervised learning. Unsupervised learning seeks to discover intrinsic patterns in data, sometimes without known labels for each data point, while supervised learning seeks to predict a label of interest from the input data. Deep learning is a subtype of ML that has the potential to learn more informative features than engineered features, but there is difficulty in model interpretability and performance is notoriously dependent on the amount of training data available 14. No ML algorithm is universally superior to another, but the data and targets to be related motivate the choice of model 158,159. With sufficient training data, deep neural networks (DNNs) have become a leading approach to capture salient patterns within data. DNNs are universal function approximators that learn a distributed representation of given data, with deep features often describing data better than competing human-defined features 160. Though these methods are limited by the need for large training datasets and the difficulty of interpreting their learned features, they are indispensable for discovering highly informative features in clinical datasets.
Specific variants of DNNs exist for different data modalities. For example, convolutional neural networks (CNNs) learn sliding window-like kernels to detect textural patterns within images, often achieving or exceeding human performance in image classification. Some of the most popular variants, available off the shelf in modern DL frameworks, are ResNet, Inception, DenseNet, and SqueezeNet 161-164. For sequential data such as time series of lab values, recurrent neural networks (RNNs) can be the architecture of choice. The RNN uses each data point to update its understanding of the data, building an amalgamated representation that is then used to predict the outcome of interest, such as risk of disease recurrence. The most successful variants are long short-term memory (LSTM) and gated recurrent unit (GRU) networks 165,166. More recently, transformer networks 167 have demonstrably outperformed RNNs in sequential learning. Although RNNs and transformers have not yet been widely applied in oncology, preliminary studies of RNNs and transformers for longitudinal medical event prediction have yielded promising results 76-78,168. For high-dimensional data such as transcriptomic profiles, the attention gating mechanisms (methods to identify which data elements are most relevant) inherent in deep highway networks 79 have helped identify salient features amidst potentially uninformative background 80.
In histological imaging, similar computational models have advanced biomarker identification, particularly from hematoxylin and eosin (H&E)-stained whole slide images (WSIs) 28-32, beyond the previously dominant practice of using pathologist-extracted features 33. One notable multi-center example in colorectal cancer showed that H&E WSIs contain information predictive of microsatellite instability (MSI) status as a biomarker for response to ICB34,35. However, these DL analyses suffer from poor interpretability and depend heavily on large training cohorts (depending on the task and data complexity, generally thousands of labeled examples are required for excellent, generalizable performance). Interpretable quantitative analyses of histological images also can be conducted using expert-guided cellular and tissue annotations, identifying biological features such as tumor-infiltrating lymphocytes (TILs) and other properties of the tumor microenvironment and their correlation with molecular features 36. A recent pan-cancer analysis found that annotation-guided interpretable features predict endogenous mutational processes and features of the tumor microenvironment 37, and other studies have linked biologically interpretable features with clinical outcomes 38,39,40. Deeper assessment of the tumor microenvironment is also possible through characterizing spatial niches derived from multiplexed imaging as well as spatial transcriptomics methods, which can be used to develop biomarkers for precision oncology 41,42.
Molecular features are the true targets of intervention, either directly or through synthetic lethality, and they are thus the most direct measure for predicting drug response. Examples include mutations in BRAF in melanoma 43, EGFR in NSCLC 44, ERBB2 (also known as HER2) in breast cancer 45, IDH1 in acute myeloid leukemia (AML) 46, BRCA1 or BRCA2 in ovarian 47 and prostate cancer 48 and even rare events such as neurotrophic tyrosine kinase (NTRK) fusions 49 for solid tumors, among many others. Targeted cancer therapies are continually being added to the clinical arena, for example, ongoing clinical trials of KRAS (G12C) inhibitors 50,51 and the PI3Kα-specific inhibitor targeting PIK3CA mutations 52 in lung and breast cancer, respectively. Higher-order genomic properties such as tumor mutational burden (TMB) 53, endogenous mutational processes such as MSI 54 and homologous recombination deficiency (HRD), and large-scale features such as whole genome duplication 55 are also clinically meaningful. In a recent study, VÖhringer et al. 56 present an algorithm (TensorSignatures) to characterise transcription-associated mutagenesis in seven cancer types. Copy number signatures from low-pass whole genome sequencing 57 and integrated ML models across single nucleotide variant (SNV) and structural variant scales 58 have also effectively stratified patients into prognostic subgroups. Both studies find that patients with HRD tumors have better prognosis, but further granularity is needed to better resolve clinically meaningful subgroups. Emerging spatial genomics techniques 59,60,61 and complementary clinical and imaging modalities are opportunities to enrich these data and refine prognostication.
Multimodal machine learning
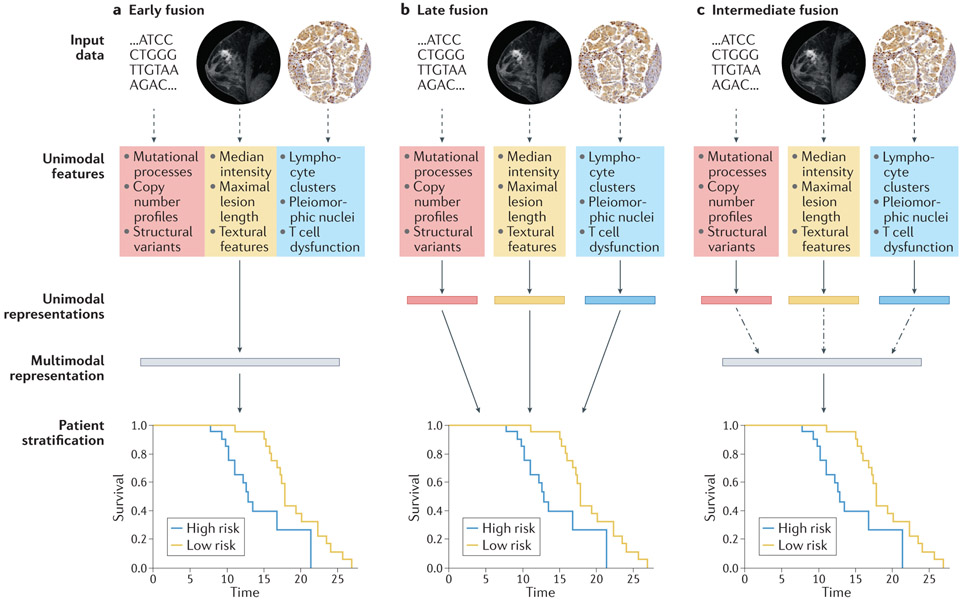
We suggest that such unimodal models across radiology, histopathology, molecular, and clinical domains discussed above will become the building blocks of integrated multimodal models (Figure 2). A major design choice for multimodal approaches is the extent to which each data input should be modeled before encoding joint representations (Figure 3). In early fusion architectures, features are simply concatenated at the outset and used to train a single model (Figure 3a). At the other extreme, late fusion architectures model unimodal data fully individually, and then aggregate learned parameters or derived scores (Figure 3b). Intermediate fusion architectures develop a representation of each modality and then model intermodal interactions before joint modeling (Figure 3c). Most multimodal architectures have more parameters to fit than their unimodal counterparts, making them prone to overfitting (learning to represent the training data too exactly, resulting in an ungeneralizable model), which paradoxically can result in worse performance in the supervised learning [G] setting 62. One mechanism to address this is incorporating the estimated generalization error in the training objective, using techniques such as gradient blending, a technique to weight each unimodal contribution to the overall loss based on its estimated generalization error 62. A related design choice in multimodal machine learning is the complexity of the constituent unimodal models. Although overparameterized DL models can outperform traditional ML, their performance is highly dependent on the size of the training dataset. This data size requirement often precludes DL application in biomedical multimodal studies, where missingness of individual data modalities, and requirement of laborious—often costly— curation of multiple data modalities limits studies to the very small data regime, defined loosely as ~5000 or fewer data points 63. This makes ML on engineered features an essential approach in the field and suggests that studies with resource constraints requiring very large cohorts, such as those in cancers with high heterogeneity, or those where a single modality overwhelmingly carries the important discriminative features, may opt for a unimodal study.
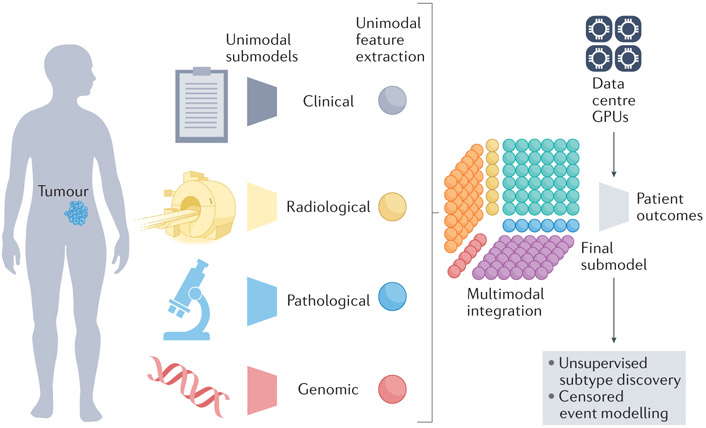
Figure 2. Multimodal models integrate features across modalities.
Sub-models extract unimodal features from each data modality. Next, a multimodal integration step generates intermodal features—a Tensor Fusion Network (TFN) is indicated here 86. A final sub-model infers patient outcomes. GPU, graphics processing unit.
Figure 3. Design choices for multimodal models with genomic, radiological, and histopathological data.
Solid arrows indicate stages with learnable parameters (linear or otherwise), dashed arrows indicate stages with no learnable parameters, and dashed and dotted arrows indicate the option for learnable parameters, depending on model architecture. (a) In early fusion, features from disparate modalities are simply concatenated at the outset. (b) In late fusion, each set of unimodal features is separately and fully processed to generate a unimodal score before amalgamation by a classifier or simple arithmetic. (c) In intermediate fusion, unimodal features are initially processed separately prior to a fusion step, which may or may not have learnable parameters, and subsequent analysis of the fused representation. All schemata shown are for deep learning (DL) on engineered features: for convolutional neural networks (CNNs) directly on images, unimodal features and unimodal representations are synonymous. For linear machine learning (ML) on engineered features, no representations are learned between features and stratification. The magnetic resonance imaging (MRI) images were obtained from The Cancer Imaging Archive (TCIA). Histology images were obtained from the Stanford Tissue Microarray database, ref. 155.
Preliminary uses of multimodal machine learning to stratify patients
Multimodal patient stratification using complementary multi-omics cancer data is well developed 64-69. The Cancer Genome Atlas (TCGA) catalogues of genomic, transcriptomic, epigenomic and proteomic data enabled integrated, multimodal inference. For example, integrating bulk transcriptomics, microRNA (miRNA) sequencing, and promoter methylation status with early fusion autoencoders [G] showed enhanced ability to stratify patients with hepatocellular carcinoma by OS 65. A similar approach identified distinct survival subtypes in the majority of TCGA cancer types, outperforming existing stratification methods 66. Joint dimensionality reduction [G] techniques, such as integrative non-negative matrix factorization, learn unsupervised representations of multi-omic profiles for downstream association with outcomes and biomarkers 70. The Clinical Proteomic Tumor Analysis Consortium (CPTAC) provides another public catalogue of correlated genomic and proteomics data across a diverse number of sites of cancers, with overlap with TCGA. As experimental and computational techniques advance, these data will more completely characterize the molecular state of patients’ disease 71,72, yet they still only capture a fraction of the informative data. Several multi-omic models also incorporate traditional clinical features 73,74. For example, dimensionality reduction, early fusion (Figure 3), and a deep Cox Proportional Hazards (CPH) model [G] to integrate multi-omics with age and hormone receptor status stratified patients with breast cancer by OS more accurately than unimodal models 74. Adding additional modalities paradoxically failed to increase performance, with most clinico-genomic models in the study slightly underperforming the genomic model alone, except when TMB and copy number burden were integrated 74. Further work is needed to determine when and why adding particular modalities is useful. CPH models also are limited by their assumption of linear dependence on each variable and challenges with handling tied samples (when events occur at the same time). Deep binned time survival 75 overcomes these limitations by discretizing follow-up times and predicts risk of NSCLC recurrence from 30 clinical and histopathological features. Recurrent neural networks [G] (RNNs) and transformers, leading methods for time series prediction [Box 1], have not yet been widely applied in oncology, but have been shown to accurately predict clinical events from multimodal serological, radiomic, and clinical data 76-78
Although under-developed relative to clinical and ‘omics integration, multimodal models including histopathology imaging features have recently emerged. One such model uses deep highway networks [Box 1] to integrate H&E images with mRNA-sequencing (mRNA-seq) and miRNA-seq data to learn the importance of individual genomic features rather than perform a priori dimensionality reduction 79, embedding the individual data modalities in the same, shared information space by minimizing the similarity loss. The model achieves a concordance index [G] (c-Index) of 0.78 to stratify patients by OS80 and is robust to missingness, but it conceptually encourages mutual information, potentially at the expense of complementary information gained via fusion methods (Figure 3), though this remains to be tested in a head-to-head comparison. Similarly, Imaging-AMARETTO 81, a framework developed on TCGA glioma data, advances associations between imaging phenotypes and molecular multi-omics, but it does not integrate information explicitly for prognostication. Other examples of multimodal ML studies using histopathology include cellular morphological features and mRNA-seq data integration in NSCLC 82, combined histological and gene expression features in breast cancer 83, and histopathological and genomic features in glioma using genomic survival CNNs84 and tensor fusion networks (TFNs) 85. TFNs are intermediate fusion architectures using the outer product of deep unimodal embeddings 86, which enables the model to learn intermodal dynamics and outperform models based only on grade and molecular subtype (c-Index 0.83 vs 0.78) or any individual modality 85. It also outperforms simpler multimodal models, such as genomic survival CNNs (c-Index 0.83 vs 0.78) 85. In general, these studies demonstrate that multimodal integration with histopathological imaging improves outcome predictions and stratification over unimodal and molecular methods alone.
Few multimodal models include radiological imaging. However, a model to diagnose breast cancer using digital mammography and diffusion contrast-enhanced MRI achieved an AUROC [G] (area under the receiver operating characteristic curve) of 0.87, higher than the respective unimodal AUROC values of 0.74 and 0.78 87. Another study found that the combination of deep features from histological imaging and engineered features from MRI outperformed unimodal classifiers for stratification of brain tumor subtypes 88. MRI radiomic features also refine survival stratification beyond IDH1 mutational status and World Health Organization (WHO) classifications alone, demonstrating the potential of multi-scale information to improve stratification 89. Multiple kernel [G] learning has been used on small, noisy datasets to integrate clinical factors with MRI- and PET-derived imaging features 90,91. PET imaging is a particularly promising area for multimodal integration, providing spatial profiles of metabolic activity 92. Similarly, MRI sequences such as dynamic contrast enhanced images depicting vasculature and diffusion weighted images, whose voxel [G] intensities are influenced by cellularity, provide rich physical profiles with potentially complementary prognostic information. Despite the shortage of multimodal works incorporating radiology, preliminary results are promising 78,93,94.
Promising methodological frontiers for multimodal integration
Multimodal ML in the medical setting is most limited by the disparity between data availability and amount of data needed to fit multimodal models. Hence, many methodological frontiers involve increasing robustness to overfitting and dealing rationally with missingness. For example, transfer learning in unimodal models involves pre-training a model on a large, tangentially related dataset and then fine-tuned on the actual dataset of interest, which is typically small. Some example datasets used are ImageNet 95, a database of more than 14 million labeled images used to train image classification algorithms for two-dimensional CNNs, and Kinetics, a curated collection of approximately 650,000 YouTube videos depicting human actions for three-dimensional CNNs (reported in a preprint 96). However, recent evidence shows that small models without pre-training, such as ResNet-50, for small medical imaging datasets can perform comparably to pre-trained large models 63. This is consistent with the hypothesis that the benefits of pre-training for small medical imaging datasets are related to low-level feature reuse and feature-independent weight scaling 63. It remains an open question whether pre-training multimodal fusion models can combat overfitting through similar weight scaling of the parameters involved in fusing unimodal representations. Both prospective clinical trials and highly curated retrospective cohorts often have low numbers of patients, highlighting the importance of studying how to use DL techniques appropriately to discover patient strata in the very small data regime.
One of the root causes of data scarcity is the need for extensive annotation: tumors need to be localized on CT scans or H&E images, and survival outcomes typically require manual review of medical records. Harnessing data at scale requires reducing this burden of annotation, especially in multimodal studies. Automated annotation approaches could provide solutions. For example, RetinaNet, an object detection CNN, has been used to localize lung nodules on CT, enabling use of 42,290 CT cases for training 97. Analogously, an ML-based model to automatically delineate representative tumor tissue from colorectal carcinoma histology slides enabled training on 6,406 specimens 35. Weakly supervised learning (WSL) also helps reduce the burden of annotation by using informative-yet-imperfect labels for the training dataset. While weak labels may be incomplete, inexact, or inaccurate 98, WSL applications in computational pathology have resulted in robust models to infer genomic alterations 31 and diagnose cancer 99. Weaknesses of this approach include the absence of a ground-truth dataset (a dataset with expert annotations that are exactly correct and can be treated as the gold standard) for model evaluation when all labels are inexact or inaccurate and its dependence on large dataset sizes. Active learning is a form of machine learning that solicits precise labels for targeted instances, selected using either informativeness or representativeness of an instance 98. For example, it can be used to prioritize expert annotations in real time for pathology tissue-type labeling (Figure 4). These strategies are essential in clinical contexts, where most data elements possess only weak labels, and are a leading strategy to learn robust models from large, information-poor datasets. Therefore, WSL is a useful strategy to augment annotations, dramatically increasing the size and robustness of usable multimodal datasets for clinical oncology.
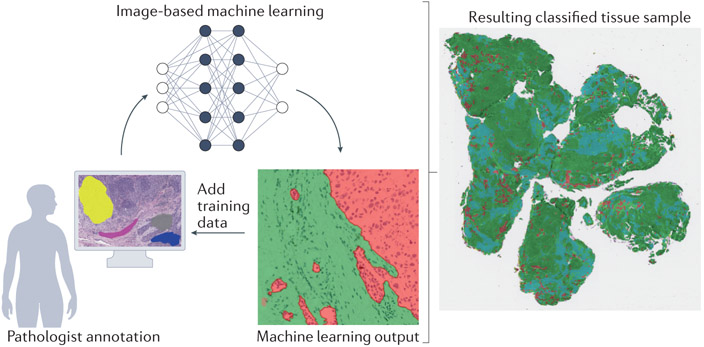
Figure 4. Active learning reduces the burden of annotation.
In active (‘human in the loop’) learning, a pathologist first annotates small training areas representing tissue areas (for example, tumor, stroma and lymphocytes). Next, a machine learning classifier is trained from these expert annotations. Finally, the resulting labeled sample can be examined for misclassified regions, and the pathologist adds targeted additional training areas. This process is repeated until the classification is accurate and can be applied to multiple samples.
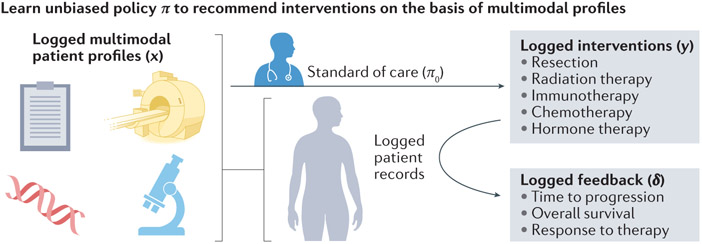
As more such datasets become annotated and integrated, oncology will benefit from multimodal recommender systems [G] , analogous to inferring cancer drug response based on unimodal gene expression data 100. Retrospective observational studies contain no matched controls, which biases training data and requires methods such as counterfactual ML [G] to learn accurate recommendation policies from logged interventions and resultant outcomes 101. In oncology, a counterfactual recommender system (Figure 5) would learn policies to recommend future therapies for new patients based on historical patient records of administered treatments, patient contexts (for example, a pre-treatment CT scan and H&E-stained biopsy), and survival outcomes 101,102. In general, this is not currently possible because patient data are not accessible and annotated at the scale required, but such methods have great potential as datasets are assembled and prospective data collection methods improve.
Figure 5. Recommender systems could learn from retrospective data to assist in clinical decision-making.
Logged healthcare data comprises multimodal patient contexts x, interventions y based on the standard of care (π0), and feedback δ based on the outcome of the intervention. Learning from such data is challenging because of the lack of two-arm design and the biased data based on the changing standard of care. Counterfactual recommender systems learn theoretically guaranteed unbiased policies from these data. Then, the validated policy π can be applied prospectively to support physicians’ management decisions.
Finally, unsupervised learning [Box 1] continues to develop in general, with potential to both facilitate discovery of new cancer phenotypes and probe multimodal associations. For example, deep probabilistic canonical correlation analysis jointly learns parameters for two DNNs and a transformation to embed them in the same information space, all with Bayesian inference [G] suitable for small datasets 103. This method is especially well suited for probing the mutual information to generate hypotheses for experimental biology, such as genomic drivers of cellular morphological heterogeneity. At the patient level, an unsupervised Bayesian topic model has been applied to learn multimodal topics that stratify patients by risk of mortality 104 and in deriving mutational process activities in genomic datasets 58. Surprisingly, progress in this area demonstrates statistical power across feature spaces from data measuring signals at vastly disparate scales (for example, histological–genomic, or radiomic–molecular). We therefore anticipate that generative methods have potential to discover new phenotypes and to generate hypotheses to guide experimental biology.
Challenges with multimodal data
The challenges inherent in multimodal integration of clinical cancer data fall into three broad categories: data engineering and curation, ML methods, and data access and governance provisions. These challenges extend to both retrospective studies seeking to discover biomarkers from standard-of-care data and prospective studies focused on bespoke or advanced data types. The field also shares two broad categories of challenges with unimodal ML studies in medicine, which are interpreting results and ensuring their reproducibility. Here, we describe these five categories of challenges along with potential solutions to address them.
Data availability
Perhaps the greatest challenge in multimodal machine learning is data scarcity. Data acquired during the standard of care are not structured in a research-ready format: stained tissue specimens typically must be manually located and scanned, and radiological images are stored in the picture archiving and communication system (PACS) 105 with limited clinical annotation. The modalities are typically organized with different patient identifiers, complicating alignment. A related challenge is spatial colocalization, which is especially important for studying biological correlation of multimodal features. Small datasets such as Ivy Glioblastoma Atlas Project (IvyGAP) 106 richly profile the genomics and multi-scale tumoral architecture of patients with matched clinical outcomes and represent the promise of spatial colocalization. To achieve this at scale, image-guided biopsies or 3D-printed molds based on tumor morphology 107,108 are possible solutions, but challenges remain before these approaches are scaled for prospective research.
The general limits of using healthcare records to conduct research are widely discussed 109: one challenge is unobserved patient outcomes, which can be handled for time-to-event analysis but requires excluding patients for categorical outcomes. Another major bottleneck is retrospective chart review, or manually reviewing patient records to extract specific features into spreadsheets. It is error prone and variable, and repeated review is often required to capture new clinical events 110,111. Efforts are underway to build models to automatically codify clinical information from unstructured text, and ontologies such as Observational Health Data Sciences and Informatics (OHDSI)112 and American Association for Cancer Research project Genomics Evidence Neoplasia Information Exchange (AACR project GENIE)1 structure disparate clinical elements to facilitate retrospective research. These models ought to be extended to incorporate additional data modalities.
As structured data emerge, data lakes [G] are a scalable solution to organize original data and track their use during subsequent analysis 113. Data lake technologies unify siloed data and accommodate both known and unforeseen file types. Cost-effective data lake storages are readily available from commercial vendors (for example, Amazon S3) as well as open source products (for example, Delta Lake 114). The ensuant technical challenges vary depending on whether the data lake is set up in the cloud, on premises, or in a hybrid solution 115. There are specific challenges when applying data lakes for biomedical research, such as the stripping of protected health information (PHI) to protect patient privacy and facilitate inter-institutional sharing.
Such cross-institutional data sharing is essential to promote and test model generalizability. Leading platforms include the database of Genotypes and Phenotypes (dbGaP), the European Genome-phenome Archive (EGA), The Cancer Imaging Archive (TCIA), the Genomic Data Commons (GDC), and other resources in the National Cancer Institute (NCI) Cancer Research Data Commons. However, beyond matched genomic data and H&E WSIs of TCGA and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), public resources contain only small patient cohorts with multiple data modalities.
Major causes of this public data scarcity include the logistical challenges of anonymizing data and institutional privacy policies. Federated learning [G] is a potential solution116. Depending on the choice of model, federated learning can require novel training approaches 117 but enables training on multi-institutional cohorts without data having to leave local networks.
Data integration and analysis
As integrated datasets mature, challenges will shift to data analysis. Complete data on all patients of a study of interest is rare, and this missingness complicates multimodal data integration. Most traditional multivariate models, such as Cox models, cannot handle this directly and thus require either exclusion of patients without all data modalities or overly simplistic interpolation (for example, by median). Both of these strategies fail to harness all available data to train effective models. To circumvent this, one simple solution is to use late fusion (Figure 3b), where each unimodal model can be trained separately to infer the outcome of interest, which can then be integrated. Bayesian approaches 118 also offer analytical solutions for missingness.
Data modeling will also be complicated by institution-specific biases in the data, such as staining and scanning particularities in histopathology 119-121, scanner parameters in MRI, and differing ontologies in clinical data. Preprocessing techniques in MRI 122 and H&E 123,124 address this heterogeneity, and with large cohorts, DL is somewhat robust to noise 28,125, but such heterogeneity is a major reason that AI systems fail when trialed in the clinic 126. An additional complexity in multimodal studies is that unimodal biases are likely to be correlated. For example, biasing factors such as MRI manufacturers and H&E staining artifacts likely differ more between institutions than within an institution. This will make it more challenging to model general intermodal relationships, motivating greater cross-institutional data representation and potentially motivating the development of methods that explicitly model these multimodal biases or normalise against them. The decision to acquire one data modality may also be based on another modality, which necessitates either limiting multimodal input to a single time point or accounting for these dependencies during time series modeling. Different modalities with different levels of heterogeneity may require different training dataset sizes—in this case, training the overall model may involve pre-training the unimodal sub-model using the larger unimodal cohort.
Another analytical challenge is overfitting. Multimodal ML is more prone to overfitting because, in most cases, multimodal datasets are smaller and multimodal models have more parameters to fit. Traditional ML models enable investigators to calculate the necessary dataset size for a tolerable generalization error before analysis. Black box models such as DNNs do not offer such analytical forms. Instead, target dataset size is decided empirically by comparing performance when the model is trained on different proportions of the full data set 35,99. Some evidence suggests that early fusion strategies can perform comparably to unimodal results using less training data 127, but in general, highly parameterized fusion models are likely to require more training data to fit the additional parameters.
Hence, in many settings, multimodal approaches cannot yet fully harness the performance benefits of DL. The most important response to this is to advance clinical data collection to assemble large datasets and better support methods development and benchmarking (see subsection below ‘Data availability’). Meanwhile, smaller datasets curated at single institutions require less complex models to avoid spurious results due to overfitting. Each unimodal model can thus be formulated using ML on engineered features, such as radiomic features from MRI and nuclear morphology features from H&E. One major drawback is the need for laborious annotation, such as segmentation on MRI and tissue type delineation on H&E, which can be reduced using WSL and active learning (see subsection above ‘Promising methodological frontiers for multimodal integration’). For all model types, cross-validation and external testing cohorts are critical to demonstrate generalizability. This is further complicated by the domain specificity of each unimodal component: a model trained on CT would not be expected to accurately interpret MRI, and vice versa. Repurposing unimodal components for integration into new combinations of modalities is likely to reduce the training burden. Furthermore, genomic features such as mutated driver genes or active mutational signatures can often be derived from multiple modalities, such as whole exome sequencing (WES) or whole genome sequencing (WGS), and these deterministic features are general enough to be used despite their modality of origin (provided that the inference of these features is accurate).
With respect to infrastructure, multimodal analytic workflows present hardware and software challenges. Centralised data lakes and workflow management tools minimize duplicated computation, such as image pre-processing, among multiple investigators’ workflows. Computational needs also differ during different parts of the workflow, with a much higher demand during model training than during cohort curation. This is especially true for multimodal models such as TFNs, which generate intermodal representations that scale exponentially with the number of data modalities. Elastic cloud computing resources and the distributed data parallelism [G] of modern DL-based frameworks handle these computational bursts appropriately, but the use of off-premises cloud computing requires robust de-identification of patient data, data security certifications, and measures to control data ingestion and egress costs.
Reproducibility
Reproducibility and benchmarking are major challenges in AI, with many published biomedical AI studies failing to provide source code, test data, or both 128. Several recent seminal works do not provide source code, claiming that internal code dependencies prevent code sharing and that textual descriptions are sufficient to reproduce the results 97,129,130. However, a recent investigation of one of these studies130 found that significant information needed to actually reproduce the study was missing, greatly reducing the impact and ability of the field at large to scrutinize 131 and improve upon it. To foster transparency, scientific reproducibility, and measurable progress, investigators should be encouraged to deposit new multimodal architectures and preprocessing regimens in standardized repositories such as modelhub.ai (reported in a preprint)132. Furthermore, to promote benchmarking and multicenter validation, journals should require investigators to make available published deidentified datasets on public platforms (see subsection above ‘Data availability’). Beyond center-specific confounders, the clinical environment has unpredictable effects on model performance, often leading to substantial performance decrements 133.
Hence, prospective clinical validation is the most relevant measure of a model’s performance 134. This is because directly comparing clinical outcomes with and without the AI system, where both arms are exposed to the inherent noise such as varying image quality and user error, provides an objective, quantitative assessment of a model’s value. SPIRIT-AI and CONSORT-AI are consensus guidelines for AI in clinical trial protocols and reports, respectively, that extend the SPIRIT and CONSORT guidelines for randomized clinical trials 134-136. In broad terms, these guidelines improve reporting transparency and ensure that readers can evaluate practical factors that may impact AI system performance in clinical contexts, such as required training, error handling, and output data format.
Balancing the need for interpretability with empiric efficacy
The nature of DL architectures creates a limiting paradox. While often outperforming standard, interpretable models, users are left to explain improved results without the benefit of drawing from model assumptions encoded in more traditional approaches such as hierarchical Bayes. We argue that investigators should seek to understand learned models from biological and clinical perspectives in order to realise rational multi-modal implementation. Depending on the goals of a study, understanding a model is arguably as important as improving its predictive capacity and will lead to greater mechanistic insight and testable hypotheses. For example, post-hoc explanation methods, which seek to interpret model predictions in terms of input feature values, have been applied to probe medical algorithms 137. However, post-hoc explanations are prone to misinterpretation and cannot supplant true interpretability 138 to elucidate a mechanism or generate hypotheses for experimental biology. Yet when the main purpose of an algorithm is to improve patient outcomes, understanding models mechanistically at the expense of denying patients empirically improved quality of life is unethical. Many empirically beneficial medical interventions, such as general anesthesia, have incompletely understood mechanisms 139. Hence, the most important threshold for using these models in the clinic is the same as for a drug: robust, prospective, multi-center empiric evidence of benefit for patients and an understanding of cases in which the model fails. Given our limited understanding of black-box models, pilot studies must demonstrate that the model is effective and equitable for all patient subpopulations it will encounter before deployment at scale 140.
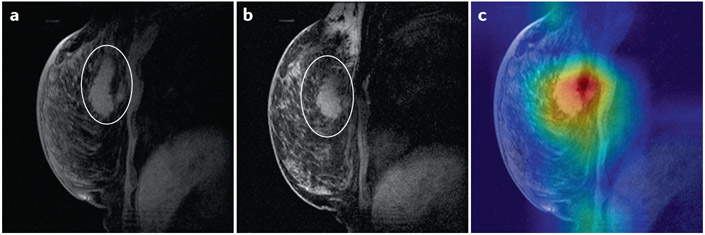
Truly causal models are a frontier of AI research, and in the future such models will be highly valuable in this field 141. Less challenging than interpretability, explicability is also useful for black-box models. For example, class activation maps (CAM) 142 (Figure 6) depict which parts of the image are most important for the model to arrive at its decision. The saliency, or dependence of the output on a specific region, is shown for prediction of response to chemotherapy in Figure 6c. This technique is limited by seeking explicability rather than interpretability 138, but it can be useful to rule out obviously spurious determinants of model output. For example, if the CAM in Figure 6c showed highest saliency in the area outside the breast, it would raise serious concern about the validity of the model. Lucid is another method for explicability which uses the learned model to generate example images for each class 143. For example, it has been applied to visualize what a CNN is looking for in breast H&E images to distinguish tumor from benign tissue 144. For DNNs with definable input variables, layer-wise relevance propagation [G] (LRP) is widely used and has been applied to clinical data 137. However, these methods were developed for unimodal ML, and interpreting multimodal ML is more challenging. Future work must quantify the relative contribution of each modality and their interactions. Uninformative feature counterfactuals also have been used to probe feature importance with guaranteed false discovery rates 145, and such a method for example, might similarly quantify the performance of a modality in a late fusion architecture. Yet feature importance is only an early step toward interpretability: probing a model with potentially informative data counterfactuals (for example, “How would the inferred genomic subtype change if the tumor texture were more coarsely heterogeneous on CT?”) would further our understanding of black-box multimodal models 141,145.
Figure 6. Class activation maps highlight the image areas most important for the model to make a decision.
Magnetic resonance imaging (MRI) images of the breast (A) before neoadjuvant chemotherapy with region of interest circled by a radiologist, (B) after neoadjuvant chemotherapy with region of interest circled by a radiologist, and (C) before neoadjuvant chemotherapy with class activation mapping by a neural network trained to predict response to therapy. Warmer colors indicate higher saliency. The images were obtained from The Cancer Imaging Archive (TCIA)156,157.
Data governance and stewardship
Progress will require appropriate data governance and stewardship. Patient consent lies at the core of appropriate use of data and dictates terms of use as stipulated by institutional review boards. Beyond patient consent, high quality, curated and annotated datasets require the expertise and domain knowledge of clinician scientists or clinical fellows. As such, terms of use for these valuable datasets are likely to be set by those who invested the expertise and time required for curation. Success will therefore depend heavily on collaborative models coupling expert clinical annotations with the expertise of data scientists for advanced analyses 146. Furthermore, cross-departmental coordination, access provisions, and governance structures will be required to achieve large scale multimodal data integration. We argue that open data models are the most productive approaches to fully leverage data for discovery and promote reproducibility. This has been demonstrated in the cancer genomics community with TCGA, and community data standards promoting multi-institutional clinical data integration, such as AACR Project GENIE, are now gaining traction 147. Moreover, as the clinical journey for a patient with cancer plays out over time, technology systems and governance structures to capture relevant events and new data in real-time will enhance efforts for data integration and computational discovery. Effective stewardship plans, including accuracy of data, collaborative access provisions, imposition of data standards, and longitudinal data updates, are therefore critical to managing and deploying appropriate use of data for large scale multimodal data integration.
Perspectives
Multimodal cancer biomarker discovery occurs at the interface of clinical oncology, ML research, and data engineering, which typically operate separately. To advance the field, collaborative research programs must unify and promote clear communication among these stakeholders through platform design, model development, and the publication lifecycle 148. These programs will enable clinical investigators to ask questions centering on patient stratification and ultimately produce predictive models by integrating multimodal data. A team science approach with appropriately shared attribution of credit and agreed-upon data stewardship provisions is essential for progress.
The main roadblock to progress in this field is the lack of usable data. Advances in multimodal ML methods have been impressive in other fields, such as sentiment analysis [G] 86,149-154 , with large benchmark datasets, but the largest multimodal oncological dataset, the TCGA, contains limited data modalities and only a few hundred patients per cancer type. This data scarcity largely prevents investigators from using advanced data-hungry models and, critically, hampers benchmarking of new methods in the field, required for rational development of multi-model biomarkers.
Institutional datasets must be assembled and shared, but current data infrastructure typically necessitates months of laborious extraction and annotation before analysis begins. This is perhaps the most well-known issue of conducting ML research for healthcare applications, and a general solution is not imminent. To address this in specific cases, imposed structures on certain notes and full-time data curators have hastened chart review. Automatic annotation strategies relying on WSL and active learning conserve scarce expert annotations and have begun to reduce annotation burdens for large imaging cohorts. Until these fundamental challenges are addressed, multimodal ML models must often operate in the very small data regime. Simple ML models should be used in place of DL methods for small cohorts. DL models should be used judiciously for tasks with large statistical sample size and with strategies to combat overfitting, such as gradient blending, early stopping, data augmentation, and weight decay [G]. Investigators must be wary of spurious results due to institutional biases and small sample sizes, with cross-validation, retrospective external validation, prospective validation, and clinical trials serving as key measures to assess algorithm effectiveness.
Ultimately, as biomedical data infrastructures develop, the goal of this approach is to refine cancer prognosis and rational management by integrating multiple data modalities. Genomic biomarkers have improved upon traditional staging and have begun to implement personalised cancer care, promoting targeted therapies. We predict new classes of multimodal biomarkers will further harness information content from various sources, thereby leading to improved predictive models for therapeutic response. Validated models will be deployed to the electronic medical record, providing near-real-time risk stratification and recommendations for individual patients for clinicians to integrate with other factors to inform management. While we focused on genomics, histology, radiomics and clinical outcomes in this Perspective, we expect additional measurements such as the microbiome, metabolic analytes, longitudinal cell free DNA analysis, and deep immune profiling will become integrated as informative determinants of clinical trajectories. In summary, we project that as data access challenges are overcome, multimodal computational techniques will play important roles in clinicians’ decisions around disease management. Developing multimodal ML methods, usefully logging and annotating patient data, and advancing data engineering infrastructures are outstanding hurdles that remain in the field. As these challenges are met, the field is poised for a reimagined class of rational, multimodal biomarkers and predictive tools to refine evidence-based cancer care and precision oncology.
This Perspective proposes that data from multiple modalities including molecular diagnostics, radiological and histological imaging and codified clinical data should be integrated by multimodal machine learning models to advance the prognosis and treatment management of patients with cancer.
Acknowledgments
We thank Drs. Nicole Rusk and Wesley Tansey for helpful comments on the manuscript. SPS is supported by the Nicholls-Biondi endowed chair in Computational Oncology and the Susan G. Komen Scholars program. KMB is supported by the National Cancer Institute of the National Institutes of Health under award number F30CA257414, the Jonathan Grayer Fellowship of Gerstner Sloan Kettering Graduate School of Biomedical Sciences, and a Medical Scientist Training Program Grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program. MSK MIND is generously supported by Cycle for Survival. All authors are supported by the NIH/NCI Cancer Center Support Grant P30 CA008748.
Glossary box
- Artificial intelligence (AI)
A broad field of computer science concerned with developing computational tools to carry out tasks historically requiring human-level intelligence.
- AUROC (area under the receiver operating characteristic curve)
Measures the ability of a binary classifier to separate the populations of interest. It describes the increase in true positive rate relative to the increase in false positive rate over the range of score thresholds chosen to separate the two classes. The highest value obtainable is 1, and random performance is associated with a value of 0.5.
- Autoencoders
Unsupervised neural network architectures trained to represent data in a lower dimensional space. It is a form of lossy compression (reducing the size of data representations, but with some loss of information) that can be used to uncover latent structure in the data or reduce computational needs before further analysis.
- Bayesian inference
A statistical method that refers to the application of Bayes’ Theorem in determining the updated probability of a hypothesis given new information. Bayesian inference allows the posterior probability to be calculated given the prior probability of a hypothesis and a likelihood function.
- Biomarkers
Measurements which indicate a biological state. Cancer biomarkers can be categorized into diagnostic (disease progression), predictive (treatment response), and prognostic (survival).
- Concordance index (c-Index)
Generalizes the AUROC to measure the ability of a model to separate censored data. As with the AUROC, the baseline value for a model with arbitrary predictions is 0.5, and the ceiling value for a perfect prediction model is 1.0.
- Convolutional neural networks (CNNs)
A form of deep neural network (DNNs) typically used to analyze images. CNNs are named for their use of convolutions, a mathematical operation involving the input data and a smaller matrix known as a kernel. This parameter sharing reduces the number of parameters to be learned and encourages the learning of features which are invariant to image shifts.
- Counterfactual machine learning
A set of techniques for machine learning based on the paradigm of modeling situations that did not factually occur. These techniques are often deployed for interpretable models or to learn from biased logged data. For example, a counterfactual analysis could involve using a model developed to predict a disease outcome using a set of measurements to predict scenarios where the input measurements are perturbed to study their causal relationship. This paradigm has also been harnessed to learn unbiased recommenders from logged data, such as user purchases on online marketplaces, despite changes in how products are recommended over time and the lack of a controlled experimental setup.
- Cox proportional hazards (CPH) model
A regression model used to associate censored temporal outcomes, such as time to survival, and potential predictor variables, such as age or cancer stage. It is the most common method to evaluate prognostic variables in survival analyses of patients with cancer.
- Deep Learning
(DL) Comprises a class of machine learning methods based on artificial neural networks (ANNs), which use multiple non-linear layers to derive progressively higher-order features from data.
- Data lakes
Store relational and non-relational data from a vast pool of raw data. The structure of the data or schema is not defined when data is captured. Different types of analytics on data like structured query language (SQL) queries, big data analytics, full text search, real-time analytics, and machine learning can be used to uncover insights.
- Data parallelism
The approach of performing a computing task in parallel utilizing multiple processors. It focuses on distributing data across various cores and enabling simultaneous sub-computations.
- Deep neural network (DNN)
A form of deep learning, namely artificial neural networks with more than one hidden layer between the input and output layers.
- Federated learning
A training strategy wherein the model to be trained is passed around among institutions instead of centrally amalgamating data. Each institution then updates the model parameters based on the local dataset. This strategy enables multi-institutional model training without data sharing among institutions.
- Kernel
A similarity function often used to transform input data implicitly into a form more suitable for machine learning tasks. For example, a two-dimensional pattern-based kernel could be used to identify the presence of specific shapes in an image, and a one-dimensional Gaussian kernel could be used to impute a smoothed trendline based on noisy data points.
- Layer-wise relevance propagation (LRP)
One of the most prominent techniques in explainable machine learning. LRP decomposes the network's output score into the individual contributions of the input neurons using model parameters (i.e., weights) and neuron activations.
- Machine learning (ML):
A type of artificial intelligence which aims to discover patterns in data which are not explicitly programmed. ML models typically use a dataset for pattern discovery, known as ‘training’, to make predictions on unseen data, known as ‘inference’.
- Recommender systems
Aim to predict relevant items to users by building a model from past behavior. In precision medicine, recommender systems can be used to predict the preferred treatment for a disease based on multiple patient measurements.
- Recurrent neural networks (RNNs)
A form of deep neural network optimized for time series data. An RNN analyzes each element of the input sequence in succession and updates its representation of the data based on previous elements.
- Sentiment analysis
A field seeking to characterize human emotional states from text, images, and sounds by the use of machine learning models.
- Supervised learning
A machine learning paradigm, which aims to elucidate the relationship between input data variables and predefined classes (‘classification’) or continuous labels (‘regression’) of interest. By contrast, unsupervised learning aims to identify patterns in a dataset without the use of such labels or classes.
- Voxel
The volume element is defined by the x, y and z coordinates in 3D space used in medical imaging modalities. Its dimensions are given by the pixel, together with the thickness of the slice.
- Weight decay
A regularization strategy to improve the generalizability of models whereby high estimated values of model parameters are penalized despite marginal increases in accuracy on the training set.
Footnotes
Competing Interests
S.P.S is a shareholder and consultant to Canexia Health Inc. K.M.B., P.K., R.V. and J.G. declare no competing interests.
References
- 1.AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 7, 818–831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasan N et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors. Science 366, 714–723 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razavi P et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 34, 427–438.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonsson P et al. Genomic Correlates of Disease Progression and Treatment Response in Prospectively Characterized Gliomas. Clin. Cancer Res 25, 5537–5547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soumerai TE et al. Clinical Utility of Prospective Molecular Characterization in Advanced Endometrial Cancer. Clin. Cancer Res 24, 5939–5947 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui M & Zhang DY Artificial intelligence and computational pathology. Lab. Invest 101, 412–422 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen SY et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Cristiano S et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klupczynska A et al. Study of early stage non-small-cell lung cancer using Orbitrap-based global serum metabolomics. J. Cancer Res. Clin. Oncol 143, 649–659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helland T et al. Serum concentrations of active tamoxifen metabolites predict long-term survival in adjuvantly treated breast cancer patients. Breast Cancer Res. 19, 125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo P et al. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma: Luo, Yin, et al. Hepatology 67, 662–675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medina-Martínez JS et al. Isabl Platform, a digital biobank for processing multimodal patient data. BMC Bioinformatics 21, 549 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhinder B, Gilvary C, Madhukar NS & Elemento O Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. 11, 900–915 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosny A, Parmar C, Quackenbush J, Schwartz LH & Aerts HJWL Artificial intelligence in radiology. Nat. Rev. Cancer 18, 500–510 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bera K, Schalper KA, Rimm DL, Velcheti V & Madabhushi A Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nature Reviews Clinical Oncology 16, 703–715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutman DA et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 267, 560–569 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zwanenburg A et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 295, 328–338(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun R et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 19, 1180–1191 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Rizzo S et al. Radiomics of high-grade serous ovarian cancer: association between quantitative CT features, residual tumour and disease progression within 12 months. Eur. Radiol 28, 4849–4859 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Pisapia JM et al. Predicting pediatric optic pathway glioma progression using advanced magnetic resonance image analysis and machine learning. Neurooncol Adv 2, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang K et al. Residual Convolutional Neural Network for the Determination of IDH Status in Low- and High-Grade Gliomas from MR Imaging. Clin. Cancer Res 24, 1073–1081(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Wang Y, Yu J, Guo Y & Cao W Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Scientific Reports 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu C-F et al. Machine Learning-Based Radiomics for Molecular Subtyping of Gliomas. Clin. Cancer Res 24, 4429–4436 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Wang S et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur. Respir. J 53, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khosravi P, Lysandrou M, Eljalby M & Li Q A Deep Learning Approach to Diagnostic Classification of Prostate Cancer Using Pathology–Radiology Fusion. J. Magn. Reson (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosny A et al. Deep learning for lung cancer prognostication: A retrospective multicohort radiomics study. PLoS Med. 15, e1002711 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajpurkar P et al. AppendiXNet: Deep Learning for Diagnosis of Appendicitis from A Small Dataset of CT Exams Using Video Pretraining. Sci. Rep 10, 3958 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khosravi P, Kazemi E, Imielinski M, Elemento O & Hajirasouliha I Deep Convolutional Neural Networks Enable Discrimination of Heterogeneous Digital Pathology Images. EBioMedicine 27, 317–328 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coudray N et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nature Medicine vol. 24 1559–1567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu Y et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nature Cancer 1, 800–810 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Kather JN et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nature Cancer 1, 789–799 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding K et al. Feature-Enhanced Graph Networks for Genetic Mutational Prediction Using Histopathological Images in Colon Cancer, in Medical Image Computing and Computer Assisted Intervention – MICCAI 2020 294–304 (Virtual, 2020). [Google Scholar]
- 33.Rutledge WC et al. Tumor-infiltrating lymphocytes in glioblastoma are associated with specific genomic alterations and related to transcriptional class. Clin. Cancer Res 19, 4951–4960 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kather JN et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med 25, 1054–1056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Echle A et al. Clinical-Grade Detection of Microsatellite Instability in Colorectal Tumors by Deep Learning. Gastroenterology 159, 1406–1416.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saltz J et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep. 23, 181–193.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diao JA et al. Human-interpretable image features derived from densely mapped cancer pathology slides predict diverse molecular phenotypes. Nat. Commun 12, 1613 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corredor G et al. Spatial Architecture and Arrangement of Tumor-Infiltrating Lymphocytes for Predicting Likelihood of Recurrence in Early-Stage Non-Small Cell Lung Cancer. Clin. Cancer Res 25, 1526–1534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abduljabbar K et al. Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nat. Med 26, 1054–1062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong J et al. Machine-based morphologic analysis of glioblastoma using whole-slide pathology images uncovers clinically relevant molecular correlates. PLoS One 8, e81049 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao A, Barkley D, França GS & Yanai I Exploring tissue architecture using spatial transcriptomics. Nature 596, 211–220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis SM et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat. Methods 1–16 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Flaherty KT et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl. J. Med 367, 107–114 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Maemondo M et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med 362, 2380–2388 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Slamon DJ et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med 344, 783–792 (2001). [DOI] [PubMed] [Google Scholar]
- 46.DiNardo CD et al. Durable Remissions with Ivosidenib in IDH1-Mutated Relapsed or Refractory AML. N. Engl. J. Med 378, 2386–2398 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Mirza MR et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N. Engl. J. Med 375, 2154–2164 (2016). [DOI] [PubMed] [Google Scholar]
- 48.de Bono J et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med 382, 2091–2102 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Drilon A et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N. Engl. J. Med 378, 731–739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Canon J et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Hallin J et al. The KRASG12C Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov. 10, 54–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andrá F et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N. Engl.J. Med 380, 1929–1940 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Samstein RM et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet 51, 202–206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le DT et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Priestley P et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 575, 210–216 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.VÖhringer H, Van Hoeck A, Cuppen E & Gerstung M Learning mutational signatures and their multidimensional genomic properties with TensorSignatures. Nat. Commun 12,3628(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Macintyre G et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat. Genet 50, 1262–1270 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Funnell T et al. Integrated structural variation and point mutation signatures in cancer genomes using correlated topic models. PLoS Comput. Biol 15, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y et al. High-Spatial-Resolution Multi-Omics Sequencing via Deterministic Barcoding in Tissue. Cell 183, 1665–1681.e18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maniatis S et al. Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis. Science 364, 89–93 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Payne AC et al. In situ genome sequencing resolves DNA sequence and structure in intact biological samples. Science 371, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang W, Tran D & Feiszli M What makes training multi-modal classification networks hard? in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 12695–12705 (Virtual, 2020). [Google Scholar]
- 63.Raghu M, Zhang C, Kleinberg J & Bengio S Transfusion: Understanding Transfer Learning for Medical Imaging, in Advances in Neural Information Processing Systems 32 (NeurIPS 2019) (Vancouver, BC, Canada, 2019). [Google Scholar]
- 64.Zhang L et al. Deep Learning-Based Multi-Omics Data Integration Reveals Two Prognostic Subtypes in High-Risk Neuroblastoma. Front. Genet 9, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chaudhary K, Poirion OB, Lu L & Garmire LX Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res 24,1248–1259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramazzotti D, Lai A, Wang B, Batzoglou S & Sidow A Multi-omic tumor data reveal diversity of molecular mechanisms that correlate with survival. Nat. Commun. 9, 4453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poirion OB, Chaudhary K & Garmire LX Deep Learning data integration for better risk stratification models of bladder cancer, in 197–206 (San Francisco, CA, USA, 2018). [PMC free article] [PubMed] [Google Scholar]
- 68.Žitnik M & Zupan B Survival regression by data fusion. Systems Biomedicine 2, 47–53 (2014). [Google Scholar]
- 69.Cancer Genome Atlas Research Network et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med 372, 2481–2498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cantini L et al. Benchmarking joint multi-omics dimensionality reduction approaches for the study of cancer. Nat. Commun 12, 124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stuart T & Satija R Integrative single-cell analysis. Nat. Rev. Genet 20, 257–272 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Hasin Y, Seldin M & Lusis A Multi-omics approaches to disease. Genome Biol. 18, 83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun D, Wang M & Li A A multimodal deep neural network for human breast cancer prognosis prediction by integrating multi-dimensional data. IEEE/ACM Trans. Comput. Biol. Bioinform (2018). [DOI] [PubMed] [Google Scholar]
- 74.Huang Z et al. SALMON: Survival Analysis Learning With Multi-Omics Neural Networks on Breast Cancer. Front. Genet 10, 166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee B et al. DeepBTS: Prediction of Recurrence-free Survival of Non-small Cell Lung Cancer Using a Time-binned Deep Neural Network. Sci. Rep 10, 1952 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choi E, Bahadori MT, Schuetz A, Stewart WF & Sun J Doctor AI: Predicting Clinical Events via Recurrent Neural Networks, in Proceedings of the 1st Machine Learning for Healthcare Conference 301–318 (Boston, MA, USA, 2016). [PMC free article] [PubMed] [Google Scholar]
- 77.Tomašev N et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature 572, 116–119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang J et al. MIA-Prognosis: A Deep Learning Framework to Predict Therapy Response, in Medical Image Computing and Computer Assisted Intervention – MICCAI 2020 211–220 (Virtual, 2020). [Google Scholar]
- 79.Srivastava RK, Greff K & Schmidhuber J Highway Networks, in Advances in Neural Information Processing Systems 28 (NIPS 2015) (Montreal, Canada, 2015). [Google Scholar]
- 80.Cheerla A & Gevaert O Deep learning with multimodal representation for pancancer prognosis prediction. Bioinformatics 35, i446–i454 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gevaert O et al. Imaging-AMARETTO: An Imaging Genomics Software Tool to Interrogate Multiomics Networks for Relevance to Radiography and Histopathology Imaging Biomarkers of Clinical Outcomes. JCO Clin Cancer Inform 4, 421–435 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu X et al. Imaging-genetic data mapping for clinical outcome prediction via supervised conditional Gaussian graphical model, in 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (Shenzhen, Guangdong, China, 2016). [Google Scholar]
- 83.Popovici V et al. Joint analysis of histopathology image features and gene expression in breast cancer. BMC Bioinformatics 17, 209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mobadersany P et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. U. S. A 115, E2970–E2979 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen RJ et al. Pathomic Fusion: An Integrated Framework for Fusing Histopathology and Genomic Features for Cancer Diagnosis and Prognosis. IEEE Trans. Med. Imaging PP, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zadeh A, Chen M, Poria S, Cambria E & Morency L-P Tensor Fusion Network for Multimodal Sentiment Analysis, in Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing (Copenhagen, Denmark, 2017). [Google Scholar]
- 87.Yuan Y, Giger ML, Li H, Bhooshan N & Sennett CA Multimodality computer-aided breast cancer diagnosis with FFDM and DCE-MRI. Acad. Radiol 17, 1158–1167 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chan H-W, Weng Y-T & Huang T-Y Automatic Classification of Brain Tumor Types with the MRI Scans and Histopathology Images, in Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries 353–359 (Quebec City, QC, Canada, 2017). [Google Scholar]
- 89.Rathore S et al. Radiomic MRI signature reveals three distinct subtypes of glioblastoma with different clinical and molecular characteristics, offering prognostic value beyond IDH1. Sci. Rep 8, 5087 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Donini M et al. Combining heterogeneous data sources for neuroimaging based diagnosis: re-weighting and selecting what is important. Neuroimage 195, 215–231 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gonen M & Alpaydin E Multiple Kernel Learning Algorithms. Journal of Machine Learning Research 12, 2211–2268 (2011). [Google Scholar]
- 92.Gillies RJ , Kinahan PE & Hricak H Radiomics: Images Are More than Pictures, They Are Data. Radiology 278, 563–577 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duanmu H et al. Prediction of Pathological Complete Response to Neoadjuvant Chemotherapy in Breast Cancer Using Deep Learning with Integrative Imaging, Molecular and Demographic Data, in Medical Image Computing and Computer Assisted Intervention – MICCAI 2020 242–252 (Virtual, 2020). [Google Scholar]
- 94.Bhattacharya I et al. CorrSigNet: Learning CORRelated Prostate Cancer SIGnatures from Radiology and Pathology Images for Improved Computer Aided Diagnosis, in Medical Image Computing and Computer Assisted Intervention – MICCAI 2020 315–325 (Virtual, 2020). [Google Scholar]
- 95.Deng J et al. ImageNet: A large-scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition (2009). [Google Scholar]
- 96.Kay W et al. The Kinetics Human Action Video Dataset. Preprint at http://arxiv.org/abs/1705.06950 (2017). [Google Scholar]
- 97.Ardila D et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med 25, 954–961 (2019). [DOI] [PubMed] [Google Scholar]
- 98.Zhou Z-H A brief introduction to weakly supervised learning. Natl Sci Rev 5, 44–53 (2018). [Google Scholar]
- 99.Campanella G et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med 25, 1301–1309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Suphavilai C, Bertrand D & Nagarajan N Predicting Cancer Drug Response using a Recommender System. Bioinformatics 34, 3907–3914 (2018). [DOI] [PubMed] [Google Scholar]
- 101.Joachims T, Swaminathan A & de Rijke M Deep Learning with Logged Bandit Feedback, in Proceedings of the International Conference on Learning Representations (ICLR) (Vancouver, BC, Canada, 2018). [Google Scholar]
- 102.Lee JS et al. Synthetic lethality-mediated precision oncology via the tumor transcriptome. Cell 184, 2487–2502.e13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gundersen G, Dumitrascu B, Ash JT & Engelhardt BE End-to-end training of deep probabilistic CCA for joint modeling of paired biomedical observations, in Proceedings of The 35th Uncertainty in Artificial Intelligence Conference 945–955 (Tel Aviv, Israel, 2020). [Google Scholar]
- 104.Li Y et al. Inferring multimodal latent topics from electronic health records. Nat. Commun 11, 2536 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choplin RH, Boehme JM 2nd & Maynard CD Picture archiving and communication systems: an overview. Radiographics 12, 127–129 (1992). [DOI] [PubMed] [Google Scholar]
- 106.Puchalski RB et al. An anatomic transcriptional atlas of human glioblastoma. Science 360, 660–663 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weigelt B et al. Radiogenomics Analysis of Intratumor Heterogeneity in a Patient With High-Grade Serous Ovarian Cancer. JCO Precis Oncol 3, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jiménez-Sánchez A et al. Unraveling tumor-immune heterogeneity in advanced ovarian cancer uncovers immunogenic effect of chemotherapy. Nat. Genet 52, 582–593 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hersh WR et al. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med. Care 51, S30–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Allison JJ et al. The Art and Science of Chart Review. Jt. Comm. J. Qual. Improv 26, 115–136(2000). [DOI] [PubMed] [Google Scholar]
- 111.Vassar M & Holzmann M The retrospective chart review: important methodological considerations. J. Educ. Eval. Health Prof 10, 12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hripcsak G et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud. Health Technol. Inform 216, 574–578(2015). [PMC free article] [PubMed] [Google Scholar]
- 113.Stein B & Morrison A The enterprise data lake: Better integration and deeper analytics. PwC Technology Forecast: Rethinking integration 1, 18 (2014). [Google Scholar]
- 114.Armbrust M et al. Delta lake: high-performance ACID table storage over cloud object stores, in vol. 13 3411–3424 (Virtual, 2020). [Google Scholar]
- 115.Zagan E & Danubianu M Cloud DATA LAKE: The new trend of data storage, in 2021 3rd International Congress on Human-Computer Interaction, Optimization and Robotic Applications (HORA) 1–4 (Ankara, Turkey, 2021). [Google Scholar]
- 116.Rieke N et al. The future of digital health with federated learning, npj Digital Medicine 3, 119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Andreux M, Manoel A, Menuet R, Saillard C & Simpson C Federated Survival Analysis with Discrete-Time Cox Models, in FL-ICML 2020 : International Workshop on Federated Learning for User Privacy and Data Confidentiality in Conjunction with ICML 2020 (Virtual, 2020). [Google Scholar]
- 118.Lin J-H & Haug PJ Exploiting missing clinical data in Bayesian network modeling for predicting medical problems. J. Biomed. Inform 41, 1–14 (2008). [DOI] [PubMed] [Google Scholar]
- 119.Khan A, Atzori M, Otálora S, Andrearczyk V & Müller H Generalizing convolution neural networks on stain color heterogeneous data for computational pathology, in Medical Imaging 2020: Digital Pathology 113200R (Houston, TX, USA, 2020). [Google Scholar]
- 120.Glatz-Krieger K, Spornitz U, Spatz A, Mihatsch MJ & Glatz D Factors to keep in mind when introducing virtual microscopy. Virchows Arch. 448, 248–255 (2006). [DOI] [PubMed] [Google Scholar]
- 121.Janowczyk A, Basavanhally A & Madabhushi A Stain Normalization using Sparse AutoEncoders (StaNoSA): Application to digital pathology. Comput. Med. Imaging Graph. 57, 50–61(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lacroix M et al. Correction for Magnetic Field Inhomogeneities and Normalization of Voxel Values Are Needed to Better Reveal the Potential of MR Radiomic Features in Lung Cancer. Front. Oncol 10, 43 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Macenko M et al. A method for normalizing histology slides for quantitative analysis, in 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro (Boston, MA, USA, 2009). [Google Scholar]
- 124.Srinidhi CL, Ciga O & Martel AL Deep neural network models for computational histopathology: A survey. Med. Image Anal 67, 101813 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hu Z, Tang A, Singh J, Bhattacharya S & Butte AJ A robust and interpretable end-to-end deep learning model for cytometry data. Proc. Natl. Acad. Sci. U. S. A 117, 21373–21380 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kleppe A et al. Designing deep learning studies in cancer diagnostics. Nat. Rev. Cancer 21, 199–211 (2021). [DOI] [PubMed] [Google Scholar]
- 127.Lopez K, Fodeh SJ, Allam A, Brandt CA & Krauthammer M Reducing Annotation Burden Through Multimodal Learning. Frontiers in Big Data 3, 19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gundersen OE & Kjensmo S State of the art: Reproducibility in artificial intelligence, in Thirty-second AAAI conference on artificial intelligence (New Orleans, LA, USA, 2018). [Google Scholar]
- 129.Courtiol P et al. Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat. Med 25, 1519–1525 (2019). [DOI] [PubMed] [Google Scholar]
- 130.McKinney SM et al. Addendum: International evaluation of an AI system for breast cancer screening. Nature 586, E19 (2020). [DOI] [PubMed] [Google Scholar]
- 131.Haibe-Kains B et al. Transparency and reproducibility in artificial intelligence. Nature 586, E14–E16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hosny A et al. ModelHub.AI: Dissemination Platform for Deep Learning Models. Preprint at http://arxiv.org/abs/1911.13218 (2019). [Google Scholar]
- 133.Beede E et al. A Human-Centered Evaluation of a Deep Learning System Deployed in Clinics for the Detection of Diabetic Retinopathy, in Proceedings of the 2020 CHI Conference on Human Factors in Computing Systems 1–12 (Honolulu, HI, USA (cancelled), 2020). [Google Scholar]
- 134.Cruz Rivera S et al. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Nat. Med 26, 1351–1363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Moher D et al. CONSORT 2010 explanation and elaboration: updated guidelines for reporting parallel group randomised trials. BMJ 340, c869 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu X et al. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat. Med 26,1364–1374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lauritsen SM et al. Explainable artificial intelligence model to predict acute critical illness from electronic health records. Nat. Commun 11, 3852 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rudin C Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nature Machine Intelligence 1, 206–215(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pavel MA, Petersen EN, Wang H, Lerner RA & Hansen SB Studies on the mechanism of general anesthesia. Proc. Natl. Acad. Sci. U. S. A 117, 13757–13766 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang F, Kaushal R & Khullar D Should Health Care Demand Interpretable Artificial Intelligence or Accept “Black Box” Medicine? Ann. Intern. Med 172, 59–60 (2020). [DOI] [PubMed] [Google Scholar]
- 141.Castro DC, Walker I & Glocker B Causality matters in medical imaging. Nat. Commun 11, 3673 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhou B, Khosla A, Lapedriza A, Oliva A & Torralba A Learning deep features for discriminative localization, in Proceedings of the IEEE conference on computer vision and pattern recognition 2921–2929 (Las Vegas, NV, USA, 2016). [Google Scholar]
- 143.Olah C et al. The Building Blocks of Interpretability. Distill 3, (2018). [Google Scholar]
- 144.Graziani M, Andrearczyk V & Müller H Visualizing and interpreting feature reuse of pretrained CNNs for histopathology. in (Dublin, Ireland, 2019). [Google Scholar]
- 145.Burns C, Thomason J & Tansey W Interpreting Black Box Models via Hypothesis Testing, in FODS ’20: Proceedings of the 2020 ACM-IMS on Foundations of Data Science Conference 47–57 (Virtual, 2019). [Google Scholar]
- 146.Donoghue MTA, Schram AM, Hyman DM & Taylor BS Discovery through clinical sequencing in oncology. Nature Cancer 1, 774–783 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kehl KL et al. Assessment of Deep Natural Language Processing in Ascertaining Oncologic Outcomes From Radiology Reports. JAMA Oncol 5, 1421–1429 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cosgriff CV, Stone DJ, Weissman G, Pirracchio R & Celi LA The clinical artificial intelligence department: a prerequisite for success. BMJ Health Care Inform 27, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zadeh A et al. Memory Fusion Network for Multi-view Sequential Learning, in Proceedings of the Thirty-Second AAAI Conference on Artificial Intelligence (AAAI-18) (New Orleans, LA, USA, 2018). [Google Scholar]
- 150.Zadeh A, Liang PP, Poria S, Cambria E & Morency L-P Multimodal Language Analysis in the Wild: CMU-MOSEI Dataset and Interpretable Dynamic Fusion Graph, in Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers) 2236–2246 (Melbourne, Australia, 2018). [Google Scholar]
- 151.Zadeh A et al. Multi-attention Recurrent Network for Human Communication Comprehension, in Proc. Conf AAAI Artif Intell. vol. 2018 5642–5649 (New Orleans, LA, USA, 2018). [PMC free article] [PubMed] [Google Scholar]
- 152.Kumar A, Srinivasan K, Cheng W-H & Zomaya AY Hybrid context enriched deep learning model for fine-grained sentiment analysis in textual and visual semiotic modality social data. Inf. Process. Manag 57, 102141 (2020). [Google Scholar]
- 153.Liang PP, Zadeh A & Morency L-P Multimodal Local-Global Ranking Fusion for Emotion Recognition, in Proceedings of the 20th ACM International Conference on Multimodal Interaction 472–476 (Boulder, CO, USA, 2018). [Google Scholar]
- 154.Liu Z et al. Efficient Low-rank Multimodal Fusion with Modality-Specific Factors, in Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Long Papers), 2247–2256 (Melbourne, Australia, 2018). [Google Scholar]
- 155.Marinelli RJ et al. The Stanford Tissue Microarray Database. Nucleic Acids Res. 36, D871–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Clark K et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26, 1045–1057 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Newitt D & Hylton N Single site breast DCE-MRI data and segmentations from patients undergoing neoadjuvant chemotherapy. The Cancer Imaging Archive (2016). [Google Scholar]
- 158.Wolpert DH The Lack of A Priori Distinctions Between Learning Algorithms. Neural Comput. 8, 1341–1390 (1996). [Google Scholar]
- 159.Wolpert DH & Macready WG No Free Lunch Theorems for Optimization. IEEE Trans. Evol. Comput 1, (1997). [Google Scholar]
- 160.LeCun Y, Bengio Y & Hinton G Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 161.He K, Zhang X, Ren S & Sun J Deep Residual Learning for Image Recognition, in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (Las Vegas, NV, USA, 2016). [Google Scholar]
- 162.Iandola FN et al. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5MB model size. Preprint at http://arxiv.org/abs/1602.07360 (2016). [Google Scholar]
- 163.Szegedy C, Vanhoucke V, Ioffe S, Shlens J & Wojna Z Rethinking the Inception Architecture for Computer Vision, in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (Las Vegas, NV, USA, 2016). [Google Scholar]
- 164.Huang G, Liu Z, van der Maaten L & Weinberger KQ Densely Connected Convolutional Networks, in 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (Honolulu, HI, USA, 2016). [Google Scholar]
- 165.Cho K et al. Learning Phrase Representations using RNN Encoder-Decoder for Statistical Machine Translation, in Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP) (Doha, Qatar, 2014). [Google Scholar]
- 166.Hochreiter S & Schmidhuber J Long short-term memory. Neural Comput. 9, 1735–1780(1997). [DOI] [PubMed] [Google Scholar]
- 167.Vaswani A et al. Attention Is All You Need, in 31st Conference on Neural Information Processing Systems (NIPS 2017) (Long Beach, CA, USA, 2017). [Google Scholar]
- 168.Lee G, Kang B, Nho K, Sohn K-A & Kim D MildInt: Deep Learning-Based Multimodal Longitudinal Data Integration Framework. Front. Genet 10, 617 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Perhaps the greatest challenge in multimodal machine learning is data scarcity. Data acquired during the standard of care are not structured in a research-ready format: stained tissue specimens typically must be manually located and scanned, and radiological images are stored in the picture archiving and communication system (PACS) 105 with limited clinical annotation. The modalities are typically organized with different patient identifiers, complicating alignment. A related challenge is spatial colocalization, which is especially important for studying biological correlation of multimodal features. Small datasets such as Ivy Glioblastoma Atlas Project (IvyGAP) 106 richly profile the genomics and multi-scale tumoral architecture of patients with matched clinical outcomes and represent the promise of spatial colocalization. To achieve this at scale, image-guided biopsies or 3D-printed molds based on tumor morphology 107,108 are possible solutions, but challenges remain before these approaches are scaled for prospective research.
The general limits of using healthcare records to conduct research are widely discussed 109: one challenge is unobserved patient outcomes, which can be handled for time-to-event analysis but requires excluding patients for categorical outcomes. Another major bottleneck is retrospective chart review, or manually reviewing patient records to extract specific features into spreadsheets. It is error prone and variable, and repeated review is often required to capture new clinical events 110,111. Efforts are underway to build models to automatically codify clinical information from unstructured text, and ontologies such as Observational Health Data Sciences and Informatics (OHDSI)112 and American Association for Cancer Research project Genomics Evidence Neoplasia Information Exchange (AACR project GENIE)1 structure disparate clinical elements to facilitate retrospective research. These models ought to be extended to incorporate additional data modalities.
As structured data emerge, data lakes [G] are a scalable solution to organize original data and track their use during subsequent analysis 113. Data lake technologies unify siloed data and accommodate both known and unforeseen file types. Cost-effective data lake storages are readily available from commercial vendors (for example, Amazon S3) as well as open source products (for example, Delta Lake 114). The ensuant technical challenges vary depending on whether the data lake is set up in the cloud, on premises, or in a hybrid solution 115. There are specific challenges when applying data lakes for biomedical research, such as the stripping of protected health information (PHI) to protect patient privacy and facilitate inter-institutional sharing.
Such cross-institutional data sharing is essential to promote and test model generalizability. Leading platforms include the database of Genotypes and Phenotypes (dbGaP), the European Genome-phenome Archive (EGA), The Cancer Imaging Archive (TCIA), the Genomic Data Commons (GDC), and other resources in the National Cancer Institute (NCI) Cancer Research Data Commons. However, beyond matched genomic data and H&E WSIs of TCGA and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), public resources contain only small patient cohorts with multiple data modalities.
Major causes of this public data scarcity include the logistical challenges of anonymizing data and institutional privacy policies. Federated learning [G] is a potential solution116. Depending on the choice of model, federated learning can require novel training approaches 117 but enables training on multi-institutional cohorts without data having to leave local networks.