Abstract
Objective
The prognostication of metastatic pancreatic adenocarcinoma (mPDAC) patients remains uncertain, mainly based on carbohydrate antigen 19-9 (CA19-9), with limited utility. Circulating tumour DNA (ctDNA) has been suggested as a prognostic factor, but its added value has been poorly explored. The objective was to determine whether ctDNA is an independent factor for the prognostication of mPDAC.
Design
Translational study based on two prospective collections of plasma samples of mPDAC patients naïve for chemotherapy. One used as a test series and the other as validation series coming from two randomised trials (Prodige 35 and Prodige 37). CtDNA was assessed by digital droplet PCR targeting two methylated markers (HOXD8 and POU4F1) according to a newly developed and validated method. Univariate and multivariate analyses were performed according to ctDNA status.
Results
Of 372 plasma samples available, 354 patients were analyzed for survival. In the validation series, 145 of 255 patients were found ctDNA positive (56.8%), Median PFS and OS were 5.3 and 8.2 months in ctDNA-positive and 6.2 and 12.6 months in ctDNA-negative patients, respectively. ctDNA positivity was more often associated with young age, high CA19-9 level and neutrophils lymphocytes ratio. In multivariate analysis including these previous markers, ctDNA was confirmed as an independent prognostic marker for PFS (adjusted hazard ratio (HR) 1.5, CI 95% [1.03–2.18], p = 0.034) and OS (HR 1.62, CI 95% [1.05–2.5], p = 0.029).
Conclusions
In this first ctDNA assessment in a large series of mPDAC derived from clinical trials, ctDNA was detectable in 56.8% of patients and confirmed as an independent prognostic marker.
Subject terms: Prognostic markers, Cancer screening, Pancreatic cancer
Introduction
Pancreatic Adenocarcinoma (PDAC) is a leading cause of cancer-related mortality worldwide [1]. As in many cancers, surgery remains the only curative treatment. Because of aggressive growth, early dissemination and lack of early symptoms, 80% of patients are diagnosed at late clinical stages [2]. Despite recent improvements with new chemotherapy protocols such as FOLFIRINOX or gemcitabine plus nab-paclitaxel, patients prognosis remains very poor [3–5]. Carbohydrate Antigen 19-9 (CA19-9) is the only currently available blood test for diagnosis, prognosis, monitoring and surveillance in PDAC [6]. However, its utility is limited by low specificity, influenced in particular by bilirubin level and sialyl Lewis blood group.
Measuring tumour-specific alterations in blood nucleic acids offers an interesting approach. Detection of circulating tumour DNA (ctDNA) using somatic acquired genetic alterations has produced interesting results to predict response to treatment, bring prognostic information and monitor disease recurrence in a wide range of cancers [7]. In several recent small retrospective series of PDAC, ctDNA was detectable with a frequency varying according to tumour stage and technics used for its detection from 26 to 87.5% [8] and suggested to be an independent prognostic marker whatever disease stage [9–11]. In these publications, the presence of detectable ctDNA was related to tumour burden, and its early decrease seemed able to predict response to chemotherapy in advanced tumours [9–11]. However, data on ctDNA prognostic value remain limited, coming from retrospective or prospective series most often with a limited number of patients, mixing non-metastatic and metastatic patients and patients not always treated with current standard chemotherapy regimens. In addition, the potential added value of ctDNA quantification for prognostication in PDAC has been poorly explored to date.
Based on computational analysis of the TCGA database, we identified two specific methylated biomarkers in PDAC, HOXD8 and POU4F1, and used them to validate the independent prognostic value of ctDNA. As compared to previously published papers on this hot topic [8–11], we propose here to test ctDNA in a large series of 372 metastatic pancreatic cancer patients included in one prospective series and two randomised phase II trials with long follow-up and high-quality annotated data to efficiently evaluate the weight of this variable in the prognostication of metastatic cancer patients.
Patients and methods
Patients
Test cohort
From January 2011 to June 2018, plasmas of all consecutive patients with histologically proven metastatic PDAC, eligible for first-line chemotherapy, were prospectively collected in the oncology department at Pitié-Salpêtrière hospital in Paris. Blood samples were collected just before the first cycle of chemotherapy. All patients signed an informed consent form approved by the ethics committee (CPP Ile-de-France 2014/59NICB). The monitoring data collected in this prospective database have been described previously [12]. Blood samples (9 mL) were withdrawn from a central catheter and placed in EDTA tubes. The collected samples were centrifuged at 1620 × g for 15 min at 4 °C within 3 h of blood draw. Plasma was stored at −80 °C until further use.
Validation cohort
The validation cohort is composed of patients included in two prospective randomised phase II trials conducted by the PRODIGE collaborative intergroup.
PRODIGE 35 (PANOPTIMOX) trial [13] was a multicenter prospective randomised trial (NCT02352337) that compared three arms in the first line in patients with metastatic PDAC: 6 months FOLFIRINOX (arm A), 4 months FOLFIRINOX followed by LV5FU2 maintenance treatment for controlled patients, and treatment reintroduction at disease progression (arm B) or a sequential treatment alternating gemcitabine and FOLFIRI.3 every 2 months (arm C). Blood samples were collected at inclusion and sent within 24 h to the central laboratory. Samples were centrifuged at 1620 × g for 15 min at 4 °C. Plasma was stored at −80 °C until further use.
PRODIGE 37 (FIRGEMAX) trial [14] was a multicenter prospective randomised trial (NCT 02827201) that compared two arms in the first line in patients with metastatic PDAC: 2 months alternating nab-paclitaxel/gemcitabine and FOLFIRI.3 (arm A) or nab-paclitaxel + gemcitabine (arm B). Blood samples were collected at inclusion and sent within 24 h to the central laboratory. Samples were centrifuged at 1620 × g for 15 min at 4 °C. Plasma was stored at −80 °C until further use.
ctDNA analysis
See eMethods online for the description of (i) plasma sample preparation, storage, DNA extraction and quantification; (ii) Assessment of circulating tumour DNA and (iii) next-generation sequencing (NGS).
Methylation marker development and validation
HOXD8 and POU4F1 methylation markers were selected from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/). The selected region contained CpGs that were significantly differentially methylated between tumour and healthy pancreas tissues. The CpGs containing region was selected from a list of 485,577 CpGs annotated with genes, including their methylation level in healthy and tumour tissue, the difference in methylation level between healthy and tumour tissue (the fold change), correlated with the tumour and normal cells count in cancer patients. Analysis of DNA methylation of HOXD8 and POU4F1 in tumour and non-tumour tissue (matched adjacent non-tumour tissue) from pancreatic cancer patients was validated by ddPCR (n = 19 tumour tissues and n = 17 matched adjacent non-tumour tissue). Samples were selected with at least one detectable mutation in KRAS, TP53 or SMAD4 (mutated allelic frequency from 2.9 to 71.3%) when sequenced by NGS using colon lung panel v2 (Thermo Fisher). One adjacent non-tumour tissue appeared mutated in TP53 (4.3%, 83.1% for the matched tumour tissue) and was thus excluded from further analyses. Compared to non-tumour tissue, DNA methylation in tumour tissue was significantly increased (p = 0.0005 for POU4F1 and p = 0.0001 for HOXD8, Wilcoxon paired test, n = 17). The sensitivity was 79%, and the specificity of 100% when considering both markers simultaneously. When analyzing plasma ccfDNA from healthy individuals (n = 12) by ddPCR for the presence of the two methylated markers, none were detected as positive for both markers and one sample presented low positivity with one maker (HOXD8, three positive droplets [lower than LOD] corresponding to 0.4% of methylated DNA).
Statistical analysis
Statistical analyses were performed using R software 3.6.3 Studio. A p value ≤ 0.05 was considered significant. The demographic characteristics of patients were compared by the Chi-square or Fisher’s exact test. Continuous data were analyzed with the independent-samples t-test. Survival rates were calculated using the Kaplan–Meier method. Progression-free survival (PFS) was measured from the date of randomisation/enrolment in the study to the first documented radiological progression (RECIST V1.1), or death, whichever occurred first. Patients lost to follow-up were censored at the last follow-up visit. Overall survival (OS) was defined as the time from randomisation/enrolment in the study to the date of death from any cause. Association between outcomes and baseline characteristics was assessed with Cox proportional hazard models, and hazard ratio (HR) with CI 95% are provided. Parameters with p < 0.05 in univariate analysis were entered into a final multivariable Cox regression. All variables included in the multivariate Cox model were tested for proportional hazards assumption by the function cox.zph of the survival R package.
Method description has been performed in order to maximise compliance to digital dMIQE2020 guidelines (see eMethods online and eTables 1 to 3) [15].
Results
Study population
Three hundred and seventy-two patients were enrolled in this study. One hundred and ten patients in the test cohort and 262 in the validation cohort (179 in PRODIGE 35 and 83 in PRODIGE 37, respectively).
ctDNA detection by Met-ddPCR assay and comparison of Met-ddPCR and BPER NGS analyses
The test cohort is an extension of an already published pancreatic cancer series [12]. Forty-eight additional patients with metastatic disease were added to the 62 patients from the previous publication to reach the final number of 110 mPDAC in our test cohort. This series allowed us to validate the quantitative analysis of ctDNA by detecting the methylation of two markers (HOXD8 and POU4F1). For 61 samples, we performed both next-generation sequencing with a panel of 22 genes including KRAS, TP53 and SMAD4 analyzed with BPER method and Met-ddPCR. The kappa coefficient between the two methods was 0.93 (CI 95% [0.83–1], eFig. 1). There was a significant correlation between the ctDNA concentration estimated by the measurement of the maximum variant allelic frequency (i.e. the allelic frequency of the more represented mutated gene) in NGS assay and the ratio of methylated ctDNA to non-methylated DNA in ddPCR assays (R2 = 0.52, p < 0.0001, eFig. 1).
Analysis of the test cohort
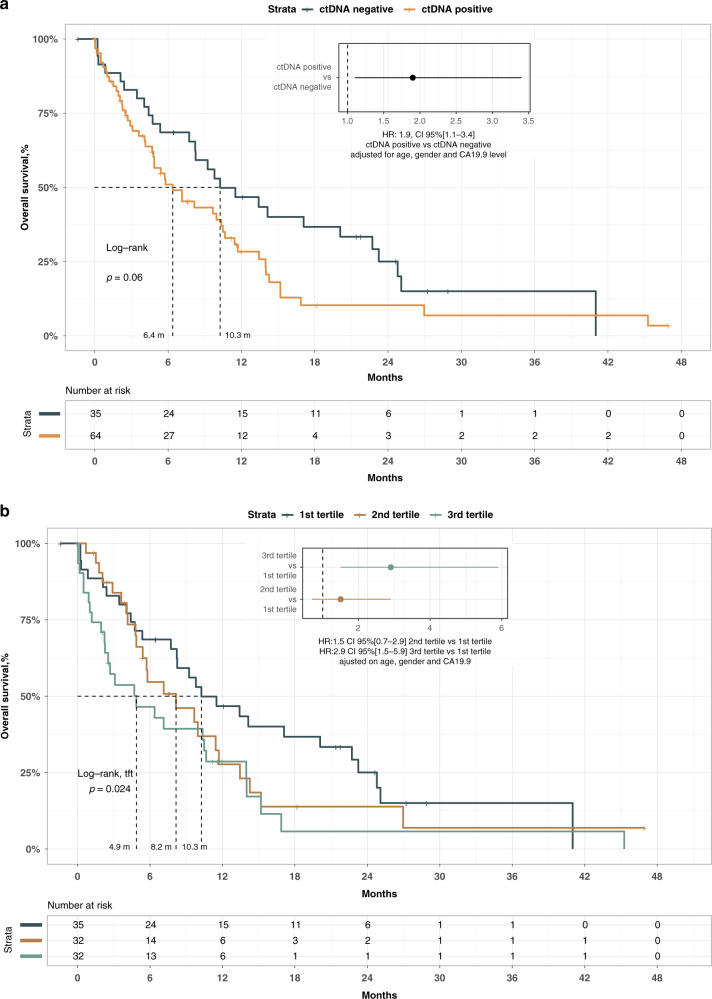
Using the Met-ddPCR assay, we were able to determine ctDNA status in 102 patients of the 110 patients of the test cohort (92.7%). The mean and standard deviation of free plasma DNA concentration was 101.4 ± 294.3 ng/mL in the test cohort. CtDNA was considered positive in 65 out the 102 patients (63.7%) with a mean methylated fraction of 7.8% ± 12.8. Among the clinical variables including age, gender, WHO performance status, CA19-9 concentration (as a continuous variable or dichotomised according to the median), the number of metastatic sites or the presence of liver metastases, the only variable associated with the positivity of ctDNA was the presence of liver metastasis (eTable 4). Indeed 96% of the patients with liver metastases had a positive ctDNA, whereas only 56% of those without liver metastasis had a positive ctDNA (p < 0.001). Analysis of OS showed a poor prognosis for patients with positive ctDNA. After adjustment on age gender and CA19-9 concentration, a hazard ratio (HR) of 1.9 (CI 95% [1.1–3.4]) was observed (Fig. 1a). Based on the fact that our Met-ddPCR assay allowed for quantitative analysis of ctDNA, we divided the patients according to tertiles and observed a significant association between ctDNA quantity and OS (Log-rank test for trends p-value = 0.024 Fig. 1B). The adjusted HR was 2.9 (CI 95% [1.5–5.9]) between patients with a ctDNA concentration belonging to the upper tertile compared to the patients with a ctDNA concentration belonging to the lower tertile after adjustment on age, gender and CA19-9.
Fig. 1. Test cohort.
a Overall survival according to ctDNA status in test cohort. b Overall survival according to ctDNA quantification (patient divided into tertiles).
Analysis of the validation cohort
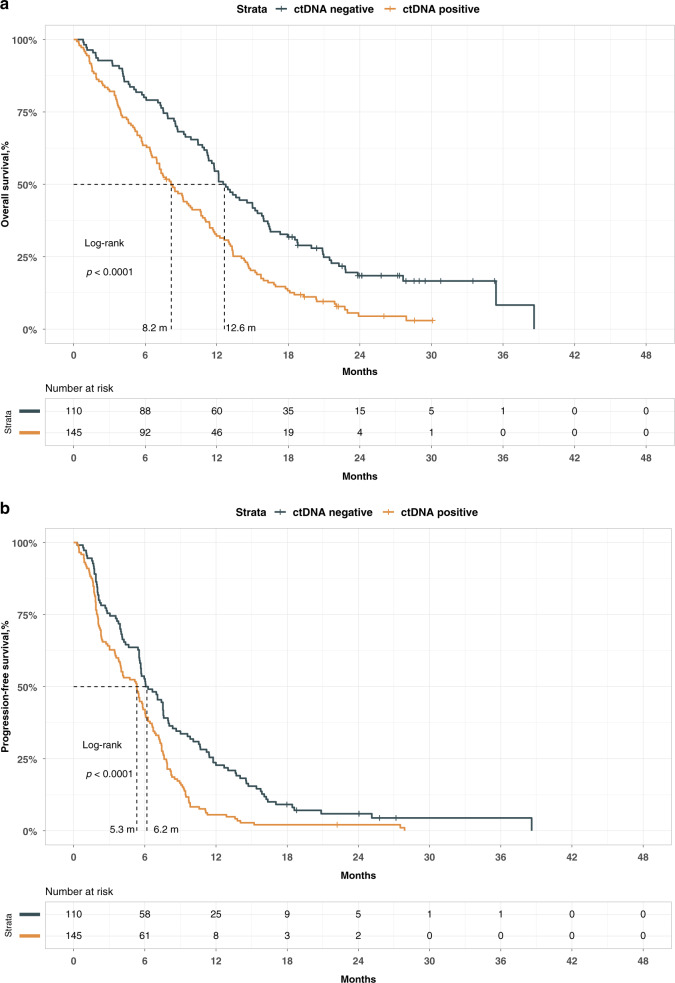
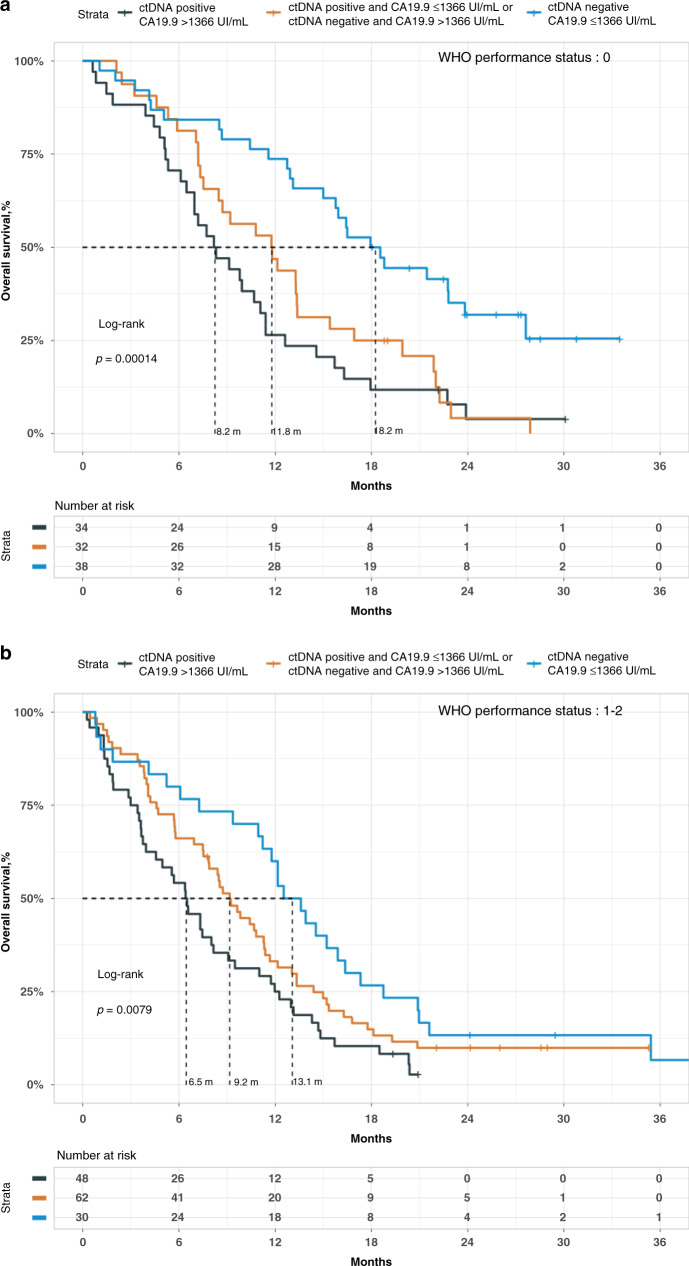
We pooled the samples of two randomised trials: PRODIGE 35 and PRODIGE 37, conducted during the same period. A plasma sample was available for 262 out of the 400 patients included in the two trials. The determination of ctDNA status was done in 255 patients out of the 262 for whom a plasma sample was available (97.3%). The clinical, biological and survival data of these 255 patients were not different from those without ctDNA determination, as shown in eTable 5, eFig. 2A, B). The mean and standard deviation of free plasma DNA concentration was 111.5 ± 273 ng/mL. CtDNA was considered positive in 145 out of 255 patients (56.9%) of the validation cohort with a mean methylated fraction of 6.3% ± 20.7. The clinical and biological characteristics of patients according to their ctDNA status are reported in Table 1. The presence of ctDNA was significantly associated with younger age, WHO performance status of 2+, high levels of CA19-9, elevated neutrophils lymphocytes ratio and the presence of liver metastases. We performed a univariate analysis for OS and PFS (eTable 6 and Fig. 2a, b). The presence of ctDNA was associated with a worse prognosis both on OS and PFS. The median OS was 8.2 versus 12.6 months in ctDNA-positive and -negative patients, respectively (HR = 2.26, CI 95% [1.56–3.27], p < 0.001). Similarly, the median PFS was 5.3 versus 6.2 months in ctDNA-positive and -negative patients, respectively (HR = 1.76, CI 95% [1.24–2.50], p < 0.001). ctDNA concentration was associated positively with a worse prognostic when the variable was considered as a continuous variable (HR = 2.94, CI 95% [1.4–6.0] and HR = 1.81, CI 95% [0.86–3.82] for OS and PFS, respectively). According to the number of patients with negative ctDNA, we divided our population into three groups according to the median and the 3rd quartile value of ctDNA concentration. We confirmed the significant association between ctDNA concentration and survivals. These results were observed both for OS and PFS (eFig. 3A, B). Finally, we performed multivariate analysis for OS and PFS. To test the impact of the addition of ctDNA in the prediction of survival, two models were built: the first one included factors showing a prognostic value in univariate analysis (reduced model) and the second one added the ctDNA dichotomised as absence or presence of ctDNA to the first one (full model). Both models were stratified on centers and studies. We tested the two models and showed that the full model fitted better than the reduced model using a robust likelihood ratio test (LR = 3.0, p = 0.04, LR = 4.8, p = 0.01) for OS and PFS, respectively. In the full model, ctDNA remained a significant prognostic factor for OS and PFS (Tables 2, 3) with HR of 1.62 (CI 95% [1.05–2.50]) and 1.50 (CI 95% [1.03–2.18]), respectively. We also built a full model using the five treatment arms as a stratification variable instead of centers and studies. The ctDNA remained an independent prognostic factor (HR 1.40, CI 95% [1.02–1.90] and HR 1.50, CI 95% [1.10–2.05] for OS and PFS, respectively). Furthermore, we investigated in ctDNA-positive patients the impact on survival of the investigational arms as compared to FOLFIRINOX in PRODIGE 35 and NAB-PACLITAXEL/GEMCITABINE in PRODIGE 37, and we found that there is no significant improvement of survival in any of the investigational arms. Finally, to measure the added value of cDNA status in predicting survival, we first compared the C index of two models with and without cDNA. For this purpose, we used the most parsimonious OS multivariate model that included WHO and CA19-9 status. The ctDNA status significantly increased the C index of the model from 0.636 to 0.671 (z-score = 1.87, p < 0.05), showing the usefulness of adding the ctDNA status in the OS model, which is highlighted in Fig. 3a, b. Next, we focused on the 19 patients with a WHO performance status of 0 and a CA19-9 ≤ 40 IU/mL. Of these, 12 were ctDNA negative, and 7 were ctDNA positive. In this small group of patients, the presence of ctDNA was highly prognostic, with an HR of 4.77 (CI 95% [1.32–17.36]), confirming the additional prognostic information of ctDNA.
Table 1.
Clinical and biological characteristics of the patients according to their ctDNA status.
| Characteristic | ctDNA negative N = 110a | ctDNA positive N = 145a | P-valueb |
|---|---|---|---|
| Age | 67 (61, 71) | 64 (56, 68) | <0.001 |
| Gender | 0.8 | ||
| Female | 47 (43%) | 58 (40%) | |
| Male | 63 (57%) | 87 (60%) | |
| WHO Score | 0.10 | ||
| 0 | 54 (49%) | 55 (38%) | |
| 1–2 | 56 (51%) | 90 (62%) | |
| CA19.9 UI/mL | 708 (76, 3167) | 3154 (165, 20,000) | 0.001 |
| Unknown | 2 | 9 | |
| CA19.9 median UI/mL | |||
| ≤1366 | 68 (63%) | 54 (40%) | <0.001 |
| >1366 | 40 (37%) | 82 (60%) | |
| Unknown | 2 | 9 | |
| NLRc | <0.001 | ||
| <3.2 | 73 (84%) | 54 (67%) | |
| ≥3.2 | 37 (16%) | 90 (33%) | |
| Unknown | 0 | 1 | |
| Liver metastasis | 76 (69%) | 138 (95%) | <0.001 |
| Number of metastatic sites | 0.5 | ||
| 1 | 68 (62%) | 83 (57%) | |
| ≥2 | 42 (38%) | 62 (43%) |
Bold values indicate statistical significance p < 0.05.
aStatistics presented: median (IQR); n (%).
bStatistical tests performed: Wilcoxon rank-sum test; Chi-square test of independence.
cNLR Neutrophils Lymphocytes ratio.
Fig. 2. Validation cohort.
a Overall survival according to ctDNA status. b Progression-free survival according to ctDNA status.
Table 2.
Multivariate analysis of overall survival in the validation cohort.
| Characteristic | HRa | 95% CIa | P-value |
|---|---|---|---|
| Age | 0.98 | 0.96–1.01 | 0.13 |
| WHO Performance status | |||
| 0 | Ref | ||
| 1–2 | 1.88 | 1.27, 2.80 | 0.002 |
| ctDNA | |||
| ctDNA negative | Ref | ||
| ctDNA positive | 1.62 | 1.05, 2.50 | 0.029 |
| CA19.9_median | |||
| ≤1366 | ref | ||
| >1366 | 1.70 | 1.17, 2.49 | 0.006 |
| NLRc | |||
| <3.2 | ref | ||
| ≥3.2 | 1.08 | 0.73, 1.60 | 0.7 |
Bold values indicate statistical significance p < 0.05.
Analysis was performed after stratification on studies and centers.
aHR Hazard ratio.
bCI confidence interval.
cNLR neutrophils lymphocytes ratio.
Table 3.
Multivariate analysis of progression-free survival in the validation cohort.
| Characteristic | HRa | 95% CIb | P-value |
|---|---|---|---|
| WHO Performance status | |||
| 0 | Ref | ||
| 1–2 | 1.43 | 0.98, 2.08 | 0.061 |
| ctDNA | |||
| ctDNA negative | Ref | ||
| ctDNA positive | 1.50 | 1.03, 2.18 | 0.034 |
| CA19.9_median | |||
| ≤1366 | Ref | ||
| >1366 | 1.36 | 0.96, 1.92 | 0.085 |
Bold values indicate statistical significance p < 0.05.
Analysis was performed after stratification on studies and centers.
aHR Hazard ratio.
bCI confidence interval.
Fig. 3. Overall survival according to CA.19.9 concentration and ctDNA status.
a Patients with a WHO performance status of 0. b Patients with a WHO performance status of 1 or 2.
Discussion
This study demonstrates that ctDNA is an independent prognostic marker for OS and PFS in mPDAC patients. We confirmed the significant association between ctDNA concentration and survival. Moreover, this is the first large-scale validation of the prognostic value of ctDNA in two randomised controlled trials.
CtDNA detection rate was 63.7% (CI 95% [53.6–73.8]) and 56.8% (CI 95% [50.5–63]) in the test and in the validation cohorts, respectively. This rate increased to 78.3% and 64.5% when only patients with liver metastases were considered in the test and validation cohorts. The prevalence of ctDNA in mPDAC observed in the literature ranges from 29.3 to 86.1% [8–12, 16–22]. These variations may be explained by different tumour volumes [10, 21], the presence or absence of liver metastases [10, 23], or other performance methods used for the ctDNA characterisation.
Different methods have been developed to assess the presence of ctDNA in pancreatic cancers, including ddPCR and optimised highly sensitive NGS methods. These two methods have specific advantages and drawbacks: (i) ddPCR is a relatively low-cost and fast method allowing accurate quantification of ctDNA but with the need of an a priori knowledge of the genetic alteration(s) present in the tumour tissue; (ii) NGS is a costly method allowing a semi-quantitative detection of ctDNA measurement, agnostic of tumour mutations but requiring complex informatics support for development and analysis [24]. The two methods give similar results when compared [12]. The frequency of KRAS gene mutations in PDAC has made them prime targets for identifying ctDNA. Nevertheless, 5–10% of PDAC are KRAS negative, and the variety of different possible RAS mutations limits the pertinence of ddPCR for ctDNA detection based on the search of these mutations [25]. To combine the best of the two technics, we developed universal markers of ctDNA based on hypermethylated cancer-type specific markers, which allow the use of the ddPCR method [26].
DNA methylation in promotor of specific genes is an early key epigenetic change observed in various cancers during oncogenesis [27]. Changes in the DNA methylation pattern can be considered as a promising biomarker, and identification of specific methylation biomarkers could be a substitute for genes mutations to identify ctDNA [26, 28, 29]. We validated two specific PDAC methylated markers (HOXD8 / POU4F1). These markers have been identified using in silico data mining of the TCGA. Their sensitivity and specificity have been tested using DNA extracted from PDAC tissues and compared to the counterpart of non-tumour tissues (see Material and methods section). The sensitivity was 79%, and the specificity was 100% when we considered a positive sample with both makers. Several other methylated markers have been reported in PDAC. These markers have been applied for PDAC detection or for discriminating benign from malignant pancreatic disease (for review) [30]. The detection of ctDNA using POU4F1 and HOXD8 epigenetic modifications show a strong correlation with the optimised NGS-based method (eFig. 1) [12]. The main advantage of this method is the cost-effectiveness and the absence of the need to characterise tumours before performing a search for ctDNA. In addition, since the detection of KRAS mutations by ddPCR requires several assays to be exhaustive, our technique based on epigenetic changes takes advantage that is limited to one triplex assay and not dependent on complex multiplex kits.
The prognostic role of ctDNA in advanced PDAC has been evaluated in many studies, but this is, to our knowledge, the first large-scale exploration of the prognostic value of ctDNA in PDAC, with validation cohort from two homogenous randomised control trials with prospective data. The presence of ctDNA was associated with a worse prognosis both on OS and PFS with HR of 1.62 (CI 95% [1.05–2.50]) and 1.50 (CI 95% [1.03–2.18]), respectively. Previous studies reported similar results with a large meta-analysis (18 articles–1243 patients), the prognostic value of ctDNA in advanced PDAC on OS, as on PFS (HR 2.57, CI 95% [1.95–3.38] and HR = 2.31, CI 95% [1.47–3.64] respectively) was evidenced [9]. Our study integrates different prognostic factors in the multivariate analyses as the WHO performance status, the ratio of neutrophils lymphocytes ratio [31], all factors having shown their prognostic role in mPDAC and which potentially explain the difference observed in hazard ratio.
The quantitative analysis of ctDNA seems to bring prognostic information beyond the presence or the absence of ctDNA. We confirmed and precise the data already published in rare series. In a retrospective cohort including 94 patients, a significant association between ctDNA (dichotomised at the median) was associated with poor patient survival (HR 4.35, CI 95% [1.85–10.24]; 6.3 versus 11.7 months, p = 0.001) [32]. Likewise, ctDNA MAF assessed as a continuous variable has also been reported to be an independent predictor of OS (HR = 1.05, CI 95% [1.01–1.09], p = 0.005) [10] in a multivariate analysis including CA19-9 as co-factor. Our results are consistent with these previous reports. We confirmed the significant association between ctDNA concentration, considered as a continuous or categorised variable, and both OS and PFS (HR 2.2 and HR 1.5, respectively, ctDNA level superior to 3rd quartile vs. ctDNA inferior or equal to median).
Of course, some limitations could be pointed out in this exploratory analysis, such as the lower fractions of methylated DNA in the validation cohort compared to the test one (means 6.3% and 7.8%, respectively). This could be explained by the fact that EDTA tubes from the validation cohort were processed within 24 h due to the plasma centralisation in the two randomised trials. The delay imposed by the transport could induce increasing leukocytes DNA contamination. Also, despite careful collection and storage of samples and the use of highly sensitive methods to detect genetic changes, some alterations may not have been detected due to limited plasma DNA amounts or very low methylation levels. The presentation of our results using multiple multivariable models could also be a limitation of our study. However, all models tested showed the independent prognostic value of methylation markers in PDAC, compared to other clinical and biological known prognostic factors.
Finally, another important point of the described work is its ability to be easily generalised to hospital molecular biology laboratories. Compared to other studies that used technologies based on highly sensitive optimised NGS, we describe a strategy compatible with clinical practices using a relatively low number of ddPCR assays. The developed methodology cumulates several advantages such as high sensitivity, shorter turn-around time, straightforward data interpretation (i.e. do not require complex bioinformatics analyses) and reduced technical costs.
Conclusion
In conclusion, this large series confirms that the ctDNA is an independent prognostic factor of OS and PFS in patients with metastatic PDAC. Moreover, we validated a Met-ddPCR assay using HOXD8 and POU4F1, two specific methylated PDAC markers. We confirmed here that the ctDNA status and its level are key tools to highlight prognostic subgroups of patients in this tumour. These results argue for patients’ stratification on ctDNA status for further randomised trials.
Supplementary information
Acknowledgements
DP, SWR, JBB, VT and PLP are inventors of HOXD8 and POU4F1 methylated markers and owners of the European Patent n°19305695.9-1111 “Detection of hypermethylated genes for diagnosing pancreatic cancer” (Faculté Paris Descartes, INSERM, CNRS, AP-HP, IDF INNOV’).
Author contributions
DP: Conceptualisation, Methodology, Formal analysis, investigation, Writing Original Draft Review and Editing, Project administration; SWR: Conceptualisation, Methodology, Resources; JT: Investigation, Writing Original draft Review and Editing, Supervision; LD: investigation, Writing Review and Editing, Project administration; MP: Methodology, Investigation, Resources; JDL: Methodology, Investigation, Resources; KLM: Methodology, Investigation, Data curation; CM: Investigation, Resources; YR: Investigation, Resources; JMP: Investigation, Resources; SD: Investigation, Resources; MD; HB: Validation, Resources; AdR: Formal analysis, Software, Data curation; JBB: Conceptualisation, Methodology, investigation, Writing Review and Editing, Project administration, Funding acquisition, Visualisation; VT: Conceptualisation, Methodology, Formal analysis, Writing Review and Editing, Project administration, Funding acquisition; PLP: Conceptualisation, Formal analysis, Software, Writing Original Draft Review and Editing, Project administration, Funding acquisition.
Funding
This work was supported by the Ministère de l’Enseignement Supérieur et de la Recherche, the Université de Paris, the Centre National de la Recherche Scientifique (CNRS), the Institut National de la Santé et de la Recherche Médicale (INSERM), the SIRIC CARPEM (Inca_DGOS_Inserm-12561), the (LNCC, Program “Equipe labelisée LIGUE”; no. EL2016.LNCC). Collection of plasma samples was supported by Celgene and Fédération Francophone de Cancérologie Digestive (FFCD). A part of this work has been supported by the Fondation Roche.
Data availability
PLP and DP have full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Competing interests
JT has received honoraria for speaker or advisory roles from AMGEN, Roche, MERCKKGaA, MSD, Lilly, Servier, Sanofi, Pierre Fabre, AstraZeneca, Samsung Bioepis, HallioDx. JBB has received personal fees from Amgen, AstraZeneca, Bayer, Merck Serono, Pierre Fabre, Roche, Sanofi, Servier, Shire, and non-financial support from Amgen, Merck Serono, and Roche. PLP declares consulting for or personal fees from Amgen, AstraZeneca, Biocartis, BMS, Boehringer-Ingelheim, Lilly, Merck Serono, MSD, Sanofi, Servier, Roche.
Ethics approval and consent to participate
All the patients included in the PLAPAN cohort signed an informed consent form approved by the ethics committee (CPP Ile-de-France 2014/59NICB). PRODIGE 35 PANOPTIMOX trial (NCT02352337) PRODIGE 37 FIRGEMAX (NCT 02827201) trials were approved by the ethics committee.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shufang Wang-Renault, Julien Taieb, Laetitia Dahan, Jean-Baptiste Bachet, Valérie Taly, Pierre Laurent-Puig.
Contributor Information
Valérie Taly, Email: valerie.taly@parisdescartes.fr.
Pierre Laurent-Puig, Email: pierre.laurent-puig@parisdescartes.fr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01624-2.
References
- 1.Henley SJ, Ward EM, Scott S, Ma J, Anderson RN, Firth AU, et al. Annual Report to the Nation on the Status of Cancer, part I: National cancer statistics. Cancer. 2020;126:2225–49. [DOI] [PMC free article] [PubMed]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 3.Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–25. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 4.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369:1691–703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conroy T, Hammel P, Hebbar M, Ben Abdelghani M, Wei AC, Raoul J-L, et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N Engl J Med. 2018;379:2395–406. doi: 10.1056/NEJMoa1809775. [DOI] [PubMed] [Google Scholar]
- 6.Poruk KE, Gay DZ, Brown K, Mulvihill JD, Boucher KM, Scaife CL, et al. The clinical utility of CA 19-9 in pancreatic adenocarcinoma: diagnostic and prognostic updates. Curr Mol Med. 2013;13:340–51. doi: 10.2174/1566524011313030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med. 2018;379:1754–65. doi: 10.1056/NEJMra1706174. [DOI] [PubMed] [Google Scholar]
- 8.Kim MK, Woo SM, Park B, Yoon K-A, Kim Y-H, Joo J, et al. Prognostic implications of multiplex detection of KRAS mutations in cell-free DNA from patients with pancreatic ductal adenocarcinoma. Clin Chem. 2018;64:726–34. doi: 10.1373/clinchem.2017.283721. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Zhang Y, Cheng Y, Zhang D, Zhu S, Ma X. Prognostic value of circulating cell-free DNA in patients with pancreatic cancer: a systemic review and meta-analysis. Gene. 2018;679:328–34. doi: 10.1016/j.gene.2018.09.029. [DOI] [PubMed] [Google Scholar]
- 10.Strijker M, Soer EC, de Pastena M, Creemers A, Balduzzi A, Beagan JJ, et al. Circulating tumor DNA quantity is related to tumor volume and both predict survival in metastatic pancreatic ductal adenocarcinoma. Int J Cancer. 2020;146:1445–56. doi: 10.1002/ijc.32586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugimori M, Sugimori K, Tsuchiya H, Suzuki Y, Tsuyuki S, Kaneta Y, et al. Quantitative monitoring of circulating tumor DNA in patients with advanced pancreatic cancer undergoing chemotherapy. Cancer Sci. 2020;111:266–78. doi: 10.1111/cas.14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietrasz D, Pécuchet N, Garlan F, Didelot A, Dubreuil O, Doat S, et al. Plasma circulating tumor DNA in pancreatic cancer patients is a prognostic marker. Clin Cancer Res. 2017;23:116–23. doi: 10.1158/1078-0432.CCR-16-0806. [DOI] [PubMed] [Google Scholar]
- 13.Dahan L, Phelip JM, Le Malicot K, Williet N, Desrame J, Volet J, et al. FOLFIRINOX until progression, FOLFIRINOX with maintenance treatment, or sequential treatment with gemcitabine and FOLFIRI.3 for first-line treatment of metastatic pancreatic cancer: a randomized phase II trial (PRODIGE 35-PANOPTIMOX) J Clin Oncol Am Soc Clin Oncol. 2018;36:4000–4000. doi: 10.1200/JCO.2018.36.15_suppl.4000. [DOI] [Google Scholar]
- 14.Taieb J, Rinaldi Y, Pointet A-L, Barbier E, Khemissa F, Gratet A, et al. gemcitabine plus nab-paclitaxel until progression or given sequentially with 5-fluorouracile plus irinotecan (FOLFIRI.3) for first-line treatment of metastatic pancreatic ductal adenocarcinoma (mPDAC): A randomized phase II study (PRODIGE 37-FIRGEMAX) J Clin Oncol Am Soc Clin Oncol. 2018;36:4107–4107. doi: 10.1200/JCO.2018.36.15_suppl.4107. [DOI] [Google Scholar]
- 15.dMIQE Group. Huggett JF. The Digital MIQE Guidelines Update: Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020. Clin Chem. 2020;66:1012–29. doi: 10.1093/clinchem/hvaa125. [DOI] [PubMed] [Google Scholar]
- 16.Bernard V, Kim DU, San Lucas FA, Castillo J, Allenson K, Mulu FC, et al. Circulating nucleic acids are associated with outcomes of patients with pancreatic cancer. Gastroenterology. 2019;156:108–118.e4. doi: 10.1053/j.gastro.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh N, Gupta S, Pandey RM, Chauhan SS, Saraya A. High levels of cell-free circulating nucleic acids in pancreatic cancer are associated with vascular encasement, metastasis and poor survival. Cancer Invest. 2015;33:78–85. doi: 10.3109/07357907.2014.1001894. [DOI] [PubMed] [Google Scholar]
- 18.Perets R, Greenberg O, Shentzer T, Semenisty V, Epelbaum R, Bick T, et al. Mutant KRAS circulating tumor DNA is an accurate tool for pancreatic cancer monitoring. Oncologist. 2018;23:566–72. doi: 10.1634/theoncologist.2017-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadano N, Murakami Y, Uemura K, Hashimoto Y, Kondo N, Nakagawa N, et al. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br J Cancer. 2016;115:59–65. doi: 10.1038/bjc.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z-Y, Ding X-Q, Zhu H, Wang R-X, Pan X-R, Tong J-H. KRAS mutant allele fraction in circulating cell-free DNA correlates with clinical stage in pancreatic cancer patients. Front Oncol. 2019;9:1295. doi: 10.3389/fonc.2019.01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uesato Y, Sasahira N, Ozaka M, Sasaki T, Takatsuki M, Zembutsu H. Evaluation of circulating tumor DNA as a biomarker in pancreatic cancer with liver metastasis. PLoS ONE. 2020;15:e0235623. doi: 10.1371/journal.pone.0235623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Earl J, Garcia-Nieto S, Martinez-Avila JC, Montans J, Sanjuanbenito A, Rodríguez-Garrote M, et al. Circulating tumor cells (Ctc) and kras mutant circulating free Dna (cfdna) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer. 2015;15:797. doi: 10.1186/s12885-015-1779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del ReM, Vivaldi C, Rofi E, Vasile E, Miccoli M, Caparello C, et al. Early changes in plasma DNA levels of mutant KRAS as a sensitive marker of response to chemotherapy in pancreatic cancer. Sci Rep. 2017;7:7931. doi: 10.1038/s41598-017-08297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Postel M, Roosen A, Laurent-Puig P, Taly V, Wang-Renault S-F. Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: a cancer diagnostic perspective. Expert Rev Mol Diagn. 2018;18:7–17. doi: 10.1080/14737159.2018.1400384. [DOI] [PubMed] [Google Scholar]
- 25.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47:D941–7. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garrigou S, Perkins G, Garlan F, Normand C, Didelot A, Le Corre D, et al. A study of hypermethylated circulating tumor DNA as a universal colorectal cancer biomarker. Clin Chem. 2016;62:1129–39. doi: 10.1373/clinchem.2015.253609. [DOI] [PubMed] [Google Scholar]
- 27.Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357:eaal2380. [DOI] [PMC free article] [PubMed]
- 28.Barault L, Amatu A, Siravegna G, Ponzetti A, Moran S, Cassingena A, et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut. 2018;67:1995–2005. doi: 10.1136/gutjnl-2016-313372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu R-H, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16:1155–61. doi: 10.1038/nmat4997. [DOI] [PubMed] [Google Scholar]
- 30.Brancaccio M, Natale F, Falco G, Angrisano T. Cell-free DNA methylation: the new frontiers of pancreatic cancer biomarkers’ discovery. Genes. 10.3390/genes11010014. 2019;11:14. [DOI] [PMC free article] [PubMed]
- 31.Zhou Y, Wei Q, Fan J, Cheng S, Ding W, Hua Z. Prognostic role of the neutrophil-to-lymphocyte ratio in pancreatic cancer: a meta-analysis containing 8252 patients. Clin Chim Acta. 2018;479:181–9. doi: 10.1016/j.cca.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 32.Patel H, Okamura R, Fanta P, Patel C, Lanman RB, Raymond VM, et al. Clinical correlates of blood-derived circulating tumor DNA in pancreatic cancer. J Hematol Oncol. 2019;12:130. doi: 10.1186/s13045-019-0824-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PLP and DP have full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.