Abstract
The inability to distinguish between species can be a serious problem in groups responsible for pathogen transmission. Culicoides biting midges transmit many pathogenic agents infecting wildlife and livestock. In North America, the C. variipennis species complex contains three currently recognized species, only one of which is a known vector, but limited species-specific characters have hindered vector surveillance. Here, genomic data were used to investigate population structure and genetic differentiation within this species complex. Single nucleotide polymorphism data were generated for 206 individuals originating from 17 locations throughout the United States and Canada. Clustering analyses suggest the occurrence of two additional cryptic species within this complex. All five species were significantly differentiated in both sympatry and allopatry. Evidence of hybridization was detected in three different species pairings indicating incomplete reproductive isolation. Additionally, COI sequences were used to identify the hybrid parentage of these individuals, which illuminated discordance between the divergence of the mitochondrial and nuclear datasets.
Subject terms: Molecular ecology, Population dynamics, Entomology
Introduction
Speciation is a dynamic evolutionary process through which populations segregate into independently evolving lineages over time1. When gene flow is restricted either through geographic, behavioral, or ecological isolation, the accumulation of genetic changes, through selection or local genetic drift, may lead to divergence and potentially reproductive isolation2–6. Thus, the amount of genetic differentiation and level of gene flow between closely related lineages can be used to evaluate the strength of this isolation and determine species status7. Depending on the completeness of the speciation process between lineages, it can be challenging to unambiguously identify species8. Shallow divergence and hybridization can mask both morphological and genetic differences. While the most accurate assumptions about species delimitation are derived from a multifaceted approach9,10, genomic data has become a powerful tool to investigate species boundaries11,12. Both substantial and fine-scale genetic divergence is being uncovered across many study systems, even in the absence of morphological variation13–16. Species delimitation is especially important when working with organisms responsible for pathogen transmission, as misidentifications will lead to inaccurate vector surveillance data. Culicoides Latreille (Diptera: Ceratopogonidae) biting midges are responsible for the transmission of many pathogens worldwide17,18, including bluetongue virus (BTV) and epizootic hemorrhagic disease virus (EHDV). These viruses can cause severe symptoms and death in wild and domestic ungulates and are responsible for substantial economic losses globally19,20.
In North America, one of the main BTV and EHDV vectors is Culicoides sonorensis Wirth and Jones20, which belongs to the C. variipennis species complex. When originally described, this group consisted of five subspecies21; though presently, three distinct species are recognized (C. occidentalis Wirth and Jones, C. sonorensis, and C. variipennis (Coquillet)) with C. albertensis Wirth and Jones and C. australis Wirth and Jones designated as synonyms of C. sonorensis22. Despite the current taxonomic arrangement, species identification remains difficult due to very subtle morphological differences and overall genetic similarities23,24. Additionally, the presence of cryptic species or hybridization within the complex could be further complicating proper species identification. Under laboratory conditions, C. sonorensis and C. occidentalis have been shown to hybridize25, and both C. occidentalis and C. variipennis occur sympatrically with C. sonorensis21. Hybridization with C. sonorensis in nature would represent a pathway for introgression, potentially for genes controlling vector competency26.
Geographic isolation limits gene flow between populations, and thus, life-history traits influencing dispersal ability can drastically influence the level of gene flow among populations (e.g.27,28). Species with low dispersal ability are particularly likely to exhibit highly differentiated populations resulting in the evolution of cryptic species over a limited spatial scale29. In contrast, species with high dispersal abilities are likely to maintain a high level of gene flow between populations. Studies of Culicoides species in Europe, Africa, and Australia have consistently revealed frequent gene flow between populations, even at continental scales30–33. Culicoides species have been shown to randomly disperse away from their larval habitats, up to 2 km daily34,35, and are also known to disperse via prevailing winds for hundreds of kilometers36–38. The high dispersal ability of biting midges decreases the likelihood of geographic isolation between populations, and as a consequence, may not have played a major role in the diversification of this group. Instead, ecological or behavioral isolation can allow closely related species to occur sympatrically in distinct ecological niches, and may help explain species divergence within Culicoides39,40. The high rate of dispersal, potential for hybridization, and numerous sympatric populations make the C. variipennis complex an intriguing system in which to study species delimitation and may also provide insights into the mechanisms responsible for speciation within this group.
Here, we evaluated the genetic structure of the C. variipennis complex from broad-ranging sampling locations to test the current taxonomic hypothesis of three distinct species. We used ddRadSeq to analyze 206 individuals collected from 17 sites throughout the United States and Canada. We first estimated the overall genetic similarity and population structure among these samples to delimit distinct lineages within the species complex. We then estimated the level of gene flow within and between the inferred species. As previous attempts to separate these species using common barcoding genes have been inconclusive, we sequenced a region of the COI gene to compare to the putative SNP species identifications. Additionally, we discuss the potential mechanisms controlling reproductive isolation within this species complex.
Results
SNP calling and clustering analyses
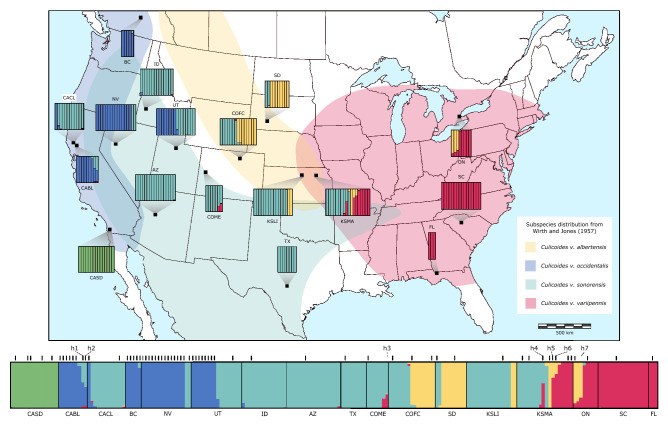
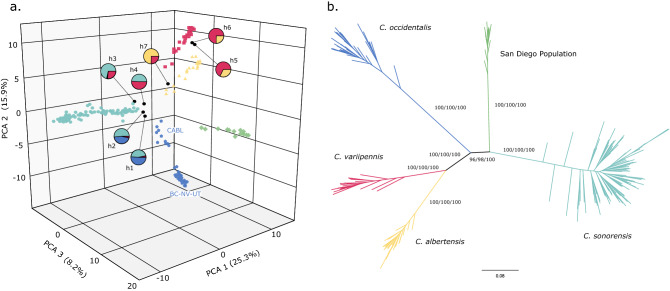
In total, 271 individuals were subjected to the ddRADseq procedure and yielded an average of 2.08 million reads per individual. During the initial filtering, 36 individuals were found to have low-quality sequences (phred score of less than 25) and were removed from the dataset. Additionally, 29 individuals were found to have more than 75% missing data and were also removed. The final dataset included 206 individuals from 17 sites and contained 3612 SNPs. The population structure inferred by fastSTRUCTURE that best explains the data was K = 5. Structure plots showing K = 3–6 can be found in Figure S1. At K = 5, most individuals (86%) were unambiguously assigned to one group (98–100% assignment score; Fig. 1). Consistent with these results, the principal component analysis (PCA) and discriminant analysis of principal components (DAPC) grouped these individuals into five main clusters (Figs. 2a & S2). The main difference being that the PCA further segregated one cluster (blue, Fig. 2a) into two separate groups; east and west of the Sierra Nevada mountain range. Further support for the same five clusters was found in the maximum likelihood trees, with a high level of support from each approximation method (Figs. 2b & S3).
Figure 1.
Geographic distribution and structure plots for each collection site (black squares) overlaid on the historical distribution of the species described in Wirth and Jones 1957. The fastSTRUCTURE results are for 206 individuals inferred by 3612 SNPs and assuming five populations (K = 5). The vertical bars within each collection site represents an individual, with each color representing a cluster. The putative species identity of each cluster are as follows: Culicoides occidentalis (blue), C. sonorensis (teal), C. albertensis (yellow), C. variipennis (red), and an unidentified population in San Diego, CA (CASD) (green). The black bars above the overall structure plot indicates an individual for which the COI gene was also sequenced. The individuals inferred to be hybrids are labeled h1-7. This map was created using Inkscap v.1.1 (https://inkscape.org/).
Figure 2.
(a) A 3D representation of the principal component analysis (PCA) of all individuals included in the study. Each color represents the cluster inferred from the structure analysis; C. albertensis (yellow), C. occidentalis (blue), C. sonorensis (teal), C. variipennis (red), and the unidentified San Diego population (green). Hybrids (h1–h7) are designated with a black circle and their inferred parental ancestry is depicted with pie charts. The geographic locations of the two C. occidentalis clusters are labeled next to each grouping (see Table 1 for abbreviation). (b) Unrooted maximum likelihood phylogenetic tree based on 199 individuals inferred from 3612 SNPs (the hybrids were removed here but are included in Fig. S3.). Clade colors also represent the clusters inferred from the structure analysis. Support values written on the branches: rapid bootstrap (%)/SH-aLRT support (%)/ultrafast bootstrap support (%). For clarity, the values within each cluster are not shown.
Inter- and intra-species population genetics
The geographic distributions of these clusters closely align with the distributions of the species (then subspecies) described in Wirth & Jones (1957) (Fig. 1), and recent morphological analyses of individuals from this study supports the species level designation of these clusters41. For the remainder of the manuscript, we will refer to each cluster by its corresponding species name. Culicoides occidentalis was located in Western North America, C. sonorensis in the Western and Southern United States (U.S.), C. albertensis in the Midwest U.S. and Ontario, C. variipennis in the Eastern U.S. and Ontario, and a fifth genetic group suggesting the occurrence of an additional, undescribed cryptic species in San Diego, CA. Notably, eight of the 17 sites had more than one species in sympatry, and one site had three species. At four sites, seven individuals were assigned to two genetic groups with an assignment score of ~ 50% for three individuals (scores = 45, 47 and 41%) and of ~ 25% for four individuals (scores = 34, 31, 25 and 24%), which suggests the occurrence of putative F1 or other types of hybrids (e.g., F2 or backcrosses). Interestingly, these hybrids were from three different species pairings (C. sonorensis X C. occidentalis; C. sonorensis X C. variipennis; and C. albertensis X C. variipennis). These hybrid individuals also stood out in the PCA, as they segregated between their parental clusters (Fig. 2a), as well as at the base of each parental branch in the phylogenetic tree (Fig. S3). In addition to these seven hybrids, 20 individuals had a secondary assignment score between 3 and 21%, signifying potential introgression between those pairings. However, we are less confident in STRUCTURE’s ability to identify this level of ancestry as other factors can also lead to mixed assignments.
The seven putative hybrids were excluded from the dataset used to calculate the intraspecies summary statistics (rearranged by cluster), which resulted in the isolation of 566 SNPs after more stringent filtering was applied. The mean FST between species was 0.7147 (0.6541–0.7470), roughly 9 times higher than the mean FST between the populations (i.e., localities) within each species (see below; Tables 2 & S1). Similarly, both the aR and LKC values of intra-individual genetic distance show a low level of divergence/high level of similarity within each species, including the San Diego population (Table S2). The four species-specific datasets were used to calculate the interspecies summary statistics as well as test for isolation by distance (IBD). These datasets contained 22 individuals of C. albertensis from four sites (3423 SNPs), 36 individuals of C. occidentalis from four sites (2714 SNPs), 97 individuals of C. sonorensis from seven sites (2357 SNPs), and 29 individuals of C. variipennis from four sites (2960 SNPs). The expected and observed heterozygosity, FIS, and number of private alleles for each species are reported in Table S2. No species-level dataset was created for the San Diego species, as this species was uncovered in a single locality.
Table 2.
Mean pairwise FST within and between species.
| Species | C. albertensis | C. occidentalis | C. sonorensis | C. variipennis |
|---|---|---|---|---|
| C. albertensis |
0.055 (− 0.009 to 0.116) |
– | – | – |
| C. occidentalis | 0.707 |
0.411 (0.143–0.704) |
– | – |
| C. sonorensis | 0.709 | 0.730 |
0.029 (0.006–0.069) |
– |
| C. variipennis | 0.654 | 0.747 | 0.730 |
0.026 (− 0.006 to 0.045) |
| San Diego pop | 0.714 | 0.719 | 0.706 | 0.734 |
The between species FST values (below diagonal) were calculated using 566 SNPs and the within-species values (on diagonal) is the mean FST calculated from individual species-specific datasets (see Table S4).
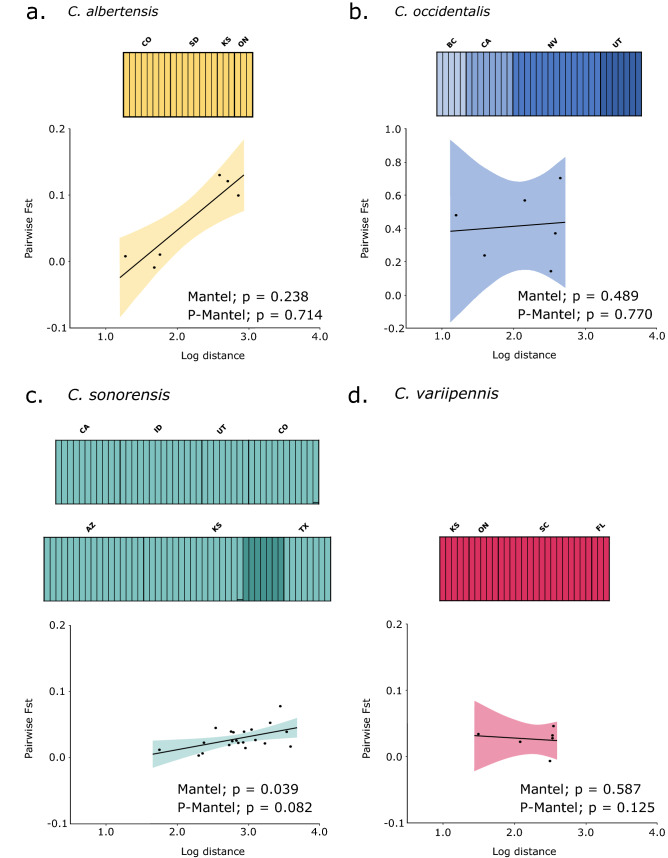
When examining each species individually, C. albertensis, had no evidence of population structure (K = 1), and had low genetic differentiation among locations (mean FST = 0.054) (Fig. 3a; Table 2). Although there does seem to be a pattern of IBD, this was found to not be significant in this species (Mantel test, P = 0.238; Partial Mantel test, P = 0.714). The low number of locations sampled potentially limits the statistical power of these correlations. The results obtained for C. occidentalis showed much more divergence compared to the other species, with populations being strongly differentiated from each other (mean FST = 0.411) (Table 2). Additionally, the fastSTRUCTURE analysis suggested that each location of C. occidentalis sampled is distinct (K = 4) (Fig. 3b). While no IBD was found (Mantel test, P = 0.489; Partial Mantel test, P = 0.770), there seems to be a considerable amount of geographic isolation among populations of this species, with pairwise FST values ranging from 0.14 to 0.70 (Table S4). Additionally, significant levels of dissimilarity were found between individuals from California and those from the other three populations (Fig. S4). In contrast, low genetic differentiation among locations were found for C. sonorensis (mean FST = 0.029), with varying levels of support for IBD in this species (Mantel test, P = 0.039; Partial Mantel test, P = 0.082) (Fig. 3c; Table 2). For this reason, the individuals from Colorado were combined into a single location, as were the individuals from Kansas. The fastSTRUCTURE analysis suggested the occurrence of population structure in C. sonorensis (K = 2), with some individuals from Kansas belonging to a distinct group, though these were not highly divergent from any other C. sonorensis location (Table S4). Individuals of C. variipennis exhibited no evidence of population structure (K = 1) or of IBD (Mantel test, P = 0.587; Partial Mantel test, P = 0.125) (Fig. 3d). Consistently, almost no genetic differentiation was found among locations of this species (mean FST = 0.026) (Table 2).
Figure 3.
For each species, an independent SNP dataset was used to calculate the most suitable K using fastSTRUCTURE with the inferred clusters denoted by varying shades. A Mantel and partial Mantel (P-Mantel) test was used to test for IBD (shown as pairwise FST by log geographic distance) for each species in Genepop. The individuals from San Diego, CA are not included here as they were only found in a single population.
Haplotype network
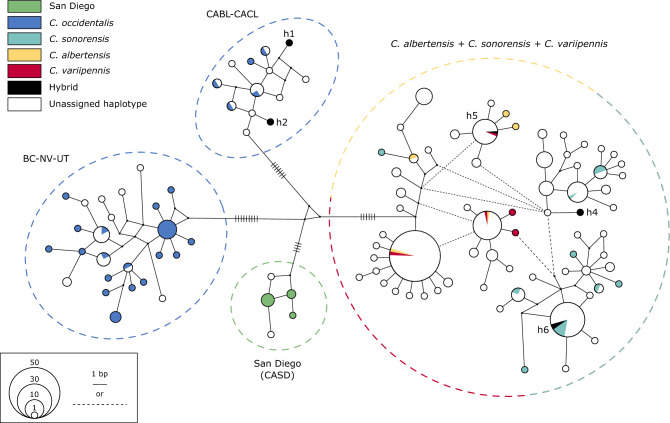
In total, 285 midges were included in the analysis of a 546 bp region of the COI gene. Four distinct haplogroups were identified with substantial genetic divergence between groups (p-distance = 2.99–3.30%) and little divergence within groups (p-distance = 0.25–0.86%; Fig. 4; Table 3). Consistent with the SNP datasets, C. occidentalis formed a divergent haplogroup, separated from the rest of its range. The mean percent divergence between the two C. occidentalis groups (2.99%) was similar to its divergence from the other species (3.01–3.30%). The San Diego population also clustered as a distinct group, with a similar level of divergence from the other species (3.01–3.03%). Interestingly, C. albertensis, C. sonorensis, and C. variipennis were not separated from each other, and in some cases, C. albertensis and C. variipennis shared identical haplotypes (Fig. 4, S5). Furthermore, these three species exhibit a mean percent divergence between individuals (0.80%) similar to the divergence observed among individuals within C. occidentalis (Table 3). Other than the grouping of C. occidentalis in California, there was no geographic clustering observed.
Figure 4.
A haplotype network inferred by a median-joining method, using 285 mitochondrial (mt) DNA sequences of the C. variipennis complex from 27 states in the U.S., as well as British Columbia and Ontario, Canada. The size of each circle represents the frequencies of the haplotype and the length of the lines connecting the circles corresponds to number of bp differences. Note that the dotted black lines also represent a single bp change. The 67 sequences obtained in the present study (see Fig. 1) are colored according the clusters assigned from the structure analysis. The four main groups of haplotypes (see Results) are circled.
Table 3.
Mean percent divergence (p-distance) within and between species clusters based on the COI gene (ranges listed in parentheses).
| Clade | occ (CABL) | occ (BC-NV-UT) | San Diego pop | alb-son-var |
|---|---|---|---|---|
| occ (CABL) |
0.48 (0.00–0.73) |
– | – | – |
| occ (BC-NV-UT) |
3.99 (3.20–5.49) |
0.86 (0.00–1.65) |
– | – |
| San Diego pop |
3.01 (2.38–4.21) |
3.66 (2.75–4.76) |
0.25 (0.00–0.66) |
– |
| alb-son-var |
3.30 (2.75–5.12) |
3.76 (3.30–6.04) |
3.03 (2.38–4.21) |
0.80 (0.00–2.74) |
Based on overall similarity, C. occidentalis was split into two groups (CABL; and BC-NV-UT) and C. albertensis, C. sonorensis, and C. variipennis were grouped into a single clade (alb-son-var).
Discussion
Our study provides valuable insights into the population genetics of the C. variipennis species complex and highlights the presence of potential cryptic species. For most of the species examined, minimal genetic divergence was observed across locations, suggesting the maintenance of gene flow even over large geographic distances. The only exception was C. occidentalis, which showed a high level of geographic isolation, as well as two distinct COI haplogroups. We confirmed that mitochondrial data is not reliable to differentiate three out of the five species, due to the lack of segregation between the mitochondrial haplotypes of C. albertensis, C. sonorensis, and C. variipennis. This stands in stark contrast to their clear differentiation and high level of divergence inferred from the SNP data. Though a substantial amount of divergence exists between all five species, low levels of hybridization, and potentially introgression, are present in sympatric populations. While we do not know the fitness of these hybrids, but this could suggest that strong post-zygotic isolation barriers may have yet to evolve in this group. Thus, pre-zygotic isolation through either ecological or behavioral segregation is a possible mechanism maintaining divergence within this complex. With a considerable amount of geographic overlap between some species (Fig. 1), each sympatric population is potentially experiencing a set of unique selective pressures to maintain species boundaries.
The high degree of genetic differentiation between clusters inferred by the SNP data supports the current species groupings of the C. variipennis complex (C. occidentalis, C. sonorensis, and C. variipennis), as well as raising C. albertensis and a cryptic species in San Diego, California to species status (Fig. S5). While this putative new species was only collected in San Diego, its true distribution could extend well into Mexico. While clear divergence was observed in the SNP data, the mitochondrial data showed a different pattern of divergence. Culicoides albertensis, C. sonorensis, and C. variipennis have a considerable amount of genome-wide differentiation (Fig. 1); however, there was no clear differentiation of the COI gene (Fig. 4). In fact, several individuals of C. albertensis and C. variipennis shared identical haplotypes. Multiple studies have shown a high degree of genetic similarity in mtDNA between C. sonorensis and C. variipennis23,24,42, though it was proposed that this was due to misidentifications. As all of the individuals included in our mitochondrial haplotype analysis from the current study were identified to species using the SNP data, this lack of mitochondrial separation must have an underlying biological cause. This finding can result from historical introgression with “leaky” pre-zygotic isolation, or semipermeable species boundaries, which have been shown to produce mitochondrial introgression without detectable nuclear DNA introgression in some taxa43,44. This is likely due to the fact that the mitochondrial genome is independent of the nuclear genome and thus unlinked to the genes contributing to reproductive isolation45. However, we cannot rule out that other possibilities could have caused this discordance, such as recent speciation and incomplete lineage sorting or selection13,46. Regardless, it appears that the evolution of the mitochondrial genome is not congruent with the species tree of the C. variipennis complex. Notably, the SNP phylogenetic tree shows that C. occidentalis, and not C. sonorensis, is the sister taxa of C. albertensis and C. variipennis (Fig. S5). This suggests that the mtDNA similarity between C. albertensis, C. sonorensis and C. variipennis could stem from ongoing hybridization and introgression, rather than incomplete lineage sorting.
Little to no IBD or structure was found within C. albertensis, C. sonorensis, and C. variipennis indicating a substantial amount of connectivity among localities of these species (Fig. 3a,c,d). The number of populations inferred by fastSTRUCTURE for C. sonorensis was K = 2; however, a mean pairwise FST of 0.0287 suggests that a high amount of gene flow exists between all locations. This could be an artifact of the propensity of delta K inferring two populations47 or from a high level of relatedness among individuals from KS (Fig. S6). Interestingly, although no IBD was found in C. occidentalis, each location of this species clustered as a distinct population (Fig. 3b). The lack of IBD is therefore not indicative of a single, genetically homogeneous population, but rather stems from high levels of divergence between populations regardless of their geographic distances. Focusing sampling efforts on each of these species will surely permit robust landscape genetic approaches to gain understanding of the evolutionary forces driving population structure in this group. This will allow studying whether Culicoides population structure is characterized by uniform or discontinuous isolation by distance, as well as, isolation by adaptation (IBA) or by environment (IBE)48.
The strong genetic divergence between the C. occidentalis from California and the other populations was observed in both the SNP and mtDNA data (Tables 2, 3, Fig. 4). It is possible that this may represent a further cryptic species with a dispersal barrier created by the Sierra-Nevada mountain range (Fig. S4). This high level of differentiation within C. occidentalis could be due to geographic isolation alone; however, endosymbionts have also been shown to significantly increase mitochondrial diversity in the presence of geographic structure49,50. Naturally occurring endosymbionts have been found in Culicoides midges, including C. sonorensis51,52, and recently, a Cardinium sp. was linked to mitochondrial divergence in C. imicola53. Further screening is needed to determine the diversity and abundance of endosymbionts infecting Culicoides midges, though the possibility remains that they could be playing a role in the phylogeographical structure of C. occidentalis if they are causing incompatibility between populations. Additionally, patchiness of the specialized larval habitat of C. occidentalis, not present in the other members of the C. variipennis complex, could create isolation between populations, as well as reduce the number of individuals within each population. A small effective population size with little to no immigration would allow for a strong effect from drift54. While the populations of C. occidentalis outside of California were less diverged from one another, the lowest pairwise FST values between these populations were still greater than the highest pairwise values observed within any other species, consistent with the findings of Holbrook et al. (2000) (Table 2).
Similar to other species of Culicoides30,32,33,55, high values of the inbreeding coefficient (FIS) were observed in all species investigated in this study (Table S2). Although these previous studies have suggested that the observed high FIS are an artifact from a large number of null alleles, the consistent reporting of these findings across various species using several types of molecular markers lends support to the hypothesis that high inbreeding has a biological origin in this genus. High levels of inbreeding and heterozygote deficiencies are common among mosquitoes56–58, even when using markers with a low level of null alleles59,60. Goubert et al. (2016) considered the typical Aedes albopictus population as “a network of interconnected breeding sites, each with a high level of inbreeding”. Although we cannot rule out all other possibilities, our results strongly suggest that some aspects of the reproductive biology of Culicoides induce inbreeding within populations. High FIS and low FST between populations can stem from high levels of migration between populations (i.e., homogenizing allele frequencies at large scale), followed by matings with close relatives within populations (i.e., increasing homozygosity without altering allelic frequencies). It is also possible that our sampling approaches (single night trapping) led to capturing cohorts of closely related individuals.
Low levels of hybridization were found in some sympatric populations involving several different species pairings. Under laboratory conditions, mating between C. sonorensis and C. occidentalis can produce viable offspring for at least six generations, though the hatch rate of the progeny is dependent on the species of the mother25. A cross of female C. sonorensis and male C. occidentalis only yields a 7% hatch rate whereas the reciprocal cross yields a 75% hatch rate. This asymmetrical hybrid viability is likely caused by cytonuclear incompatibility61,62, though endosymbionts have also been shown to cause reproductive incompatibility63. Upon secondary contact of closely related species, and in the absence of post-zygotic reproductive isolation, the production of unfit hybrids can induce the rapid evolution of premating barriers2,64–66. In most populations however, C. sonorensis females are unlikely to come across C. occidentalis males due to differences in mating behavior. Conversely, C. occidentalis females do come into contact with C. sonorensis males, who do not appear to have mate discrimination67, and will likely attempt to mate with these heterospecific females. As there are demographic disparities (population size and structure) between these two species, as well as viable offspring produced from this cross, rampant hybridization and asymmetric introgression would be detrimental to C. occidentalis68. Strong selection against hybridization can maintain species boundaries, but as two of the ten C. occidentalis collected from Borax Lake in California (CABL) appeared to be F1 hybrids (Fig. 1), another mechanism, potentially differences in the larval habitat or mating behavior, appears to be limiting directional introgression from C. sonorensis.
Culicoides occidentalis females lay their eggs in highly saline environments (up to 88.0 parts per thousand (ppt))69, whereas C. sonorensis eggs will not hatch in water with salinity over 20.0 ppt70. However, ecological exclusion via the larval habitat would only limit introgression if the hybrids were inviable in highly saline environments, which does not appear to be the case. The difference in mating behavior between these two species may be a more likely mechanism by which the detrimental effects of hybridization are diminished. Culicoides occidentalis mates at the larval habitat while C. sonorensis mates at or near a host22,71. Even if a C. occidentalis female mated with a C. sonorensis male, she would return to the high saline pools to lay her eggs and these hybrid offspring would have a high chance of only backcrossing within the C. occidentalis lineage. While only two C. occidentalis x C. sonorensis hybrids were tested in this study, both had C. occidentalis mothers (Fig. 4), providing evidence that this scenario takes place in nature. However, this type of isolation would not explain how C. sonorensis and C. variipennis maintain species boundaries in sympatry as they share a larval habitat. Further studies are needed to determine the mechanisms behind reproductive isolation within this group.
The C. variipennis complex is one of many vector groups in which species delimitation can be challenging46,72–76; however, species identification is an integral part of vector surveillance. The species status of these group members has implications for vector surveillance, as any ambiguity in identification will lead to unreliable data. For example, while C. albertensis and C. sonorensis occur in sympatry, only C. sonorensis is reported as a vector species77. The addition of the non-vector species when conducting serological surveys could lead to a severe underestimation of the infection rate within the vector species. As BTV and EHDV are expanding northward into eastern Canada78, it has been suggested that the dispersal of C. sonorensis into new areas could be to blame for this incursion42. Specimens assigned to C. sonorensis by Jewiss-Gaines et al. (2017) were included in the present study and cluster instead with C. albertensis (“ON”, Fig. 1). Thus, there are likely alternative reasons for the range expansion of these viruses, including an unidentified vector species outside of the C. variipennis complex, such as C. stellifer79,80. Molecular tools for accurate species-level delimitation within this complex is sorely needed for proper vector surveillance. Additionally, the detection of hybridization between a non-vector and vector species may be evidence of recent speciation, but it also highlights a potential path of introgression for genes controlling vector competency81,82.
Our study shows that using a population genomic approach to analyze sibling species can identify species-level divergence, fine-scale genetic structuring within species, and uncover the existence of hybrids and cryptic species in Culicoides. Radiation within the C. variipennis complex occurred despite the long-range dispersal capabilities of biting midges as well as hybridization between sympatric species. This does not preclude historical geographic isolation; however, we believe that behavioral and ecological isolation may have shaped evolution within this group or is at least maintaining the current species boundaries. Significant geographic isolation was only found between populations of C. occidentalis, but more sampling is needed to determine if the lack of gene flow between California and the other populations represents an incipient speciation event or IBD. Additionally, focusing efforts in these various hybrid zones may provide a better understanding of the evolution of reproductive isolation in this group. Cryptic species have been reported in a number of other Culicoides species complexes83 and the analyses presented here could help to identify these putative species. Delimiting the species in these complexes, will not only aid in vector surveillance efforts, but continued study of the speciation of closely related vector and non-vector species could produce valuable evolutionary insights into vector competency.
Materials and methods
Sample collection and sequencing
Culicoides midges were collected from 17 sites across the United States and Canada (Table 1). Specimens were collected either as pupae and reared to adulthood, or as adults using CDC light traps baited with CO2 and UV light (Bioquip 2836BQ). Individuals morphologically assigned to the C. variipennis complex were sorted out from the by-catch and stored in 95% ethanol at -80 °C. Total DNA was extracted from individuals using a Puregene extraction protocol (Gentra Systems, Inc., D-5500A) with the addition of glycogen (ThermoFisher, R0561) to increase yields. DNA was only extracted only from females as their larger body size (compared to the males) produced sufficient amount of DNA for next-gen sequencing. The DNA quality was checked using gel electrophoresis and DNA concentration was measured using a Qubit 3.0 fluorometer and a Qubit dsDNA HS assay kit (Invitrogen, Q33230). A total of 300–400 ng of DNA per sample was sent to Floragenex, Inc. for library preparation using the protocol from Truong et al. (2012). DNA was digested using the restriction enzymes MseI and PstI. After PCR amplification, the samples in each plate were pooled and sequenced on a lane of single-end 100 bp sequencing on a HiSeq4000 at the University of Oregon Genomics Facility, Eugene, OR.
Table 1.
Collection site information and numbers of individuals retained for the SNP analyses.
| Country | State/Province | Lat | Long | Collection date | Collection method | N | Abbreviation |
|---|---|---|---|---|---|---|---|
| Canada | British Columbia | 49.3065 | − 119.6323 | 5/7/2019 | Pupal rearing | 5 | BC |
| USA | California | 39.0245 | − 122.8515 | 8/14/2018 | Pupal rearing | 12 | CACL |
| USA | California | 38.9811 | − 122.6731 | 8/14/2018 | Pupal rearing | 9 | CABL |
| USA | California | 32.5522 | − 117.0628 | 11/7/2014 | Light trap | 15 | CASD |
| USA | Idaho | 43.7065 | − 116.4236 | 8/19/2014 | Light trap | 14 | ID |
| USA | Nevada | 40.0521 | − 118.4681 | 7/29/2013 | Light trap | 17 | NV |
| USA | Arizona | 34.5792 | − 112.4258 | 7/21/2010 | Light trap | 17 | AZ |
| USA | Utah | 40.7844 | − 112.1090 | 9/10/2018 | Light trap | 16 | UT |
| USA | South Dakota | 43.7438 | − 101.9509 | 8/6/2018 | Light trap | 10 | SD |
| USA | Colorado | 40.6560 | − 104.9878 | 8/8/2019 | Light trap | 15 | COFC |
| USA | Colorado | 39.0546 | − 108.5170 | 7/16/2013 | Light trap | 7 | COME |
| USA | Kansas | 38.8793 | − 98.4481 | 9/25/2018 | Pupal rearing | 16 | KSLI |
| USA | Kansas | 39.2234 | − 96.5906 | 7/17/2018 | Light trap | 18 | KSMA |
| USA | Texas | 29.9515 | − 99.6010 | 7/29/2017 | Light trap | 8 | TX |
| Canada | Ontario | 43.2167 | − 79.9500 | 7/5/2013 | Light trap | 8 | ON |
| USA | South Carolina | 34.3080 | − 81.7550 | 7/23/2014 | Light trap | 16 | SC |
| USA | Florida | 30.4782 | − 84.6401 | 8/27/2018 | Light trap | 3 | FL |
Raw sequence filtering and processing
Raw sequence quality was first assessed using FastQC v.0.11.9 and MultiQC v.1.784,85, and then reads were filtered and processed using Stacks v.2.386. Reads with a phred score below 25 were removed as well as individuals with a > 75.0% missing data. Next, reads were aligned to the C. sonorensis genome87 (Accession: PRJEB19938) using the Burrows-Wheeler Aligner (BWA-mem)88. Finally, aligned reads were run through the reference-based pipeline of Stacks. Filtering options were set to only include loci found in at least half of the sampling locations (-p 8) and those occurring in at least 50% of individuals within those sites (-r 0.5)89. The minimum allele frequency was set to 0.05 to protect against potential sequencing errors90, and only the first SNP per locus was kept to minimize linkage disequilibrium between SNPs from influencing population structure and phylogenetic analyses. All subsequent file reformatting was done with PGDSpider v.2.1.1.591.
Clustering analysis
Population structure in the overall dataset was evaluated using fastSTRUCTURE v.1.04, with Structure_threader utilized to parallelize distinct runs of K92,93. Models were fitted with the number of genetic clusters (K) set to range from 1 to 10. The most suitable value of K was selected using the chooseK.py function from the fastSTRUCTURE package which selects the model that maximizes the marginal likelihood of the data. Using the output from fastSTRUCTURE and Distruct v.2.3 (http://distruct2.popgen.org), a bar plot was created where each individual is represented by a vertical line divided into K colored segments with the length of each segment being proportional to the estimated membership in each of the inferred K groups. A map of the structuring at each collection site was created using Inkscap v.1.1 (https://inkscape.org/). The clustering of individuals into the distinct genetic groups was also visualized using a principal component analysis (PCA) and a discriminant analysis of principal components (DAPC). The most likely number of genetic groups was inferred by the find.clusters algorithm for the PCA and the optimal number of principal components to inform the DAPC was defined using the function optim.a.score. Both were performed in R94 through the adegenet package95.
Any individual with more than 25% of their loci grouping with a second cluster in the fastSTRUCTURE analysis was marked as a hybrid and removed from the phylogenetic analysis, due to the uncertainty of assigning them to a given species. Maximum likelihood phylogeny among individuals was run using RAxML v.8.2.1296. An acquisition bias correction was applied to the likelihood calculations as alignments were solely composed of SNPs, with each invariant site removed through Phrynomics (https://github.com/bbanbury/phrynomics)97. The GTR + G nucleotide substitution model was used for each search. A rapid bootstrap analysis and search for the best-scoring maximum likelihood tree was executed using the extended majority rule-based bootstopping criterion to achieve a sufficient number of bootstrap replicates98. Additionally, to cross-validate our results, a second phylogeny was inferred in W-IQ-Tree version 1.6.1299, using the TVM + F + G4 substitution model determined by ModelFinder100,101. Branch support was calculated using 1000 ultrafast bootstraps102 and a Shimodaira–Hasegawa like approximate likelihood-ratio test (SH-aRLT)102,103.
To measure the amount of divergence between genetic clusters, a new SNP dataset was generated with individuals grouped by cluster rather than locality. Additionally, to measure the amount of divergence within each genetic cluster, four cluster-specific datasets (grouped by site) were also generated. SNPs were obtained from these new datasets using the same processing methods above except with more stringent filtering parameters. Only SNPs that occurred in at least 75% of the clusters or collection sites and at least half of the individuals within those groups were included. Genetic diversity estimates (FIS, HE, and HO) and population differentiation (pairwise FST) were calculated for each species dataset using Genepop v.4.7.0104. Population differentiation was not calculated for the new species found in San Diego, as only a single population of this species was uncovered. Rousset’s distance aR105 and Loiselle’s kinship coefficient (LKC)106 were calculated respectively with SPAGeDi v.1.5107. Geographic distances among localities were calculated as both Euclidean and anisometric distances and a Mantel test and a Partial Mantel test were preformed to test for isolation-by-distance (IBD)108. Tests for areas of significant genetic dissimilarity among individuals using aR were implemented in MAPI using 1000 replications109.
Mitochondrial sequencing and haplotype network
Mitochondrial DNA haplotypes were obtained from a subset of 67 individuals from the five genetic clusters. PCR reactions were performed using a Taq-Pro COMPLETE kit (Denville Scientific, CB4065-4) targeting a partial region of the COI gene with the Lep50 primer set from Folmer et al. (1994) and the thermocycler profile from Herbert et al. (2003). PCR products were cleaned using an EXOSAP-IT kit (ThermoFisher, 78201.1.ML) and prepared for sequencing using a BigDye Terminator v.3.1 Cycle Sequencer Kit (Applied Biosystems, 4337454). Sanger sequencing was done using an Applied Biosystems 3500 Genetic Analyzer. Chromatograms were cleaned and aligned using the software Geneious v.9.1110.
A haplotype network analysis was conducted using the 67 COI sequences obtained in this study combined with 218 C. variipennis complex sequences previously collected from 25 states across the U.S.111. Sequences were aligned in MEGA v.10.1.8112 and trimmed to ensure all sequences contained identical lengths. A median-joining analysis was performed using NETWORK v.5.0.1.0113. Specimens collected in this study were assigned a color based on the results from the SNP clustering analyses while the remaining samples were left unassigned. All individuals were used to calculate the mean uncorrected p-divergence between and within the different groupings inferred from the haplotype network using MEGA.
Supplementary Information
Acknowledgements
The authors thank Art Borkent, Adam Jewess-Gaines, Dustin Swanson, Bonnie Ryan, Nadja Mayerle, multiple vector control agencies, USDA-APHIS-Wildlife Services, and several landowners for access to property or assisting with collecting specimens used in this study. Funding was provided by a USDA Non-Assistance Cooperative Agreement: 58-3020-9-007 and the Urban Entomology Endowment fund at Texas A&M University.
Author contributions
P.S., M.M., L.C., and E.V. planned and designed the study, P.S., M.H., and L.C. collected samples, P.S., P.E., and A.B. carried out lab work and analyzed the data, P.S. and P.E. produced the figures, P.S. led writing; M.H., P.E., A.B., M.M., L.C., and E.V. contributed to drafting and editing the manuscript, M.M., L.C., and E.V. provided supervision, P.S., L.C., and E.V. contributed to procuring funds.
Data availability
The data reported in this study will be deposited in the Open Science Framework database upon acceptance, https://osf.io (10.17605/OSF.IO/E3Z72). Mitochondrial sequences obtained in the current study have been deposited under Genbank Accession Numbers OL604713—OL604779.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Phillip Shults, Email: phillip.shults@gmail.com.
Lee W. Cohnstaedt, Email: lee.cohnstaedt@usda.gov
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-05856-x.
References
- 1.De Queiroz K. Species concepts and species delimitation. Syst. Biol. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- 2.Coyne JA, Orr HA. Speciation. Sinauer Associates Inc; 2004. [Google Scholar]
- 3.Endler JA. Gene flow and population differentiation: studies of clines suggest that differentiation along environmental gradients may be independent of gene flow. Science. 1973;179:243–250. doi: 10.1126/science.179.4070.243. [DOI] [PubMed] [Google Scholar]
- 4.Mayr E. Systematics and the Origin of Species, from the Viewpoint of a Zoologist. Harvard University Press; 1999. [Google Scholar]
- 5.Richardson JL, Urban MC, Bolnick DI, Skelly DK. Microgeographic adaptation and the spatial scale of evolution. Trends Ecol. Evol. 2014;29:165–176. doi: 10.1016/j.tree.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Nosil P. Ernst Mayr and the integration of geographic and ecological factors in speciation. Biol. J. Lin. Soc. 2008;95:26–46. doi: 10.1111/j.1095-8312.2008.01091.x. [DOI] [Google Scholar]
- 7.Kisel Y, Barraclough TG. Speciation has a spatial scale that depends on levels of gene flow. Am. Nat. 2010;175:316–334. doi: 10.1086/650369. [DOI] [PubMed] [Google Scholar]
- 8.Leliaert F, et al. DNA-based species delimitation in algae. Eur. J. Phycol. 2014;49:179–196. doi: 10.1080/09670262.2014.904524. [DOI] [Google Scholar]
- 9.Carstens BC, Pelletier TA, Reid NM, Satler JD. How to fail at species delimitation. Mol. Ecol. 2013;22:4369–4383. doi: 10.1111/mec.12413. [DOI] [PubMed] [Google Scholar]
- 10.Schlick-Steiner BC, et al. Integrative taxonomy: a multisource approach to exploring biodiversity. Annu. Rev. Entomol. 2010;55:421–438. doi: 10.1146/annurev-ento-112408-085432. [DOI] [PubMed] [Google Scholar]
- 11.Capblancq T, Mavárez J, Rioux D, Després L. Speciation with gene flow: evidence from a complex of alpine butterflies (Coenonympha, Satyridae) Ecol. Evol. 2019;9:6444–6457. doi: 10.1002/ece3.5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedraza-Marrón CdR, et al. Genomics overrules mitochondrial DNA, siding with morphology on a controversial case of species delimitation. Proc. R. Soc. B. 2019;286:20182924. doi: 10.1098/rspb.2018.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinojosa JC, et al. A mirage of cryptic species: genomics uncover striking mitonuclear discordance in the butterfly Thymelicus sylvestris. Mol. Ecol. 2019;28:3857–3868. doi: 10.1111/mec.15153. [DOI] [PubMed] [Google Scholar]
- 14.Nygren A, et al. A mega-cryptic species complex hidden among one of the most common annelids in the North East Atlantic. PLoS ONE. 2018;13:e0198356. doi: 10.1371/journal.pone.0198356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thielsch A, Knell A, Mohammadyari A, Petrusek A, Schwenk K. Divergent clades or cryptic species? Mito-nuclear discordance in a Daphnia species complex. BMC Evol. Biol. 2017;17:1–9. doi: 10.1186/s12862-017-1070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eyer PA, Hefetz A. Cytonuclear incongruences hamper species delimitation in the socially polymorphic desert ants of the Cataglyphis albicans group in Israel. J. Evol. Biol. 2018;31:1828–1842. doi: 10.1111/jeb.13378. [DOI] [PubMed] [Google Scholar]
- 17.Borkent, A. Biology of Disease Vectors. 2nd edn, i–xxiii + 1–785 (Elsevier Academic Press, 2004).
- 18.Mellor P, Boorman J, Baylis M. Culicoides biting midges: their role as arbovirus vectors. Annu. Rev. Entomol. 2000;45:307–340. doi: 10.1146/annurev.ento.45.1.307. [DOI] [PubMed] [Google Scholar]
- 19.Rushton J, Lyons N. Economic impact of Bluetongue: a review of the effects on production. Veterinaria italiana. 2015;51:401–406. doi: 10.12834/VetIt.646.3183.1. [DOI] [PubMed] [Google Scholar]
- 20.Tabachnick WJ. Culicoides vriipennis and Bluetongue-Virus eidemiology in the United States. Annu. Rev. Entomol. 1996;41:23–43. doi: 10.1146/annurev.en.41.010196.000323. [DOI] [PubMed] [Google Scholar]
- 21.Wirth WW, Jones RH. The North American Subspecies of Culicoides variipennis (Diptera, Heleidae) U. S. Dep. Agric. Tech. Bull. 1957;1170:1–35. [Google Scholar]
- 22.Holbrook FR, et al. Sympatry in the Culicoides variipennis Complex (Diptera: Ceratopogonidae): a Taxonomic Reassessment. J. Med. Entomol. 2000;37:65–76. doi: 10.1603/0022-2585-37.1.65. [DOI] [PubMed] [Google Scholar]
- 23.Hopken MW. Pathogen Vectors at the Wildlife-Livestock Interface: Molecular Approaches to Elucidating Culicoides (Diptera: Ceratopogonidae) Biology. University of Colorado; 2016. [Google Scholar]
- 24.Shults P. A Study of the Taxonomy, Ecology, and Systematics of Culicoides Species (Diptera: Ceratopogonidae) Including those Associated with Deer Breeding Facilities in Southeast Texas. Texas A&M University; 2015. [Google Scholar]
- 25.Velten RK, Mullens BA. Field morphological variation and laboratory hybridization of Culicoides variipennis sonorensis and C. v. occidentalis (Diptera:Ceratopogonidae) in southern California. J. Med. Entomol. 1997;34:277–284. doi: 10.1093/jmedent/34.3.277. [DOI] [PubMed] [Google Scholar]
- 26.Fontaine MC, et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 2015;347:1258522. doi: 10.1126/science.1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolnick DI, Otto SP. The magnitude of local adaptation under genotype-dependent dispersal. Ecol. Evol. 2013;3:4722–4735. doi: 10.1002/ece3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- 29.Pante E, et al. Species are hypotheses: avoid connectivity assessments based on pillars of sand. Mol. Ecol. 2015;24:525–544. doi: 10.1111/mec.13048. [DOI] [PubMed] [Google Scholar]
- 30.Jacquet S, et al. Colonization of the Mediterranean basin by the vector biting midge species Culicoides imicola: an old story. Mol. Ecol. 2015;24:5707–5725. doi: 10.1111/mec.13422. [DOI] [PubMed] [Google Scholar]
- 31.Onyango MG, et al. Genotyping of whole genome amplified reduced representation libraries reveals a cryptic population of Culicoides brevitarsis in the Northern Territory, Australia. BMC Genomics. 2016;17:769. doi: 10.1186/s12864-016-3124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onyango MG, et al. Delineation of the population genetic structure of Culicoides imicola in East and South Africa. Parasit. Vectors. 2015;8:660. doi: 10.1186/s13071-015-1277-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mignotte A, et al. High dispersal capacity of Culicoides obsoletus (Diptera: Ceratopogonidae), vector of bluetongue and Schmallenberg viruses, revealed by landscape genetic analyses. Parasit. Vectors. 2021;14:1–14. doi: 10.1186/s13071-020-04522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanders CJ, Carpenter S. Assessment of an immunomarking technique for the study of dispersal of Culicoides biting midges. Infect. Genet. Evol. 2014;28:583–587. doi: 10.1016/j.meegid.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 35.Kluiters G, Swales H, Baylis M. Local dispersal of palaearctic Culicoides biting midges estimated by mark-release-recapture. Parasit. Vectors. 2015;8:86. doi: 10.1186/s13071-015-0658-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ducheyne E, et al. Quantifying the wind dispersal of Culicoides species in Greece and Bulgaria. Geospat. Health. 2007;10:177–189. doi: 10.4081/gh.2007.266. [DOI] [PubMed] [Google Scholar]
- 37.Purse BV, et al. Climate change and the recent emergence of bluetongue in Europe. Nat. Rev. Microbiol. 2005;3:171–181. doi: 10.1038/nrmicro1090. [DOI] [PubMed] [Google Scholar]
- 38.Jacquet S, et al. Range expansion of the Bluetongue vector, Culicoides imicola, in continental France likely due to rare wind-transport events. Sci. Rep. 2016 doi: 10.1038/srep27247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rundle HD, Nosil P. Ecological speciation. Ecol. Lett. 2005;8:336–352. doi: 10.1111/j.1461-0248.2004.00715.x. [DOI] [Google Scholar]
- 40.Wang IJ, Bradburd GS. Isolation by environment. Mol. Ecol. 2014;23:5649–5662. doi: 10.1111/mec.12938. [DOI] [PubMed] [Google Scholar]
- 41.Shults, P. A Study of Culicoides Biting Midges in the Subgenus Monoculicoides: Population Genetics, Taxonomy, Systematics, and Control. Ph.D. thesis, Texas A&M University (2021).
- 42.Jewiss-Gaines A, Barelli L, Hunter FF. First records of Culicoides sonorensis (Diptera: Ceratopogonidae), a known vector of bluetongue virus, Southern Ontario. J. Med. Entomol. 2017;54:757–762. doi: 10.1093/jme/tjw215. [DOI] [PubMed] [Google Scholar]
- 43.Chan KM, Levin SA. Leaky prezygotic isolation and porous genomes: rapid introgression of maternally inherited DNA. Evolution. 2005;59:720–729. doi: 10.1111/j.0014-3820.2005.tb01748.x. [DOI] [PubMed] [Google Scholar]
- 44.Harrison RG. Hybrid zones: windows on evolutionary process. Oxf. Surv. Evol. Biol. 1990;7:69–128. [Google Scholar]
- 45.Harrison RG. Animal mitochondrial DNA as a genetic marker in population and evolutionary biology. Trends Ecol. Evol. 1989;4:6–11. doi: 10.1016/0169-5347(89)90006-2. [DOI] [PubMed] [Google Scholar]
- 46.Després L. One, Two or More Species? Mitonuclear Discordance and Species Delimitation. Molecular ecology. 2019;28(17):3845–3847. doi: 10.1111/mec.15211. [DOI] [PubMed] [Google Scholar]
- 47.Janes JK, et al. The K= 2 conundrum. Mol. Ecol. 2017;26:3594–3602. doi: 10.1111/mec.14187. [DOI] [PubMed] [Google Scholar]
- 48.De Meester L, Vanoverbeke J, Kilsdonk LJ, Urban MC. Evolving perspectives on monopolization and priority effects. Trends Ecol. Evol. 2016;31:136–146. doi: 10.1016/j.tree.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 49.Ballard JWO, Chernoff B, James AC. Divergence of mitochondrial DNA is not corroborated by nuclear DNA, morphology, or behavior in Drosophila simulans. Evolution. 2002;56:527–545. doi: 10.1111/j.0014-3820.2002.tb01364.x. [DOI] [PubMed] [Google Scholar]
- 50.Behura S, Sahu S, Mohan M, Nair S. Wolbachia in the Asian rice gall midge, Orseolia oryzae (Wood-Mason): Correlation between host mitotypes and infection status. Insect Mol. Biol. 2001;10:163–171. doi: 10.1046/j.1365-2583.2001.00251.x. [DOI] [PubMed] [Google Scholar]
- 51.Covey H, et al. Cryptic Wolbachia (Rickettsiales: Rickettsiaceae) detection and prevalence in Culicoides (Diptera: Ceratopogonidae) midge populations in the United States. J. Med. Entomol. 2020;57:1262–1269. doi: 10.1093/jme/tjaa003. [DOI] [PubMed] [Google Scholar]
- 52.Pagès N, Muñoz-Muñoz F, Verdún M, Pujol N, Talavera S. First detection of Wolbachia-infected Culicoides (Diptera: Ceratopogonidae) in Europe: Wolbachia and Cardinium infection across Culicoides communities revealed in Spain. Parasit. Vectors. 2017;10:582. doi: 10.1186/s13071-017-2486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pilgrim J, et al. Cardinium symbiosis as a potential confounder of mtDNA based phylogeographic inference in Culicoides imicola (Diptera: Ceratopogonidae), a vector of veterinary viruses. Parasit. Vectors. 2021;14:100. doi: 10.1186/s13071-020-04568-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hare MP. Prospects for nuclear gene phylogeography. Trends Ecol. Evol. 2001;16:700–706. doi: 10.1016/S0169-5347(01)02326-6. [DOI] [Google Scholar]
- 55.Onyango MG, et al. Assessment of population genetic structure in the arbovirus vector midge, Culicoides brevitarsis (Diptera: Ceratopogonidae), using multi-locus DNA microsatellites. Vet. Res. 2015;46:108. doi: 10.1186/s13567-015-0250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fonseca DM, Smith JL, Kim H-C, Mogi M. Population genetics of the mosquito Culex pipiens pallens reveals sex-linked asymmetric introgression by Culex quinquefasciatus. Infect. Genet. Evol. 2009;9:1197–1203. doi: 10.1016/j.meegid.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goubert C, Minard G, Vieira C, Boulesteix M. Population genetics of the Asian tiger mosquito Aedes albopictus, an invasive vector of human diseases. Heredity. 2016;117:125–134. doi: 10.1038/hdy.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lehmann T, et al. Microgeographic structure of Anopheles gambiae in western Kenya based on mtDNA and microsatellite loci. Mol. Ecol. 1997;6:243–253. doi: 10.1046/j.1365-294X.1997.00177.x. [DOI] [PubMed] [Google Scholar]
- 59.Chapuis M-P, Estoup A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2006;24:621–631. doi: 10.1093/molbev/msl191. [DOI] [PubMed] [Google Scholar]
- 60.Manni M, et al. Molecular markers for analyses of intraspecific genetic diversity in the Asian Tiger mosquito, Aedes albopictus. Parasit. Vectors. 2015;8:1–11. doi: 10.1186/s13071-015-0794-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arntzen JW, Jehle R, Bardakci F, Burke T, Wallis GP. Asymmetric viability of reciprocal-cross hybrids between Crested and Marbled Newts (Triturus cristatus and T. marmoratus) Evolution. 2009;63:1191–1202. doi: 10.1111/j.1558-5646.2009.00611.x. [DOI] [PubMed] [Google Scholar]
- 62.Gibeaux R, et al. Paternal chromosome loss and metabolic crisis contribute to hybrid inviability in Xenopus. Nature. 2018;553:337. doi: 10.1038/nature25188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008;6:741. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 64.Servedio MR, Kirkpatrick M. The effects of gene flow on reinforcement. Evolution. 1997;51:1764–1772. doi: 10.1111/j.1558-5646.1997.tb05100.x. [DOI] [PubMed] [Google Scholar]
- 65.Howard, D. J. Reinforcement: origin, dynamics, and fate of an evolutionary hypothesis. Hybrid zones and the evolutionary process, 46–69 (1993).
- 66.Yukilevich R. Asymmetrical patterns of speciation uniquely support reinforcement in Drosophila. Evolution. 2012;66:1430–1446. doi: 10.1111/j.1558-5646.2011.01534.x. [DOI] [PubMed] [Google Scholar]
- 67.Downes JA. The Culicoides variipennis complex: a necessary re-alignment of nomenclature (Diptera: Ceratopogonidae) Can. Entomol. 1978;110:63–69. doi: 10.4039/Ent11063-1. [DOI] [Google Scholar]
- 68.Toews DP, Brelsford A. The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 2012;21:3907–3930. doi: 10.1111/j.1365-294X.2012.05664.x. [DOI] [PubMed] [Google Scholar]
- 69.Smith H, Mullens BA. Seasonal activity, size, and parity of Culicoides occidentalis (Diptera: Ceratopogonidae) in a coastal southern California salt marsh. J. Med. Entomol. 2003;40:352–355. doi: 10.1603/0022-2585-40.3.352. [DOI] [PubMed] [Google Scholar]
- 70.Linley J. The effect of salinity on oviposition and egg hatching in Culicoides variipennis sonorensis (Diptera: Ceratopogonidae) J. Am. Mosq. Control Assoc. 1986;2:79–82. [PubMed] [Google Scholar]
- 71.Gerry AC, Mullens BA. Response of Male Culicoides variipennis sonorensis (Diptera: Ceratopogonidae) to carbon dioxide and observations of mating behavior on and near cattle. J. Med. Entomol. 1998;35:239–244. doi: 10.1093/jmedent/35.3.239. [DOI] [PubMed] [Google Scholar]
- 72.Nolan DV, et al. Rapid diagnostic PCR assays for members of the Culicoides obsoletus and Culicoides pulicaris species complexes, implicated vectors of bluetongue virus in Europe. Vet. Microbiol. 2007;124:82–94. doi: 10.1016/j.vetmic.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 73.Sebastiani F, et al. Molecular differentiation of the Old World Culicoides imicola species complex (Diptera, Ceratopogonidae), inferred using random amplified polymorphic DNA markers. Mol. Ecol. 2001;10:1773–1786. doi: 10.1046/j.0962-1083.2001.01319.x. [DOI] [PubMed] [Google Scholar]
- 74.Carlson D. Identification of mosquitoes of Anopheles gambiae species complex A and B by analysis of cuticular components. Science. 1980;207:1089–1091. doi: 10.1126/science.7355276. [DOI] [PubMed] [Google Scholar]
- 75.Palacios G, et al. Characterization of the Sandfly fever Naples species complex and description of a new Karimabad species complex (genus Phlebovirus, family Bunyaviridae) J. Gen. Virol. 2014;95:292. doi: 10.1099/vir.0.056614-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rivas G, Souza N, Peixoto AA. Analysis of the activity patterns of two sympatric sandfly siblings of the Lutzomyia longipalpis species complex from Brazil. Med. Vet. Entomol. 2008;22:288–290. doi: 10.1111/j.1365-2915.2008.00742.x. [DOI] [PubMed] [Google Scholar]
- 77.Wilson WC, et al. Current status of bluetongue virus in the Americas. Bluetongue. 2009;10:197–220. doi: 10.1016/B978-012369368-6.50013-7. [DOI] [Google Scholar]
- 78.Allen SE, et al. Epizootic Hemorrhagic Disease in White-Tailed Deer, Canada. Emerg. Infect. Dis. 2019;25:832–834. doi: 10.3201/eid2504.180743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McGregor BL, et al. Field data implicating Culicoides stellifer and Culicoides venustus (Diptera: Ceratopogonidae) as vectors of epizootic hemorrhagic disease virus. Parasit. Vectors. 2019;12:258. doi: 10.1186/s13071-019-3514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shults P, Ho A, Martin EM, McGregor BL, Vargo EL. Genetic diversity of Culicoides stellifer (Diptera: Ceratopogonidae) in the Southeastern United States compared with sequences from Ontario, Canada. J. Med. Entomol. 2020;57:1324–1327. doi: 10.1093/jme/tjaa025. [DOI] [PubMed] [Google Scholar]
- 81.Mallet J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 82.Ciota AT, Chin PA, Kramer LD. The effect of hybridization of Culex pipiens complex mosquitoes on transmission of West Nile virus. Parasit. Vectors. 2013;6:1–4. doi: 10.1186/1756-3305-6-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meiswinkel R, Gomulski L, Delécolle J, Goffredo M, Gasperi G. The taxonomy of Culicoides vector complexes-unfinished business. Vet. Ital. 2004;40:151–159. [PubMed] [Google Scholar]
- 84.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics (Oxford, England) 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Andrews, S. Babraham bioinformatics-FastQC a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc (2010).
- 86.Rochette NC, Rivera-Colón AG, Catchen JM. Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol. Ecol. 2019;28:4737–4754. doi: 10.1111/mec.15253. [DOI] [PubMed] [Google Scholar]
- 87.Morales-Hojas R, et al. The genome of the biting midge Culicoides sonorensis and gene expression analyses of vector competence for bluetongue virus. BMC Genomics. 2018;19:624. doi: 10.1186/s12864-018-5014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pante E, et al. Use of RAD sequencing for delimiting species. Heredity. 2015;114:450–459. doi: 10.1038/hdy.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Benestan LM, et al. Conservation genomics of natural and managed populations: building a conceptual and practical framework. Mol. Ecol. 2016;25:2967–2977. doi: 10.1111/mec.13647. [DOI] [PubMed] [Google Scholar]
- 91.Lischer HE, Excoffier L. PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics (Oxford, England) 2012;28:298–299. doi: 10.1093/bioinformatics/btr642. [DOI] [PubMed] [Google Scholar]
- 92.Pina-Martins F, Silva DN, Fino J, Paulo OS. Structure_threader: An improved method for automation and parallelization of programs structure, fastStructure and MavericK on multicore CPU systems. Mol. Ecol. Resour. 2017;17:e268–e274. doi: 10.1111/1755-0998.12702. [DOI] [PubMed] [Google Scholar]
- 93.Raj A, Stephens M, Pritchard JK. Variational Inference of Population Structure in Large SNP Datasets. bioRxiv. 2013;10:001073. doi: 10.1534/genetics.114.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.http://www.R-project.org/ (2013).
- 95.Jombart, Thibaut, and Caitlin Collins. A tutorial for discriminant analysis of principal components (DAPC) using adegenet 2.0. 0. London: Imperial College London, MRC Centre for Outbreak Analysis and Modelling (2015).
- 96.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Leaché AD, Banbury BL, Felsenstein J, De Oca AN-M, Stamatakis A. Short tree, long tree, right tree, wrong tree: New acquisition bias corrections for inferring SNP phylogenies. Syst. Biol. 2015;64:1032–1047. doi: 10.1093/sysbio/syv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pattengale ND, Alipour M, Bininda-Emonds OR, Moret BM, Stamatakis A. How many bootstrap replicates are necessary? J. Comput. Biol. 2010;17:337–354. doi: 10.1089/cmb.2009.0179. [DOI] [PubMed] [Google Scholar]
- 99.Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 30. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 104.Rousset F. genepop’007: a complete re‐implementation of the genepop software for Windows and Linux. Molecular ecology resources. 2010;8(1):103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 105.Rousset F. Genetic differentiation between individuals. J Evol Biol. 2000;13:58–62. doi: 10.1046/j.1420-9101.2000.00137.x. [DOI] [Google Scholar]
- 106.Loiselle BA, Sork VL, Nason J, Graham C. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae) Am. J. Bot. 1995;82:1420–1425. doi: 10.1002/j.1537-2197.1995.tb12679.x. [DOI] [Google Scholar]
- 107.Hardy, O. & Vekemans, X. SPAGeDi 1.5. A program for Spatial Pattern Analysis of Genetic Diversity. User's manual http://ebe.ulb.ac.be/ebe/SPAGeDi_files/SPAGeDi_1.5_Manual.pdf. Université Libre de Bruxelles, Brussells, Belgium.[Google Scholar] (2015).
- 108.Jay F, Sjödin P, Jakobsson M, Blum MG. Anisotropic isolation by distance: the main orientations of human genetic differentiation. Mol. Biol. Evol. 2013;30:513–525. doi: 10.1093/molbev/mss259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Piry S, et al. Mapping Averaged Pairwise Information (MAPI): a new exploratory tool to uncover spatial structure. Methods Ecol. Evol. 2016;7:1463–1475. doi: 10.1111/2041-210X.12616. [DOI] [Google Scholar]
- 110.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxford, England) 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hopken, M. W. Pathogen Vectors at The Wildlife-Livestock Interface: Molecular Approaches to Elucidating Culicoides (Diptera: Ceratopogonidae) Ph.D. thesis, Colorado State University (2016).
- 112.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this study will be deposited in the Open Science Framework database upon acceptance, https://osf.io (10.17605/OSF.IO/E3Z72). Mitochondrial sequences obtained in the current study have been deposited under Genbank Accession Numbers OL604713—OL604779.