Abstract
Although non-coding RNAs have long been considered as non-functional “junk” RNAs, accumulating evidence in the past decade indicates that they play a critical role in pathogenesis of various cancers. In addition to their biological significance, the recognition that their expression levels are frequently dysregulated in multiple cancers have fueled the interest for exploiting their clinical potential as cancer biomarkers. In particular, microRNAs (miRNAs), a subclass of small non-coding RNAs that epigenetically modulate gene-transcription, have become one of the most well-studied substrates for the development of liquid biopsy biomarkers for cancer patients. The emergence of high-throughput sequencing technologies has enabled comprehensive molecular characterisation of various non-coding RNA expression profiles in multiple cancers. Furthermore, technological advances for quantifying lowly expressed RNAs in the circulation have facilitated robust identification of previously unrecognised and undetectable biomarkers in cancer patients. Here we summarise the latest progress on the utilisation of non-coding RNAs as non-invasive cancer biomarkers. We evaluated the suitability of multiple non-coding RNA types as blood-based cancer biomarkers and examined the impact of recent technological breakthroughs on the development of non-invasive molecular biomarkers in cancer.
Subject terms: Biomarkers, Cancer
Introduction
The term ‘liquid biopsy’ refers to sampling and analysis of non-solid biological tissues such as blood, saliva, urine and other bodily fluids for a multitude of clinical applications, including disease diagnostics and post-treatment surveillance and monitoring [1]. Unlike the conventional tissue biopsy (tissue), a liquid biopsy is generally non-invasive (e.g. saliva, urine, stool) or minimally invasive (e.g. blood), and can be used to assess disease and/or health status of organs that are otherwise difficult to access. Over the last two decades, circulating nucleic acids, cell-free DNA (cfDNA) in particular, have been utilised clinically as a liquid biopsy source in multiple diseases including cancers [2–6]. Although identification of genetic alterations residing within the circulating tumour-derived DNA (ctDNA) shed by cancer cells can be detected in circulation, the challenges lie in the scarcity of well-established and recognisable mutations, as well as dearth of mutational burden in early vs. later stages of cancer, that can be easily identified in cfDNA [7]—thus, highlighting one of the limitations for the use of ctDNA as a biomarker source for non-invasive, and early detection of cancers. In contrast to analysis of the mutational burden in ctDNA, other molecular alterations including transcriptional profiles in cell-free RNA (cfRNA), as well epigenetic changes such as DNA methylation, are dynamic events that are known to occur at early stages in carcinogenesis. In particular, non-coding RNAs (ncRNAs) such as microRNAs (miRNAs) have been extensively investigated as a potential source for the development of non-invasive cancer biomarkers. Here we have summarised the latest progress on the major types of ncRNAs as substrates for the development of non-invasive cancer biomarker. Furthermore, we evaluated the suitability of ncRNAs as a liquid biopsy platform vis-à-vis other conventional approaches such as cfDNA. In addition, we summarised the recent technological advances for the quantification of ncRNAs and how these technological refinements will impact the development of non-invasive non-coding RNA-based cancer biomarkers in the near future.
Potential clinical applications of non-coding RNA-based liquid biopsy biomarkers in cancer
Currently, majority of ncRNA-based liquid biopsy biomarker candidates are investigated for diagnostic and screening purposes [4, 8]. Since many of ncRNAs are frequently overexpressed in cancers, they are plausibly suitable for monitoring of cancer progression and recurrence as well. Accordingly, several studies have demonstrated the utility of circulating ncRNAs, including miRNAs and lncRNAs, for cancer monitoring [9, 10]. Furthermore, considering that tissue-based profiling of various ncRNA types in multiple cancers have clarified the functional roles of these novel ncRNAs in cancers [11–13], there is an increasing research interest to understand the molecular profiles of these novel ncRNAs in the circulation for potential blood-based cancer biomarker discovery. In addition, there are multiple ncRNA-based therapeutics, primarily those targeting miRNAs, which are currently being tested in preclinical and early-stage clinical trials [14]. Therefore, it is possible that circulating ncRNAs could be used as companion diagnostic biomarkers for upcoming ncRNA-based therapeutic trials. Although further studies are necessary to evaluate the suitability of ncRNAs in these clinical applications, there is an increasing optimism for the utilisation of ncRNAs in the development of cancer biomarkers.
Non-coding RNA biomarker types
To date, the majority of non-coding RNA (ncRNA) types have been identified in the cell-free component of the blood and many of these RNA types have already been assessed as potential biomarker species in multiple cancers. In general, ncRNAs are divided into two major categories based on their size: small ncRNAs (<200 nucleotides) and long ncRNAs (lncRNA) (>200 nucleotides). In particular, small ncRNAs can be further divided into multiple sub-categories including: microRNAs (miRNAs), transfer RNAs (tRNAs), piwi-interacting RNAs (piRNAs), transcription initiating RNAs (tiRNAs), and enodogenous small interfering RNAs (endo-siRNAs) [15, 16]. In contrast, despite the recognition of pivotal functional role of lncRNAs, these typically fall under a single category, and have not been optimal biomarker substrates due to their stability and degradation concerns. Although many groups have tried to subcategorise lncRNAs based on their genomic location (intergenic, intragenic, bidirectional and enhancers) and the sense in which they are expressed (sense and antisense), these subcategories remain ambiguous and require further evaluations. Although ncRNAs were initially considered as “junk”, accumulating evidence over the past two decades have highlighted their functional importance in various diseases, including cancer. As we improve our understanding for the functional significance of ncRNAs in various biological processes, there is an increasing desire to simultaneously exploit their potential clinical utility for the development of the next-generation of non-invasive cancer biomarkers. Although molecular functionality of different types of ncRNAs may differ significantly, the process by which these biomarkers can be used in the clinic will be quite uniform (Fig. 1). In this section, we will review the potential of various types of ncRNAs as sources for the development of liquid biopsy-based cancer biomarker.
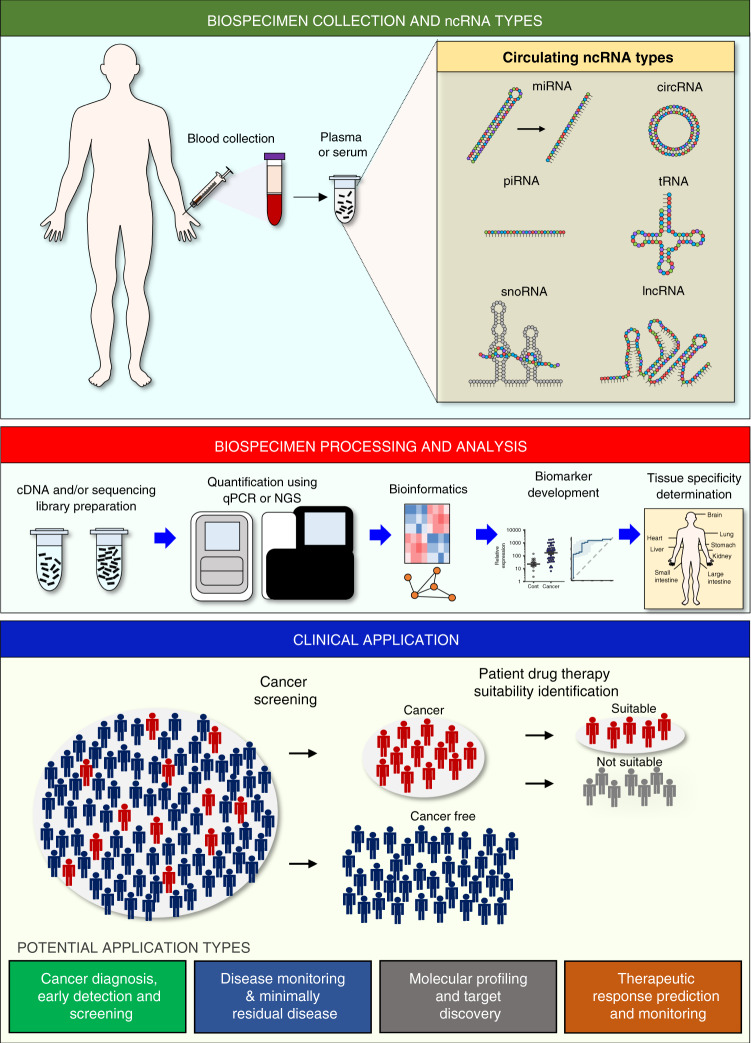
Fig. 1. An overview of blood-based noncoding RNA cancer biomarker development.
(Top) Biospecimen collection and serum/plasma processing. Various types of noncoding RNAs can be found in cell free component of peripheral blood. (Middle) Sample processing and data analysis. cDNA and sequencing libraries are prepared from serum/plasma samples. Targets can be quantified by qPCR or next generation sequencing (NGS). Data are subsequently analysed and bioinformatic processes are utilised. Genes that are differentially expressed between cancer patients and healthy controls are identified (for cancer diagnostic biomarkers) and diagnostic classifiers are developed, and tissue and cancer specificities are assessed. (Bottom) Clinical applications of ncRNA biomarkers. The biomarkers can be utilised in various applications including: Cancer screening and identifying patients that are most likely to respond to a therapeutic agent.
MicroRNAs
microRNAs (miRNAs) are endogenous single-stranded nucleotides, 18–25 in length, that bind to 3′-untranslated regions of a target gene(s), which in turn might regulate multiple cellular processes through modulation of RNA translation. To date, miRNAs are by far the most studied ncRNAs in cancer and are certainly the most well-studied RNA source as liquid biopsies. Due to their high abundance and inherent stability in a variety of bodily fluids (e.g. blood, urine, stool and saliva), miRNAs have been regarded as one of the most promising non-invasive biomarker sources [8, 17, 18]. In view of the fact that the biomarker suitability of miRNAs as non-invasive cancer biomarkers and specific circulating cancer-associated miRNA candidates have been extensively covered previously [4, 8, 14, 19, 20], in this article, we will focus primarily on the overall feasibility of miRNAs as a potential liquid biopsy cancer biomarkers. To date, the most well-recognised cancer-associated circulating miRNAs have been miR-21 and miR-155. Both miRNAs have been shown to act as oncogenes or “onco-miRs” in multiple cancers and are frequently overexpressed in a multitude of cancers [21, 22]. Therefore, not surprisingly, miR-21 and miR-155 are reported to be consistently upregulated in the circulation of cancer patients in multiple cancers types [23–27]. However, the disease specificity of these oncogenic miRNAs remain questionable as both miR-21 and miR-155 have been linked to inflammation and have also been shown to be elevated in a variety of inflammatory diseases [28]. Therefore, one of the key questions for miRNAs as a potential non-invasive biomarker source is whether they possess sufficient disease and tissue-specificity. In addition, with the majority of miRNAs being expressed in most tissues, those that are transcriptionally suppressed in cancer tissues are generally not suitable as blood-based cancer diagnostic biomarkers. While there are several key challenges for miRNAs as a liquid biopsy source, their abundance and tissue stability remain attractive characteristics as a potential resource for the development of non-invasive biomarkers for cancer patients.
Long non-coding RNAs
Long non-coding RNAs (lncRNAs) belong to the groups of ncRNAs that are longer than 200 nucleotides and are recognised to play major roles in multiple biological processes [29]. Furthermore, several lncRNAs appear to have functional roles in the pathogenesis of various cancers, primarily through their ability to regulate key cancer-associated transcriptional activators [30]. In particular, lncRNAs such as HOTAIR, MALAT1 and PVT1 have been shown to be consistently dysregulated in multiple cancers and their oncogenic properties have been validated in subsequent studies [31–33]. While there are more than 60,000 lncRNAs identified from human tumours and cancer cell lines, a large majority of them have yet to be annotated and their functional roles remain unclear [34]. Nevertheless, emerging evidence indicates that lncRNAs are detectable in circulation as cell-free RNAs and appear to be a promising source for non-invasive cancer biomarkers [35]. Moreover, several well-established cancer-associated lncRNAs such as UCA1, MALAT1 and H19 have been shown to be overexpressed in serum and plasma of cancer patients [36–39]. Although further studies are required to determine whether these lncRNAs are truly derived from cancers and can be used as robust cancer biomarkers, identification of key oncogenic circulating RNAs in cancer patients is encouraging. Interestingly, multiple studies have identified the presence of lncRNAs in extracellular vesicles (EVs) in human serum/plasma samples [40–42], suggesting that it is possible for a subset of lncRNAs to be secreted into the circulation inside tumour derived EVs. Collectively, lncRNAs appear to be a promising cancer liquid biopsy source and we need to pay close attention to upcoming studies to fully evaluate their potential.
Circular RNAs
Circular RNAs (circRNA) are a novel class of endogenous ncRNAs that were discovered in RNA viruses in the 1970s, but have recently gained recognition as another class of ncRNAs that are functionally involved in oncogenesis. CircRNAs are RNAs with a loop like structure and possess covalently closed loops with neither 5′ or 3′ polarities nor polyadenylated tails [43, 44]. The unique structure and properties of circRNAs prevent exonuclease digestion and increase their stability in the circulation. Despite that, at present time, not all the biological functions of circRNAs have been elucidated and they are primarily known to act as miRNA sponges in mediating their gene regulatory potential [45, 46]. CircRNAs can be generated from both protein-coding and non-coding genes and some of these can also be derived from putative oncogenes that are typically overexpressed in cancers [47, 48]. To date, several circRNAs have been identified as potential non-invasive cancer biomarker candidates in multiple cancer types [49–52]. In addition, circRNAs have been identified in EVs, such as exosomes [53]. Interestingly, RNA sequencing of exosomal circRNAs revealed that the majority of the ones contained within the exosomes appear to be derived from protein-coding exons [53].
One of the major challenges of adopting circRNAs as a source for liquid biopsy in cancer patients is the difficulty associated with profiling circRNAs using conventional RNA-sequencing technologies. While the classic short-read paired-end RNA sequencing can identify back-spliced junctions of circRNAs, it requires an effective methodology that can profile non-polyadenylated (non-polyA) transcripts [46, 54]. Nevertheless, several groups have optimised sequencing library steps to further improve the quantification of circRNAs; such approaches include utilisation of RNase to digest linear mRNAs [54], use of Ribo-Zero to enrich for circRNAs while depleting rRNAs [55], and utilisation of exome capture probes [56]. These RNA-sequencing modifications have significantly improved the quantification of circRNAs in cancers. Considering that circulating circRNAs have the properties that are highly suitable for liquid biopsy research, there is a great degree of enthusiasm for exploring their potential as non-invasive cancer biomarker candidates.
Piwi-interacting RNAs
Piwi-interacting RNAs (piRNAs) are another class of small ncRNAs that are 26–31 nucleotides in length, and quite comparable in size to miRNAs. However, piRNAs are distinctly different from miRNA counterparts because they lack the sequence conservation present in miRNAs. Furthermore, unlike miRNAs and siRNAs, which are converted from double-stranded RNA precursors into small RNAs by RNase enzymes, piRNAs are derived from an alternative biosynthesis process [57]. Functionally, piRNAs interact with piwi proteins to form RNA-protein complexes that induce epigenetic transcriptional and post-transcriptional gene silencing. In multiple cancers, a large number of dysregulated piRNAs in cancer tissues have already been reported [58, 59]. Accordingly, previous studies have shown that there is a subset of piRNAs which act as either tumour suppressors or oncogenes [60–62]. As a source for cancer liquid biopsy, piRNAs are highly regarded considering that they have similar properties to miRNAs. piRNAs are highly stabile in the circulation and resistant to ribonuclease-mediated degradation. Already several studies have shown that dysregulation of piRNAs in the circulation correlates with cancer diagnosis [63–65]. Overall, piRNAs are a fascinating resource for development as liquid biomarkers in cancer and other diseases, and it will be important to understand how these ncRNAs are released into the circulation by cancers and how this process differs from that for miRNAs.
Transfer RNAs
Transfer RNAs (tRNA) are RNA-derived adaptive molecules that act as a physical link between RNA and the amino acid sequence of proteins. Functionally, tRNAs are known to transport amino acids to ribosomes, a protein synthetic machinery during protein translation. In the circulation tRNAs are typically found as small fragmented forms called “transfer RNA-derived small RNAs (tsRNAs)”, which are further divided into two categories: tRNA fragments (tRFs) and tRNA-derived stress-induced RNAs (tiRNAs). While tRFs are derived from mature or primary tRNAs that are typically 14–30 nt in length, tiRNAs are generated by specific cleavage at the mature tRNA anticodon loop of over 31 nt in length following stress or starvation [66]. tRFs are present in most organisms and are one of the most abundant family of short ncRNAs. tRNAs have been speculated to have higher turnover rates in cancers than normal tissues and their expression levels have been shown to be elevated in cancers under stress [67, 68]. Accordingly, several studies have identified elevation of several tRNAs in serum and plasma as well as urine of cancer patients [69, 70]. Moreover, a study that examined the serum RNA-sequencing profiles of small RNAs showed that the expression of serum tRFs intimately associated with clinicopathologic characteristics in patients with breast cancer [71]. In addition, a recent study identified several exosomal tRFs that are highly enriched in the circulation of liver cancer patients compared to that of healthy controls [72]. Despite the role of tRNAs in cancer needs further clarification, these accumulating evidences suggest that tRNAs could potentially serve as novel cancer diagnostic biomarkers.
Other types of non-coding RNAs-based liquid biopsies
A liquid biopsy is defined as a simple and non-invasive alternative to classical surgical (tissue) biopsies. Considering that blood is the most frequently studied liquid biopsy type, we have focused primarily on blood-based cancer biomarkers in this review. However, the potential of cell-free ncRNAs have been examined in other liquid biopsy substrates including saliva, urine and stool; and these alternative liquid biopsy types may have substantial advantages over blood-based biomarkers depending on the cancer types. As expected, the expression levels of various miRNAs have been extensively evaluated in saliva, urine and stool in multiple cancers [73–75]. While disease specificity of oncogenic miRNAs remains a major hurdle for establishing clinically relevant miRNA-based cancer biomarkers in these liquid biopsy types, the stability and abundance of miRNAs in these sources make them highly attractive cancer biomarker candidates. Currently, the second most evaluated ncRNA source for cancer liquid biopsies are lncRNAs, with several oncogenic lncRNAs that have been detected in saliva [76], urine [77] and stool [78] of cancer patients. Moreover, molecular profiling of piRNAs and circRNAs have been conducted in human saliva [79], demonstrating the feasibility of quantifying these ncRNA types in saliva. Although, the evaluation of ncRNA types as potential non-invasive cancer biomarkers in these liquid biopsy types, excluding miRNA and lncRNA, remain limited, there is an anticipation of increasing utilisation of these novel ncRNAs in multiple liquid biopsy types in the near future.
Comparison of non-coding RNAs to other non-invasive biomarker sources
While there are various types of molecular liquid biopsy sources, their suitability as cancer biomarkers is highly dependent on the specific biomarker types. Based on the current knowledge, we have summarised various molecular sources for liquid biopsy in cancer patients and have highlighted their advantages and disadvantages compared to other types of ncRNAs. The most relevant molecular biomarkers are shown in Fig. 2 and key characteristics of each biomarker sources are also summarised in Table 1.
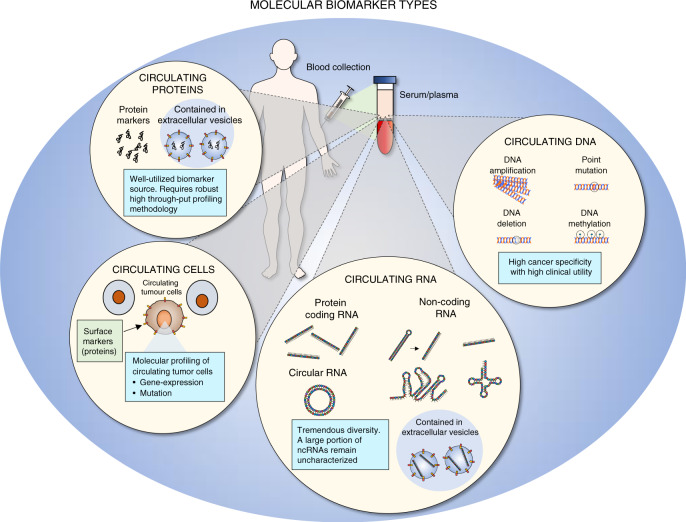
Fig. 2. Molecular biomarker types in cancer.
A list of molecular biomarker types. (Right) Circulating DNA: DNA biomarkers can range from mutation, amplification, deletion and DNA methylation. (Centre-bottom) Circulating RNA: Circulating RNA is primarily divided into coding RNA (messenger RNA) and non-coding RNA (such as miRNAs and lncRNAs). RNAs can also exist as circular RNAs (both variants of coding and non-coding RNAs exist). In addition, RNAs are present in extracellular vesicles such as exosomes. (Centre-left) Circulating tumour cells: Circulating tumour cells can be identified using surface proteins. Furthermore, once isolated, genomic and transcriptomic profile can be examined. (Left) Protein markers have been widely utilised as cancer biomarkers. Proteins exist as free forms or within extracellular vesicles. Improvements in high-through put protein profiling may rejuvenate blood-based cancer protein biomarkers.
Table 1.
Suitability of various molecular sources for developing liquid biopsy biomarkers in cancer.
| Type | Abundance | Stability | Tissue specificity | Functional info availability | Comprehensive profiling feasibility | Present in EV | Published studies* |
|---|---|---|---|---|---|---|---|
| RNA | |||||||
| Non-coding RNA | |||||||
| miRNA | High | High | Intermediate | intermediate | High | High | High (1420) |
| lncRNA | Low | Low | Unclear | Low | Low | Intermediate | Low (135) |
| Circular RNA | Low | High | Unclear | Low | Low | Low | Low (29) |
| piRNA | High | High | Unclear | Low | High | High | Low (8) |
| tRNA | High | High | Unclear | Low | High | High | Low (3) |
| Messenger RNA | Low | Low | High | High | Intermediate | Intermediate | Low (113) |
| DNA | |||||||
| Mutation | Low | Intermediate | Low | intermediate | Intermediate | Low | High (979) |
| Methylation | Intermediate | Intermediate | Intermediate | intermediate | Intermediate | Low | Intermediate (387) |
| Circulating tumour cell | Very low | High | High | Intermediate | Intermediate | No | High (1582) |
| Protein | High | High | High | High | Low | High | High (15,020) |
*Note: Total number of cancer biomarker articles that are published in Pubmed as of 9/13/2021. Pubmed Search Strategy: (serum[ti] OR plasma[ti] OR “non-invasive”[ti] OR “noninvasive”[ti] OR “circulating”[ti] OR “cell free”[ti] OR “liquid biops*“[ti]) AND “Neoplasms”[Mesh] AND “Biomarkers”[Mesh] AND “MicroRNAs”[Mesh]. For other ncRNAs, “MicroRNAs”[Mesh] was replaced by “RNA, Long Noncoding”[Mesh], “RNA, Circular”[Mesh], “RNA, Transfer”[Mesh], “RNA, Messenger”[Mesh], “Mutation”[Mesh], “DNA Methylation”[Mesh], “Neoplastic Cells, Circulating”[Mesh], “Proteins”[Mesh] and (piRNA OR piwi interacting RNA). Reviews, meta-analysis and unrelated publications were excluded from the final count.
Circulating tumour DNA
Circulating tumour DNA (CtDNA) is a tumour-derived fragment of DNA that can be found in the circulation. In most cases, the main analyte of ctDNA is gene mutations that are associated with cancers. ctDNA is highly specific to tumours and studies have shown that it is highly suitable for monitoring of cancer recurrence once the specific mutations have been identified from the tumours [80, 81]. Furthermore, several studies have demonstrated the robustness of ctDNA for identifying late stage cancers [7]. However, the overall clinical application for the use of ctDNA as an early cancer diagnostic biomarker has been limited, primarily because of the fact that the burden of sporadic mutations is invariably quite low in pre-cancerous lesions and even early stage cancers—rendering the sensitivity of any ctDNA based assay a suspect [82]. Therefore, even if the diagnostic resolution of ctDNA quantification improves in the future, this fundamental biological issue will likely be a limiting factor for ctDNA as a cancer diagnostic biomarker source. In comparison, RNA-based liquid biopsy sources are highly dynamic and known to be dysregulated in early stage cancers; hence, implicating their better suitability as early diagnostic cancer biomarkers. Collectively, it is important to understand the molecular characteristics of each biomarker source and evaluate their suitability when developing any specific type of clinical biomarker.
Circulating RNAs
Among RNA-based liquid biopsy sources, both ncRNAs and messenger RNAs (mRNAs) have been investigated extensively in the past. However, even though gene-expression profiling is highly popular in cancer tissue samples, circulating miRNAs are by far the most well-studied liquid biopsy sources in non-invasive cancer biomarkers [4]. This is primarily due to the higher abundance of miRNAs in the circulation compared to mRNAs. In blood, RNase and RNA degrading enzymes are highly abundant, which led to the initial misconception that perhaps cell free RNAs (cfRNAs) are not present in serum and plasma. One of the key mechanisms by which cfRNAs evaded RNase digestion was discovered in 1970s, where a proportion of circulating cfRNAs were shown to be protected from RNase-mediated degradation via their encapsulation within EVs [83]. Subsequent studies have shown that EVs are highly stable from freeze-thaw process and moderate temperature heating [84, 85], thus providing highly protective environment for RNAs inside these tumour-derived vesicles. While EVs are one of the main sources of cfRNAs, certain types of RNAs can be found outside of EVs. miRNAs have been shown to form a stable complex with argonaute (Ago) protein and exist outside of EVs [86, 87]. This was confirmed in a recent study that a high level of miRNAs can be detected outside of EVs, but very few mRNAs were expressed outside of the EVs [88]. Collectively, to date the abundance of miRNAs in the circulation have made them the most well-characterised cfRNA source and subsequently availability for plethora of preexisting datasets are helpful for various stages of cancer biomarker development from target discovery to biomarker validation [89].
Proteins
Circulating protein-based biomarkers are perhaps the most well investigated non-invasive cancer biomarker source and several protein markers are already being utilised clinically as indirect indicators of cancer diagnosis [90]. Since proteins are translated from mRNAs, they are generally regarded to have a similar expression profile to that of the counterpart mRNAs. With recent advances in DNA and RNA sequencing technology platforms, the feasibility of evaluating whole genome and whole transcriptome have improved dramatically. However, one of the major challenges for quantifying proteins in the circulation is the difficulty associated with accurate quantification of the whole proteome. Considering that unbiased transcriptomic and genomic profiling approaches for identifying biomarker candidates have become more pedestrian, it is important to develop approaches that could provide an equivalent quantification platform for a comprehensive proteome profiling. While there are new technologies that allow multiplexing of up to 5000 proteins simultaneously, protein profiling remains limited relative to genomic and transcriptomic approaches.
DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA residues. It is one of the most common and well-characterised epigenetic processes where gene-expression is regulated without a permanent alteration in the DNA sequence itself [16, 91, 92]. Circulating methylated DNA has been evaluated as a potential cancer biomarker source for close to two decades and have been recognised as a promising cancer biomarker source [92, 93]. In this regard, a plethora of data exist for specific DNA methylation sites that are associated with cancers. In particular, SEPT9 is a gene that is frequently methylated in colorectal cancer and circulating methylated SEPT9 has been identified as a potential diagnostic biomarker candidate in multiple cancers [94–96]. Furthermore, recently differentially methylated genes from colorectal cancer tissue samples were quantified in systemic circulation using targeted bisulfite sequencing [97]. This study utilised a large set of circulating methylated genes derived from bisulfite sequencing to develop methylated-gene diagnostic classifiers using machine learning approaches for colorectal cancer.
Intriguingly, emerging evidence indicate that the origin for cfDNA and cfRNA are distinctively different. A recent study showed that only a small fraction of exosomes contain genomic DNA [98]. Considering that the most circulating RNAs exist predominantly within EVs [88], the differences in the origin of circulating DNA and RNA should contribute to the abundance of cancer derived nucleic acids in the circulation. Several studies have postulated that the primary source of cfDNA to be apoptotic cells and bodies [99, 100]. Therefore, understanding the underlying mechanisms by which cancer cells release DNAs and RNAs into circulation will be crucial for the development of non-invasive cancer biomarkers in the future.
Recent advances in circulating ncRNA quantification techniques
Molecular profiling approaches
One of the most significant recent advancements within the field of diagnostic cancer biomarkers is the emergence of high-throughput technological platforms such as next-generation sequencing (NGS) which allows comprehensive quantification of whole genome or transcriptome. There are two key factors that contributed to a large number of groups adapting this technological platform for non-invasive cancer biomarker development. First, wide availability of tissue-based sequencing databases has provided researchers insights into the molecular characteristics of each type of cancer. For most cancer types, there are multiple publicly available large-scale datasets such as TCGA [101–104] and these datasets permit an accurate assessment for the identification of genes that are consistently dysregulated in a specific cancer. Furthermore, based on the cancer tissue expression levels, it is possible to estimate whether the targeted genes can be detected in the circulation. Second, the application of NGS technology for liquid biopsy biomarker development has evolved significantly in the recent years. Over the past decade, the cost of NGS has reduced significantly, allowing sequencing of a larger number of clinical biospecimens, as well as the ability to generate datasets with higher depth of coverage. The latter is particularly important for the development of molecular non-invasive cancer biomarkers, as most of the molecular signals from cancers are typically low in the systemic circulation. Therefore, the opportunity to generate higher resolution data for low expressing transcripts improves the ability to detect rare cancer-derived transcripts in circulation. Furthermore, the emergence of single-cell sequencing technologies has markedly improved the resolution of low-input sequencing. The improvements in low-input sequencing allows accurate quantification of transcriptome for the samples with limited starting nucleic acid template, such as plasma and serum. Collectively, these technological advancements will allow more researchers to conduct comprehensive assessment of molecular profiles in the circulation.
Systematic studies which have utilised molecular profiling of ncRNAs for cancer biomarker development remain limited. There are several publications that have utilised tissue profiling data to identify potential ncRNA biomarker targets and subsequently validated these cancer-associated ncRNA signatures in blood (e.g. plasma and serum) cohorts. A recent study utilised multiple tissue sequencing datasets to identify highly overexpressed miRNAs in gastric cancer tissues and evaluated the robustness of a gastric cancer associated miRNA signature in multiple cohorts [105]. A similar approach was utilised in ovarian cancer, where multiple ovarian cancer tissue and serum datasets were used to establish a miRNA-based diagnostic signature for ovarian cancer and subsequently validated in a prospective collected serum samples [106]. In addition, multiple tissue datasets were used to identify miRNAs that are associated with pancreatic cancer patients with poor prognosis and the prognostic miRNA signature was validated in a serum cohort [107]. Collectively, these studies highlight the effectiveness of utilising tissue datasets for identification of candidates for blood-based cancer biomarkers.
Bioinformatics approaches
One of the major challenges in developing any cancer biomarker is overcoming the inherent heterogeneity associated with cancers. Molecular profiling of various types of cancer tissues indicate that there is no consensus single gene biomarker that can accurately distinguish cancer from healthy tissues. In order to overcome limited sensitivity and specificity of individual genes, multiple biomarkers have been combined to develop robust gene signatures for cancer identification. Nevertheless, analysing a large set of sequencing data is complex and challenging. One of the leading strategies to overcome disease heterogeneity is utilisation of innovative bioinformatic algorithms. Machine learning encompasses a set of computational techniques that are widely utilised to simplify large numbers of measurements into lower-dimensional outputs that are more interpretable [108]. Utilisation of such artificial intelligence-based machine learning approach provides significant advantage over conventional single biomarker or small panel approach for developing cancer biomarkers. Not surprisingly, machine learning algorithms have been increasingly utilised in liquid biopsy biomarker development in a wide range of diseases including cancer [108]. While the utilisation of machine learning approaches will undoubtfully become a more commonplace in the future, there are several fundamental hurdles that need to overcome in order to unleash the full potential of this approach. In particular, the quality of datasets dictates the performance of how well machine learning approaches can be utilised to develop biomarkers. Considering that cancer-derived molecular signals in the circulation are low, it is crucial to improve the overall resolution of the circulating molecular signals in the future.
Functional roles of extracellular vesicle encapsulated noncoding RNAs
Although EVs have been initially considered as a cellular waste disposal mechanism with little biological importance, currently they are being recognised as one of the key cell-cell communication mechanisms that transfer proteins, mRNAs and ncRNAs to target cells [109]. Delivery of genetic material via EVs can be achieved through multiple mechanisms including receptor-ligand interactions, direct fusion of membranes and endocytic internalisation [110]. Once EVs are internalised, the horizontal genetic transfer takes place, sending their content to the cytoplasm of target cells via endosomal fusion. Interestingly, a recent characterisation of extracellular RNA in both healthy individuals and cancer patients revealed that RNA content of EVs were primarily miRNAs and piRNAs (40% respectively), while mRNAs occupied only 2% of total EV content [111]. Therefore, based on the high content of non-coding RNAs in the EVs, they are likely to play an important functional role in EVs.
Currently, one of the most provocative, but well supported, biological hypotheses to date involving EV genetic transfer is promotion of distant cancer metastasis through the formation of a premetastatic niche [112]. Using a rodent model of metastasis, a previous study showed that a subgroup of cancer cell-derived exosomes was taken up by specific organs, altering the molecular profile of the host cells. Subsequently these cancer-derived exosomes redirected metastasis of tumour cells that normally lack the capacity to metastasise to a specific organ. While further validations are needed to confirm these findings, this study brought EV-facilitated genetic material transfer to the forefront of cancer research. Furthermore, the functional role of EV-facilitated RNA transfer was demonstrated in a rodent model of breast cancer [113]. Collectively, these studies highlight the importance of EVs in cancer. Considering that these particles can be found in the circulation and they contain high portion of non-coding RNAs, EVs appear to have an enormous potential to be implemented into cancer biomarker development.
Future challenges
Cancer specificity of ncRNAs
When developing a circulating non-invasive biomarker, specificity for a particular disease and/or organ provides an additional assurance and indicated robustness of that marker for that disease. The biomarkers that are highly specific to a specific type of cancer will reduce the false positive rates, thus making them more informative for clinical diagnosis. In contrast, there are a group of genes that are expressed highly specifically to distinct organs. If these organ-specific genes are dysregulated in cancer, it is possible to identify the location of cancer. Many of cancer mutations, such as KRAS mutations, are rarely found in healthy individuals, thus making them highly cancer-specific. However, since these mutations can occur in multiple cancer types, identification of these mutations in the circulation is insufficient to pinpoint the location of cancer. To date, most of the well-recognisable putative miRNA biomarkers have limited specificity to cancer. However, isoforms of miRNAs (isomiRs) have been postulated to be produced constitutively in human tissues and their expression are dependent on tissue and disease types [114, 115]. The analysis of isomiRs in multiple TCGA datasets showed a subset of such ncRNAs exhibit high tissue specificity [116]. Although the quantification of isomiRs by qPCR is difficult due to their sequence similarity with the (parent) reference sequence, these can be accurately quantified using RNA-sequencing. Considering that RNA sequencing is becoming highly utilised in the biomarker research, we will soon find out the effectiveness of isomiRs as a non-invasive cancer biomarker source.
In addition, accumulating studies indicate that some EVs that are released from specific organs maintain molecular features that are tissue-specific. Previous studies have identified a subset of exosomes with surface proteins that are tissue-specific [117, 118]. Furthermore, several exosomal surface markers that are specific to cancers have also been identified [119, 120]. Therefore, it is possible to measure ncRNA content of tissue-specific or cancer-specific exosomes to further improve cancer specificity as biomarkers. While, a subset of exosomes will be scarce in the circulation, combining the molecular features of exosomes and ncRNA expression inside the exosomes has an enormous potential.
Quantification issues
One of the key issues for the quantification of circulating ncRNAs is the limitations that are associated with normalisation of qPCR-based quantification. This issue has been well documented for circulating miRNAs and remains to be an important issue [8]. Currently, two major protocols have been utilised for the normalisation of miRNAs. First is the use of synthetic spiked-in controls such as cel-miR-39 or ath-miR-159a to normalise the expression of miRNAs in the circulation. This methodology works particularly well, if the exogenous controls are added in a standardised manner. However, the method is limited for older retrospectively collected samples as RNAs will undergo progressive degradation even if they are stored in a −80 °C freezer. In contrast, the use of endogenous controls to normalise the expression levels of miRNAs is also limited since there are no consensus endogenous normalisation controls. While endogenous controls such as miR-16, miR-451 and U6 are used commonly as the endogenous controls to calculate the expression levels of target miRNAs in the circulation, these markers have been shown to be dysregulated in some cancers, thus making them less ideal normalisation controls [121].
One of the potential methodologies to overcome these normalisation issues is to utilise NGS platform to quantify ncRNAs. Typically, RNA-sequencing data is normalised using a different strategy compared to that of qPCR. The most RNA-sequencing data is normalised by counts or reads per million (CPM or RPM). This means that the expression levels of genes are calculated as a relative abundance of all genes that are detected. Furthermore, due to the differences in the length of genes contribute to the expression of genes (i.e. longer genes are more likely to generate more RNA fragments), the expression levels can be expressed as transcripts per million (TPM). TPM is a normalisation method similar to that of CPM and RPM, but normalises the output according to the length of each genes. Collectively, these normalisation methodologies for RNA-sequencing are less likely to be influenced by the fluctuations associated with conventional normalisation methods. Nevertheless, there are issues associated with sequencing-based normalisation methodologies such as not accounting for patients who have higher overall levels of circulating RNAs. Therefore, further assessment is needed to evaluate the best normalisation methodologies for the quantification of circulating transcripts.
Conclusions
In this review, we summarised the current developments in the ncRNAs as a source for the development of non-invasive biomarkers for cancer patients. Although not a long time ago, miRNAs were the only ncRNAs that were viewed as a potential blood-based cancer biomarker source, currently other types of ncRNAs are fast emerging as potential blood-based cancer biomarker sources. Even though hypothesis driven methods have identified several potential cancer diagnostic biomarkers, utilisation of comprehensive molecular profiling allows for a hypothesis-independent biomarker discovery process and such an unbiased approach should result in the identification of novel cancer biomarkers. Although it is still early days for implementing high-throughput profiling methods into non-invasive biomarker development, the evaluation of circulating ncRNA profiles in cancers will provide exciting information for the research community and hopefully will contribute to the development of clinically game-changing non-invasive cancer biomarkers as we embark on enhanced precision oncology efforts.
Acknowledgements
Not applicable
Author contributions
The review article was conceived and wrote by ST and AG.
Funding information
The present work was supported by the grants CA72851, CA181572, CA184792, CA187956 and CA202797 from the National Institute of Health (NIH) to AG.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472–84. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 2.Ignatiadis M, Lee M, Jeffrey SS. Circulating tumor cells and circulating tumor DNA: challenges and opportunities on the path to clinical utility. Clin Cancer Res. 2015;21:4786–4800. doi: 10.1158/1078-0432.CCR-14-1190. [DOI] [PubMed] [Google Scholar]
- 3.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–37. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 4.Toiyama Y, Okugawa Y, Fleshman J, Richard Boland C, Goel A. MicroRNAs as potential liquid biopsy biomarkers in colorectal cancer: A systematic review. Biochim Biophys Acta Rev Cancer. 2018;1870:274–82. doi: 10.1016/j.bbcan.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wada, Y, Shimada, M, Murano, T, Takamaru, H, Morine, Y, Ikemoto, T et al. A liquid biopsy assay for noninvasive identification of lymph node metastases in T1 colorectal cancer. Gastroenterology. 2021;161:151–62.e1. [DOI] [PMC free article] [PubMed]
- 6.Campos-Carrillo A, Weitzel JN, Sahoo P, Rockne R, Mokhnatkin JV, Murtaza M, et al. Circulating tumor DNA as an early cancer detection tool. Pharm Ther. 2020;207:107458. doi: 10.1016/j.pharmthera.2019.107458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shigeyasu K, Toden S, Zumwalt TJ, Okugawa Y, Goel A. Emerging Role of MicroRNAs as Liquid Biopsy Biomarkers in Gastrointestinal Cancers. Clin Cancer Res. 2017;23:2391–9. doi: 10.1158/1078-0432.CCR-16-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326–48. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 10.Volovat SR, Volovat C, Hordila I, Hordila DA, Mirestean CC, Miron OT, et al. MiRNA and LncRNA as Potential Biomarkers in Triple-Negative Breast Cancer: A Review. Front Oncol. 2020;10:526850. doi: 10.3389/fonc.2020.526850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dvinge H, Git A, Gräf S, Salmon-Divon M, Curtis C, Sottoriva A, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–82. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 12.Niknafs YS, Han S, Ma T, Speers C, Zhang C, Wilder-Romans K, et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat Commun. 2016;7:12791. doi: 10.1038/ncomms12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16:129. doi: 10.1186/s12943-017-0696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toden S, Zumwalt TJ, Goel A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875:188491. doi: 10.1016/j.bbcan.2020.188491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12:136–49. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung G, Hernandez-Illan E, Moreira L, Balaguer F, Goel A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020;17:111–30. doi: 10.1038/s41575-019-0230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, et al. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomark Prev. 2010;19:1766–74. doi: 10.1158/1055-9965.EPI-10-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okugawa Y, Toiyama Y, Goel A. An update on microRNAs as colorectal cancer biomarkers: where are we and what’s next? Expert Rev Mol Diagn. 2014;14:999–1021. doi: 10.1586/14737159.2014.946907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed Res Int. 2015;2015:125094. doi: 10.1155/2015/125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brase JC, Wuttig D, Kuner R, Sültmann H. Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer. 2010;9:306. doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 22.Tili E, Croce CM, Michaille JJ. miR-155: on the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28:264–84. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- 23.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849–59. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abue M, Yokoyama M, Shibuya R, Tamai K, Yamaguchi K, Sato I, et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int J Oncol. 2015;46:539–47. doi: 10.3892/ijo.2014.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Wang M, Lin G, Sun S, Li X, Qi J, et al. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS ONE. 2012;7:e47003. doi: 10.1371/journal.pone.0047003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu R, Liao J, Yang M, Shi Y, Peng Y, Wang Y, et al. Circulating miR-155 expression in plasma: a potential biomarker for early diagnosis of esophageal cancer in humans. J Toxicol Environ Health A. 2012;75:1154–62. doi: 10.1080/15287394.2012.699856. [DOI] [PubMed] [Google Scholar]
- 27.Shi C, Yang Y, Xia Y, Okugawa Y, Yang J, Liang Y, et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut. 2016;65:1470–81. doi: 10.1136/gutjnl-2014-308455. [DOI] [PubMed] [Google Scholar]
- 28.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–8. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 29.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–63. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Gao Y, Wang X, Li L, Zhang J, Zhang L, et al. Circulating lncRNAs as noninvasive biomarkers in bladder cancer: A diagnostic meta-analysis based on 15 published articles. Int J Biol Markers. 2020;35:40–48. doi: 10.1177/1724600820926685. [DOI] [PubMed] [Google Scholar]
- 36.Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng ZK, Pang C, Yang Y, Duan Q, Zhang J, Liu WC. Serum long noncoding RNA urothelial carcinoma-associated 1: A novel biomarker for diagnosis and prognosis of hepatocellular carcinoma. J Int Med Res. 2018;46:348–56. doi: 10.1177/0300060517726441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Q, Su Y, He X, Zhao W, Wu C, Zhang W, et al. Plasma long non-coding RNA MALAT1 is associated with distant metastasis in patients with epithelial ovarian cancer. Oncol Lett. 2016;12:1361–6. doi: 10.3892/ol.2016.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–9. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yousefi H, Maheronnaghsh M, Molaei F, Mashouri L, Reza Aref A, Momeny M, et al. Long noncoding RNAs and exosomal lncRNAs: classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene. 2020;39:953–74. doi: 10.1038/s41388-019-1040-y. [DOI] [PubMed] [Google Scholar]
- 41.Zang X, Gu J, Zhang J, Shi H, Hou S, Xu X, et al. Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 2020;11:215. doi: 10.1038/s41419-020-2409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lane, JS, Hoff, DV, Cridebring, D & Goel, A. Extracellular vesicles in diagnosis and treatment of pancreatic cancer: current state and future perspectives. Cancers (Basel) 2020;12:1530. [DOI] [PMC free article] [PubMed]
- 43.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–8. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–6. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–8. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 46.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 47.Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, et al. Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer Res. 2017;77:2339–50. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–19. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 49.Hang D, Zhou J, Qin N, Zhou W, Ma H, Jin G, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7:2783–91. doi: 10.1002/cam4.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin J, Cai D, Li W, Yu T, Mao H, Jiang S, et al. Plasma circular RNA panel acts as a novel diagnostic biomarker for colorectal cancer. Clin Biochem. 2019;74:60–68. doi: 10.1016/j.clinbiochem.2019.10.012. [DOI] [PubMed] [Google Scholar]
- 51.Chen J, Cui L, Yuan J, Zhang Y, Sang H. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem Biophys Res Commun. 2017;494:126–32. doi: 10.1016/j.bbrc.2017.10.068. [DOI] [PubMed] [Google Scholar]
- 52.Li Z, Zhou Y, Yang G, He S, Qiu X, Zhang L, et al. Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin Chim Acta. 2019;492:37–44. doi: 10.1016/j.cca.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–4. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giannoukos G, Ciulla DM, Huang K, Haas BJ, Izard J, Levin JZ, et al. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol. 2012;13:R23. doi: 10.1186/gb-2012-13-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Wu YM, et al. The Landscape of Circular RNA in Cancer. Cell. 2019;176:869–81. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Czech B, Hannon GJ. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci. 2016;41:324–37. doi: 10.1016/j.tibs.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Dou M, Song X, Dong Y, Liu S, Liu H, et al. The emerging role of the piRNA/piwi complex in cancer. Mol Cancer. 2019;18:123. doi: 10.1186/s12943-019-1052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weng W, Li H, Goel A. Piwi-interacting RNAs (piRNAs) and cancer: Emerging biological concepts and potential clinical implications. Biochim Biophys Acta Rev Cancer. 2019;1871:160–9. doi: 10.1016/j.bbcan.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505:353–9. doi: 10.1038/nature12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe T, Lin H. Posttranscriptional regulation of gene expression by Piwi proteins and piRNAs. Mol Cell. 2014;56:18–27. doi: 10.1016/j.molcel.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weng W, Liu N, Toiyama Y, Kusunoki M, Nagasaka T, Fujiwara T, et al. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol Cancer. 2018;17:16. doi: 10.1186/s12943-018-0767-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mai D, Zheng Y, Guo H, Ding P, Bai R, Li M, et al. Serum piRNA-54265 is a new biomarker for early detection and clinical surveillance of human colorectal cancer. Theranostics. 2020;10:8468–78. doi: 10.7150/thno.46241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qu A, Wang W, Yang Y, Zhang X, Dong Y, Zheng G, et al. A serum piRNA signature as promising non-invasive diagnostic and prognostic biomarkers for colorectal cancer. Cancer Manag Res. 2019;11:3703–20. doi: 10.2147/CMAR.S193266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu X, Wang C, Deng H, Qing C, Liu R, Liu S, et al. Exosomal piRNA profiling revealed unique circulating piRNA signatures of cholangiocarcinoma and gallbladder carcinoma. Acta Biochim Biophys Sin (Shanghai) 2020;52:475–84. doi: 10.1093/abbs/gmaa028. [DOI] [PubMed] [Google Scholar]
- 66.Jia Y, Tan W, Zhou Y. Transfer RNA-derived small RNAs: potential applications as novel biomarkers for disease diagnosis and prognosis. Ann Transl Med. 2020;8:1092. doi: 10.21037/atm-20-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–42. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 69.Gehrke CW, Kuo KC, Waalkes TP, Borek E. Patterns of urinary excretion of modified nucleosides. Cancer Res. 1979;39:1150–3. [PubMed] [Google Scholar]
- 70.Lakings DB, Waalkes TP, Borek E, Gehrke CW, Mrochek JE, Longmore J, et al. Composition, associated tissue methyltransferase activity, and catabolic end products of transfer RNA from carcinogen-induced hepatoma and normal monkey livers. Cancer Res. 1977;37:285–92. [PubMed] [Google Scholar]
- 71.Dhahbi JM, Spindler SR, Atamna H, Boffelli D, Martin DI. Deep Sequencing of Serum Small RNAs Identifies Patterns of 5’ tRNA Half and YRNA Fragment Expression Associated with Breast Cancer. Biomark Cancer. 2014;6:37–47. doi: 10.4137/BIC.S20764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu L, Li J, Gong Y, Wu Q, Tan S, Sun D, et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol Cancer. 2019;18:74. doi: 10.1186/s12943-019-1000-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoshino I. The usefulness of microRNA in urine and saliva as a biomarker of gastroenterological cancer. Int J Clin Oncol. 2021;26:1431–40. doi: 10.1007/s10147-021-01911-1. [DOI] [PubMed] [Google Scholar]
- 74.Li L, Wang A, Cai M, Tong M, Chen F, Huang L. Identification of stool miR-135b-5p as a non-invasive diaognostic biomarker in later tumor stage of colorectal cancer. Life Sci. 2020;260:118417. doi: 10.1016/j.lfs.2020.118417. [DOI] [PubMed] [Google Scholar]
- 75.Duran-Sanchon S, Moreno L, Gómez-Matas J, Augé JM, Serra-Burriel M, Cuatrecasas M, et al. Fecal microRNA-based algorithm increases effectiveness of fecal immunochemical test-based screening for colorectal cancer. Clin Gastroenterol Hepatol. 2021;19:323–30. doi: 10.1016/j.cgh.2020.02.043. [DOI] [PubMed] [Google Scholar]
- 76.Xie Z, Chen X, Li J, Guo Y, Li H, Pan X, et al. Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget. 2016;7:25408–19. doi: 10.18632/oncotarget.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhan Y, Du L, Wang L, Jiang X, Zhang S, Li J, et al. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol Cancer. 2018;17:142. doi: 10.1186/s12943-018-0893-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gharib E, Nazemalhosseini-Mojarad E, Baghdar K, Nayeri Z, Sadeghi H, Rezasoltani S, et al. Identification of a stool long non-coding RNAs panel as a potential biomarker for early detection of colorectal cancer. J Clin Lab Anal. 2021;35:e23601. doi: 10.1002/jcla.23601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221–30. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vidal J, Muinelo L, Dalmases A, Jones F, Edelstein D, Iglesias M, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol. 2017;28:1325–32. doi: 10.1093/annonc/mdx125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kruger S, Heinemann V, Ross C, Diehl F, Nagel D, Ormanns S, et al. Repeated mutKRAS ctDNA measurements represent a novel and promising tool for early response prediction and therapy monitoring in advanced pancreatic cancer. Ann Oncol. 2018;29:2348–55. doi: 10.1093/annonc/mdy417. [DOI] [PubMed] [Google Scholar]
- 82.Ruiz-Banobre J, Goel A. Genomic and epigenomic biomarkers in colorectal cancer: from diagnosis to therapy. Adv Cancer Res. 2021;151:231–304. doi: 10.1016/bs.acr.2021.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stroun M, Anker P, Beljanski M, Henri J, Lederrey C, Ojha M, et al. Presence of RNA in the nucleoprotein complex spontaneously released by human lymphocytes and frog auricles in culture. Cancer Res. 1978;38:3546–54. [PubMed] [Google Scholar]
- 84.Sokolova V, Ludwig AK, Hornung S, Rotan O, Horn PA, Epple M, et al. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf B Biointerfaces. 2011;87:146–50. doi: 10.1016/j.colsurfb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 85.Kalra H, Adda CG, Liem M, Ang CS, Mechler A, Simpson RJ, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. 2013;13:3354–64. doi: 10.1002/pmic.201300282. [DOI] [PubMed] [Google Scholar]
- 86.Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 2011;8:1149–57. doi: 10.4161/rna.8.6.17665. [DOI] [PubMed] [Google Scholar]
- 87.Fuji T, Umeda Y, Nyuya A, Taniguchi F, Kawai T, Yasui K, et al. Detection of circulating microRNAs with Ago2 complexes to monitor the tumor dynamics of colorectal cancer patients during chemotherapy. Int J Cancer. 2019;144:2169–80. doi: 10.1002/ijc.31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Enderle D, Spiel A, Coticchia CM, Berghoff E, Mueller R, Schlumpberger M, et al. Characterization of RNA from Exosomes and Other Extracellular Vesicles Isolated by a Novel Spin Column-Based Method. PLoS ONE. 2015;10:e0136133. doi: 10.1371/journal.pone.0136133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen, Y, Wu, T, Zhu, Z, Huang, H, Zhang, L, Goel, A et al. An integrated workflow for biomarker development using microRNAs in extracellular vesicles for cancer precision medicine. Semin Cancer Biol 2021;74:134–55. [DOI] [PubMed]
- 90.Chatterjee SK, Zetter BR. Cancer biomarkers: knowing the present and predicting the future. Future Oncol. 2005;1:37–50. doi: 10.1517/14796694.1.1.37. [DOI] [PubMed] [Google Scholar]
- 91.Goel A, Boland CR. Epigenetics of colorectal cancer. Gastroenterology. 2012;143:1442–60. doi: 10.1053/j.gastro.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015;149:1204–25. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lam K, Pan K, Linnekamp JF, Medema JP, Kandimalla R. DNA methylation based biomarkers in colorectal cancer: A systematic review. Biochim Biophys Acta. 2016;1866:106–20. doi: 10.1016/j.bbcan.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 94.Toyota M, Ho C, Ahuja N, Jair KW, Li Q, Ohe-Toyota M, et al. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307–12. [PubMed] [Google Scholar]
- 95.Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317–25. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–32. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 97.Luo, H, Zhao, Q, Wei, W, Zheng, L, Yi, S, Li, G et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med.2020;12:eaax7533. [DOI] [PubMed]
- 98.Yokoi A, Villar-Prados A, Oliphint PA, Zhang J, Song X, De Hoff P, et al. Mechanisms of nuclear content loading to exosomes. Sci Adv. 2019;5:eaax8849. doi: 10.1126/sciadv.aax8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lui YY, Chik KW, Chiu RW, Ho CY, Lam CW, Lo YM. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin Chem. 2002;48:421–7. [PubMed] [Google Scholar]
- 100.Lui YY, Woo KS, Wang AY, Yeung CK, Li PK, Chau E, et al. Origin of plasma cell-free DNA after solid organ transplantation. Clin Chem. 2003;49:495–6. doi: 10.1373/49.3.495. [DOI] [PubMed] [Google Scholar]
- 101.Cancer Genome Atlas Research, N., Analysis Working Group: Asan, U., Agency, B. C. C., Brigham, Women’s, H., Broad, I. et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–75. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Network CGAR. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Izumi D, Zhu Z, Chen Y, Toden S, Huo X, Kanda M, et al. Assessment of the diagnostic efficiency of a liquid biopsy assay for early detection of gastric cancer. JAMA Netw Open. 2021;4:e2121129. doi: 10.1001/jamanetworkopen.2021.21129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kandimalla R, Wang W, Yu F, Zhou N, Gao F, Spillman M, et al. OCaMIR-A noninvasive, diagnostic signature for early-stage ovarian cancer: a multi-cohort retrospective and prospective study. Clin Cancer Res. 2021;27:4277–86. doi: 10.1158/1078-0432.CCR-21-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kandimalla, R, Shimura, T, Mallik, S, Sonohara, F, Tsai, S, Evans, DB et al. Identification of serum miRNA signature and establishment of a nomogram for risk stratification in patients with pancreatic ductal adenocarcinoma. Ann Surg. 2020; 10.1097/sla.0000000000003945. [DOI] [PMC free article] [PubMed]
- 108.Ko J, Baldassano SN, Loh PL, Kording K, Litt B, Issadore D. Machine learning to detect signatures of disease in liquid biopsies - a user’s guide. Lab Chip. 2018;18:395–405. doi: 10.1039/c7lc00955k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 110.Tian T, Wang Y, Wang H, Zhu Z, Xiao Z. Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. J Cell Biochem. 2010;111:488–96. doi: 10.1002/jcb.22733. [DOI] [PubMed] [Google Scholar]
- 111.Yuan T, Huang X, Woodcock M, Du M, Dittmar R, Wang Y, et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci Rep. 2016;6:19413. doi: 10.1038/srep19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–35. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nabet BY, Qiu Y, Shabason JE, Wu TJ, Yoon T, Kim BC, et al. Exosome RNA Unshielding Couples Stromal Activation to Pattern Recognition Receptor Signaling in Cancer. Cell. 2017;170:352–66. doi: 10.1016/j.cell.2017.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Loher P, Londin ER, Rigoutsos I. IsomiR expression profiles in human lymphoblastoid cell lines exhibit population and gender dependencies. Oncotarget. 2014;5:8790–802. doi: 10.18632/oncotarget.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Telonis AG, Loher P, Jing Y, Londin E, Rigoutsos I. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res. 2015;43:9158–75. doi: 10.1093/nar/gkv922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Telonis AG, Magee R, Loher P, Chervoneva I, Londin E, Rigoutsos I. Knowledge about the presence or absence of miRNA isoforms (isomiRs) can successfully discriminate amongst 32 TCGA cancer types. Nucleic Acids Res. 2017;45:2973–85. doi: 10.1093/nar/gkx082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteom: MCP. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vallabhajosyula P, Korutla L, Habertheuer A, Yu M, Rostami S, Yuan CX, et al. Tissue-specific exosome biomarkers for noninvasively monitoring immunologic rejection of transplanted tissue. J Clin Invest. 2017;127:1375–91. doi: 10.1172/JCI87993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–21. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Castillo J, Bernard V, San Lucas FA, Allenson K, Capello M, Kim DU, et al. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann Oncol. 2018;29:223–9. doi: 10.1093/annonc/mdx542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xiang M, Zeng Y, Yang R, Xu H, Chen Z, Zhong J, et al. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun. 2014;454:210–4. doi: 10.1016/j.bbrc.2014.10.064. [DOI] [PubMed] [Google Scholar]