Abstract
Background
Targeted sequencing of circulating tumour DNA (ctDNA) is a promising tool to monitor dynamic changes in the variant allele frequencies (VAF) of genomic alterations and predict clinical outcomes in patients with advanced urothelial carcinoma (UC).
Methods
We performed targeted sequencing of 182 serial ctDNA samples from 53 patients with advanced UC.
Results
Serial ctDNA-derived metrics predicted the clinical outcomes in patients with advanced UC. Combining serial ctDNA aggregate VAF (aVAF) values with clinical factors, including age, sex, and liver metastasis, improved the performance of prognostic models. An increase of the ctDNA aVAF by ≥1 in serial ctDNA samples predicted disease progression within 6 months in 90% of patients. The majority of patients with aVAFs ≤0.7 in three consecutive ctDNA samples achieved durable clinical responses (≥6 months).
Conclusions
Serial ctDNA analysis predicts disease progression and enables dynamic monitoring to guide precision medicine in patients with advanced UC.
Subject terms: Molecular medicine, Urological cancer
Introduction
Urothelial carcinoma (UC) is known to have a high tumour mutational burden (TMB) [1, 2]. Using whole-exome sequencing of paired primary-metastatic tumour samples, we previously discovered a significant degree of clonal heterogeneity and continuous evolution in patients with advanced UC [3]. However, biopsies of metastatic lesions are invasive and are associated with potential complications. Tumour cells shed small fragments of circulating tumour DNA (ctDNA) which are 120–200 base pairs in length, into the circulation [4, 5]. Several methods have been developed to interrogate the ctDNA in plasma obtained from cancer patients [4, 6, 7]. UC is among the cancer types with the highest detection rates of ctDNA (range 75–90%) [8]. The value of ctDNA has been explored in the clinically localised stages of UC using targeted sequencing, personalised tumour-specific ddPCR, or tumour-informed multiplexed PCR assays to predict recurrence or progression to invasive disease [9–11]. A cross-sectional analysis of genomic alterations (GAs) in ctDNA samples from 369 patients with advanced UC was previously published [8]. Longitudinal monitoring of ctDNA has demonstrated promising prognostic and predictive values in patients with various cancer types [4, 12–16]. Exploratory analyses from early phase clinical trials revealed that changes in ctDNA VAF were associated with survival outcomes and response to targeted therapeutics and immune checkpoint blockade in patients with UC [17–20]. In the current study, we evaluated the utility of serial ctDNA measurements to predict clinical progression and map the evolutionary trajectories of advanced UC.
Methods
Patients and clinical samples
We evaluated a cohort of 53 patients with advanced UC from two institutions (Weill Cornell Medicine and the Comprehensive Cancer Centers of Nevada). Analysis of plasma samples using Guardant360® was approved by the Western Institutional Review Board (Protocol No. 20152817). A total of 182 serial individual plasma samples were collected, and ctDNA was analysed using the Guardant360® NGS assay (Guardant Health, Redwood City, CA). The testing was performed in a CLIA-certified, CAP-accredited laboratory and included analysis of 73 genes for single-nucleotide variants (SNVs), insertion-deletion mutations (indels), fusions, and amplifications [21] (Supplementary Table 1). Clinical data, including age, presence of visceral or liver metastasis, prior lines of treatment, response status to active anti-cancer treatment, and overall survival, were collected. Overall survival was measured from the time of diagnosis of invasive urothelial cancer to death or last follow-up. Non-progressive disease (non-PD) was defined as stable disease, partial response, or complete response based on restaging scans done within four weeks of the corresponding ctDNA sample. Scans were assessed at each study site using RECIST v1.1 criteria by the study investigators. ctDNA samples with corresponding radiographic scans outside the four-week window were excluded from the final analysis.
Sequencing and analysis
A total of 182 serial individual plasma samples were collected, and ctDNA was analysed using the Guardant360® NGS commercial assay (Guardant Health, Redwood City, CA). The testing was performed in a CLIA-certified, CAP-accredited laboratory and included analysis of up to 73 genes for single-nucleotide variants (SNVs), insertions-deletion mutations (indels), fusions, and amplifications. The methods of the Guardant360® test have been previously described in detail [21]. Briefly, patient samples are collected in two 10 ml Streck Cell-Free DNA blood collection tubes and sent to Guardant Health for analysis. Cell-free DNA is extracted from the plasma aliquots. Extracted cfDNA is enriched by hybrid capture (Agilent Technologies), pooled, and sequenced using paired-end synthesis (NextSeq 500 or HiSeq 2500). The average depth of coverage is 15,000×, and sequencing reads are mapped to hg19/GRCh37. A computational pipeline was trained and validated for variant detection with a detection limit of 0.04% for SNVs and 0.02% for indels. Six patients had their FFPE tumour samples undergo whole-exome sequence (WES) as previously described [7, 22]. Select FFPE tumours were tested using Oncomine (143 genes) and Caris Molecular Intelligence (592 genes) targeted sequencing panels [23, 24] (Supplementary Notes). We used several sources to curate a list of therapeutically actionable genomic alterations, including the COSMIC cancer census list https://cancer.sanger.ac.uk/cosmic and https://www.mycancergenome.org/. In addition, we used two precision oncology databases, PMKB (https://pmkb.weill.cornell.edu/) [25] and OncoKB (https://www.oncokb.org/) [26], to identify oncogenic actionable GAs with strong clinical evidence in patients with UC, i.e., levels 1–3 in OncoKB and tier 1 in PMKB. We included GAs that were annotated as oncogenic or likely oncogenic in OncoKB.
Statistical analysis
The comparison of categorical variables was performed using Fisher’s exact test. The comparison of continuous variables among responders and non-responders was performed using the non-parametric Wilcoxon rank-sum (Mann–Whitney) test. The ctDNA aggregate VAF (aVAF) was defined as the sum of VAF of all identified genomic alterations in a given sample. The ctDNA delta aVAF was calculated by subtracting the ctDNA aVAF of the second sample from the ctDNA aVAF of the previously collected sample within a three-month period. Figures and analyses were produced using GraphPad Prism 8.3. Overall survival was estimated from the time of diagnosis to the time of death or loss to follow-up. Overall survival was analysed using Kaplan–Meier curves. The equality of two survivor functions was tested using the Log-rank test, and the hazard ratio was reported using the Mantel–Haenszel method. Lollipops of genomic alterations were produced using the ProteinPaint tool (https://pecan.stjude.cloud/proteinpaint).
Results
Patient characteristics and study design
The clinical characteristics of patients in our cohort are outlined in Table 1 and Supplementary Table 2. The median age was 69 years (range 38–88). 39 (74%) were men. A median of 3 (range 1–10) ctDNA samples per patient were collected. Distant metastases were found in 39 patients (74%), and 24 patients (45%) had visceral metastases. The median number of treatment lines per patient was 2 (range 0-6). A total of 36/53 (68%) patients received first-line platinum-based chemotherapy (Table 1). The median overall survival was 70 months from the time of the initial diagnosis.
Table 1.
Baseline patient characteristics.
| N (%) | |
|---|---|
| Sex | |
| Male | 39 (74%) |
| Female | 14 (26%) |
| Median Age, years (range) | 69 (38–88) |
| Location | |
| Bladder | 48 (91%) |
| Upper tract | 5 (9%) |
| Highest Stage (AJCC 8th edition) | |
| Stage ≤3 | 12 (23%) |
| Stage 4 | 41 (77%) |
| Lymph node metastasis | |
| Yes | 40 (76%) |
| No | 13 (24%) |
| Distant metastasis | |
| Yes | 39 (74%) |
| No | 14 (26%) |
| Visceral metastasis | |
| Yes | 24 (45%) |
| No | 29 (55%) |
| Liver metastasis | |
| Yes | 15 (28%) |
| No | 38 (72%) |
| Lung metastasis | |
| Yes | 17 (32%) |
| No | 36 (68%) |
| Non-UC active malignancy | |
| Localised Prostate Cancer (GS ≤ 7) | 4 (7.5%) |
| CLL | 1 (1.9%) |
| Low-grade NHL | 1 (1.9%) |
| Breast Cancer | 1 (1.9%) |
| None | 1 (1.9%) |
| 46 (87%) | |
| Primary tumour removed | |
| Yes | 32 (60%) |
| No | 21 (40%) |
| Response to first-line therapy | |
| Non-PD | 22 (49%) |
| PD | 23 (51%) |
| NA | 8 |
| Vital status at last follow-up | |
| Live | 36 (68%) |
| Dead | 17 (32%) |
UC urothelial carcinoma, GS Gleason score, CLL chronic lymphocytic leukaemia, NHL non-Hodgkin lymphoma, PD progressive disease, Non-PD non-progressive disease, NA no available data.
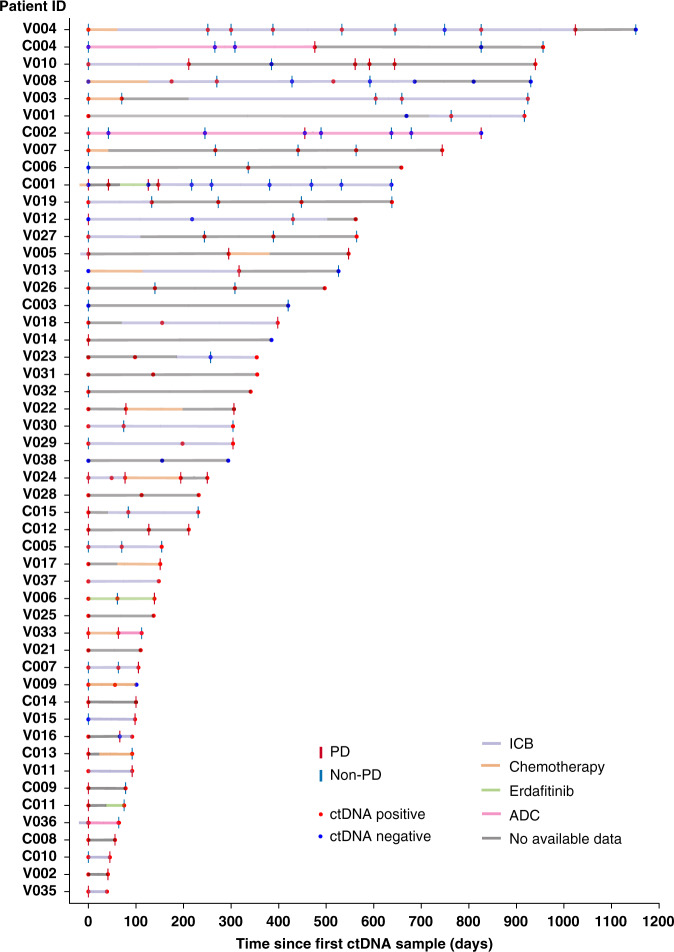
The study included 53 patients diagnosed with advanced UC, including patients with nodal and distant metastases. A total of 182 serial individual plasma samples were collected for ctDNA analysis between 2015 and 2020 (Fig. 1). An individual plasma sample was considered ctDNA positive if one or more GAs were identified. Using this definition, 76% (138/182) of all plasma samples were ctDNA positive (Fig. 1). The most common active therapies received proximate to ctDNA collection were immune checkpoint blockade (ICB) (35%), antibody–drug conjugates (ADC) (9.3%), and platinum-based (8.2%) (Fig. 1). The median follow-up period following the collection of the first ctDNA sample was 185 (range 42–1151) days. During the follow-up period, 45 PD and 82 non-PD events were documented on radiographic scans within 4 weeks from collecting the respective ctDNA sample (Fig. 1). Overall, 462 different GAs were identified in 58 genes in 138/182 (75.8%) of ctDNA samples. The landscape of ctDNA genomic alterations (GAs) identified in patients with advanced UC is described in Supplementary Notes and Supplementary Fig. 1.
Fig. 1. Swimmer plot showing the disease course in each patient, treatment, ctDNA status, and radiographic response.
Red dots represent positive ctDNA samples, blue dots represent a negative ctDNA samples, vertical red lines represent PD, and vertical blue lines represent non-PD. ICB: immune checkpoint blockade, ADC: antibody–drug conjugates.
Serial ctDNA sampling identifies additional actionable GAs
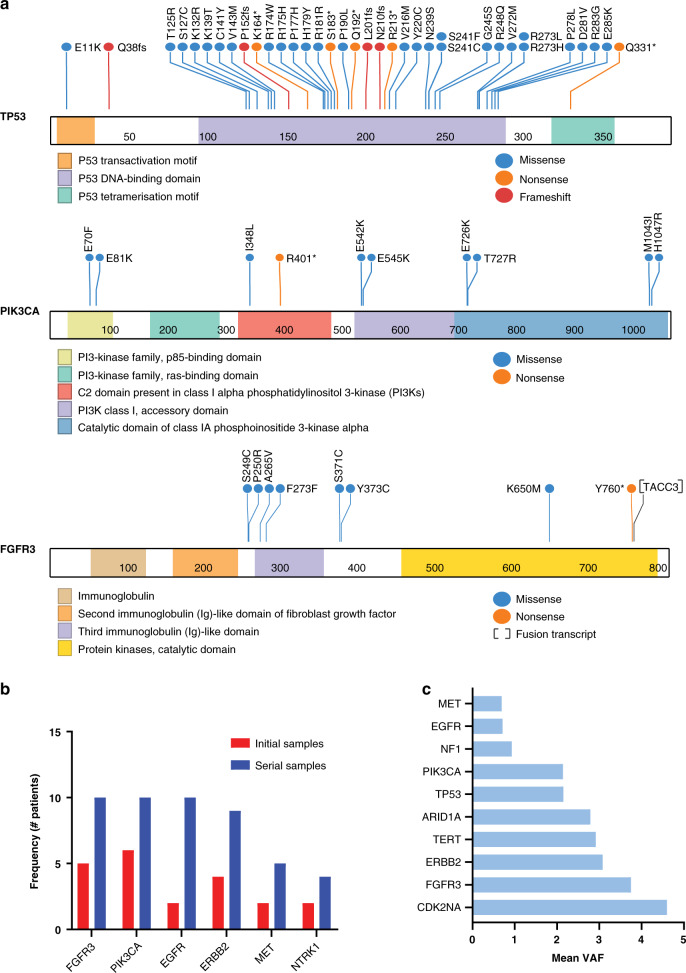
The genes most commonly harbouring GAs were TP53 (64%), BRCA1/2 (22.6%), PIK3CA, EGFR, and ERBB2 (each in 19% of patients) (Fig. 2a). FGFR3 GAs were identified in 10 patients (19%, two patients with FGFR3-TACC3 fusions, and 8 with SNVs, including two novel mutations P250R, and Y760*), that were not previously reported in the COSMIC database (Fig. 2a and Supplementary Table 3). Actionable GAs with US FDA-approved targeted agents were identified in 15 genes. The median frequency of patients harbouring actionable GAs in each gene was 7.5% (range 2–19%) (Supplementary Table 4). Frequently co-occurring actionable GAs were observed (Supplementary Table 5). ERBB2 GAs frequently co-occurred with either PIK3CA (11.3%) or ARID1A GAs (9.4%). We defined actionable oncogenic GAs in patients with advanced UC as those from tier 1 or 2 in our Precision Medicine Knowledgebase PMKB [25] or levels 1–3 in the OncoKB database [26]. We identified 19 GAs in 6 genes, including FGFR3, ERBB2, BRCA2, TSC1, HRAS, and NTRK1, affecting 30% (16/53) of patients (Supplementary Fig. 2). Patients with UC harbouring these GAs were shown to respond to the corresponding targeted agents in clinical studies [17, 25–28]. In addition, somatic ctDNA GAs in the BRCA1/2 and ATM homologous recombination repair genes were identified in 32% (17/53) of patients. Using the OncoKB [26] and Findlay et al. [29] functional annotations, six patients harboured pathogenic mutations in BRCA1/2 and ATM, and the remaining variants were of unknown significance.
Fig. 2. Serial ctDNA captures actionable genomic alterations.
a Lollipop plots showing the location and type of mutations identified in ctDNA samples of patients with UC in the TP53, PIK3CA, and FGFR3 genes. b ctDNA analysis of serial samples from 53 patients with advanced UC detected significantly more actionable GAs than analysis of a single sample from each patient. Vertical red bars represent the frequency of patients harbouring actionable GAs from a single ctDNA sample, and vertical blue bars represent the frequency of patients harbouring actionable GAs from serial ctDNA samples. c The mean ctDNA VAF across serial samples for commonly altered genes.
Serial ctDNA sampling identified more patients with actionable GAs than single timepoint ctDNA sampling at a frequency of 31/53 (58.5%) vs. 14/53 (26.4%) patients with any actionable GAs (Fisher’s p = 0.02) and a median of 10 vs. 3 patients per each actionable gene, paired rank p = 0.003)(Fig. 2b). This suggests that using single snapshot ctDNA testing to select patients for clinical trials based on their ctDNA molecular profiles may miss a significant number of patients with actionable GAs.
Among the top 10 commonly altered genes, CDKN2A, FGFR3, ERBB2 GAs were associated with the highest mean ctDNA aVAF across all serial samples (Fig. 2c), indicating that tracking GAs in these genes should be prioritised in ctDNA-based strategies to monitor patients receiving the corresponding targeted therapies. We noted that 120/462 of the identified GAs had very low frequency (VAF ≤ 0.2). To test whether the low-frequency GAs are UC-related, generated by clonal hematopoiesis of indeterminate potential (CHIP) or sequencing artifacts, we performed the following analyses: first, we manually reviewed these GAs and confirmed that none were previously reported in a CHIP database [30], second, we analysed the dynamic changes of the VAFs of these low abundance GAs in serial ctDNA samples. We postulated that the ctDNA delta aVAF changes of UC-associated low abundance GAs would track the clinical response status. Indeed, seven out of nine GAs followed the clinical response status (Supplementary Fig. 3). These data suggest that very low-frequency GAs are potentially informative and should not be excluded from serial ctDNA analyses.
Initial and serial ctDNA measurements are prognostic
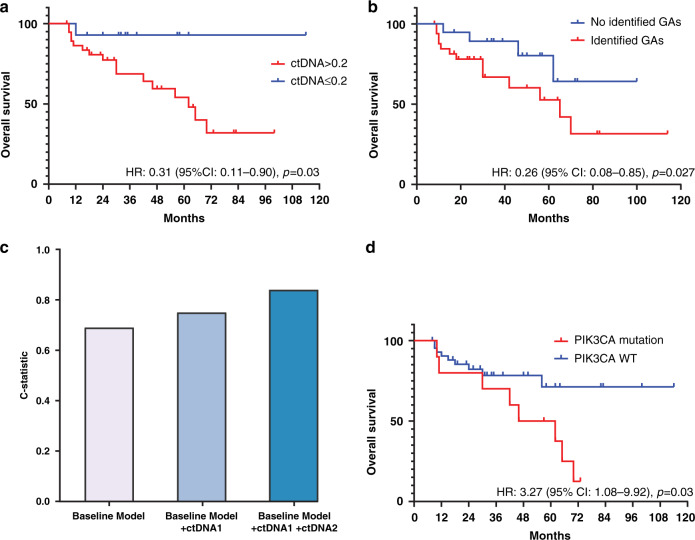
We hypothesised that an initial low ctDNA aVAF and subsequent clearance over serial samples would be associated with improved survival outcomes. We found that patients with lower initial ctDNA aVAF (≤0.2) had longer overall survival (hazard ratio (HR): 0.31, 95% CI: 0.11–0.90, p = 0.03) (Fig. 3a). Higher ctDNA aVAF level was associated with shorter overall survival (HR: 1.07, 95% CI: 1.03–1.10, p < 0.0001). Moreover, a higher number of GAs in the initial sample of each patient was associated with shorter overall survival (HR: 1.12, 95% CI: 1.02–1.24, p = 0.025). These associations remained significant in multivariate regression analyses after adjusting for age at diagnosis, sex, liver metastasis, and the number of prior treatment lines (Supplementary Table 6), suggesting that the ctDNA aVAF and the number of identified GAs are independent prognostic factors in this setting. Clearance of ctDNA (i.e., no detectable GAs) in any serial sample (n = 19/53) was associated with longer overall survival (median OS: not reached vs. 65 months). In a multivariate regression analysis adjusting for age, sex, and liver metastasis, ctDNA clearance at any time point was independently associated with a significantly reduced hazard of death (HR: 0.26, 95% CI: 0.08–0.85, adjusted p = 0.027) (Fig. 3b).
Fig. 3. ctDNA measurements are prognostic.
a Kaplan–Meier overall survival curves showing better overall survival (OS) in patients with ctDNA aVAF ≤0.2. b Patients achieving clearance of their ctDNA aVAF at any time point had better overall survival. c Addition of ctDNA aVAF and ctDNA delta aVAF improved the accuracy of the multivariate model for the prediction of overall survival. The baseline model included age, sex, and liver metastasis. d Kaplan–Meier overall survival curve showing a worse OS of patients having PIK3CA genomic alterations.
To evaluate whether the ctDNA aVAF value and ctDNA delta aVAF between two serial ctDNA samples (‘Methods’) added value to the radiographic assessment of progression, we compared the predictive power of survival models including baseline clinical characteristics along with ctDNA aVAF at the initial sample (ctDNA1), and the second sample (ctDNA2). The baseline clinical model included age, sex, and liver metastasis. Using these clinical characteristics in multivariate cox regression analysis yielded an accuracy of 0.65 Harrel’s C-statistic in predicting overall survival consistent with previously published models [31, 32]. The addition of ctDNA1 and ctDNA2 to the baseline prognostic model improved the accuracy of multivariate regression models of overall survival (Harrel’s C-statistic improved from 0.65 to 0.84) (Fig. 3c), suggesting an added prognostic value for including ctDNA aVAF values across serial samples. We tested the performance of different ctDNA-derived parameters to predict radiographic progression. The area under the curve (AUC) was 0.71–0.84, supporting the predictive value of serial ctDNA-derived parameters (Supplementary Notes and Supplementary Fig. 4a–c).
Somatic ctDNA GAs identified in BRCA1/2 and ATM (n = 17/53), were not significantly associated with overall survival in univariate (HR = 1.02, p = 0.9) or multivariate analysis (HR = 1.2, p = 0.7). Patients with detectable PIK3CA ctDNA GAs at any time point (n = 10/53) had shorter overall survival in univariate analysis (HR: 3.27, 95% CI:1.08–9.92, p = 0.03) (Fig. 3d) and multivariate analysis (HR: 2.87, p = 0.038) (Supplementary Table 6). Consistent with the previously described association between ARID1A-mutations in UC tumours and response to anti-PD-L1 therapy [33], we identified seven patients with ARID1A ctDNA GAs who received immune checkpoint inhibitors as salvage therapy. Three out of the seven patients achieved a durable clinical response to immune checkpoint inhibitors (range 8–31 months).
Serial ctDNA changes predict treatment response
We hypothesised that the aVAF in individual ctDNA samples correlates with the radiographic cancer burden. We limited our analysis to 127 ctDNA samples collected within 4 weeks before or after restaging scans. The mean interval between imaging and ctDNA sampling was 12 days. Overall, there were 45 PD and 82 non-PD events in our cohort.
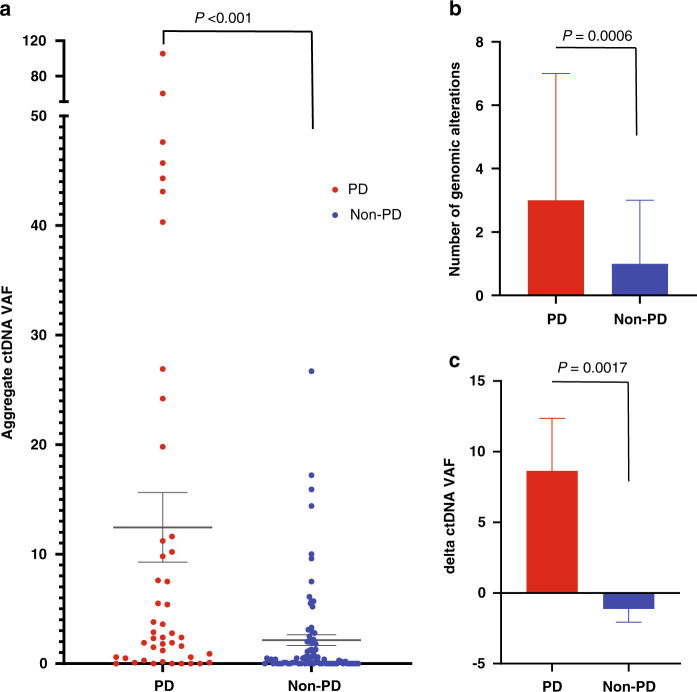
Positive ctDNA samples were significantly more likely to be associated with PD events than non-PD events (odds ratio (OR) 3.02, 95% CI: 1.20–8.72, p = 0.02). Samples with higher ctDNA aVAF (>median for the cohort) were significantly more likely to be associated with PD events (OR: 4.05, 95% CI: 1.89–9.11, p = 0.0005) within the same time window. The mean ctDNA aVAF was significantly increased at PD events compared to non-PD events (12.31 vs. 2.10, p < 0.0001) (Fig. 4a). The median number of GAs per ctDNA sample was significantly higher in PD vs. non-PD events (3 vs. 1, p = 0.0006) (Fig. 4b), suggesting the emergence of new genomic alterations with progression. We hypothesised that the ctDNA delta aVAF defined as the difference between ctDNA aVAF in two consecutive samples collected within a three-month interval correlates with UC progression. We found that ctDNA delta aVAF between two consecutive ctDNA samples increased significantly in PD events and decreased in non-PD events (8.64 compared to −1.12, p = 0.0017) (Fig. 4c). Patients with a ctDNA delta aVAF ≤0 (unchanged or decreased) were more significantly likely to have non-PD events on their subsequent scans (OR: 4.27 (95% CI: 1.80–10.38, p = 0.0006).
Fig. 4. ctDNA dynamics reflect radiographic disease burden.
a The mean ctDNA aVAF increased significantly in samples corresponding to PD events compared to scans showing non-PD events (PD events = 45, non-PD events = 82). b The number of total genomic alterations per sample showed a significant decrease in ctDNA samples corresponding to non-PD events compared to samples corresponding to PD events. c The ctDNA delta aVAF was significantly increased with PD events and decreased in non-PD events.
We posited that ctDNA aVAF reduction reflects radiographic volumetric reduction of disease burden. We found that patients with scans showing complete or partial response had significant mean ctDNA delta aVAF reduction compared to scans with stable disease (SD) or progression (PD) (ctDNA delta aVAF was −0.28 for CR, −10.41 for PR, 0.51 for SD, and 8.64 for PD, p = 0.011). In a subgroup of patients receiving ICB at the time of ctDNA collection, maintaining a ctDNA delta aVAF ≤0.9 (the third quartile) was significantly associated with achieving non-PD events (45/52 (87%) compared to 9/22 (40%) PD events (p = 0.006).
Serial ctDNA predicts clinical outcomes in patients with urothelial cancer
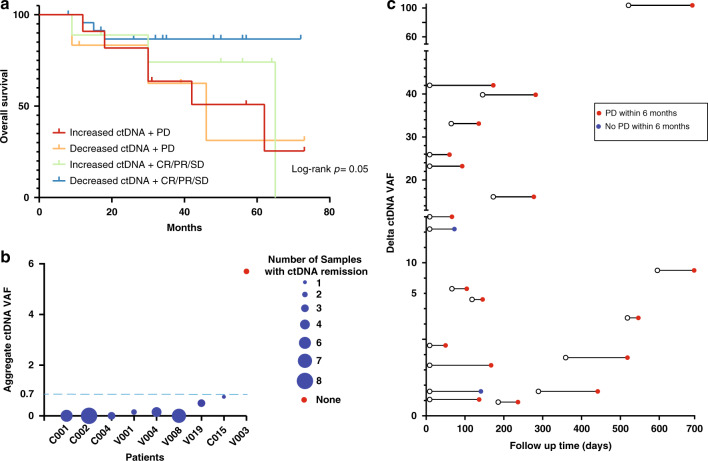
To test the added value of serial ctDNA on top of radiographic response assessment in predicting overall survival, we stratified the patients into four categories based on ctDNA delta aVAF (increased vs. decreased) and radiographic response (PD vs. non-PD). We found that using ctDNA delta aVAF improved the stratification of patients into risk subgroups that correlated with overall survival. The subgroup of patients with both decreased ctDNA delta aVAF and non-PD had the longest overall survival (Log-rank p = 0.05) (Fig. 5a).
Fig. 5. Serial ctDNA predicts clinical outcomes.
a The addition of ctDNA status to the clinical response status stratified prognosis into four risk groups. b Durable clinical responses were associated with continued ctDNA remission in serial ctDNA samples (defined as less than 0.7). The dashed line indicates the threshold for ctDNA remission. The size of each circle indicates the number of consecutive samples with ctDNA remission during the clinical response period for each patient. c Follow-up of patients (n = 20) with ctDNA delta aVAF increase ≥1 showed that 90% (18/20) developed PD events within 6 months. Each line represents one patient. Lines with red dots indicate the occurrence of a PD event within 6 months, and lines with blue dots indicate no PD events occurring within the 6-months follow-up period.
In 8/9 patients who achieved a durable radiographic response of six months, the ctDNA aVAF was ≤0.7 in a median of three successive samples (Fig. 5b). These observations indicate that continuous molecular ctDNA remission by ctDNA aVAF measurement in three serial ctDNA samples is associated with durable clinical responses.
To test the predictive value of the increase in ctDNA aVAF occurring before clinical progression, we identified 20 patients with ctDNA delta aVAF above the third quartile (>0.9) with one or more subsequent radiographic scans within 6 months. 18/20 patients achieved PD event with a median lead time of 92 days over the imaging (Fig. 5c). These data suggest that an increase in ctDNA aVAF by 1 or more is predictive of disease progression.
We provide several molecular case studies to demonstrate how serial ctDNA measurements can provide insights into the clonal evolutionary trajectories of UC patients during treatment (Supplementary Notes and Supplementary Fig. 5). Concordance analysis between genomic alterations detected by ctDNA and tissue sequencing was provided in Supplementary Notes and Supplementary Table 7.
Discussion
Serial ctDNA measurements potentially capture subclonal dynamics and their respective evolutionary trajectories during UC progression or in response to treatment. Before this study, it was unclear whether longitudinal monitoring of serial ctDNA samples has additional prognostic and predictive benefits for patients with advanced UC. We show that serial ctDNA monitoring identified patients who were more likely to achieve durable clinical responses. On the other hand, an increase in ctDNA VAF ≥1 predicted disease progression within 6 months.
Our study provides direct evidence supporting the feasibility of tracking genomic alterations in serial ctDNA samples. We tested the association of several serial ctDNA-derived metrics with clinical outcomes, including the number of GAs identified per ctDNA sample, the aVAF in a single ctDNA sample, and the delta aVAF between successive ctDNA samples. We show that serial sampling detected more patients with actionable GAs emerging during progression compared to the analysis of ctDNA at a single time point. For example, serial sampling identified six additional patients with FGFR 2/3 alterations (13 vs. 7). These alterations would not have been detected in the initial ctDNA samples. In addition, 30% of patients harboured at least one actionable GA (defined as tier 1–2 in PMKB or level 1–3 in OncoKB). For instance, three patients harboured potentially actionable TSC1 E1044fs and the HRAS G12S mutations in our cohort. Patients with the nonsense TSC1 and missense HRAS mutations have been reported to have higher responses to everolimus and tipifarnib, respectively [27, 34]. This suggests a potentially critical role for serial ctDNA sampling to identify actionable targets.
A previous study by Christensen et al. showed that the detection of ctDNA was highly prognostic in patients with localised UC [10]. In this study, we show that ctDNA aVAF ≤0.2 was associated with longer overall survival in patients with advanced UC. This is consistent with the data by Vandekerkhove et al. [16] showing that patients with advanced UC having ctDNA in the first quartile had longer overall survival. We show that patients with advanced UC and undetectable GAs at any timepoint in their clinical course achieved longer overall survival, suggesting that ctDNA persistence is an adverse prognostic factor. In addition, Our findings are consistent with the recent results from the phase III IMvigor010 trial, which showed that patients who achieved ctDNA clearance after adjuvant atezolizumab had longer disease-free and overall survival [20].
Several models were developed to predict prognosis in patients with advanced UC. These models often rely on clinical factors without including molecular data. This applies to the Memorial Sloan-Kettering Cancer Center (MSKCC) risk-score model [35] and another four-parameter model developed by Apolo et al. [36]. The c-statistic for these models ranges from 0.64 to 0.67 [36]. In the current study, we demonstrate that adding the ctDNA aVAF significantly improved the performance of the baseline model, including age, sex, and liver metastasis for predicting survival [31, 32]. As ctDNA is minimally invasive and easily obtained, incorporating it into the initial patient stratification will potentially improve prognostic accuracy.
Our results also support using the ctDNA aVAF change as a tool to predict response to therapy [15, 37]. We showed that each time a patient progressed clinically, the ctDNA delta aVAF increased by a mean of 8.64 from the previous sample, and each time a patient achieved clinical response, the ctDNA delta aVAF decreased by a mean of 1.12. This is consistent with an earlier observation showing a significant decrease in the mean post-treatment VAF in patients with advanced UC responding to durvalumab [18]. Notably, a ctDNA delta aVAF increase ≥1 heralded disease progression within 6 months in most patients. This information potentially enables oncologists to change management earlier to avoid treatment failure.
Our study provides evidence that monitoring the molecular UC burden, defined by ctDNA aVAF, potentially adds value to the conventional imaging-based assessments of disease burden. In the clinically localised setting, the presence of ctDNA as a dichotomous outcome (detectable vs. none) was previously linked to cancer recurrence [10, 15, 38]. In the metastatic setting, dynamic ctDNA-derived metrics that quantify the depth and the durability of responses to treatment are needed to predict progression events. A recent study by Avanzini et al. developed a mathematical model of tumour evolution and ctDNA shedding to predict the imaging-based tumour size in patients with lung cancer, inferring that the death of each tumour cell death sheds 0.014% of ctDNA into the bloodstream [39]. The extent of ctDNA shedding from tumour cells potentially varies in different cancer types and the clinical stage. More precise models to predict tumour size from the quantity of ctDNA and a better understanding of the factors contributing to ctDNA shedding are needed in patients with advanced UC.
Limitations of our study include the heterogeneity of the treatments received during ctDNA sample collection and the variability in the timing of sampling and radiographic assessments. However, it is important to note that our data reflect real-world practices and highlight how incorporating longitudinal ctDNA analysis could yield additional prognostic value to radiographic assessment regardless of treatment. Mutations with low VAF may have originated from other mutational processes unrelated to UC, such as CHIP. However, we showed that these mutations expand significantly with UC progression, suggesting they were most likely UC-related. The number of serial samples varied between patients, and matched tumour tissue sequencing was unavailable for most patients. While the Guardant360® assay is a tumour-only sequencing assay, the Guardant computational pipeline can distinguish between somatic and germline variants [21]. Other limitations inherent to our retrospective study design include the lack of a randomised control group and the potential selection and confounding biases.
In conclusion, our study shows that serial ctDNA monitoring predicts clinical outcomes, augments radiographic assessment of clinical response, and provides insights into the clonal evolutionary trajectories in patients with advanced UC. Minimally-invasive serial ctDNA testing is a promising option for real-time monitoring of treatment effectiveness, predicting outcomes, and guiding adaptive therapy paradigms. Future prospective studies are needed to confirm the value of serial ctDNA dynamics for predicting radiographic progression.
Supplementary information
Acknowledgements
BMF was supported by the Department of Defense CDMRP grant (CA160212), a STARR Cancer Consortium grant (I14-0047), and the Gellert Family-John P. Leonard, MD Research Scholarship in Hematology and Medical Oncology. This work was also supported by a Conquer Cancer Foundation Long Term International Fellowship Award (KSS) and the Englander Institute for Precision Medicine at WCM (OE, BMF). Guardant performed sequencing and initial mutational testing of ctDNA samples. We thank Duy Nguyen for his assistance with editing the text.
Author contributions
Initiation and design of the study: KSS, KSP, RN, PG, NJV and BMF. Subject enrollment, sample, and clinical data collection: KSS, DV, YC, JT, STT, AMM, CNS, DMN, JMM, OE, GPS, PV, NJV and BMF. Sample sequencing: KSP and RN. Statistical, and bioinformatic analyses: KSS and OE. Supervision of research: NV and BMF. Writing of the first draft of the manuscript: KSS, OE, GPV, PG, NV and BMF. All authors contributed to the writing and editing of the revised manuscript and approved the manuscript.
Funding
Not applicable.
Data availability
Data access and responsibility: NJV and BMF had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Data sharing policy: Data are available for bona fide researchers upon request from the authors.
Competing interests
KSS, DMV, YC and JT: No competing interests. AMM: Honoraria—ASCO, Consulting or advisory role—EISAi; Exelixis; Janssen. KSP and RN: Employment—Guardant Health Inc. CNS: Consulting or advisory role—Astellas Pharma; AstraZeneca; Bayer; Genzyme; Immunomedics; Incyte; Medscape; Merck; MSD; Pfizer; Roche; UroToday. STT: Consulting or advisory role—Abbvie; Amgen; Astellas Pharma; Bayer; Clovis Oncology; Dendreon; Endocyte; Genentech; Immunomedics; Janssen; Karyopharm Therapeutics; Medivation; Pfizer; QED Therapeutics; Sanofi; Tolmar, Research funding—Abbvie (Inst); Amgen (Inst); Astellas Pharma (Inst); AstraZeneca (Inst); AVEO (Inst); Bayer (Inst); Boehringer Ingelheim (Inst); Bristol-Myers Squibb (Inst); Clovis Oncology (Inst); Dendreon (Inst); Endocyte (Inst); Exelixis (Inst); Genentech (Inst); Immunomedics (Inst); Inovio Pharmaceuticals (Inst); Janssen (Inst); Karyopharm Therapeutics (Inst); Lilly (Inst); Medivation (Inst); Merck (Inst); Millennium (Inst); Newlink Genetics (Inst); Novartis (Inst); Progenics (Inst); Rexahn Pharmaceuticals (Inst); Sanofi (Inst); Stem CentRx (Inst), Travel, Accommodations, expenses—Amgen; Immunomedics; Sanofi. DMN: Consulting or advisory role—Roche/Genentech, Research funding—Boehringer Ingelheim (Inst); Novartis (Inst); Zenith Epigenetics (Inst). JMM: Research funding—Personal genome diagnostics, Travel, accommodations, expenses—Personal genome diagnostics. OE: Stock and other ownership interests—OneThree Biotech; Owkin; Volastra Therapeutics. GPS: Honoraria—UpToDate, Consulting or advisory role—Astellas Pharma; AstraZeneca; Bicycle Therapeutics; Bristol-Myers Squibb; Eisai; EMD Serono; Exelixis; Genentech; Janssen; Merck; Pfizer; Seattle Genetics, Speakers’ Bureau - Medscape; Onclive; Physicans’ Education Resource; Research to practice, research funding—AstraZeneca (Inst); Janssen (Inst); Sanofi (Inst), Travel, Accommodations, expenses—Bristol-Myers Squibb, Other relationship—Astellas Pharma; AstraZeneca; Bavarian Nordic; Bristol-Myers Squibb; Debiopharm Group; Elsevier; QED Therapeutics. PG: Consulting or advisory role—AstraZeneca; Bayer; Bristol-Myers Squibb; Clovis Oncology; Driver, Inc; EMD Serono; Exelixis; Foundation Medicine; Genzyme; GlaxoSmithKline; HERON; Janssen; Merck; Mirati Therapeutics; Pfizer; QED Therapeutics; Roche; Seattle Genetics, Research funding—Bavarian Nordic (Inst); Bristol-Myers Squibb (Inst); Clovis Oncology (Inst); Debiopharm Group (Inst); Immunomedics (Inst); Pfizer (Inst). NJV: Employment—US Oncology, Stock and Other Ownership interests—Caris Life Sciences, Honoraria—Novartis; Pfizer; UpToDate, Consulting or Advisory role—Astellas Pharma, AstraZeneca; Bayer; Boehringer Ingelheim; Caris Life Sciences; Clovis Oncology; Corvus Pharmaceuticals; Eisai; Genentech/Roche; Janssen Oncology; Merck; Modra Pharmaceuticals; Pfizer; Tolero Pharmaceuticals, Speakers’ Bureau—Bayer; Bristol-Myers Squibb; Clovis Oncology; Genentech/Roche; Sanofi; Seattle Genetics/Astellas, Research funding—Endocyte (Inst); Merck (Inst); Suzhou Kintor Pharmaceuticals (Inst); US Oncology (Inst), Expert testimony—Novartis, Travel, accommodations, expenses—AstraZeneca/MedImmune; Bayer/Onyx; Exelixis; Genentech/Roche; Pfizer; Sanofi/Aventis; US Oncology. BMF: Honoraria—Digital Science Press, Consulting or advisory role—QED therapeutics, Immunomedics, Merck, Seattle Genetics. Patent royalties Immunomedics/Gilead Research funding—Eli Lilly.
Ethics approval and consent to participate
The study was approved by the Western Institutional Review Board (Protocol No. 20152817).
Consent to publish
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nicholas J. Vogelzang, Email: nicholas.vogelzang@usoncology.com
Bishoy Morris Faltas, Email: bmf9003@med.cornell.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01648-8.
References
- 1.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder. Cancer Cell. 2017;171:540–556.e25. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sailer V, Eng KW, Zhang T, Bareja R, Pisapia DJ, Sigaras A, et al. Integrative molecular analysis of patients with advanced and metastatic cancer. JCO Precis Oncol. 2019;3:1–12. doi: 10.1200/PO.19.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faltas BM, Prandi D, Tagawa ST, Molina AM, Nanus DM, Sternberg C, et al. Clonal evolution of chemotherapy-resistant urothelial carcinoma. Nat Genet. 2016;48:1490–9. doi: 10.1038/ng.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Razavi P, Li BT, Brown DN, Jung B, Hubbell E, Shen R, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med. 2019;25:1928–37. doi: 10.1038/s41591-019-0652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24–224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS ONE. 2015;10:1–27. doi: 10.1371/journal.pone.0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agarwal N, Pal SK, Hahn AW, Nussenzveig RH, Pond GR, Gupta SV, et al. Characterization of metastatic urothelial carcinoma via comprehensive genomic profiling of circulating tumor DNA. Cancer. 2018;124:2115–24. doi: 10.1002/cncr.31314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birkenkamp-Demtröder K, Nordentoft I, Christensen E, Høyer S, Reinert T, Vang S, et al. Genomic alterations in liquid biopsies from patients with bladder cancer. Eur Urol. 2016;70:75–82. doi: 10.1016/j.eururo.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Christensen E, Birkenkamp-Demtröder K, Sethi H, Shchegrova S, Salari R, Nordentoft I, et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J Clin Oncol. 2019;37:1547–57. doi: 10.1200/JCO.18.02052. [DOI] [PubMed] [Google Scholar]
- 11.Powles TB, Assaf ZJ, Davarpanah N, Hussain M, Oudard S, Gschwend JE, et al. Clinical outcomes in post-operative ctDNA-positive muscle-invasive urothelial carcinoma (MIUC) patients after atezolizumab adjuvant therapy. Ann Oncol. 2020;31:S1417. doi: 10.1016/j.annonc.2020.10.486. [DOI] [Google Scholar]
- 12.Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, et al. Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clin Cancer Res. 2015;21:4586–96. doi: 10.1158/1078-0432.CCR-15-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel KM, Van Der Vos KE, Smith CG, Mouliere F, Tsui D, Morris J, et al. Association of plasma and urinary mutant DNA with clinical outcomes in muscle invasive bladder cancer. Sci Rep. 2017;7:1–12. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grivas P, Lalani A-KA, Pond GR, Nagy RJ, Faltas B, Agarwal N, et al. Circulating tumor DNA alterations in advanced urothelial carcinoma and association with clinical outcomes: a pilot study. Eur Urol Oncol. 2020;3:695–9. doi: 10.1016/j.euo.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Hilke FJ, Muyas F, Admard J, Kootz B, Nann D, Welz S, et al. Dynamics of cell-free tumour DNA correlate with treatment response of head and neck cancer patients receiving radiochemotherapy. Radiother Oncol. 2020;151:182–9. doi: 10.1016/j.radonc.2020.07.027. [DOI] [PubMed] [Google Scholar]
- 16.Vandekerkhove G, Lavoie J, Annala M, Murtha AJ, Sundahl N, Walz S, et al. Plasma ctDNA is a tumor tissue surrogate and enables clinical-genomic stratification of metastatic bladder cancer. Nat Commun. 2021;12:184. doi: 10.1038/s41467-020-20493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powles T, Carroll D, Chowdhury S, Gravis G, Joly F, Carles J, et al. An adaptive, biomarker-directed platform study of durvalumab in combination with targeted therapies in advanced urothelial cancer. Nat Med. 2021;27:793–801. doi: 10.1038/s41591-021-01317-6. [DOI] [PubMed] [Google Scholar]
- 18.Raja R, Kuziora M, Brohawn PZ, Higgs BW, Gupta A, Dennis PA, et al. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin Cancer Res. 2018;24:6212–22. doi: 10.1158/1078-0432.CCR-18-0386. [DOI] [PubMed] [Google Scholar]
- 19.Sundahl N, Vandekerkhove G, Decaestecker K, Meireson A, De Visschere P, Fonteyne V, et al. Randomized phase 1 trial of pembrolizumab with sequential versus concomitant stereotactic body radiotherapy in metastatic urothelial carcinoma. Eur Urol. 2019;75:707–11. doi: 10.1016/j.eururo.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Powles T, Assaf ZJ, Davarpanah N, Banchereau R, Szabados BE, Yuen KC, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature. 2021;595:432–7. doi: 10.1038/s41586-021-03642-9. [DOI] [PubMed] [Google Scholar]
- 21.Odegaard JI, Vincent JJ, Mortimer S, Vowles JV, Ulrich BC, Banks KC, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res. 2018;24:3539–49. doi: 10.1158/1078-0432.CCR-17-3831. [DOI] [PubMed] [Google Scholar]
- 22.Vosoughi A, Zhang T, Shohdy KS, Vlachostergios PJ, Wilkes DC, Bhinder B, et al. Common germline-somatic variant interactions in advanced urothelial cancer. Nat Commun. 2020;11:6195. doi: 10.1038/s41467-020-19971-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang L, Fernandes H, Zia H, Tavassoli P, Rennert H, Pisapia D, et al. The cancer precision medicine knowledge base for structured clinical-grade mutations and interpretations. J Am Med Inform Assoc. 2017;24:513–9. doi: 10.1093/jamia/ocw148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakravarty D, Gao J, Phillips S, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;1:1–16. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221–221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loriot Y, Necchi A, Park SH, Garcia-Donas J, Huddart R, Burgess E, et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N. Engl J Med. 2019;381:338–48. doi: 10.1056/NEJMoa1817323. [DOI] [PubMed] [Google Scholar]
- 29.Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–22. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bick AG, Weinstock JS, Nandakumar SK, Fulco CP, Bao EL, Zekavat SM, et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature. 2020;586:763–8. doi: 10.1038/s41586-020-2819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nassar AH, Mouw KW, Jegede O, Shinagare AB, Kim J, Liu C-J, et al. A model combining clinical and genomic factors to predict response to PD-1/PD-L1 blockade in advanced urothelial carcinoma. Br J Cancer. 2020;122:555–63. doi: 10.1038/s41416-019-0686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khaki AR, Li A, Diamantopoulos LN, Miller NJ, Carril-Ajuria L, Castellano D, et al. A new prognostic model in patients with advanced urothelial carcinoma treated with first-line immune checkpoint inhibitors. Eur Urol Oncol. 2021;4:464–72. doi: 10.1016/j.euo.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge Z, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24:556–62. doi: 10.1038/s41591-018-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HW, Sa JK, Gualberto A, Scholz C, Sung HH, Jeong BC, et al. A phase II trial of tipifarnib for patients with previously treated, metastatic urothelial carcinoma harboring hras mutations. Clin Cancer Res. 2020;26:5113–9. doi: 10.1158/1078-0432.CCR-20-1246. [DOI] [PubMed] [Google Scholar]
- 35.Bajorin DF, Dodd PM, Mazumdar M, Fazzari M, McCaffrey JA, Scher HI, et al. Long-term survival in metastatic transitional-cell carcinoma and prognostic factors predicting outcome of therapy. J Clin Oncol. 1999;17:3173–81. doi: 10.1200/JCO.1999.17.10.3173. [DOI] [PubMed] [Google Scholar]
- 36.Apolo AB, Ostrovnaya I, Halabi S, Iasonos A, Philips GK, Rosenberg JE, et al. Prognostic model for predicting survival of patients with metastatic urothelial cancer treated with cisplatin-based chemotherapy. J Natl Cancer Inst. 2013;105:499–503. doi: 10.1093/jnci/djt015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bratman SV, Yang SYC, Iafolla MAJ, Liu Z, Hansen AR, Bedard PL, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer. 2020;1:873–81. doi: 10.1038/s43018-020-0096-5. [DOI] [PubMed] [Google Scholar]
- 38.Birkenkamp-Demtröder K, Christensen E, Nordentoft I, Knudsen M, Taber A, Høyer S, et al. Monitoring treatment response and metastatic relapse in advanced bladder cancer by liquid biopsy analysis. Eur Urol. 2018;73:535–40. doi: 10.1016/j.eururo.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 39.Avanzini S, Kurtz DM, Chabon JJ, Moding EJ, Hori SS, Gambhir SS, et al. A mathematical model of ctDNA shedding predicts tumor detection size. Sci Adv. 2020;6:1–10. doi: 10.1126/sciadv.abc4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data access and responsibility: NJV and BMF had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Data sharing policy: Data are available for bona fide researchers upon request from the authors.