Abstract
Background
The benefit of alpelisib in hormone-receptor-positive (HR+) metastatic breast cancer patients provided clinical evidence for the increasing importance of PIK3CA testing. We performed a comparison of liquid biopsy and tissue-based detection of PIK3CA mutations.
Materials and methods
PIK3CA hotspot mutation analysis using a high-resolution SiMSen-Seq assay was performed in plasma from 93/99 eligible patients with HR+/HER2− breast cancer. Additionally, mFAST-SeqS was used to estimate the tumour fractions in plasma samples. In 72/93 patients, matched tissue was available and analysed using a customised Ion Torrent panel.
Results
PIK3CA mutations were detected in 48.6% of tissue samples and 47.3% of plasma samples, with identical PIK3CA mutation detected in 24/72 (33.3%) patients both in tissue and plasma. In 10 (13.9%) patients, mutations were only found in plasma, and in 6 (8.3%) patients, PIK3CA mutations found in tissue were not detectable in ctDNA. In 49/93 plasma samples without detectable PIK3CA mutations, 22 (44.9%) samples had elevated tumour fractions, implying true negative results.
Conclusion
SiMSen-Seq-based detection of PIK3CA mutations in plasma shows advantageous concordance with the tissue analyses. A combination with an untargeted approach for detecting ctDNA fractions may confirm a negative PIK3CA result and enhance the performance of the SiMSen-Seq test.
Subject terms: Oncology, Breast cancer
Introduction
Breast cancer (BC) remains the leading cause of cancer-related death in the global female population [1]. Despite the substantial progress in systemic treatment, metastatic disease remains incurable, but novel treatment options offer the chance for prolonged disease stabilisation [2]. However, particularly for the treatment of hormone-receptor (HR)-positive disease, substantial progress in systemic treatment has been made. After the introduction of CDK4/6 inhibitors, targeted signal-transduction pathways have been established in the meantime [3]. Phosphatidylinositol 3-kinases (PI3Ks) are among the most important targets in this regard, apparently mediating the conversion of phosphatidylinositol 4,5- bisphosphate (PI2) to phosphatidylinositol 3,4,5-triphosphate (PIP3) [4]. The PI3K/AKT/mTOR pathway’s ensuing upregulation plays a critical role in essential processes of disease progression, including cell growth, proliferation, metabolism and invasion [5].
In breast cancer, the PI3K/AKT/mTOR signaling pathway can be altered by different mechanisms. In approximately a third of early breast cancers activating mutations in the helical or kinase domain of PIK3CA can be found (45% in luminal A, 29% in luminal B, 39% in HER2-enriched and 9% in the basal carcinomas, respectively) [6, 7]. Similar rates of mutations can be found in metastatic breast cancer, although fewer data are available [6].
Recent data suggest that PIK3CA alterations have an adverse impact on the prognosis of HR-positive disease and may play a role in endocrine resistance [8–10]. Several drugs targeting the PI3 kinase pathway have been tested, but to date, a clinically meaningful benefit of adding an α-specific PI3K inhibitor (alpelisib) with an acceptable tolerability profile was only demonstrated in the SOLAR-1 phase III trial [11]. In this trial, only a minor proportion of patients was pretreated with CDK4/6 inhibitors, but the benefit of PI3K inhibition in this patient’s subset was confirmed in the phase II BYLieve study [12]. With these data, the importance of the PIK3CA encoding for the α-isoform of the catalytic subunit of the class IA PI3K kinase as a biomarker was postulated; the best testing approach, however, remains elusive. The stability of mutations throughout the disease was only recently addressed by Mosele et al. [9], and prospective collection of metastatic tissue is ongoing in several large clinical trials such as AURORA (Aiming to Understand the Molecular Aberrations in Metastatic Breast Cancer, NCT02102165) [13, 14]. As tissue from metastatic sites may not always be readily available, interest in liquid biopsy techniques has grown. In the SOLAR-1 trial, in parallel to the tissue-based detection, cell-free DNA was analysed for the presence of PIK3CA mutations. Along with the approval of alpelisib, FDA has approved the companion diagnostic test, therascreen® PIK3CA RGQ PCR Kit (QIAGEN Manchester, Ltd.), to select patients who have PIK3CA mutations in tumour tissue specimens and/or in circulating tumour DNA (ctDNA) isolated from plasma specimens.
Therefore, our study’s primary aim was to compare plasma-based detection of PIK3CA hotspot mutations with the tissue-based detection from the most recent tumour tissue.
Materials and methods
Patients and sample collections
Between February 2017 and June 2021, 99 locally advanced or metastatic breast cancer patients were recruited for this prospective observational study at the Division of Oncology, Department of Internal Medicine, Medical University of Graz, Austria. One patient who had received a bone marrow transplantation and three patients who had triple-positive or triple-negative tumours were excluded from the study (Fig. 1). The study was approved by the ethics committee of the Medical University Graz (ethical approval number 21–227 ex 09/10), and written informed consent was obtained from all patients. All analyses were carried out considering the guidelines for good scientific practice as officially required from the Medical University of Graz. Clinical data and histopathological tumour characteristics were collected from clinical and pathological records in an electronic database system. Blood samples were obtained either at first diagnosis of metastases, during varying lines of treatment, and/or at a further moment of progression/development of new metastases, along with a planned introduction of a new line of systemic treatment. The most recent tissue was used for tissue analyses, either metastatic tissue, or if metastatic tissue was not available the primary tumour tissue was analysed.
Fig. 1. CONSORT diagram for the study.
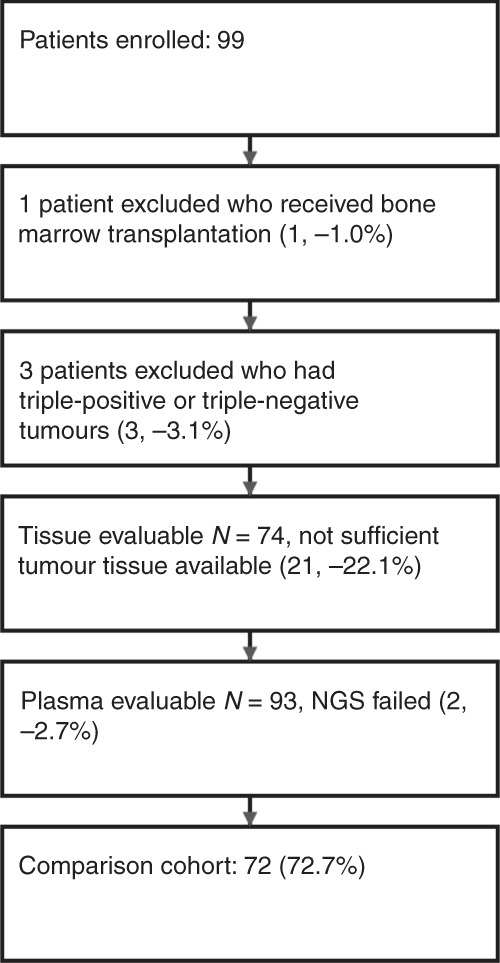
The numbers of patients and samples included in the study and reasons for the exclusion are shown.
Preparation of plasma DNA
Isolation of plasma DNA was performed as previously described [15]. Briefly, whole blood (18 mL) was collected in PAXgene Blood ccfDNA Tubes (Qiagen, Hilden, Germany). Plasma was isolated from blood samples by centrifugation at 200 × g for 10 min, followed by 1600 × g for 10 min with brake and acceleration powers set to slow. The supernatant was then removed and centrifuged again at 1600 × g for an additional 10 min. Plasma was then stored in 2 mL tubes at −80 °C. DNA was isolated from 2 mL plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations or using the QIASymphony PAXgene Blood ccfDNA Kit (Qiagen, Hilden, Germany). Qubit dsDNA HS Assay Kit (Life Technologies, Vienna, Austria) was used for the quantification of plasma DNA.
High-resolution screening for PIK3CA mutations in plasma
To screen for PIK3CA mutation, the SiMSen-Seq method (simple, multiplexed, PCR-based barcoding of DNA for sensitive mutation detection using sequencing) was used [16]. This technology is based on molecular barcoding of individual DNA template strands to track all sequencing reads back to a single original template and correct for PCR errors during library preparation. To assess the most common hotspots (n = 11), three assays covering exons 7, 9 and 20 were designed and validated as described in ref. [16] (see Table S1, Table S2 and Supplementary methods). After a comprehensive evaluation of the assay performance using SeraCare reference materials and control samples, we set the limit of detection (LOD) to 0.25%. In addition, samples were only considered PIK3CA mutated if ≥5 consensus reads carried the mutation (for details, see supplementary material, Supplementary Figs. S1 and S2).
For the analysis of plasma samples, an average input of 26.3 ng (range, 8.5–103.8 ng) cfDNA per sample was used for library preparation. After purification with a 0.8-fold volume of AMPure XP Beads (Beckman Coulter, Brea, CA, USA), libraries were quantified with qPCR using the QIAseq Library Quant Assay (QIAGEN, Hilden, Germany). Libraries were sequenced on an Illumina MiSeq, or NextSeq using 150 bp single-end reads aiming for 10 million reads per sample. Barcoded SiMSen-Seq data were analysed using Debarcer [16]. After consensus read generation, we obtained an average coverage of 2185x (range 49–17227x) for all interrogated positions. The molecular coverage of the SiMSen-Seq assay significantly correlated with the amount of input DNA used for sequencing (Spearman’s rho = 0.66, p < 0.001; Supplementary Fig. S3). The presence of mutations was reported as variant allele frequency (VAF, percentage of alternate reads relative to the total number of reads at a given position).
The raw sequencing datasets have been deposited at the European Genome-phenome Archive (EGA; http://www.ebi.ac.uk/ega/) under study accession number EGAS00001004940 as following dataset: PIK3CA SiMSen-Seq data of plasma samples.
mFAST-SeqS
LINE1-amplicon libraries for mFAST-SeqS were prepared as previously described [17]. Briefly, 1 ng of plasma DNA was amplified with Phusion Hot Start II Polymerase in eight PCR cycles using target-specific L1 primers. The 10 µl of the purified (AMPure Beads, Beckman Coulter, Brea, CA, USA) PCR products were used for the second PCR in which Illumina-specific adaptors and indices were added. L1 amplicon libraries were pooled equimolarly and sequenced on an Illumina MiSeq or NextSeq, generating 150 bp single reads or 76 bp paired-end reads aiming for 100,000 reads per sample. Sequence reads were aligned to the hg19 genome, and reads with a mapping quality >15 were counted per chromosome arm using an in-house script. Normalised read counts were compared to a control population (n = 35) using z-score statistics by subtracting the mean and dividing by the standard deviation to assess over- and under-representation of LINE1-sequences. Because little or no reads aligned to the short arms of acrocentric chromosomes 13p, 14p, 15p, 21p and 22p, these were omitted from the analysis. Finally, all chromosome arm-specific z-scores were squared and summed up, resulting in a genome-wide z-score, which acts as a surrogate for tumour fraction [18].
Tissue-based analyses
DNA was extracted from macrodissected tumour areas of unstained FFPE sections. Adequate tumour cell content was verified from HE stained parallel sections. Extraction was performed using the Maxwell 16 instrument (Promega, Wisconsin, USA) and the Maxwell RSC DNA FFPE Kit (Promega, CatNr: AS1450) or the Ion AmpliSeq Direct FFPE DNA Kit (Thermo Fisher Scientific, Massachusetts, USA). DNA was quantified by Picogreen fluorescence, and typically, 40 ng DNA were used for library preparation. NGS libraries were prepared using a custom-designed AmpliSeqHD panel covering the following mutation hotspots: EGFR (L858), ERBB2 (S310, L755, V762, V777, P780), ERBB3 (V104, E928), KRAS (G12, G13), PTEN (T319), ESR1 (E207, K303, E380, Y537, D538), PIK3CA (E103, N345, C420, E542, E545, Q546, E726, H1047, G1049), AKT1 (E17), TP53 (R175, H179, R213, Y220, R248, R273). The panel includes all mutations evaluated in the SOLAR-1 trial [11]. Molecular barcoding enables a detection threshold of 0.1% mutant allele. Sequencing was performed on an Ion S5XL benchtop sequencer (Thermo Fisher Scientific) to a length of 200 bp. We used the open-source Ion Torrent software (Thermo Fisher Scientific, open source, GPL, https://github.com/iontorrent/) for initial data analysis. Briefly, this included base calling, alignment to the reference genome (hg19) using the TMAP mapper and variant calling by a modified diBayes approach taking the flow space information into account. Called variants were deconvoluted with regards to molecular barcodes and annotated using open-source software ANNOVAR [19] and SnpEff [20]. All coding, nonsynonymous mutations were further evaluated and visually inspected in IGV (http://www.broadinstitute.org/igv/), and variant calls resulting from technical read errors or sequence effects were excluded from the result. This data are available on request from the authors.
Statistical analyses
All statistical analyses were performed with Stata 16.1 (Stata Corp., Houston, TX, USA) and Graphpad. Continuous variables were reported as medians [25–75th percentile] and count data as absolute frequencies (%). The distribution of baseline variables between patients with or without a PIK3CA mutation (binary variable) was assessed with rank-sum tests (continuous variables) and χ2, or with Fisher’s exact tests (categorical variables). The agreement between plasma and tissue was evaluated with a kappa score and 95% CIs. The exact McNemar’s test for comparing paired sample proportions was used to assess the statistical significance of differences between plasma and tissue results. The correlation between z-scores and mutant PIK3CA allele fractions was determined by the Spearman rank correlation coefficient. In an explorative analysis, we associated PIK3CA mutation status with progression-free survival using Kaplan–Meier estimators, log-rank tests and Cox proportional hazards regression. Figures were generated with Stata or GraphPad Prism (v9).
Results
Patient characteristics
Ninety-nine patients were enrolled, and 95 advanced or metastatic breast cancer patients with a median age of 65.4 years [25–75th percentile: 54.9–74.7] were analysed in this study. Table 1 shows the main patients’ characteristics. The majority of patients were female (95.8%), and all patients had HR+/HER2− tumours (100%). The most frequent metastatic site was bone (66.3%), followed by lymph nodes (31.6%), lung (24.2%), liver (22.1%) and pleura (10.5%). Fifty patients (52.6%) had multiple metastatic sites, and most patients (80.0%) received first or second-line palliative treatment at study inclusion. Seventy-four tissue samples and ninety-three plasma samples were available for PIK3CA mutation analysis (Fig. 1). For the comparison cohort of 72 paired tissue and plasma samples, the median time interval between tissue and plasma sample collection was 5.5 months [25–75th percentile: 0.7–34.7 months].
Table 1.
Baseline characteristics of the study population (N = 95).
| Total cohort N = 95 | |
|---|---|
| Age at inclusion (years) | 65.4 (54.9–74.7) |
| Female gender | 91 (95.8%) |
| Histological type | |
| IDC/NST | 59 (62.1%) |
| ILC | 23 (24.2%) |
| Other or not reported | 13 (13.7%) |
| Molecular subtype | |
| HR+/HER2− | 95 (100.0%) |
| Bone metastases | 63 (66.3%) |
| Lung metastases | 23 (24.2%) |
| Liver metastases | 21 (22.1%) |
| Pleura metastases | 10 (10.5%) |
| Lymph node metastases | 30 (31.6%) |
| Number of metastatic sites | |
| 0–1 sitesa | 45 (47.4%) |
| ≥2 sites | 50 (52.6%) |
| Treatment line at inclusion | |
| ≤2 lines | 76 (80.0%) |
| >2 lines | 19 (20.0%) |
| PIK3CA mutation detected in plasma (N = 93)b | 44 (47.3%) |
| PIK3CA mutation detected in tissue (N = 74)c | 36 (48.6%) |
| Source of tissue (N = 74) | |
| Metastasis | 45 (60.8%) |
| Primary tumour | 29 (39.1%) |
| Interval between tissue and blood sample collection (months) | 7.7 (0.7–40.5) |
Data are medians [25–75th percentile] for continuous data and absolute frequencies (%) for count data.
aIncludes one patient with locally advanced disease.
bNGS failed in two plasma samples.
cNo sufficient tumour material was available for NGS testing in 21 cases.
PIK3CA mutations and z-scores in patients with evaluable plasma samples (N = 93)
Of all 93 patients with evaluable plasma samples, 47.3% (44/93) had detectable PIK3CA mutations. In detail, 48 PIK3CA mutations were detected, and in four samples, the co-occurrence of two PIK3CA mutations was observed (Table 2). Double or compound PIK3CA mutations are described in cis on the same allele; they are supposed to increase PI3K activity and enhance downstream signaling, cell proliferation and tumour growth [21]. The VAF of detected PIK3CA mutations in plasma ranged from 0.27 to 49.85%, with a median of 1.67%. The most frequent mutation reported in plasma samples was H1047R (32.6%), followed by E542K (9.7%) and E545K (7.5%).
Table 2.
Patients with double PIK3CA mutations detected in plasma.
| Patient ID | First mutation in plasma (AF %) | Second mutation in plasma (AF %) | First mutation in tissue (%MAF) | Second mutation in tissue (%MAF) | Δ time (months) |
|---|---|---|---|---|---|
| B211 | E542K (12.76) | H1047R (0.33) | E542K (28.57) | None detected | 20.3 |
| B235 | E542K (1.34) | H1047R (0.77) | None detected | None detected | 22.6 |
| B259 | E542K (1.71) | H1047R (4.42) | E542K (13.52) | None detected | 60.3 |
| B387 | E545K (16.36) | E542K (9.87) | E545K (62.09) | E542K (26.66) | 17.6 |
Metastatic sites, including bone, liver, lung, pleura and lymph nodes, were not significantly associated with PIK3CA mutated ctDNA. Multiple metastases were numerically (56.3% versus 37.8%) but not statistically associated with PIK3CA mutations (χ2 test p = 0.075). Finally, there was no difference between the line of treatment and the presence of PIK3CA mutations in plasma.
The median z-score resulting from mFAST-SeqS analyses was 2.13 [25–75th percentile: 1.53–4.18]. Based on a previously established cut-off at ≥3, which depending on the amount and amplitude of copy number alterations, correlates to a tumour fraction of 5–10% [22], 33/93 (35.5%) patients had elevated z-scores. We found no statistically significant correlation between z-scores and mutant PIK3CA allele frequencies (Spearman’s rho = −0.11, p = 0.285). Surprisingly, z-scores were higher in samples where no PIK3CA mutation was detected (median 2.43, range 0.85–69.68) than PIK3CA mutated samples (median 1.82, range 0.75–16.59; Wilcoxon rank-sum test p = 0.042; Supplementary Fig. S4). 22/49 (44.9%) of patients with no detectable plasma PIK3CA mutation presented z-scores ≥3, indicating a high tumour content. Since ctDNA but no PIK3CA mutation could be detected in these samples, these results can be considered true negatives for the assessed PIK3CA mutations. In contrast, for 27/49 (55.1%) patients with low (<3) z-scores and, therefore, low tumour fractions in plasma, the presence of PIK3CA mutations below the LOD cannot be entirely excluded. In these cases, tissue-based PIK3CA analysis might be indicated. Taken together, in 27 out of 93 patients (29.0%), the PIK3CA status was negative with a z-score < 3 based on ctDNA analyses. For these 27 patients, 20 tissue samples were evaluable for PIK3CA testing, and PIK3CA mutations were detected only in 4/20 (20.0%) of these patients.
PIK3CA mutations in patients with evaluable tissue samples (N = 74)
PIK3CA mutations were detected in 48.6% (36/74) of archival tumour tissue samples (29 primary, 45 metastatic). In one tumour sample, two PIK3CA mutations were observed (Table 2). VAFs ranged from 4.36 to 72.93% (median 28.64%). The most frequent PIK3CA mutation detected in tissue was H1047R (23.0%), followed by E545K (13.5%), E542K (9.5%) and H1047L (2.7%). The location and number of metastases, as well as treatment lines, were not associated with PIK3CA mutations in tissue samples.
Concordance of PIK3CA mutation status in plasma and tissue
Comparative analyses of plasma and tissue were performed on samples from 72 patients for the 11 PIK3CA hotspot mutations covered by both assays (Supplementary Fig. S5). We found a moderate agreement between tissue and plasma samples, with a Cohen kappa = 0.58 (95% CI, 0.426–0.651) and an overall concordance rate of 72.2% (52/72, Table 3). Overall concordance was similar if only metastatic tissue samples (32/45, 71.1%) or only primary tissue samples (20/27, 74.1%) were considered. In detail, 24/72 patients (33.3%) had the same PIK3CA alteration in plasma and tissue, while no PIK3CA mutation in either sample type was found in 28/72 patients (38.9%). Discordant results (20/72, 27.8%; Table 4) included 10 (13.9%) patients with a PIK3CA mutation in plasma only and six patients (8.3%) with a PIK3CA mutation in tissue only. In addition, in four patients (5.6%), a different PIK3CA mutation was detected in the plasma sample compared to the tissue sample. The overall comparison showed no statistical difference between the results of tissue and plasma (McNemar’s test p = 0.317). For concordant samples, the PIK3CA VAF in ctDNA were generally higher (median 2.78, range 0.57–17.70) than for the discordant samples (median 0.97, range 0.27–2.97; Wilcoxon rank-sum test p = 0.001), where mutations were only detected in plasma (Supplementary Fig. S6). Z-scores were similar in discordant samples (median 2.09, range 0.77–7.90) compared to concordant samples (median 2.12, range 0.85–69.68; Wilcoxon rank-sum test p = 0.475). Within the 20 discordant samples, 6 had z-scores ≥3. Finally, the time interval between plasma and tissue collection did not differ significantly and ranged from −9.9 to 139.2 months (median 1.9 months) for discordant samples and from −14.8 to 229.3 months (median 9.6 months) for concordant samples (Wilcoxon rank-sum test p = 0.119).
Table 3.
Summary of PIK3CA mutation concordance data comparing plasma with tissue samples (N = 72).
| No | % | |
|---|---|---|
| Total concordance (no alteration or identical alteration) | 52 | 72.2 |
| No alterations found in plasma and tissue | 28 | 38.9 |
| Identical alterations found in plasma and tissue | 24 | 33.3 |
| Total discordant samples | 20 | 27.8 |
| Different alterations found in plasma and tissue | 4 | 5.6 |
| Plasma only | 10 | 13.9 |
| Tissue only | 6 | 8.3 |
| Total number of alterations found in tissue | 35 | – |
| Total number of alterations found in plasma | 42 | – |
| Samples with double mutations in tissue | 1 | 1.4 |
| Samples with double mutations in plasma | 4 | 5.6 |
Table 4.
Discordant PIK3CA mutation results (N = 20) between plasma and tissue samples.
| Patient ID | Mutation(s) in plasma (AF %) | z-score | Mutation in tissue (MAF %) | Source of tissue | Time interval between plasma and tissue collectiona |
|---|---|---|---|---|---|
| B176 | H1047R (0.27) | 7.39 | ND | Metastasis | −11 days |
| B195 | H1047R (2.97) | 4.18 | ND | Metastasis | 6 days |
| B200 | H1047R (0.98) | 1.75 | ND | Metastasis | 4.4 months |
| B201 | E542K (0.46) | 2.11 | H1047R (21.1) | Metastasis | 22 days |
| B205 | H1047R (0.72) | 7.90 | ND | Metastasis | 14 days |
| B207 | H1047R (0.97) | 3.30 | ND | Metastasis | −3.8 months |
| B209 | H1047R (1.17) | 1.22 | ND | Primary | 6.6 months |
| B212 | ND | 3.19 | E545K (30.7) | Primary | −9.9 months |
| B218 | ND | 3.40 | E542K (23.7) | Metastasis | 16.1 months |
| B235 | E542K (1.34), H1047R (0.77) | 1.69 | ND | Primary | 22.6 months |
| B242 | ND | 2.07 | E545K (55.8) | Metastasis | 10.1 months |
| B275 | ND | 2.51 | H1047R (25.6) | Primary | 2.2 months |
| B285 | H1047R (0.31) | 1.81 | ND | Metastasis | −4.6 months |
| B296 | H1047R (0.58) | 0.77 | H1047L (30.5) | Metastasis | 2.3 months |
| B303 | H1047R (0.42) | 0.81 | E542K (29.5) | Metastasis | 7 days |
| B304 | H1047R (1.08) | 2.20 | ND | Primary | 139.2 months |
| B307 | H1047R (0.41) | 1.51 | H1047L (25.9) | Primary | 88.5 months |
| B341 | ND | 2.43 | E545K (30.9) | Primary | 20 days |
| B350 | H1047R (0.47) | 1.29 | ND | Metastasis | 1.8 months |
| B354 | ND | 0.94 | H1047R (15.9) | Metastasis | 119.0 months |
ND no mutation detected.
aNegative numbers mean that plasma samples were analysed at an earlier date than tissue samples.
Overall in the comparison cohort (N = 72), 38 patients had PIK3CA mutations (52.8%) detectable in plasma samples, compared to 34 patients with PIK3CA mutations in tissue samples (47.2%). In four plasma samples, double PIK3CA mutations were detected, while only one double mutation occurred in tumour tissue samples. H1047R was the most frequent mutation found in both plasma and tissue samples (34.7% in plasma, 22.2% in tissue), followed by E545K (8.3% in plasma, 12.5% in tissue), E542K (8.3% in both plasma and tissue) and C420R (1.4% in both plasma and tissue). The H1047L variant was detected in two tissue samples (2.8%) but not in plasma samples.
Focusing only on the question of whether we were able to identify patients qualifying for alpelisib treatment based on the detection of any of the 11 PIK3CA mutations included in the SOLAR-1 trial [11], we detected at least one mutation in 28/72 patients both in tissue and plasma. In 10 patients, such mutations were found only in plasma. Among six patients with tissue PIK3CA mutations not detectable in ctDNA, four patients had low tumour fractions (z-score < 3). Comparing our results to the liquid biopsy-based results in the SOLAR-1 study, we found that the proportions of discordant plasma samples compared to wild-type tissue samples were significantly higher in our study (p < 0.001). In contrast, the proportion of discordant plasma samples compared to mutant tissue samples was considerably lower in our study (p = 0.002, Fig. 2). (https://www.accessdata.fda.gov/cdrh_docs/pdf19/P190004B.pdf) These results indicate that our SiMSen-Seq assay was more sensitive than the therascreen assay used in the SOLAR-1 study [11].
Fig. 2. Concordance of PIK3CA mutation status between ctDNA and tissue samples in our study and in the SOLAR-1 study.
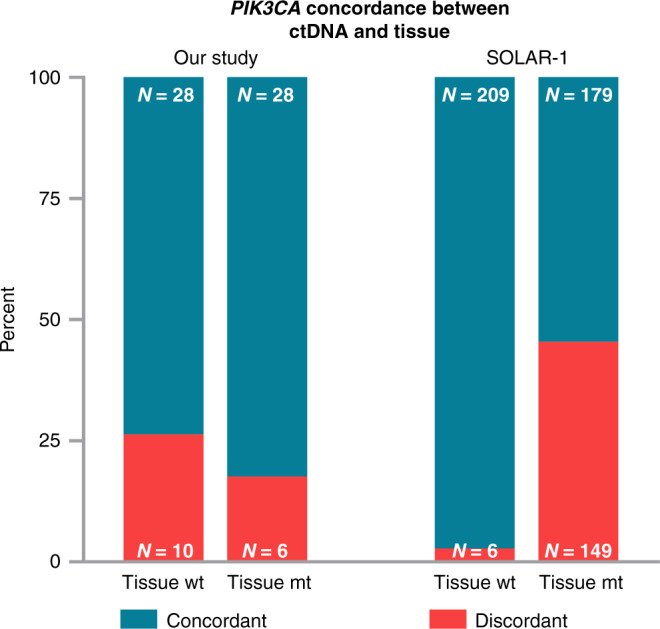
Total numbers of concordant (turquoise) and discordant (red) cases; ctDNA, circulating tumour DNA; wt, wild type; mt, mutant.
Finally, we performed an exploratory analysis assessing the association between PIK3CA mutation and progression-free survival in a subset of 47 HR+/HER2− breast cancer patients receiving CDK inhibitor treatment. In this subset of patients, the presence of a PIK3CA mutation in plasma was not significantly associated with PFS (HR 1.79, 95% CI 0.73–4.37, log-rank p = 0.197; Supplementary Fig. S7)
Discussion
This analysis with patient-paired plasma and tissue samples shows that a liquid biopsy-based approach holds promise to identify patients with the most common activating PIK3CA mutations. Using the high-resolution NGS-based SiMSen-Seq approach, PIK3CA mutations were detected non-invasively in 47% of our patients with a moderate concordance to tissue data (overall concordance rate 72%). Although digital PCR may currently be regarded as the gold standard for mutation detection at hotspots, it can be particularly challenging when analyzing multiple targets since either multiple reactions are needed, or no specific sequence information can be obtained. Although SiMSen-Seq is more expensive than digital PCR, all targets can be analysed within one reaction, and sequence information of the mutations is provided. A combination with an untargeted, mutation-independent approach for detecting ctDNA fractions might confirm a negative PIK3CA result, thereby enhancing the performance of the SiMSen-Seq test. This approach allowed the optimisation of the selection of candidate patients for PIK3CA targeted treatment.
The concept of ctDNA based detection of PIK3CA mutations as predictive biomarkers for the PI3K inhibitors was already evaluated in the phase III trials BELLE-2 and BELLE-3 with the Pan-PI3K inhibitor buparlisib [8, 23]. and in the SANDPIPER trial with the alpha-specific PI3K inhibitor taselisib [24]. Although these trials were positive, based on the toxicity and the magnitude of benefit, neither buparlisib nor taselisib have been clinically developed further in HR+/HER2− breast cancer. However, PIK3CA mutation testing based on liquid biopsy has shown promising results as a predictive biomarker for PIK3 inhibitors [25], and these data were more recently confirmed in the SOLAR-1 trial. In contrast, PIK3CA mutations in ctDNA did not predict the clinical benefit of everolimus in the BOLERO2 study. Among 550 patients included in the ctDNA analyses, median PFS was comparable in PIK3CA wild-type and mutant patients receiving everolimus [10].
The FDA approval of alpelisib includes the companion diagnostic test therascreen® PIK3CA RGQ PCR Kit [26] and liquid biopsy testing. Details on the detection of PIK3CA mutations in ctDNA in the SOLAR-1 trial have not yet been published, and only conference papers refer to the sensitivity of the liquid biopsy-based testing to detect PIK3CA mutant patients [27]. Out of 328 patients identified as positive in the tissue, in 54.9% of patients, PIK3CA mutations could be confirmed in ctDNA via the qPCR assay. These data indicate that the sensitivity of the test performed in patients included in the SOLAR-1 study was substantially lower than the sensitivity of the NGS-based SiMSen-Seq assay used in our study. Furthermore, by adding the mFAST-SeqS z-score as a surrogate marker for tumour fraction in plasma to the results of SiMSen-Seq, we could narrow down the need for tissue testing to only 27/93 (29%) patients. In 4 out of these 20 patients with evaluable tissue samples, we identified PIK3CA mutation in the tissue. This finding may be of great importance for ctDNA testing in general since the combination of mutation-specific analyses with an untargeted, fast and cost-effective assessment of tumour fractions may help data interpretation. In addition to the relevance of its one-time measurement, longitudinal assessment of tumour fractions using mFAST-SeqS sequencing harbors important prognostic information [28].
PIK3CA mutations occur on average in ~40% of HR+ patients, with a slightly higher proportion in Luminal A patients [6]. In a recent analysis of metastatic breast cancer tissue samples from the SAPHIR study, the positivity of PIK3CA mutated samples was lower in the HR+ HER2− patients, with 28%. However, recent data show a highly heterogenous mutation frequency between distinct ethnicities in the first line setting in the HR+ HER2− subgroup of patients (46% in Chinese vs 22% in US patients) [29].
Preliminary data from the AURORA demonstrated a similar overall range of PIK3CA mutations in metastatic breast cancer without providing information on the distribution with intrinsic subtypes [13]. Nevertheless, the activating PIK3CA mutations were associated with diminished sensitivity to chemotherapy and worse OS [9]. Tzanikou et al. have shown that the frequency of detection of PIK3CA mutations in liquid biopsy is higher in metastatic patients than in early stages, possibly either due to a higher proportion of mutated patients but also to higher tumour load in peripheral blood [30]. In the recently published analyses of the first 381 matched pairs of primary tumours and metastases from the AURORA study, an enrichment of PIK3CA mutations in metastasis was observed [14, 31]. Thus, it is reasonable to expect an increase in PIK3CA mutations in progressive or recurrent disease [31]. In our present study, we did not observe this increase since the most recent tissue available was used for the NGS, and discordant results were observed independently, as summarised in Table 4. Yet, it may contribute to a higher proportion of positive patients overall. Another study demonstrated that the proportion of multiple PIK3CA mutated tumours increases with metastatic disease, thereby further increasing the importance of continuous testing at the later stage [21]. Our results demonstrate that co-occurrence of mutations (e.g. double mutations) were primarily found in ctDNA, thereby substantiating the assumption that DNA may be more representative of the heterogeneity of the disease. However, with our approach, we can neither technically nor biologically confirm that these mutations occurred on the same allele. Additionally, the VAF of the second mutation was mainly low, except for one patient where the detected VAF of the second mutation not found in tissue was higher in plasma. One possible reason for the discrepancy, in this case, could be that the time difference between the primary tumour and plasma analysis was significantly longer compared to the other samples. Peripheral blood for ctDNA was also partly collected at a later advanced and/ or metastatic stage, whereas a significant proportion of the analysed tissues (61%) were from the most recently biopsied single metastatic lesion. Since compound mutations were shown to be associated with improved benefit from PIK3CA inhibitors, this fact may also be in favour of the definition of PIK3CA mutation status based on the ctDNA analyses [21].
The detection of PIK3CA mutations in plasma implies the presence of tumour-specific mutations since germline PIK3CA mutations lead to early lethality and are not expected in humans [32]. Analytical sensitivity of the assays may allow detection of very low amounts of mutated DNA, with a detection limit of 0.25% VAF in our study. However, the clinical relevance of detecting very low VAF is not well understood. There are several possibilities for low VAF, including a low tumour DNA fraction and a low proportion of PIK3CA mutated cells in the overall tumour mass. Concerning clonality, data suggest that most PIK3CA mutations are clonal or truncal mutations [33], leaving the possibility that low tumour ctDNA burden is the primary cause of low-level detection. In our study, we observed that discordant samples with low-level VAF were mainly retrieved from patients with low ctDNA burden as measured by z-scores. The correlation between PIK3CA VAF levels and clinical benefits from the alpelisib and the best time point to start the treatment is currently unknown and remains a topic for future studies.
Our study has several limitations, including the small sample size, a higher percentage of PIK3CA mutated cancers, and the focus on PIK3CA mutations only. A broader approach providing information on various important genes would be beneficial, but with a potential of decreased sensitivity for PIK3CA mutation detection compared to our present analyses. Our future studies will also include a comparison of SiMSen-Seq based detection of PIK3CA mutations with NGS technologies analyzing a larger number of genes important for treating metastatic breast cancer, as was recently performed in the PlasmaMATCH study [34]. Finally, for the clinical implementation of our SIMSen-Seq assays additional in-depth technical validation experiments are necessary to assess sensitivity and specificity. These experiments are already ongoing at our institution.
Nevertheless, the PlasmaMATCH study demonstrated groundbreaking clinical progress for selected metastatic breast cancer patients, and we believe that this study paved the way for future large clinical trials. On the other hand, we currently need to establish precise and low-cost technologies to select patients for currently available PI3K treatments, and our SiMSen-Seq based assay performed well in the current study.
In conclusion, within our study, SiMSen-Seq-based detection of PIK3CA hotspot mutations from plasma has shown promising results. In combination with an untargeted, mutation-independent approach for detecting ctDNA fractions, we believe that for samples with z-score >3 and no PIK3CA hotspot mutations, no additional tissue analyses are needed. Adding ctDNA fractions thus further increased the validity of negative plasma results. SiMSen-Seq alone and this combinatory approach for plasma analyses substantially improved the robustness of liquid biopsy-based PIK3CA mutations testing, particularly compared to the FDA-approved companion diagnostic test therascreen® PIK3CA used in the SOLAR-1 study. This approach should be validated in a larger cohort of patients and compared with larger panel NGS ctDNA sequencing methods to establish the most promising approach for selecting individual treatments for our patients.
Supplementary information
Acknowledgements
We thank Klara Balic for English proofreading. In addition, we want to acknowledge that part of this manuscript was published as a doctoral thesis.
Author contributions
CS, AT, ND and MB designed the study. RG and KK performed sequencing experiments, SJ and PR performed pathological evaluation, CS, EVK, AT, FP and MB contributed to patient care and data acquisition. ND and FP performed the statistical analysis, CS, AT, ND, MB, EH and PJJ performed interpretation of results. CS, ND, MB and FP drafted and revised the manuscript. ND, EH and MB supervised the study. All authors reviewed the manuscript and approved its final version.
Funding
This work was supported by the Christian Doppler Research Fund for Liquid Biopsies for Early Detection of Cancer led by EH supported by the Federal Ministry of Digital and Economic Affairs and by Novartis and Pfizer.
Data availability
Data are available at request from authors and were partially (SiMSen-Seq and mFAST-SeqS) deposited at the European Genome-phenome Archive (EGA; http://www.ebi.ac.uk/ega/) under study accession number EGAS00001004940 as following dataset: PIK3CA SiMSen-Seq data of plasma samples.
Ethics approval and consent to participate
The study was approved by the ethics committee of the Medical University Graz (ethical approval number 21–227 ex 09/10), and written informed consent was obtained from all patients.
Competing interests
CS received travel expenses, consulting fees and honoraria from Amgen, Astra Zeneca, Eli Lilly, Novartis, Pfizer, Roche and Samsung, SJ received honoraria from Novartis and Roche, EVK has received travel expenses and honoraria from Astra Zeneca and Pierre- Fabre, RB had a consulting role, received honoraria, research funding and travel expenses from Astra Zeneca, Daiichi, Eisai, Eli Lilly, MSD, Novartis, Pfizer, Pierre-Fabre, Puma, Roche, AT received honoraria and research funding from Astra Zeneca, BMS, Merck and Roche. KK received honoraria and advisory from Astra Zeneca, Thermo Fischer, Novartis and BMS. PR received consulting fees, honoraria and travel expenses from Diaceutics, Novartis, and Roche, PJJ has had a consulting or advisory role, received honoraria, research funding and/or travel/accommodation expenses from Abbvie, Bayer, Boehringer, Novartis, Pfizer, Servier, Roche, BMS and Celgene, HG is a full time/part-time employee by Novartis, EH Servier, has received funding from Servier, Freenome, CA and PreAnalytiX and had an advisory role for Roche. MB has received honoraria, consulting fees research funds and/or travel expenses from Amgen, Astra Zeneca, Daichii, Eli Lilly, MSD, Novartis, Pierre- Fabre, Pfizer, Roche and Samsung.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nadia Dandachi, Email: nadia.dandachi@medunigraz.at.
Marija Balic, Email: marija.balic@medunigraz.at.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01601-9.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Gennari A, Conte P, Rosso R, Orlandini C, Bruzzi P. Survival of metastatic breast carcinoma patients over a 20-year period: a retrospective analysis based on individual patient data from six consecutive studies. Cancer. 2005;104:1742–50. doi: 10.1002/cncr.21359. [DOI] [PubMed] [Google Scholar]
- 3.Ribnikar D, Volovat SR, Cardoso F. Targeting CDK4/6 pathways and beyond in breast cancer. Breast. 2019;43:8–17. doi: 10.1016/j.breast.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006;94:455–9. doi: 10.1038/sj.bjc.6602970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K pathway in human disease. Cell. 2017;170:605–35.. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–9. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 8.Di Leo A, Johnston S, Lee KS, Ciruelos E, Lonning PE, Janni W, et al. Buparlisib plus fulvestrant in postmenopausal women with hormone-receptor-positive, HER2-negative, advanced breast cancer progressing on or after mTOR inhibition (BELLE-3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2018;19:87–100. doi: 10.1016/S1470-2045(17)30688-5. [DOI] [PubMed] [Google Scholar]
- 9.Mosele F, Stefanovska B, Lusque A, Tran Dien A, Garberis I, Droin N, et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann Oncol. 2020;31:377–86.. doi: 10.1016/j.annonc.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Moynahan ME, Chen D, He W, Sung P, Samoila A, You D, et al. Correlation between PIK3CA mutations in cell-free DNA and everolimus efficacy in HR(+), HER2(−) advanced breast cancer: results from BOLERO-2. Br J Cancer. 2017;116:726–30.. doi: 10.1038/bjc.2017.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380:1929–40.. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 12.Rugo HS, Lerebours F, Ciruelos E, Drullinsky P, Ruiz-Borrego M, Neven P, et al. Alpelisib plus fulvestrant in PIK3CA-mutated, hormone receptor-positive advanced breast cancer after a CDK4/6 inhibitor (BYLieve): one cohort of a phase 2, multicentre, open-label, non-comparative study. Lancet Oncol. 2021;22:489–98.. doi: 10.1016/S1470-2045(21)00034-6. [DOI] [PubMed] [Google Scholar]
- 13.Aftimos PG, Antunes De Melo e Oliveira AM, Hilbers F, Venet D, Vingiani A, Nili Gal Yam E, et al. First report of AURORA, the breast international group (BIG) molecular screening initiative for metastatic breast cancer (MBC) patients (pts). Ann Oncol. 2019;30:iii48.
- 14.Aftimos P, Oliveira M, Irrthum A, Fumagalli D, Sotiriou C, Nili Gal-Yam E, et al. Genomic and transcriptomic analyses of breast cancer primaries and matched metastases in AURORA, the Breast International Group (BIG) molecular screening initiative. Cancer Discov. 2021;11:2796–2811.. doi: 10.1158/2159-8290.CD-20-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heitzer E, Ulz P, Belic J, Gutschi S, Quehenberger F, Fischereder K, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5:30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stahlberg A, Krzyzanowski PM, Egyud M, Filges S, Stein L, Godfrey TE. Simple multiplexed PCR-based barcoding of DNA for ultrasensitive mutation detection by next-generation sequencing. Nat Protoc. 2017;12:664–82.. doi: 10.1038/nprot.2017.006. [DOI] [PubMed] [Google Scholar]
- 17.Belic J, Koch M, Ulz P, Auer M, Gerhalter T, Mohan S, et al. Rapid identification of plasma DNA samples with increased ctDNA levels by a modified FAST-SeqS approach. Clin Chem. 2015;61:838–49. doi: 10.1373/clinchem.2014.234286. [DOI] [PubMed] [Google Scholar]
- 18.Belic J, Graf R, Bauernhofer T, Cherkas Y, Ulz P, Waldispuehl-Geigl J, et al. Genomic alterations in plasma DNA from patients with metastasized prostate cancer receiving abiraterone or enzalutamide. Int J Cancer. 2018;143:1236–48.. doi: 10.1002/ijc.31397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasan N, Razavi P, Johnson JL, Shao H, Shah H, Antoine A, et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kalpha inhibitors. Science. 2019;366:714–23.. doi: 10.1126/science.aaw9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suppan C, Brcic I, Tiran V, Mueller HD, Posch F, Auer M, et al. Untargeted assessment of tumor fractions in plasma for monitoring and prognostication from metastatic breast cancer patients undergoing systemic treatment. Cancers (Basel) 2019;11:1171. doi: 10.3390/cancers11081171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baselga J, Im SA, Iwata H, Cortes J, De Laurentiis M, Jiang Z, et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18:904–16.. doi: 10.1016/S1470-2045(17)30376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baselga J, Dent SF, Cortés J, Im Y-H, Diéras V, Harbeck N, et al. Phase III study of taselisib (GDC-0032) + fulvestrant (FULV) v FULV in patients (pts) with estrogen receptor (ER)-positive, PIK3CA-mutant (MUT), locally advanced or metastatic breast cancer (MBC): primary analysis from SANDPIPER. J Clin Oncol. 2018;36:LBA1006–LBA1006. doi: 10.1200/JCO.2018.36.18_suppl.LBA1006. [DOI] [Google Scholar]
- 25.Baselga J, Sellami D, El-Hashimy M, Dharan B, Wang A, Scheuer N, et al. Abstract A050: PIK3CA mutation status in tumor tissue and ctDNA as a biomarker for PFS in patients with HR+, HER2- ABC treated with buparlisib or placebo plus fulvestrant: results from the BELLE-2 and BELLE-3 randomized studies. Mol Cancer Ther. 2018;17:A050–A050. [Google Scholar]
- 26.Martinez-Saez O, Chic N, Pascual T, Adamo B, Vidal M, Gonzalez-Farre B, et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. 2020;22:45. doi: 10.1186/s13058-020-01284-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juric D, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo H, et al. Abstract GS3-08: Alpelisib + fulvestrant for advanced breast cancer: subgroup analyses from the phase III SOLAR-1 trial. Cancer Res. 2019;79:GS3-08-GS3. [Google Scholar]
- 28.Dandachi N, Posch F, Graf R, Suppan C, Klocker EV, Müller HD, et al. Longitudinal tumor fraction trajectories predict risk of progression in metastatic HR + breast cancer patients undergoing CDK4/6 treatment. Mol Oncol. 2020;15:2390–2400. doi: 10.1002/1878-0261.12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Davis AA, Liu X-R, Xie F, Gui X-Y, Chen Y-F, et al. Cell-free DNA comparative analysis of hormone receptor-positive, first-line metastatic breast cancer genomic landscape in the United States and China. J Clin Oncol. 2020;38:1059. doi: 10.1200/JCO.2020.38.15_suppl.1059. [DOI] [Google Scholar]
- 30.Tzanikou E, Markou A, Politaki E, Koutsopoulos A, Psyrri A, Mavroudis D, et al. PIK3CA hotspot mutations in circulating tumor cells and paired circulating tumor DNA in breast cancer: a direct comparison study. Mol Oncol. 2019;13:2515–30. doi: 10.1002/1878-0261.12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markou A, Farkona S, Schiza C, Efstathiou T, Kounelis S, Malamos N, et al. PIK3CA mutational status in circulating tumor cells can change during disease recurrence or progression in patients with breast cancer. Clin Cancer Res. 2014;20:5823–34. doi: 10.1158/1078-0432.CCR-14-0149. [DOI] [PubMed] [Google Scholar]
- 32.Bi L, Okabe I, Bernard DJ, Wynshaw-Boris A, Nussbaum RL. Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110alpha subunit of phosphoinositide 3-kinase. J Biol Chem. 1999;274:10963–8. doi: 10.1074/jbc.274.16.10963. [DOI] [PubMed] [Google Scholar]
- 33.O’Leary B, Cutts RJ, Liu Y, Hrebien S, Huang X, Fenwick K, et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 2018;8:1390–403. doi: 10.1158/2159-8290.CD-18-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner NC, Kingston B, Kilburn LS, Kernaghan S, Wardley AM, Macpherson IR, et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): a multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 2020;21:1296–308. doi: 10.1016/S1470-2045(20)30444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available at request from authors and were partially (SiMSen-Seq and mFAST-SeqS) deposited at the European Genome-phenome Archive (EGA; http://www.ebi.ac.uk/ega/) under study accession number EGAS00001004940 as following dataset: PIK3CA SiMSen-Seq data of plasma samples.


