Abstract
Head and neck squamous cell carcinoma (HNSCC) is a molecularly heterogeneous disease, with a 5-year survival rate that still hovers at ~60% despite recent advancements. The advanced stage upon diagnosis, limited success with effective targeted therapy and lack of reliable biomarkers are among the key factors underlying the marginally improved survival rates over the decades. Prevention, early detection and biomarker-driven treatment adaptation are crucial for timely interventions and improved clinical outcomes. Liquid biopsy, analysis of tumour-specific biomarkers circulating in bodily fluids, is a rapidly evolving field that may play a striking role in optimising patient care. In recent years, significant progress has been made towards advancing liquid biopsies for non-invasive early cancer detection, prognosis, treatment adaptation, monitoring of residual disease and surveillance of recurrence. While these emerging technologies have immense potential to improve patient survival, numerous methodological and biological limitations must be overcome before their implementation into clinical practice. This review outlines the current state of knowledge on various types of liquid biopsies in HNSCC, and their potential applications for diagnosis, prognosis, grading treatment response and post-treatment surveillance. It also discusses challenges associated with the clinical applicability of liquid biopsies and prospects of the optimised approaches in the management of HNSCC.
Subject terms: Head and neck cancer, Tumour biomarkers
Background
Head and neck cancer is the seventh most common malignancy worldwide with 930,000 new cases and 470,000 deaths reported in 2020, an obvious increase compared to 890,000 cases and 450,000 deaths recorded in 2018 [1]. It comprises a heterogeneous group of malignancies originating from the oral cavity, oropharynx, larynx, nasopharynx and hypopharynx. The predominant histological subtype (90%) is squamous cell carcinoma (head and neck squamous cell carcinoma (HNSCC)) originating from the mucosal epithelium of the oral cavity (OCSCC), oropharynx (OPSCC), hypopharynx and larynx (LSCC). The remaining 10% of the tumours originate from lymphocytes, cells of connective tissues or salivary glands and include lymphoma, adenocarcinoma and sarcoma [2]. HNSCC is of multifactorial aetiology, with major associated confounding factors including use of tobacco, excessive alcohol consumption and/or infection with Epstein–Barr virus (EBV) and high-risk human papillomavirus (HPV) [3, 4]. HPV-16, and to a lesser extent HPV-18, is a significant risk factor for oropharyngeal squamous cell carcinoma (OPSCC) [5]. HPV-positive OPSCC typically occurs at a younger age with no prior history of tobacco consumption and is associated with a more favourable prognosis than HPV-negative counterparts [5].
Current treatment paradigms for HNSCC includes surgical resection followed by adjuvant radiation with or without chemotherapy, depending on the disease stage and adverse pathologic feature [6]. Despite improvements in molecular diagnosis, cetuximab, a monoclonal antibody targeting epidermal growth factor receptor (EGFR), is the only Food and Drug Administration (FDA)-approved targeted therapy available for this disease [7]. However, the response rate to cetuximab monotherapy is variable. Given the limited success of targeted treatment and cytotoxic chemotherapy against locally advanced or metastatic HNSCC, the current clinical focus has turned to immunotherapy with antibodies targeting T cell inhibitory receptors that function as immune checkpoints, such as programmed death-1 (PD-1) [4, 6]. However, despite improved locoregional control and reduced treatment-related morbidity, the 5-year overall survival rate for patients with HPV-negative HNSCC has remained relatively low, in part due to failure in early diagnosis [8, 9].
While most of the HNSCC cases can be cured if detected early, survival of patients with advanced-stage disease remains ~20–60% [9], underscoring the need for an earlier diagnosis of cancer. With the advancements in high-throughput next-generation sequencing (NGS) technologies, tumour molecular profiling has become a significant and promising tool in disease diagnosis, prognosis and post-treatment surveillance. In the diagnostic setting, analysis based on conventional visual and tactile examination followed by tissue biopsy is still the gold standard for head and neck malignancies. However, the major drawbacks of this approach include biased acquisition of tissue due to intratumoral heterogeneity, pain and discomfort caused by its invasive nature, and difficulty in identifying occult regional metastasis [9]. Consequently, while tissue biopsy remains useful for primary tumour genetic profiling, over the past few years there has been a significant interest in exploring liquid biopsies as an alternative approach for molecular characterisation of HNSCC and detection of cancer-specific biomarkers that can be applied for early diagnosis, treatment planning, response monitoring and surveillance.
Given their minimally invasive and easy-to-collect nature, liquid biopsies have emerged as a promising platform for inexpensive screening, diagnosis, and monitoring of HNSCC, including for cases that require serial sample collection over time. Circulating tumour DNA (ctDNA), circulating tumour cells (CTCs) and exosomal microRNAs (miRNAs) have been shown to be present in patients’ body fluids, including plasma and saliva, and could therefore be used as a surrogate for a biopsy [10–12]. In this review article, we will summarise the current state of knowledge on liquid biopsies and their potential clinical application in head and neck malignancies as a non-invasive diagnostic, prognostic and treatment monitoring tool. Furthermore, we will discuss the significance of HPV quantification in liquid biopsy to aid treatment de-escalation in HNSCC and provide an outlook on future prospects in this rapidly emerging field.
Types of liquid biopsy biomarkers in HNSCC
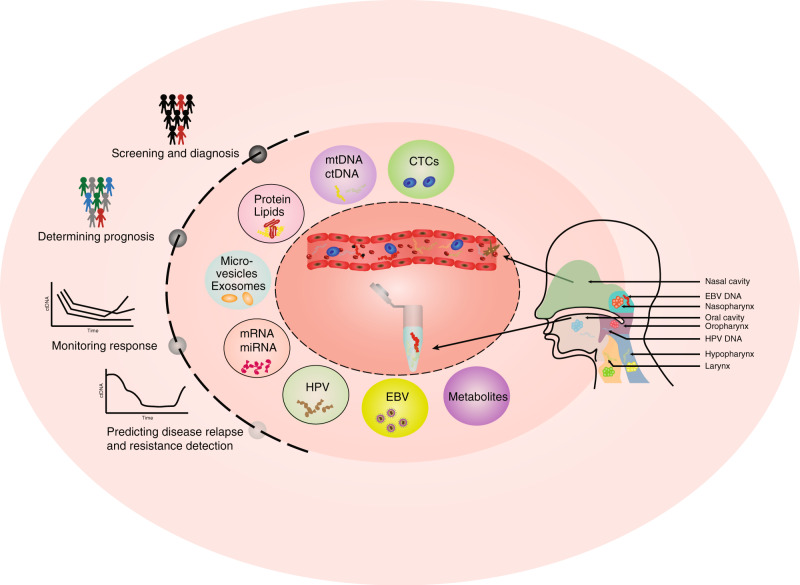
Studies of liquid biopsies in oncology have flourished over the past decade, and are rapidly gaining importance as promising non-invasive cancer biomarkers. Traditionally, the field was segregated into two major strategies: detection of free nucleic acids and isolating intact CTCs from different body fluids. More recently, additional approaches such as identifying extracellular vesicles (EVs) and targeted proteomics have emerged as alternative sources of liquid biopsy biomarkers (Fig. 1). Although a few liquid biopsy-based assays have been already approved for clinical use and many more in clinical trials, each strategy comes with its own advantages and drawbacks. The next section will describe different liquid biopsy technologies applied to patients with HNSCC and discuss their usability for clinical applications.
Fig. 1. Liquid biopsy biomarkers and clinical applications.
Schematic summary of a wide variety of blood- and saliva-based liquid biopsy biomarkers and their application for precision medicine in HNSCC.
Somatic DNA mutations based biomarkers
Somatic mutations are among the hallmarks of cancer progression that allow reliable differentiation between cancer and normal tissues. The exclusive nature of tumour-defining driver genetic alterations makes them attractive biomarkers with a theoretical specificity approaching 100% when detectable. Although HNSCC is considered to be a vastly heterogeneous disease, our group and others have revealed that HPV-negative HNSCC is largely driven by mutations in tumour suppressor genes, such as TP53 (mutated in over 80% of the cases), followed by FAT1, CDKN2A and NOTCH1 alterations (detected in ~ 20% of the tumours) [13–15]. In addition, mutations in the oncogene PIK3CA were also identified in approximately 20% of head and neck malignancies. In fact, over 80% of the patients with HPV-negative disease harbour genetic alterations in the narrow number of frequently mutated genes [16], which may serve as predictive biomarkers for detection of HNSCC despite the considerable inter-tumoral heterogeneity among HNSCC patients.
It is well established that as a tumour grows, it sheds tumour cells and DNA into various body fluids, including saliva and plasma [9, 12]. The idea that tumour-defining genetic alterations can be detected in body fluids has been proposed nearly two decades ago [17]. Over the years, multiple studies have shown that somatic mutations detected in HNSCC tissue biopsies can also be identified in matched body fluid specimens, and may act as diagnostic and predictive biomarkers in patients with HNSCC [9, 12, 17]. For example, multiplex PCR analysis of saliva DNA from 93 patients with HNSCC found tumour DNA (either somatic mutations in TP53, PIK3CA, CDKN2A, FBXW7, HRAS and NRAS or presence of HPV genes) in 100% of the patients with OCSCC, whereas detection frequency was substantially lower (47–70%) for patients with oropharyngeal, laryngeal and hypopharyngeal cancers, suggesting enrichment for tumour DNA from the oral cavity [12]. In contrast, tumour DNA was found in over 80% of the matched plasma samples available for a subset of these patients, demonstrating high detection frequency across different histological sites [12]. Of note, three of the nine patients with available “follow-up” samples were found to have tumour DNA in their saliva or plasma after surgery, but before clinical evidence of disease recurrence, supporting the feasibility of DNA-based liquid biopsies for cancer surveillance. In a more recent study, Shanmugam et al. explored the potential of saliva-based screening to identify patients who are at high risk of developing OCSCC [9]. A custom NGS panel with unique molecular identifiers was designed to cover the coding region of the seven most frequently mutated genes in OCSCC. Applying this targeted sequencing panel on DNA extracted from 121 treatment-naive OCSCC tumours and matched preoperative oral rinse specimens revealed that mutations identified in primary lesions were also detected at high concordance (93%) in saliva samples, whereas no functionally relevant mutations were detected in oral rinse DNA collected from healthy individuals [9]. To date, several studies have correlated the presence and abundance of tumour-specific DNA alterations detected in patients’ plasma and/or saliva with clinicopathological characteristics [18]. Mutation-based DNA biomarkers have several distinct advantages—unlike RNA and protein, they have no physiologic background and are not influenced by signalling changes induced during disease progression or therapy. Furthermore, unlike RNA or protein-based assays, DNA-based alterations should theoretically be found in appreciable levels only within cancer cells and not normal cells, allowing high specificity. Moreover, DNA is stable and amplifiable and may better stoichiometrically correlate with disease burden. Collectively, cancer-specific genetic mutations allow for tremendous specificity, and its use in addressing the continuing challenges in clinical oncology will grow.
Methylation-based biomarkers
Along with the mutational landscape, assessing the tumour-specific methylation signatures in body fluids has also been extensively studied over the past decade. DNA methylation is an important epigenetic mechanism of gene expression regulation and often occurs at an early stage of tumorigenesis, providing the rationale for its use as a biomarker for early detection of the disease.
Recently, a largely targeted methylation analysis of plasma circulating cell-free DNA (cfDNA) that involved 6689 participants across over 50 cancer types (including HNSCC) was performed to localise the tumour tissue of origin [19]. Using both a whole-genome bisulfite sequencing and a panel that targets >100,000 distinct methylation regions, the tissue of origin for a broad range of cancer types at metastatic and non-metastatic stages was localised with >90% accuracy, supporting the feasibility of employing methylation analysis of cfDNA for early cancer detection [19]. As an alternative to blood-based assays, several studies have focused on detecting tumour-specific DNA methylation biomarkers in saliva, an attractive matrix for early detection of head and neck cancers [20, 21]. For example, a recent study that used methylation-specific PCR to assess the promoter methylation status of p16INK4a, RASSF1A, TIMP3 and PCQAP/MED15 tumour suppressor genes in salivary DNA from OCSCC and OPSCC patients revealed that a subset of these genes was significantly hyper-methylated in patients compared to healthy controls [20]. Another study that performed a comprehensive meta-analysis to evaluate the diagnostic potential of salivary DNA methylation for detecting head and neck cancers showed a higher diagnostic accuracy of combined methylation gene panel compared to individual gene analysis [22].
While these and other studies [23–25] provide valuable insights regarding the potential application of cancer-associated DNA methylation changes in body fluids as an accurate tool for diagnosis and monitoring of head and neck malignancies, the major challenge to the discovery of biomarkers is inherent heterogeneity of methylation profiles in clinical samples and unclear origin of methylated cfDNA. This, however, does not preclude the discovery of accurate correlative biomarkers that might become extremely valuable in clinical practice.
Mitochondrial DNA-based biomarkers
Mutations in mitochondrial DNA (mtDNA) have been linked to risk, progression and treatment response of HNSCC [26–30]. Due to their clonal nature and high copy number, mitochondrial mutations could serve as powerful molecular markers for the detection of cancer cells in surgical margins, biopsies and lymph node metastasis, especially at sites where tumour involvement is not histologically apparent. Additionally, due to its circular configuration, mtDNA is thought to be more stable than genomic DNA, and may therefore be suitable for sequencing when isolated from low-quality formalin-fixed paraffin-embedded or body fluid-derived samples [31]. Over the past two decades, several groups have demonstrated that mtDNA alterations are valuable markers of head and neck tumorigenesis [32, 33]. However, only a few studies have assessed tumour-associated mitochondrial genome mutations and copy number changes in body fluids of patients with HNSCC [34–36], in part, due to insufficient sensitivity of the early sequencing platforms to detect low-prevalence heteroplasmies. Recently, our group has used an ultra-deep amplicon-based sequencing approach that covers the entire mitochondrial genome to reliably detect low-frequency tumour-associated mtDNA mutations in sputum and serum specimens collected from patients with HNSCC [31]. While these findings provide the foundation for using mitochondrial sequencing as a non-invasive cancer biomarker, additional studies in larger patient cohorts are required to further validate the accuracy of the mtDNA sequencing approach.
Exosomes and EVs as potential biomarkers in HNSCC
The EVs are actively produced by the tumour cells and released into the tumour microenvironment (TME). According to their cell compartment origin, diameter and surface protein markers, EVs can be divided into three subgroups: exosomes (40–100 nm), microvesicles (50–1000 nm) and apoptotic bodies (50–2000 nm) [37]. A complex cargo of the EVs contains DNA, messenger RNAs (mRNAs), miRNAs, long non-coding RNAs, circRNAs, proteins and metabolites. These biologically active substances are detectable in biological fluids and play a vital role in cancer progression [38]. Several techniques have been developed for the isolation of exosomes based on their biophysical properties or immune affinity [39, 40] and extensively studied as potential biomarkers for HNSCC. For example, plasma exosomes were evaluated for their levels of EMT markers (such as E-cadherin, N-cadherin and TGF-β1) by flow cytometry as a potential biomarker of post-treatment surveillance [41]. It was found that exosomes obtained at baseline had a mesenchymal profile (high levels of N-cadherin and TGF-β1) that was associated with increased tumour proliferation, migration and invasion, whereas exosomes collected post-treatment with photodynamic therapy showed epithelial phenotype [41]. In a more recent study, profiling of circulating plasma exosomes was performed in 18 HNSCC patients receiving a combination of cetuximab, ipilimumab and radiation therapy. In patients whose disease recurred, total exosome proteins, total exosome ratio, total CD3+, CD3(−)PD-L1+ and CD3+15s+ (a marker of highly differentiated and most suppressive regulatory T cells) exosomes increased from baseline levels compared to patients who remained disease-free, supporting the role of exosomes as biomarkers for post-treatment surveillance in HNSCC [42].
The tumour-specific exosomal miRNA signatures have also been studied as potential liquid biopsy biomarkers for HNSCC. Langevin et al. reported that exosomal miRNA content in saliva collected from HNSCC patients is substantially different than the microRNA-sequencing profile seen in saliva samples collected from healthy controls, providing insights on the utility of miRNA signatures as non-invasive saliva biomarker [43]. In another study, Rabinowits et al. has demonstrated that plasma exosomal miRNA mirrored the miRNA within the HNSCC tumour tissue, and tested the potential for blood-borne miRNA to identify HNSCC and to predict outcome [44]. Paralleling previous reports, patients with HNSCC exhibited miRNA profiles within circulating exosomes that were distinct from normal controls, with levels of most exosomal miRNAs being suppressed in patients responding to therapy [44]. Several other studies have explored circulating miRNA as a biomarker for early HNSCC detection, prognosis and monitoring recurrence and metastasis [45–48] underscoring that miRNA profiles within body fluid exosomes may have utility in the HNSCC diagnosis and disease monitoring.
Similar to exosomes, EVs represent an emerging liquid biopsy modality in HNSCC. Apart from promoting cell growth, proliferation and migration, EVs and their cargos (miRNAs and proteins) hold promise as potential biomarkers for cancer diagnosis and prognosis [49, 50]. Dynamic examination of circulating EVs and their contents has been shown to provide information on real-time therapeutic response as well as therapy resistance in patients with several types of cancer including HNSCC. Rodrigues-Junior et al. explored EVs as plasma-based biomarkers that prospectively segregate the outcome of HNSCC patients treated with chemoradiation therapy (CRT) [51]. Screening the EVs cargo isolated from plasma of six HNSCC patients who responded to CRT and six patients who presented incomplete response with antibody array revealed a protein signature that was able to distinguish between these two cohorts, posing the non-invasive EV evaluation as a potential tool for response assessment and guiding therapeutic decisions [51].
While these studies suggest the potential of exosomes and EV profiling in body fluids for HNSCC diagnosis and surveillance, their use in a clinical setting is associated with technical challenges in terms of isolation, purification and quantification, and demands future technological advancements to better understand the true sensitivity of this approach.
Transcriptomic biomarkers
The genetic and epigenetic alterations associated with cancer cells lead to altered gene expression patterns which can be identified long before the phenotype is manifested. Altered gene expression patterns exclusive to cancer cells can be exploited as molecular biomarkers for early detection and management of the disease, and have been studied in serum or plasma of several solid malignancies including head and neck [52, 53]. One such early study used microarray to investigate the distinct mRNA expression patterns in saliva from OCSCC patients as a potential biomarker for early cancer detection, reporting a small gene signature distinguishing OCSCCs from the controls [54]. A series of more recent pre-clinical studies using whole-transcriptome analysis, such as microarray and more recently RNA-sequencing, have demonstrated the ability to detect tumour-specific gene expression changes in both saliva and blood after cancer therapy and show their associations with tumour staging, grading and clinical outcomes [55–58]. However, none has gained traction in clinical practice. Most of the transcriptomic signatures defined by these studies are either platform-dependent, insufficiently validated in independent cohorts or have a limited sample size, resulting in high variability and limiting their translational value. Therefore, there is a need for a well-validated, platform-independent and easy-to-interpret gene expression signatures with high predictive value before transcriptomic-based liquid biopsy biomarkers could be successfully implemented as a diagnostic or prognostic tool applicable to clinical practice.
Of note, while several recent studies have also investigated circulating non-coding RNA and salivary microRNA as potential biofluid-based diagnostic and prognostic biomarkers in head and neck cancer [57, 59, 60], these results are not yet sufficiently validated.
Non-genomic molecular biomarkers
Along with genetic and epigenetic biomarkers, tumour-specific metabolites/metabolomic signatures, proteomic and lipidomic signatures have also been extensively studied as promising diagnostic and predictive biomarkers for head and neck malignancies [61, 62]. Metabolomics, an emerging field in systems biology, has been shown as a promising approach to measure global changes in metabolic profiles in response to disease or treatment via non-invasive analytes such as saliva and plasma [62–64]. For example, screening plasma samples collected from either healthy controls or patients with oesophageal squamous cell carcinoma (ESCC) before, during and after chemoradiotherapy using mass spectrometry determined global alterations in the metabolic profiles between these cohorts and identified a signature of altered plasma metabolites associated with therapeutic responses and outcomes in ESCC [65]. In a more recent study, Shahid et al. used mass spectrometry analysis to profile plasma samples collected from 62 healthy individuals, 81 tobacco snuff dippers and 91 oral cancer patients and identified stearyl alcohol and sucrose as predictive biomarkers associated with the progression of tobacco snuff dippers toward oral cancer [66].
Similar to metabolomic studies, protein and lipid profiling in saliva and plasma of patients with head and neck cancers have also been investigated as potential diagnostic and prognostic biomarkers [67–73]. Blood-based protein biomarkers are attractive targets, as they can reflect both variations in the TME as well as the host immune response [70, 71, 73]. Similarly, changes in lipid profile have also been associated with cancer progression, as lipids play a key role in the maintenance of cell integrity [68, 69].
While promising, these have obvious limitations, including questionable sensitivity and specificity, low half-life and influence by signalling changes induced during disease progression as well as therapy.
Circulating tumour cells
CTCs, passively shedding from the primary tumour or metastatic lesion into the vasculature, may provide unprecedented insights and definitive evidence of the aggressiveness of the tumour prior to detection of identifiable metastasis. In head and neck cancer, CTCs have been identified as a marker of disease severity with poor outcomes [74, 75]. The recent advances in CTC detection methods and their applications as “liquid biopsy” biomarkers for disease aggressiveness, detection of metastatic disease, monitoring treatment response and post-treatment surveillance in patients with HNSCC are summarised in a series of recent reviews [76–80], supporting their potential diagnostic and prognostic significance. For example, a recent study that evaluated the prognostic value of CTCs in relapsing non-resectable or metastatic HNSCC treated by chemotherapy and cetuximab revealed that progression-free survival was significantly lower in patients with increasing or stable CTC count [74]. In a different study, it was found that CTCs were more frequent in patients with stage IV compared to those with stage I–III cancer, whereas partial or complete response was associated with clearance of CTCs during the treatment [81]. While there is mounting evidence for using CTC counts and CTC-derived molecular signatures as prognostic and predictive biomarkers in patients with primary or recurrent/metastatic HNSCC [82–84], detection of CTCs in peripheral blood remains challenging, owing to their low number in the circulation. With rapidly evolving technologies attempted to overcome these pitfalls [74, 76, 77], CTCs and the associated genetic material will be a valuable clinical tool to improve the current treatment approach for this disease.
Viral-based biomarkers
In OPSCC, HPV status is an important prognostic factor and is predictive of prognosis to treatment [85–88]. For this reason, screening OPSCC tumour tissue for HPV is recommended by an increasing number of organisations, including the National Comprehensive Cancer Network and the College of American Pathologists. While non-invasive HPV detection is an attractive strategy, there is no standard or routine bodily fluid-based HPV-DNA testing for patients with OPSCC. A number of retrospective studies, including by members of our group, have reported that high-risk ctHPV-DNA is detectable in plasma and saliva of patients with HPV + OPSCC using real-time qPCR, suggesting its use for disease surveillance [12, 89–91]. While these studies support the clinical implication of this promising approach, detection and quantification of low abundance viral targets in a complex background of liquid biopsy using qPCR is challenging: it lacks reliable endogenous reference for normalisation of results in biofluids and when used in absolute quantification, it must rely on the use of external calibrators [92]. Ultra-sensitive droplet digital PCR (ddPCR) technology overcomes these complications and offers an advantageous approach for rapid and highly sensitive quantitative HPV genotyping in plasma specimens [93–96]. However, its clinical adoption as a routine screening practice is hindered by a relatively high cost of the reagents and challenges with performing higher-order multiplexing that enables the simultaneous identification of multiple targets in a single reaction. Due to the rapid decreases in sequencing cost and the concomitant development of the infrastructure needed to effectively apply sequencing data in clinical care, NGS is likely to become more routinely available over the next few years and may ultimately displace other HPV detection tests. However so far, this method is not widely used for HPV diagnosis, and its accuracy is yet to be confirmed [97, 98].
EBV infection is closely associated with nasopharyngeal carcinogenesis. A series of recent studies using PCR assay, targeted sequencing or serological analysis have reported the clinical relevance of plasma EBV DNA liquid biopsy as a promising biomarker for early detection, treatment response monitoring and prognostication of the clinical outcomes in patients with nasopharyngeal carcinoma (NPC) [99–105]. Furthermore, by using genome-wide methylation profiling, Lam et al. have demonstrated differential methylation patterns amongst NPC and healthy subjects, along with the changes in fractional concentration (count) and size of plasma EBV DNA [106]. Very recently, Tan et al systematically evaluated the diagnostic efficiency of plasma EBV DNA, anti-EBV antibodies and miRNA level. The analysis revealed a 76 bp fragment of the repetitive BamHI-W sequence as the most sensitive early-stage detection plasma biomarker (sensitivity 96.7 % and specificity 90–94%) and reported that EBNA1 99 bp may differentiate NPC patients with poor prognosis in an early and advanced stage of disease, thereby reflecting evidence for improved screening and monitoring of patients with NPC [107].
Application of liquid biopsy in pre-clinical and clinical settings
As summarised above, different types of circulating tumour biomarkers have been identified and studied as potential non-invasive diagnostic, prognostic and surveillance tools in patients with HNSCC. However, due to their clear tumour-specific origin, somatic DNA alterations, HPV- and CTC-based biomarkers have emerged as the most frequently studied and the most promising liquid biopsy approaches in oncology. Therefore, the following chapter will limit the discussion to these methodologies.
Early detection and screening
Tumour-specific somatic mutations, regardless of whether they are driver or passenger aberrations, offer a unique opportunity for early detection of the disease. As CTCs and ctDNA share common somatic mutations with the primary tumour, it opens a window for non-invasive early diagnosis and screening of high-risk populations. In an exploratory prospective study, saliva-based discrete genetic signature comprising alterations in PMAIP1 and PTPN1 genes correctly discriminated 100% of HNSCC patients from normal controls [108]. More recently, it was reported that mutations identified in primary tumours are highly concordant with mutations detected in ctDNA isolated from either saliva or plasma of patients HNSCC [9, 12]. Notably, the sensitivity of ctDNA detection in saliva was found to be site-specific, with higher efficiency for tumours of the oral cavity, whereas plasma was preferentially enriched for tumour DNA from the other sites of the head and neck region [12]. Given these observations, saliva samples could therefore be used as an easy-to-collect surrogate for the oral cancer biopsy. Unlike ctDNA that can be identified even at early stages of HNSCC, CTCs are more frequently detected in advanced stages of the disease [50, 109], and correlate with a poorer prognosis, locoregional relapse, and shorter disease-free survival. As such, CTCs are more likely to be used as a prognostic biomarker, rather than a platform for screening or early diagnosis of head and neck malignancies [75, 110–114].
For HNSCC subtypes with a viral aetiology, detection of the viral DNA in body fluids may provide a promising strategy for screening and early detection. Several studies have investigated the feasibility of plasma EBV as a marker for early detection of NPC [105, 115, 116], and plasma or saliva HPV-DNA as a diagnostic biomarker of HPV-positive oropharyngeal carcinoma [90, 117–121].
Taken together, DNA-based liquid biopsy biomarkers in head and neck cancer hold great promise as a non-invasive tool for early diagnosis of HNSCC and screening of high-risk populations. However, robust large-scale pre-clinical studies are required to further assess their true potential and limitations.
Predictive biomarker of response to therapy and relapse
Along with being promising diagnostic and screening biomarkers, liquid biopsies are well-suited for measuring disease burden, minimal residual disease (MRD) and predicting real-time response to therapy, as shown in several proof-of-concept studies [74, 75, 81]. An important paradigm for utilising circulating biomarkers for monitoring treatment response is that the changes in tumour volume should be reflected by changes in biomarker levels. Several studies reported that both ctDNA and CTCs in plasma and serum of HNSCC patients have prognostic significance [110, 111]. For example, in a prospective multicenter study (CIRCUTEC), Garrel et al. dynamically collected peripheral blood from patients with relapsing non-resectable or metastatic HNSCC treated by chemotherapy and cetuximab [74], with the goal to determine whether CTC level can predict progression-free survival (PFS) or overall survival (OS). It was found that median PFS was significantly lower in patients with progressively increasing or stable CTC counts [74]. In another study, Hilke et al. longitudinally analysed ctDNA dynamics in plasma and corresponding clinical outcomes in patients with locally advanced HNSCC and demonstrated a significant correlation between tumour volume and patient ctDNA prior to the start of the treatment [122]. Furthermore, a negative association of ctDNA and HPV-DNA kinetics was observed with time and treatment dosage in most patients, supporting the usage ctDNA as a potential surrogate marker for disease burden [122]. More recently, a pilot study using a targeted sequencing panel covering 61 tumour-related genes has reported that the absence of TP53 and PIK3CA mutations in plasma samples collected from patients with advanced HNSCC at different time points post surgery was associated with recurrence-free survival during over 20 months of follow-up [123]. Aligning with these observations, several recent studies have reported the potential of ctDNA as a potential tool for monitoring disease burden even at early stages, which may aid in improving prognostication and selection of treatment strategies [11, 124–127].
These findings, supported by additional evidence, indicate the potential of liquid biopsy biomarkers for real-time monitoring of early response to systemic therapies as well as predicting PFS and OS in patients with HNSCC [74, 75, 81].
Non-invasive tool to monitor therapy resistance
Cetuximab, an anti-EGFR monoclonal antibody and the only FDA-approved targeted therapy for locoregionally advanced HNSCC, has limited efficacy due to the development of resistance. Elucidating the molecular mechanisms underlying the therapy resistance may improve the clinical management of the disease. Liquid biopsies have been exploited to track molecular dynamics of tumour evolution and acquired therapy resistance in several solid malignancies, including head and neck cancer. Braig et al. were the first group that utilised plasma ctDNA to study clonal tumour evolution leading to targeted therapy resistance in patients with HNSCC [128]. Specifically, NGS of plasma DNA samples collected from 20 patients during and after cetuximab/platinum/5-fluorouracil treatment, revealed that 46% of the patients with disease progression during treatment carried acquired RAS mutations, whereas no RAS mutations were found in a subset of patients who did not progress, indicating that acquisition of RAS mutant clones significantly correlated with cetuximab resistance to HNSCC [128].
Two PD-1 inhibitors (nivolumab and pembrolizumab) have been shown to unleash antitumor immunity and produce durable clinical responses in approximately 20% of patients with platinum-refractory recurrent and/or metastatic HNSCC. In a very recent study, Abbott et al. has assessed the development of resistance mechanisms in HNSCC patients treated with nivolumab [129]. Whole-exome sequencing was performed on matched pre- and post-intervention tumour biopsy biopsies, normal control samples and plasma DNA specimens collected from 13 patients. The study reports a rapid evolution of the TME and mutational profile following therapy, with strong concordance, detected between plasma and tumour variants at each timepoint. Notably, post-therapy interrogation of cfDNA revealed dynamic changes in numerous oncogenes and clinically relevant pathways, such as ERK1/2 and MAPK, that were not observed in a solid tumour, suggesting that single-lesion biopsy of the primary tumour misses co-occurring, clinically relevant resistance alterations [129].
While these proof-of-concept studies provide evidence that liquid biopsies can provide a more comprehensive view into therapeutic response and resistance mechanisms in HNSCC, further validation in larger patient cohorts is warranted prior to advancing this concept to clinical settings.
An immunotherapeutic biomarker
Given the prognostic significance of CTC levels in HNSCC [75, 110, 114, 130], an increasing number of studies have focused on investigating the PD-L1-expressing CTCs as potential biomarkers for selecting candidates for anti-PD-1/PD-L1 therapy [131–137]. Strati et al. were the first to report the potential of PD-L1 overexpression in CTCs to predict clinical outcomes using a prospective cohort of HNSCC patients treated with induction chemotherapy [137]. PD-L1 mRNA expression was evaluated using a PCR assay at baseline and at different time points during treatment. Overexpression of PD-L1 in CTCs was associated with short PFS and OS, whereas the lack of PD-L1-overexpressing CTCs at the end of treatment was strongly associated with complete response [137]. These observations were further supported by a series of recent follow-up studies [131–137]. For example, in 2020 Economopoulou et al. investigated the prognostic value of PDL-1 and IDO1 expression in CTCs at baseline and following completion of chemoradiotherapy in patients with locally advanced HNSCC [138]. This study revealed that patients whose CTCs had concurrently decreased the expression of both PDL-1 and IDO1 after treatment showed substantially longer PFS [138]. It is important to note that the expression of immune-regulatory molecules in the CTCs do not correspond completely with that in the tumour tissue [134].
These cumulative findings suggest that the CTCs are rapidly becoming a powerful tool in cancer treatments that involve the use of immune checkpoint inhibitors; however, these concepts are still developing and their clinical implementations remain a challenge.
ctHPV-DNA: a promising biomarker for surveillance and treatment de-escalation
Over the past several decades, there has been a dramatic increase in the incidence of HPV-associated OPSCC despite a reduction in smoking-related head and neck cancer [139]. A causal association between high-risk HPV subtypes and OPSCC has been well established, with HPV-16 being the most commonly implicated strain [139]. As patients with HPV-related OPSCC show significantly higher cure rates and improved survival, determining HPV status in newly diagnosed patients is important for risk stratification and prognostication. cfHPV-DNA represents a promising non-invasive surrogate of disease burden in these patients, and several assays focusing on detection of high-risk HPV in body fluids have been developed in both academic and industrial settings [140, 141]. As mentioned above, a series of studies using quantitative PCR assays have demonstrated the potential usefulness of cfHPV-DNA for OPSCC clinical management [90, 117]. However, the reported detection rate of low-level circulating plasma HPV-DNA was modest. Novel ddPCR technologies significantly enhance the cfHPV-DNA detection rate and allow highly sensitive quantitative HPV genotyping in plasma specimens [93–96, 117], supporting its predictive and prognostic potential. Indeed, a growing body of evidence suggests that dynamic assessment of viral kinetics over time can capture changes in disease burden associated with treatment response and detect MRD in HPV-associated OPSCC [93, 95, 126, 142]. Recently, our group has developed a highly sensitive assay for plasma HPV-16/18 detection and quantification based on NGS technology, and have successfully applied it to monitor therapeutic response in HPV + OPSCC patients [98]. Prospective studies are underway to further validate this and other approaches and evaluate the kinetics of cfHPV-DNA as a predictor of response to therapy and disease recurrence prior to radiography [143].
Current treatment for locoregionally advanced HPV + OPSCC includes definitive concomitant chemoradiation or surgical resection followed by adjuvant radiation with or without chemotherapy. However, standard combined modality therapy is associated with substantial acute and long-term toxicities, which has led to an interest in developing a de-intensification treatment paradigm for HPV + OPSCC that optimises the therapeutic to toxicity ratio for patients [144, 145]. Response to induction chemotherapy represents an attractive strategy to select candidates for treatment de-escalation in HPV + OPSCC. The rationale for this strategy is based on the observation that a favourable response to induction therapy is associated with superior prognosis following subsequent definitive chemoradiation. While risk and response-adaptive de-escalated chemoradiation is a promising strategy to dynamically select patients for de-intensified definitive treatment [146, 147], improved strategies to monitor and adapt treatment response to personalise patient de-intensification is urgently needed. Reliable quantitative HPV-DNA assessment in plasma and oral rinses represent an appealing potential to dynamically monitor treatment response during induction therapy, definitive treatment and following completion of definitive therapy to monitor for disease recurrence [118, 119, 126, 148]. Currently, several pilot studies, including by our group, are set to evaluate the dynamics of quantitative cfHPV-DNA levels in combination with an established response-adaptive treatment de-intensification and provide the data needed to develop a blood-based biomarker-driven strategy. Data from such pilot studies will be invaluable in the design of a subsequent larger proof-of-concept clinical trial incorporating longitudinal assessment of HPV-DNA as a component of treatment response assessment, adaptive de-intensification and monitoring for recurrence.
Current challenges and future directions
In recent years, tremendous advancement has been made to adapt liquid biopsy as an alternative and non-invasive approach for molecular characterisation of HNSCC and detection of specific biomarkers for early diagnosis, treatment and response monitoring. Although a growing body of evidence implicates the utility of liquid biopsy to guide treatment strategies, there is no FDA-approved liquid biopsy-based biomarker for use in the clinical setting for patients with HNSCC. The major challenge that hinders the translation of liquid biopsy methodologies is the sensitivity and specificity requirements that have to be reached prior to recognising these platforms as alternative or companion assays to conventional tissue-based biopsy. The pre-analytical and analytical steps such as sample collection, storage and specimen processing quality control, along with the well-validated assay design, are critical, as errors can result in unreliable measurements and subsequently incorrect clinical decisions. The inherent heterogeneity of molecular profiles in clinical samples, difficulties associated with detection and quantification of circulating biomarkers with low half-life and stability add another twist to the already complicated field. As such, standardisation and refinement of detection techniques as well as identification and targeting multiple unique molecular identifiers will facilitate the adoption of liquid biomarker assays in routine clinical practice. As tumours of the upper aerodigestive mucosal origin can shed tumour cells, DNA and other tumour-specific analytes directly into the oral cavity, several studies have investigated the utility of saliva and oral rinse specimens as diagnostic and prognostic fluids in patients with HNSCC. However, small cohort sizes and a lack of validation limit the clinical utility of these findings.
With multiple clinical trials underway, liquid biopsies are rapidly becoming an integral platform for cancer diagnosis, prognostication and assessment of treatment efficacy. Although at present, liquid biopsy-based biomarkers are still imperfect for routine clinical practice, studies summarised in this review support their potential to effectively support clinical decision-making that may lead to patient benefit. As various approaches have been applied to overcome these challenges and improve the clinical relevance, the value of this tool in addressing the continuing challenges in clinical oncology will almost certainly increase over time.
Conclusion
The applications of liquid biopsies in head and neck cancer have emerged at an incredible rate over the past few years, due to their potential to provide valuable insights for cancer diagnosis, prognosis, treatment response and surveillance. In this context, there is growing hope that intensified efforts to improve sensitivity and specificity of these assays, along with future prospective studies set to assess dynamic changes in liquid biopsy biomarkers in large and heterogeneous patient’s populations, will enhance their adoption in a clinic and eventually improve clinical outcomes for patients with HNSCC.
Author contributions
VM, EI, AS and NA performed a literature search, analysed the data and wrote the original draft. VM, EI, NA, AS, XC, AJR, ATP, PAS, AZ and MWL developed study concepts, performed data interpretation, manuscript writing, editing and review. EI, NA and MWL acquired funding. All authors reviewed and edited the final manuscript.
Funding
This study was supported by the US National Institutes of Health (NIH) grants R01DE027809 to EI, R01DE028674 to NA and MWL and U01CA230691 to NA.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nishant Agrawal, Email: na@uchicago.edu.
Evgeny Izumchenko, Email: izumchen@uchicago.edu.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Tumban E. A current update on human papillomavirus-associated head and neck cancers. Viruses. 2019;11:922. doi: 10.3390/v11100922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beynon RA, Lang S, Schimansky S, Penfold CM, Waylen A, Thomas SJ, et al. Tobacco smoking and alcohol drinking at diagnosis of head and neck cancer and all-cause mortality: Results from head and neck 5000, a prospective observational cohort of people with head and neck cancer. Int J Cancer. 2018;143:1114–27. doi: 10.1002/ijc.31416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Prim. 2020;6:92. doi: 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elrefaey S, Massaro MA, Chiocca S, Chiesa F, Ansarin M. HPV in oropharyngeal cancer: the basics to know in clinical practice. Acta Otorhinolaryngol Ital. 2014;34:299–309. [PMC free article] [PubMed] [Google Scholar]
- 6.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 7.Astsaturov I, Cohen RB, Harari PM. EGFR-targeting monoclonal antibodies in head and neck cancer. Curr Cancer Drug Targets. 2006;6:691–710. doi: 10.2174/156800906779010191. [DOI] [PubMed] [Google Scholar]
- 8.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 9.Shanmugam A, Hariharan AK, Hasina R, Nair JR, Katragadda S, Irusappan S, et al. Ultrasensitive detection of tumor-specific mutations in saliva of patients with oral cavity squamous cell carcinoma. Cancer. 2021;127:1576–89. doi: 10.1002/cncr.33393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soda N, Rehm BHA, Sonar P, Nguyen NT, Shiddiky MJA. Advanced liquid biopsy technologies for circulating biomarker detection. J Mater Chem B. 2019;7:6670–704. doi: 10.1039/c9tb01490j. [DOI] [PubMed] [Google Scholar]
- 11.Egyud M, Sridhar P, Devaiah A, Yamada E, Saunders S, Stahlberg A, et al. Plasma circulating tumor DNA as a potential tool for disease monitoring in head and neck cancer. Head Neck. 2019;41:1351–58. doi: 10.1002/hed.25563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Springer S, Mulvey CL, Silliman N, Schaefer J, Sausen M, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med. 2015;7:293ra104. doi: 10.1126/scitranslmed.aaa8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–57. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. [DOI] [PMC free article] [PubMed]
- 16.Lawrence MS, Sougnez C, Lichtenstein L, Cibulskis K, Lander E, Gabriel SB, et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyle JO, Mao L, Brennan JA, Koch WM, Eisele DW, Saunders JR, et al. Gene mutations in saliva as molecular markers for head and neck squamous cell carcinomas. Am J Surg. 1994;168:429–32. doi: 10.1016/s0002-9610(05)80092-3. [DOI] [PubMed] [Google Scholar]
- 18.Pall AH, Jakobsen KK, Grønhøj C, von Buchwald C. Circulating tumour DNA alterations as biomarkers for head and neck cancer: a systematic review. Acta Oncol. 2020;59:845–50. doi: 10.1080/0284186X.2020.1742930. [DOI] [PubMed] [Google Scholar]
- 19.Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV, Consortium C. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 2020;31:745–59. doi: 10.1016/j.annonc.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liyanage C, Wathupola A, Muraleetharan S, Perera K, Punyadeera C, Udagama P. Promoter hypermethylation of tumor-suppressor genes p16(INK4a), RASSF1A, TIMP3, and PCQAP/MED15 in salivary DNA as a quadruple biomarker panel for early detection of oral and oropharyngeal cancers. Biomolecules. 2019;9:148. doi: 10.3390/biom9040148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim Y, Wan Y, Vagenas D, Ovchinnikov DA, Perry CF, Davis MJ, et al. Salivary DNA methylation panel to diagnose HPV-positive and HPV-negative head and neck cancers. BMC Cancer. 2016;16:749. doi: 10.1186/s12885-016-2785-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rapado-Gonzalez O, Martinez-Reglero C, Salgado-Barreira A, Muinelo-Romay L, Muinelo-Lorenzo J, Lopez-Lopez R, et al. Salivary DNA methylation as an epigenetic biomarker for head and neck cancer. Part I: a diagnostic accuracy meta-analysis. J Pers Med. 2021;11:568. doi: 10.3390/jpm11060568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schrock A, Leisse A, de Vos L, Gevensleben H, Droge F, Franzen A, et al. Free-circulating methylated DNA in blood for diagnosis, staging, prognosis, and monitoring of head and neck squamous cell carcinoma patients: an observational prospective cohort study. Clin Chem. 2017;63:1288–96. doi: 10.1373/clinchem.2016.270207. [DOI] [PubMed] [Google Scholar]
- 24.Langevin SM, Koestler DC, Christensen BC, Butler RA, Wiencke JK, Nelson HH, et al. Peripheral blood DNA methylation profiles are indicative of head and neck squamous cell carcinoma: an epigenome-wide association study. Epigenetics. 2012;7:291–9. doi: 10.4161/epi.7.3.19134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Jesus LM, Dos Reis MB, Carvalho RS, Scapulatempo Neto C, de Almeida GC, Laus AC, et al. Feasibility of methylated ctDNA detection in plasma samples of oropharyngeal squamous cell carcinoma patients. Head Neck. 2020;42:3307–15. doi: 10.1002/hed.26385. [DOI] [PubMed] [Google Scholar]
- 26.Challen C, Brown H, Cai C, Betts G, Paterson I, Sloan P, et al. Mitochondrial DNA mutations in head and neck cancer are infrequent and lack prognostic utility. Br J Cancer. 2011;104:1319–24. doi: 10.1038/bjc.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dasgupta S, Koch R, Westra WH, Califano JA, Ha PK, Sidransky D, et al. Mitochondrial DNA mutation in normal margins and tumors of recurrent head and neck squamous cell carcinoma patients. Cancer Prev Res. 2010;3:1205–11. doi: 10.1158/1940-6207.CAPR-10-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koc EC, Haciosmanoglu E, Claudio PP, Wolf A, Califano L, Friscia M, et al. Impaired mitochondrial protein synthesis in head and neck squamous cell carcinoma. Mitochondrion. 2015;24:113–21. doi: 10.1016/j.mito.2015.07.123. [DOI] [PubMed] [Google Scholar]
- 29.Sun W, Zhou S, Chang SS, McFate T, Verma A, Califano JA. Mitochondrial mutations contribute to HIF1alpha accumulation via increased reactive oxygen species and up-regulated pyruvate dehydrogenease kinase 2 in head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15:476–84. doi: 10.1158/1078-0432.CCR-08-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mithani SK, Taube JM, Zhou S, Smith IM, Koch WM, Westra WH, et al. Mitochondrial mutations are a late event in the progression of head and neck squamous cell cancer. Clin Cancer Res. 2007;13:4331–5. doi: 10.1158/1078-0432.CCR-06-2613. [DOI] [PubMed] [Google Scholar]
- 31.Schubert AD, Channah Broner E, Agrawal N, London N, Pearson A, Gupta A, et al. Somatic mitochondrial mutation discovery using ultra-deep sequencing of the mitochondrial genome reveals spatial tumor heterogeneity in head and neck squamous cell carcinoma. Cancer Lett. 2020;471:49–60. doi: 10.1016/j.canlet.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim MM, Clinger JD, Masayesva BG, Ha PK, Zahurak ML, Westra WH, et al. Mitochondrial DNA quantity increases with histopathologic grade in premalignant and malignant head and neck lesions. Clin Cancer Res. 2004;10:8512–15. doi: 10.1158/1078-0432.CCR-04-0734. [DOI] [PubMed] [Google Scholar]
- 33.Ha PK, Tong BC, Westra WH, Sanchez-Cespedes M, Parrella P, Zahurak M, et al. Mitochondrial C-tract alteration in premalignant lesions of the head and neck: a marker for progression and clonal proliferation. Clin Cancer Res. 2002;8:2260–5. [PubMed] [Google Scholar]
- 34.Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–9. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 35.Kumar M, Srivastava S, Singh SA, Das AK, Das GC, Dhar B, et al. Cell-free mitochondrial DNA copy number variation in head and neck squamous cell carcinoma: A study of non-invasive biomarker from Northeast India. Tumour Biol. 2017;39:1010428317736643. doi: 10.1177/1010428317736643. [DOI] [PubMed] [Google Scholar]
- 36.Jiang WW, Masayesva B, Zahurak M, Carvalho AL, Rosenbaum E, Mambo E, et al. Increased mitochondrial DNA content in saliva associated with head and neck cancer. Clin Cancer Res. 2005;11:2486–91. doi: 10.1158/1078-0432.CCR-04-2147. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Guo J, Yu P, Guo L, Mao X, Wang J, et al. The roles of extracellular vesicles in the development, microenvironment, anticancer drug resistance, and therapy of head and neck squamous cell carcinoma. J Exp Clin Cancer Res. 2021;40:35. doi: 10.1186/s13046-021-01840-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao J, Zhang M, Xie F, Lou J, Zhou X, Zhang L, et al. Exosomes in head and neck cancer: Roles, mechanisms and applications. Cancer Lett. 2020;494:7–16. doi: 10.1016/j.canlet.2020.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Hong CS, Funk S, Muller L, Boyiadzis M, Whiteside TL. Isolation of biologically active and morphologically intact exosomes from plasma of patients with cancer. J Extracell Vesicles. 2016;5:29289. doi: 10.3402/jev.v5.29289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li P, Kaslan M, Lee SH, Yao J, Gao ZQ. Progress in exosome isolation techniques. Theranostics. 2017;7:789–804. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theodoraki MN, Yerneni SS, Brunner C, Theodorakis J, Hoffmann TK, Whiteside TL. Plasma-derived exosomes reverse epithelial-to-mesenchymal transition after photodynamic therapy of patients with head and neck cancer. Oncoscience. 2018;5:75–87. doi: 10.18632/oncoscience.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Theodoraki MN, Yerneni S, Gooding WE, Ohr J, Clump DA, Bauman JE, et al. Circulating exosomes measure responses to therapy in head and neck cancer patients treated with cetuximab, ipilimumab, and IMRT. Oncoimmunology. 2019;8:1593805. doi: 10.1080/2162402X.2019.1593805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langevin S, Kuhnell D, Parry T, Biesiada J, Huang S, Wise-Draper T, et al. Comprehensive microRNA-sequencing of exosomes derived from head and neck carcinoma cells in vitro reveals common secretion profiles and potential utility as salivary biomarkers. Oncotarget. 2017;8:82459–74. doi: 10.18632/oncotarget.19614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rabinowits G, Taylo CG, Kloecke GH, Pate A, Hall MB, Taylor DD. Exosomal-miRNA profiles as diagnostic and prognostic biomarkers in head and neck squamous cell carcinoma (HNSCC) J Clin Oncol. 2011;29:5515. [Google Scholar]
- 45.Chang YA, Weng SL, Yang SF, Chou CH, Huang WC, Tu SJ, et al. A three-microRNA signature as a potential biomarker for the early detection of oral cancer. Int J Mol Sci. 2018;19:758. doi: 10.3390/ijms19030758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greither T, Vorwerk F, Kappler M, Bache M, Taubert H, Kuhnt T, et al. Salivary miR-93 and miR-200a as post-radiotherapy biomarkers in head and neck squamous cell carcinoma. Oncol Rep. 2017;38:1268–75. doi: 10.3892/or.2017.5764. [DOI] [PubMed] [Google Scholar]
- 47.Severino P, Oliveira LS, Andreghetto FM, Torres N, Curioni O, Cury PM, et al. Small RNAs in metastatic and non-metastatic oral squamous cell carcinoma. BMC Med Genomics. 2015;8:31. doi: 10.1186/s12920-015-0102-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haas BJ, Dobin A, Stransky N, Li B, Yang X, Tickle T, et al. STAR-Fusion: fast and accurate fusion transcript detection from RNA-Seq. bioRxiv:120295 [Preprint] 2017. Available from: 10.1101/120295.
- 49.Qu X, Li JW, Chan J, Meehan K. Extracellular vesicles in head and neck cancer: a potential new trend in diagnosis, prognosis, and treatment. Int J Mol Sci. 2020;21:8260. doi: 10.3390/ijms21218260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou S, Wang L, Zhang W, Liu F, Zhang Y, Jiang B, et al. Circulating tumor cells correlate with prognosis in head and neck squamous cell carcinoma. Technol Cancer Res Treat. 2021;20:1533033821990037. doi: 10.1177/1533033821990037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodrigues-Junior DM, Tan SS, de Souza Viana L, Carvalho AL, Lim SK, Iyer NG, et al. A preliminary investigation of circulating extracellular vesicles and biomarker discovery associated with treatment response in head and neck squamous cell carcinoma. BMC Cancer. 2019;19:373. doi: 10.1186/s12885-019-5565-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lacombe J, Brooks C, Hu CC, Menashi E, Korn R, Yang F, et al. Analysis of saliva gene expression during head and neck cancer radiotherapy: a pilot study. Radiat Res. 2017;188:75–81. doi: 10.1667/RR14707.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panta P, Venna VR. Salivary RNA signatures in oral cancer detection. Anal Cell Pathol. 2014;2014:450629. doi: 10.1155/2014/450629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;10:8442–50. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- 55.Zanotti L, Paderno A, Piazza C, Pagan E, Bignotti E, Romani C, et al. Epidermal growth factor receptor detection in serum and saliva as a diagnostic and prognostic tool in oral cancer. Laryngoscope. 2017;127:E408–14. doi: 10.1002/lary.26797. [DOI] [PubMed] [Google Scholar]
- 56.Liu GH, Zeng XJ, Wu BL, Zhao J, Pan YB. RNA-Seq analysis of peripheral blood mononuclear cells reveals unique transcriptional signatures associated with radiotherapy response of nasopharyngeal carcinoma and prognosis of head and neck cancer. Cancer Biol Ther. 2020;21:139–46. doi: 10.1080/15384047.2019.1670521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao Y, Chen XY, Lu S, Zhou C, Xu GL, Yan ZY, et al. Circulating long noncoding RNAs as biomarkers for predicting head and neck squamous cell carcinoma. Cell Physiol Biochem. 2018;50:1429–40. doi: 10.1159/000494605. [DOI] [PubMed] [Google Scholar]
- 58.Li Y, Elashoff D, Oh M, Sinha U, St John MAR, Zhou XF, et al. Serum circulating human mRNA profiling and its utility for oral cancer detection. J Clin Oncol. 2006;24:1754–60. doi: 10.1200/JCO.2005.03.7598. [DOI] [PubMed] [Google Scholar]
- 59.Diez-Fraile A, Ceulaer J, Derpoorter C, Spaas C, Backer T, Lamoral P, et al. Circulating non-coding RNAs in head and neck cancer: roles in diagnosis, prognosis, and therapy monitoring. Cells. 2020;10:48. doi: 10.3390/cells10010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fadhil RS, Wei MQ, Nikolarakos D, Good D, Nair RG. Salivary microRNA miR-let-7a-5p and miR-3928 could be used as potential diagnostic bio-markers for head and neck squamous cell carcinoma. PLoS ONE. 2020;15:e0221779. doi: 10.1371/journal.pone.0221779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song X, Yang X, Narayanan R, Shankar V, Ethiraj S, Wang X, et al. Oral squamous cell carcinoma diagnosed from saliva metabolic profiling. Proc Natl Acad Sci USA. 2020;117:16167–73. doi: 10.1073/pnas.2001395117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ishikawa S, Sugimoto M, Kitabatake K, Sugano A, Nakamura M, Kaneko M, et al. Identification of salivary metabolomic biomarkers for oral cancer screening. Sci Rep. 2016;6:31520. doi: 10.1038/srep31520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wojakowska A, Zebrowska A, Skowronek A, Rutkowski T, Polanski K, Widlak P, et al. Metabolic profiles of whole serum and serum-derived exosomes are different in head and neck cancer patients treated by radiotherapy. J Pers Med. 2020;10:229. doi: 10.3390/jpm10040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mukherjee PK, Funchain P, Retuerto M, Jurevic RJ, Fowler N, Burkey B, et al. Metabolomic analysis identifies differentially produced oral metabolites, including the oncometabolite 2-hydroxyglutarate, in patients with head and neck squamous cell carcinoma. BBA Clin. 2017;7:8–15. doi: 10.1016/j.bbacli.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu J, Chen YH, Zhang RP, Song YM, Cao JZ, Bi N, et al. Global and targeted metabolomics of esophageal squamous cell carcinoma discovers potential diagnostic and therapeutic biomarkers. Mol Cell Proteom. 2013;12:1306–18. doi: 10.1074/mcp.M112.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shahid N, Iqbal A, Siddiqui AJ, Shoaib M, Musharraf SG. Plasma metabolite profiling and chemometric analyses of tobacco snuff dippers and patients with oral cancer: Relationship between metabolic signatures. Head Neck. 2019;41:291–300. doi: 10.1002/hed.25511. [DOI] [PubMed] [Google Scholar]
- 67.Brondum L, Eriksen JG, Sorensen BS, Mortensen LS, Toustrup K, Overgaard J, et al. Plasma proteins as prognostic biomarkers in radiotherapy treated head and neck cancer patients. Clin Transl Radiat Oncol. 2017;2:46–52. doi: 10.1016/j.ctro.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patel PS, Shah MH, Jha FP, Raval GN, Rawal RM, Patel MM, et al. Alterations in plasma lipid profile patterns in head and neck cancer and oral precancerous conditions. Indian J Cancer. 2004;41:25–31. [PubMed] [Google Scholar]
- 69.Abdul Rahman M, Mohamad Haron DE, Hollows RJ, Abdul Ghani ZDF, Ali Mohd M, Chai WL, et al. Profiling lysophosphatidic acid levels in plasma from head and neck cancer patients. PeerJ. 2020;8:e9304. doi: 10.7717/peerj.9304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu S, Arellano M, Boontheung P, Wang J, Zhou H, Jiang J, et al. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res. 2008;14:6246–52. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vidotto A, Henrique T, Raposo LS, Maniglia JV, Tajara EH. Salivary and serum proteomics in head and neck carcinomas: before and after surgery and radiotherapy. Cancer Biomark. 2010;8:95–107. doi: 10.3233/CBM-2011-0205. [DOI] [PubMed] [Google Scholar]
- 72.Wang LN, Wang X, Li Y, Hou Y, Sun FY, Zhou S, et al. Plasma lipid profiling and diagnostic biomarkers for oral squamous cell carcinoma. Oncotarget. 2017;8:92324–32. doi: 10.18632/oncotarget.21289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jou YJ, Lin CD, Lai CH, Chen CH, Kao JY, Chen SY, et al. Proteomic identification of salivary transferrin as a biomarker for early detection of oral cancer. Anal Chim Acta. 2010;681:41–8. doi: 10.1016/j.aca.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 74.Garrel R, Mazel M, Perriard F, Vinches M, Cayrefourcq L, Guigay J, et al. Circulating tumor cells as a prognostic factor in recurrent or metastatic head and neck squamous cell carcinoma: the CIRCUTEC Prospective Study. Clin Chem. 2019;65:1267–75. doi: 10.1373/clinchem.2019.305904. [DOI] [PubMed] [Google Scholar]
- 75.Grisanti S, Almici C, Consoli F, Buglione M, Verardi R, Bolzoni-Villaret A, et al. Circulating tumor cells in patients with recurrent or metastatic head and neck carcinoma: prognostic and predictive significance. PLoS ONE. 2014;9:e103918. doi: 10.1371/journal.pone.0103918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kulasinghe A, Hughes BGM, Kenny L, Punyadeera C. An update: circulating tumor cells in head and neck cancer. Expert Rev Mol Diagn. 2019;19:1109–15. doi: 10.1080/14737159.2020.1688145. [DOI] [PubMed] [Google Scholar]
- 77.Perumal V, Corica T, Dharmarajan AM, Sun Z, Dhaliwal SS, Dass CR, et al. Circulating tumour cells (CTC), head and neck cancer and radiotherapy; future perspectives. Cancers. 2019;11:367. doi: 10.3390/cancers11030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Künzel J, Gribko A, Lu Q, Stauber RH, Wünsch D. Nanomedical detection and downstream analysis of circulating tumor cells in head and neck patients. Biol Chem. 2019;400:1465–79. doi: 10.1515/hsz-2019-0141. [DOI] [PubMed] [Google Scholar]
- 79.Rao VU, Arakeri G, Subash A, Bagadia RK, Thakur S, Kudpaje AS, et al. Circulating tumour cells in head and neck cancers: biological insights. J Oral Pathol Med. 2020;49:842–48. doi: 10.1111/jop.13075. [DOI] [PubMed] [Google Scholar]
- 80.Curtin J, Choi SW, Thomson PJ, Lam AK. Characterization and clinicopathological significance of circulating tumour cells in patients with oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2021. 10.1016/j.ijom.2021.05.020. [DOI] [PubMed]
- 81.Buglione M, Grisanti S, Almici C, Mangoni M, Polli C, Consoli F, et al. Circulating tumour cells in locally advanced head and neck cancer: preliminary report about their possible role in predicting response to non-surgical treatment and survival. Eur J Cancer. 2012;48:3019–26. doi: 10.1016/j.ejca.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 82.Tada H, Takahashi H, Ida S, Nagata Y, Chikamatsu K. Epithelial-mesenchymal transition status of circulating tumor cells is associated with tumor relapse in head and neck squamous cell carcinoma. Anticancer Res. 2020;40:3559–64. doi: 10.21873/anticanres.14345. [DOI] [PubMed] [Google Scholar]
- 83.Hsieh JC, Lin HC, Huang CY, Hsu HL, Wu TM, Lee CL, et al. Prognostic value of circulating tumor cells with podoplanin expression in patients with locally advanced or metastatic head and neck squamous cell carcinoma. Head Neck. 2015;37:1448–55. doi: 10.1002/hed.23779. [DOI] [PubMed] [Google Scholar]
- 84.Tinhofer I, Konschak R, Stromberger C, Raguse JD, Dreyer JH, Johrens K, et al. Detection of circulating tumor cells for prediction of recurrence after adjuvant chemoradiation in locally advanced squamous cell carcinoma of the head and neck. Ann Oncol. 2014;25:2042–7. doi: 10.1093/annonc/mdu271. [DOI] [PubMed] [Google Scholar]
- 85.Worden FP, Kumar B, Lee JS, Wolf GT, Cordell KG, Taylor JM, et al. Chemoselection as a strategy for organ preservation in advanced oropharynx cancer: response and survival positively associated with HPV16 copy number. J Clin Oncol. 2008;26:3138–46. doi: 10.1200/JCO.2007.12.7597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lassen P, Eriksen JG, Hamilton-Dutoit S, Tramm T, Alsner J, Overgaard J. Effect of HPV-associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. J Clin Oncol. 2009;27:1992–8. doi: 10.1200/JCO.2008.20.2853. [DOI] [PubMed] [Google Scholar]
- 87.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gillison M. HPV and its effect on head and neck cancer prognosis. Clin Adv Hematol Oncol. 2010;8:680–2. [PubMed] [Google Scholar]
- 89.Cao H, Banh A, Kwok S, Shi X, Wu S, Krakow T, et al. Quantitation of human papillomavirus DNA in plasma of oropharyngeal carcinoma patients. Int J Radiat Oncol. 2012;82:e351–8. doi: 10.1016/j.ijrobp.2011.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ahn SM, Chan JY, Zhang Z, Wang H, Khan Z, Bishop JA, et al. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus-related head and neck cancer. JAMA Otolaryngol Head Neck Surg. 2014;140:846–54. doi: 10.1001/jamaoto.2014.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dahlstrom KR, Li G, Hussey CS, Vo JT, Wei Q, Zhao C, et al. Circulating human papillomavirus DNA as a marker for disease extent and recurrence among patients with oropharyngeal cancer. Cancer. 2015;121:3455–64. doi: 10.1002/cncr.29538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arvia R, Sollai M, Pierucci F, Urso C, Massi D, Zakrzewska K. Droplet digital PCR (ddPCR) vs quantitative real-time PCR (qPCR) approach for detection and quantification of Merkel cell polyomavirus (MCPyV) DNA in formalin fixed paraffin embedded (FFPE) cutaneous biopsies. J Virol Methods. 2017;246:15–20. doi: 10.1016/j.jviromet.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 93.Chera BS, Kumar S, Beaty BT, Marron D, Jefferys S, Green R, et al. Rapid Clearance profile of plasma circulating tumor HPV type 16 DNA during chemoradiotherapy correlates with disease control in HPV-associated oropharyngeal cancer. Clin Cancer Res. 2019;25:4682–90. doi: 10.1158/1078-0432.CCR-19-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kacew AJ, Hanna GJ. Value and unmet needs in non-invasive human papillomavirus (HPV) testing for oropharyngeal cancer. Cancers. 2021;13:562. doi: 10.3390/cancers13030562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Veyer D, Wack M, Mandavit M, Garrigou S, Hans S, Bonfils P, et al. HPV circulating tumoral DNA quantification by droplet-based digital PCR: a promising predictive and prognostic biomarker for HPV-associated oropharyngeal cancers. Int J Cancer. 2020;147:1222–7. doi: 10.1002/ijc.32804. [DOI] [PubMed] [Google Scholar]
- 96.Isaac A, Kostiuk M, Zhang H, Lindsay C, Makki F, O’Connell DA, et al. Ultrasensitive detection of oncogenic human papillomavirus in oropharyngeal tissue swabs. J Otolaryngol Head Neck Surg. 2017;46:5. doi: 10.1186/s40463-016-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yi X, Zou J, Xu J, Liu T, Liu T, Hua S, et al. Development and validation of a new HPV genotyping assay based on next-generation sequencing. Am J Clin Pathol. 2014;141:796–804. doi: 10.1309/AJCP9P2KJSXEKCJB. [DOI] [PubMed] [Google Scholar]
- 98.Sloane H, Izumchenko E, Mattox A, Hasina R, Patel A, Jones F, et al. Ultra-sensitive detection and quantification of HPV DNA in the plasma of patients with oropharyngeal squamous cell carcinoma (OPSCC) enrolled in the OPTIMA 2 treatment de-escalation trial. J Clin Oncol. 2021;39:6048. [Google Scholar]
- 99.Fung SY, Lam JW, Chan KC. Clinical utility of circulating Epstein-Barr virus DNA analysis for the management of nasopharyngeal carcinoma. Chin Clin Oncol. 2016;5:18. doi: 10.21037/cco.2016.03.07. [DOI] [PubMed] [Google Scholar]
- 100.Chen Y, Zhao W, Lin L, Xiao X, Zhou X, Ming H, et al. Nasopharyngeal Epstein-Barr virus load: an efficient supplementary method for population-based nasopharyngeal carcinoma screening. PLoS ONE. 2015;10:e0132669. doi: 10.1371/journal.pone.0132669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang WY, Twu CW, Chen HH, Jiang RS, Wu CT, Liang KL, et al. Long-term survival analysis of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA levels. Cancer. 2013;119:963–70. doi: 10.1002/cncr.27853. [DOI] [PubMed] [Google Scholar]
- 102.Liu TB, Zheng ZH, Pan J, Pan LL, Chen LH. Prognostic role of plasma Epstein-Barr virus DNA load for nasopharyngeal carcinoma: a meta-analysis. Clin Investig Med. 2017;40:E1–12. doi: 10.25011/cim.v40i1.28049. [DOI] [PubMed] [Google Scholar]
- 103.Sengar M, Chorghe S, Jadhav K, Singh S, Laskar SG, Pai P, et al. Cell-free Epstein-Barr virus-DNA in patients with nasopharyngeal carcinoma: plasma versus urine. Head Neck. 2016;38:E1666–73. doi: 10.1002/hed.24297. [DOI] [PubMed] [Google Scholar]
- 104.Lam WKJ, Jiang P, Chan KCA, Cheng SH, Zhang H, Peng W, et al. Sequencing-based counting and size profiling of plasma Epstein-Barr virus DNA enhance population screening of nasopharyngeal carcinoma. Proc Natl Acad Sci USA. 2018;115:E5115–24. doi: 10.1073/pnas.1804184115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chan KCA, Woo JKS, King A, Zee BCY, Lam WKJ, Chan SL, et al. Analysis of plasma Epstein-Barr Virus DNA to screen for nasopharyngeal cancer. N Engl J Med. 2017;377:513–22. doi: 10.1056/NEJMoa1701717. [DOI] [PubMed] [Google Scholar]
- 106.Lam WKJ, Jiang P, Chan KCA, Peng W, Shang H, Heung MMS, et al. Methylation analysis of plasma DNA informs etiologies of Epstein-Barr virus-associated diseases. Nat Commun. 2019;10:3256. doi: 10.1038/s41467-019-11226-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tan LP, Tan GW, Sivanesan VM, Goh SL, Ng XJ, Lim CS, et al. Systematic comparison of plasma EBV DNA, anti-EBV antibodies and miRNA levels for early detection and prognosis of nasopharyngeal carcinoma. Int J Cancer. 2020;146:2336–47. doi: 10.1002/ijc.32656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sethi S, Benninger MS, Lu M, Havard S, Worsham MJ. Noninvasive molecular detection of head and neck squamous cell carcinoma: an exploratory analysis. Diagn Mol Pathol. 2009;18:81–7. doi: 10.1097/PDM.0b013e3181804b82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu K, Chen N, Wei J, Ma L, Yang S, Zhang X. Clinical significance of circulating tumor cells in patients with locally advanced head and neck squamous cell carcinoma. Oncol Rep. 2020;43:1525–35. doi: 10.3892/or.2020.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu XL, Tu Q, Faure G, Gallet P, Kohler C, Bittencourt Mde C. Diagnostic and prognostic value of circulating tumor cells in head and neck squamous cell carcinoma: a systematic review and meta-analysis. Sci Rep. 2016;6:20210. doi: 10.1038/srep20210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sun T, Zou K, Yuan Z, Yang C, Lin X, Xiong B. Clinicopathological and prognostic significance of circulating tumor cells in patients with head and neck cancer: a meta-analysis. Onco Targets Ther. 2017;10:3907–16. doi: 10.2147/OTT.S136530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang HM, Wu MH, Chang PH, Lin HC, Liao CD, Wu SM, et al. The change in circulating tumor cells before and during concurrent chemoradiotherapy is associated with survival in patients with locally advanced head and neck cancer. Head Neck. 2019;41:2676–87. doi: 10.1002/hed.25744. [DOI] [PubMed] [Google Scholar]
- 113.Jatana KR, Lang JC, Chalmers JJ. Identification of circulating tumor cells: a prognostic marker in squamous cell carcinoma of the head and neck? Fut Oncol. 2011;7:481–4. doi: 10.2217/fon.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gröbe A, Blessmann M, Hanken H, Friedrich RE, Schön G, Wikner J, et al. Prognostic relevance of circulating tumor cells in blood and disseminated tumor cells in bone marrow of patients with squamous cell carcinoma of the oral cavity. Clin Cancer Res. 2014;20:425–33. doi: 10.1158/1078-0432.CCR-13-1101. [DOI] [PubMed] [Google Scholar]
- 115.Chan KC, Hung EC, Woo JK, Chan PK, Leung SF, Lai FP, et al. Early detection of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA analysis in a surveillance program. Cancer. 2013;119:1838–44. doi: 10.1002/cncr.28001. [DOI] [PubMed] [Google Scholar]
- 116.Anh VNQ, Van Ba N, Anh DT, Ung ND, Hiep NH, Ly VT, et al. Validation of a highly sensitive qPCR assay for the detection of plasma cell-free Epstein-Barr virus DNA in nasopharyngeal carcinoma diagnosis. Cancer Control. 2020;27:1073274820944286. doi: 10.1177/1073274820944286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Damerla RR, Lee NY, You D, Soni R, Shah R, Reyngold M, et al. Detection of early human papillomavirus–associated cancers by liquid biopsy. JCO Precis Oncol. 2019;1–17. 10.1200/PO.18.00276. [DOI] [PMC free article] [PubMed]
- 118.Capone RB, Pai SI, Koch WM, Gillison ML, Danish HN, Westra WH, et al. Detection and quantitation of human papillomavirus (HPV) DNA in the sera of patients with HPV-associated head and neck squamous cell carcinoma. Clin Cancer Res. 2000;6:4171–5. [PubMed] [Google Scholar]
- 119.Zhao M, Rosenbaum E, Carvalho AL, Koch W, Jiang W, Sidransky D, et al. Feasibility of quantitative PCR-based saliva rinse screening of HPV for head and neck cancer. Int J Cancer. 2005;117:605–10. doi: 10.1002/ijc.21216. [DOI] [PubMed] [Google Scholar]
- 120.Tang KD, Vasani S, Menezes L, Taheri T, Walsh LJ, Hughes BGM, et al. Oral HPV16 DNA as a screening tool to detect early oropharyngeal squamous cell carcinoma. Cancer Sci. 2020;111:3854–61. doi: 10.1111/cas.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tang KD, Vasani S, Taheri T, Walsh LJ, Hughes BGM, Kenny L, et al. An occult HPV-driven oropharyngeal squamous cell carcinoma discovered through a saliva test. Front Oncol. 2020;10:408. doi: 10.3389/fonc.2020.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hilke FJ, Muyas F, Admard J, Kootz B, Nann D, Welz S, et al. Dynamics of cell-free tumour DNA correlate with treatment response of head and neck cancer patients receiving radiochemotherapy. Radiother Oncol. 2020;151:182–89. doi: 10.1016/j.radonc.2020.07.027. [DOI] [PubMed] [Google Scholar]
- 123.Bai J, Zhang S, Li X, Zhang Y, Liu X. Next-generation sequencing of plasma cell-free DNA for treatment monitoring in advanced head and neck cancer patients. Clin Lab. 2020;66. [DOI] [PubMed]
- 124.Meng Y, Bian L, Zhang M, Bo F, Lu X, Li D. Liquid biopsy and their application progress in head and neck cancer: focus on biomarkers CTCs, cfDNA, ctDNA and EVs. Biomark Med. 2020;14:1393–1404. doi: 10.2217/bmm-2020-0022. [DOI] [PubMed] [Google Scholar]
- 125.Khandelwal AR, Greer AH, Hamiter M, Fermin JM, McMullen T, Moore-Medlin T, et al. Comparing cell-free circulating tumor DNA mutational profiles of disease-free and nonresponders patients with oropharyngeal squamous cell carcinoma. Laryngoscope Investig Otolaryngol. 2020;5:868–78. doi: 10.1002/lio2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chera BS, Kumar S, Shen C, Amdur R, Dagan R, Green R, et al. Plasma circulating tumor HPV DNA for the surveillance of cancer recurrence in HPV-associated oropharyngeal cancer. J Clin Oncol. 2020;38:1050–8. doi: 10.1200/JCO.19.02444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schirmer MA, Beck J, Leu M, Oellerich M, Rave-Frank M, Walson PD, et al. Cell-free plasma DNA for disease stratification and prognosis in head and neck cancer. Clin Chem. 2018;64:959–70. doi: 10.1373/clinchem.2017.285668. [DOI] [PubMed] [Google Scholar]
- 128.Braig F, Voigtlaender M, Schieferdecker A, Busch CJ, Laban S, Grob T, et al. Liquid biopsy monitoring uncovers acquired RAS-mediated resistance to cetuximab in a substantial proportion of patients with head and neck squamous cell carcinoma. Oncotarget. 2016;7:42988–95. doi: 10.18632/oncotarget.8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Abbott CW, Bedi N, Zhang SV, Northcott J, Li R, Pyke RM, et al. Abstract 555: longitudinal exome-scale liquid biopsy monitoring of evolving therapeutic resistance mechanisms in head and neck squamous cell carcinoma patients receiving anti-PD-1 therapy. Cancer Res. 2021;81:555. [Google Scholar]
- 130.Cho JK, Lee GJ, Kim HD, Moon UY, Kim MJ, Kim S, et al. Differential impact of circulating tumor cells on disease recurrence and survivals in patients with head and neck squamous cell carcinomas: an updated meta-analysis. PLoS ONE. 2018;13:e0203758. doi: 10.1371/journal.pone.0203758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kulasinghe A, Perry C, Kenny L, Warkiani ME, Nelson C, Punyadeera C. PD-L1 expressing circulating tumour cells in head and neck cancers. BMC Cancer. 2017;17:333. doi: 10.1186/s12885-017-3316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kulasinghe A, Kenny L, Punyadeera C. Circulating tumour cell PD-L1 test for head and neck cancers. Oral Oncol. 2017;75:6–7. doi: 10.1016/j.oraloncology.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 133.Kulasinghe A, Kapeleris J, Kimberley R, Mattarollo SR, Thompson EW, Thiery JP, et al. The prognostic significance of circulating tumor cells in head and neck and non-small-cell lung cancer. Cancer Med. 2018;7:5910–9. doi: 10.1002/cam4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chikamatsu K, Tada H, Takahashi H, Kuwabara-Yokobori Y, Ishii H, Ida S, et al. Expression of immune-regulatory molecules in circulating tumor cells derived from patients with head and neck squamous cell carcinoma. Oral Oncol. 2019;89:34–9. doi: 10.1016/j.oraloncology.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 135.Tada H, Takahashi H, Kuwabara-Yokobori Y, Shino M, Chikamatsu K. Molecular profiling of circulating tumor cells predicts clinical outcome in head and neck squamous cell carcinoma. Oral Oncol. 2020;102:104558. doi: 10.1016/j.oraloncology.2019.104558. [DOI] [PubMed] [Google Scholar]
- 136.Tada H, Takahashi H, Kawabata-Iwakawa R, Nagata Y, Uchida M, Shino M, et al. Molecular phenotypes of circulating tumor cells and efficacy of nivolumab treatment in patients with head and neck squamous cell carcinoma. Sci Rep. 2020;10:21573. doi: 10.1038/s41598-020-78741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Strati A, Koutsodontis G, Papaxoinis G, Angelidis I, Zavridou M, Economopoulou P, et al. Prognostic significance of PD-L1 expression on circulating tumor cells in patients with head and neck squamous cell carcinoma. Ann Oncol. 2017;28:1923–33. doi: 10.1093/annonc/mdx206. [DOI] [PubMed] [Google Scholar]
- 138.Economopoulou P, Kladi-Skandali A, Strati A, Koytsodontis G, Kirodimos E, Giotakis E, et al. Prognostic impact of indoleamine 2,3-dioxygenase 1 (IDO1) mRNA expression on circulating tumour cells of patients with head and neck squamous cell carcinoma. ESMO Open. 2020;5:e000646. doi: 10.1136/esmoopen-2019-000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–301. [DOI] [PMC free article] [PubMed]
- 140.Wuerdemann N, Jain R, Adams A, Speel EJM, Wagner S, Joosse SA, et al. Cell-free HPV-DNA as a Biomarker for oropharyngeal squamous cell carcinoma—a step towards personalized medicine? Cancers. 2020;12:2997. doi: 10.3390/cancers12102997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Reder H, Taferner VF, Wittekindt C, Brauninger A, Speel EM, Gattenlohner S, et al. Plasma cell-free human papillomavirus oncogene E6 and E7 DNA predicts outcome in oropharyngeal squamous cell carcinoma. J Mol Diagn. 2020. 10.1016/j.jmoldx.2020.08.002. [DOI] [PubMed]
- 142.Hanna GJ, Supplee JG, Kuang Y, Mahmood U, Lau CJ, Haddad RI, et al. Plasma HPV cell-free DNA monitoring in advanced HPV-associated oropharyngeal cancer. Ann Oncol. 2018;29:1980–6. doi: 10.1093/annonc/mdy251. [DOI] [PubMed] [Google Scholar]
- 143.Chera BS, Kumar S, Shen C, Amdur R, Dagan R, Green R, et al. Plasma circulating tumor HPV DNA for the surveillance of cancer recurrence in HPV-associated oropharyngeal cancer. J Clin Oncol. 2020. 10.1200/JCO.19.02444. [DOI] [PMC free article] [PubMed]
- 144.Seiwert TY, Salama JK, Vokes EE. The concurrent chemoradiation paradigm-general principles. Nat Clin Pract Oncol. 2007;4:86–100. doi: 10.1038/ncponc0714. [DOI] [PubMed] [Google Scholar]
- 145.Rosenberg AJ, Vokes EE. Optimizing treatment de-escalation in head and neck cancer: current and future perspectives. Oncologist. 2020. 10.1634/theoncologist.2020-0303. [DOI] [PMC free article] [PubMed]
- 146.Vokes EE, De Souza JA, Hannigan N, Brisson RJ, Seiwert TY, Villaflor VM, et al. Response-adapted volume de-escalation (RAVD) in locally advanced head and neck cancer. Ann Oncol. 2016;27:908–13. doi: 10.1093/annonc/mdw051. [DOI] [PubMed] [Google Scholar]
- 147.Seiwert TY, Foster CC, Blair EA, Karrison TG, Agrawal N, Melotek JM, et al. OPTIMA: a phase II dose and volume de-escalation trial for human papillomavirus-positive oropharyngeal cancer. Ann Oncol. 2019;30:1673. doi: 10.1093/annonc/mdz171. [DOI] [PubMed] [Google Scholar]
- 148.Chuang AY, Chuang TC, Chang S, Zhou SY, Begum S, Westra WH, et al. Presence of HPV DNA in convalescent salivary rinses is an adverse prognostic marker in head and neck squamous cell carcinoma. Oral Oncol. 2008;44:915–9. doi: 10.1016/j.oraloncology.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]