Abstract
Four hundred ninety-nine methicillin-resistant Staphylococcus aureus (MRSA) isolates recovered from 1996 to 1998 from 22 hospitals in five countries of Latin America—Argentina, Brazil, Chile, Uruguay and Mexico—were examined for antimicrobial susceptibility and clonal type in order to define the endemic clones in those hospitals. The hybridization of ClaI restriction digests with the mecA- and Tn554-specific DNA probes combined with pulsed-field gel electrophoresis of chromosomal SmaI digests (ClaI-mecA::ClaI-Tn554::PFGE clonal types) documented not only the predominance and persistence of the Brazilian clone (XI::B::B) in Brazil (97%) and Argentina (86%) but also its massive dissemination to Uruguay (100%). Moreover, a close relative of the Brazilian clone (XI::κ::B) was highly represented in Chile (53%) together with a novel clone (47%) (II::E′::F) resistant to pencillin, oxacillin, ciprofloxacin, chloramphenicol, clindamycin, erythromycin, and gentamicin. A unique clonal type (I::NH::M) was detected in Mexico among pediatric isolates and was resistant to penicillin, oxacillin, and gentamicin only. This study clearly documented the very large capacity for geographic expansion and the persistence of the Brazilian clone, contributing not only to the increasing uniformity of the MRSA in South America but worldwide as well.
Nosocomial infections due to multidrug-resistant Staphylococcus aureus are an important health problem worldwide. Antimicrobial resistance prolongs the duration of hospitalization, thereby increasing the cost of patient care.
The Center for Molecular Epidemiology and International Network of Hospitals (CEM/NET) has been created to keep track of the movement and to identify reservoirs of major multidrug-resistant clones of S. aureus and other gram-positive pathogens (10, 38). This initiative involved a collaborative network with hospitals and health institutions of several countries that reported relatively high levels of multidrug-resistant bacteria in Europe, North America, Latin America, and Asia. Clinical isolates of methicillin-resistant S. aureus (MRSA) have been provided by collaborating national centers and deposited at two central microbiology laboratories — the Laboratory of Molecular Genetics located at the Instituto de Tecnologia Química e Biológica da Universidade Nova de Lisboa (ITQB/UNL), Oeiras, Portugal, and the Laboratory of Microbiology of the Rockefeller University—for quality control and for testing by microbiological and molecular typing techniques. Such studies have identified some internationally spread MRSA clones, i.e., the Iberian (12, 33), Brazilian (35), Pediatric (29), and New York-Tokyo clones (1, 26).
The SENTRY Antimicrobial Surveillance Program, another private network that monitors the frequency of occurrence and antimicrobial susceptibility of predominant pathogens causing infections worldwide, conducted several studies during 1997 and 1998 in Latin America (Argentina, Brazil, Chile, Colombia, Mexico, Uruguay, and Venezuela). These surveillance studies have reported a high occurrence of MRSA for commonly observed infections in this geographic region. (i) A survey of bloodstream infections carried out in 1997 showed that 21% of the 1,642 infections were attributable to S. aureus, out of which 29% were resistant to oxacillin (24). (ii) In the same year, another survey of lower respiratory tract infection of hospitalized patients with pneumonia revealed that 23% of 556 isolates collected were identified as S. aureus and resistance to oxacillin was detected in nearly 50% of these isolates (27). (iii) During 1997 and 1998, S. aureus was found as the most common (31%) etiologic agent among 885 bacterial isolates responsible for skin and soft tissue infections and 31% of the isolates were MRSA (14). Moreover, oxacillin resistance among S. aureus isolated from patients with bloodstream infections increased approximately 5% in Latin America over the 1-year period from 1997 to 1998 (11).
In a more recent study performed under the CEM/NET Initiative, Project RESIST, we have shown that marked geographic variation in multiresistance patterns exist among MRSA isolated in 6 countries of South America (Colombia, Argentina, Brazil, Chile, Mexico, and Uruguay) (32).
The molecular typing performed with the MRSA strains from Colombia has been previously reported (15). The aim of the present study was to assess the current MRSA clonal types responsible for nosocomial infection in 22 medical centers of the other five Latin American countries: Argentina, Brazil, Chile, Mexico, and Uruguay.
MATERIALS AND METHODS
Participating institutions.
Twenty-two clinical institutions from five Latin American countries—Argentina, Brazil, Chile, Uruguay, and Mexico—participated in this study. The institutions provided isolates recovered in different periods of isolation, 1, 2, or 3 years, within the period from 1996 to 1998. The names and locations of the hospitals included in this study, their specialty, their size, the number of strains recovered from each institution, and the year of collection are listed in Table 1.
TABLE 1.
Clinical institutions that provided the 499 S. aureus isolates
| Country (total no. of isolates) | City | Hospital codea | Specialtyb | No. of beds | No. of isolates | Isolation date (yr) |
|---|---|---|---|---|---|---|
| Argentina (89) | Buenos Aires | 1 | Pediatrics | 450 | 26 | 1997–1998 |
| Buenos Aires | 2 | Cardiology | 220 | 37 | 1996–1998 | |
| Buenos Aires | 3 | Community/university | 120 | 26 | 1996–1998 | |
| Brazil (162) | São Paulo | 4 | Community | 762 | 154 | 1997–1998 |
| São Paulo | 5 | – | – | 2 | 1996 | |
| São Paulo | 6 | – | – | 2 | 1996 | |
| São Paulo | 7 | – | – | 2 | 1996 | |
| São Paulo | 8 | – | – | 1 | 1996 | |
| São Paulo | 9 | – | – | 1 | 1997 | |
| Chile (118) | Santiago | 10 | Military | 184 | 36 | 1997–1998 |
| Santiago | 11 | Public/university | 795 | 21 | 1997–1998 | |
| Santiago | 12 | Public/university | 500 | 25 | 1997 | |
| Santiago | 13 | Community | 114 | 7 | 1997–1998 | |
| Santiago | 14 | Public/university | 900 | 14 | 1997 | |
| Santiago | 15 | Public/university | 600 | 5 | 1997 | |
| Puerto Montt | 16 | Public | 418 | 10 | 1997 | |
| Uruguay (102) | Montevideo | 17 | Community/university | 420 | 35 | 1997–1998 |
| Montevideo | 18 | Public | 280 | 48 | 1997–1998 | |
| Montevideo | 19 | Public | – | 16 | 1996–1997 | |
| Montevideo | 20 | Public/pediatrics | – | 2 | 1997 | |
| Young, Rio Negro | 21 | Public | – | 1 | 1996 | |
| Mexico (28) | C. de Mexico | 22 | Pediatrics | 200 | 28 | 1997–1998 |
Hospital code: 1, Hospital de Pediatría “Prof. J. P. Garrahan”; 2, Inst, Univ. de Ciencias Biomedicas, Fundación Favaloro; 3, Centro de Educacion Médica e Investigaciones Clínicas “Dr. N. Quirno”; 4, Casa de Saúde de Santa Marcelina; 5, Hospital Infantil Darcy Vargas; 6, Centro de Referência e Treinamento em AIDS; 7, Santa Casa; 8, Hospital Universitario; 9, Hospital Geral São Mateus; 10, Hospital Fach; 11, Hospital Sotero del Rio; 12, Hospital Felix Bulnes; 13, Hospital de Castro; 14, Hospital Barros Luco Trudeau; 15, Hospital, San Borja Arriaran; 16, Hospital de Puerto Montt; 17, Hospital Clinicas; 18, Hospital Pasteur; 19, Maciel Hospital; 20, HPR Children Hospital; 21, Young Hospital; 22, Hospital de Pediatria CMN Siglo XXI.
–, data not available.
Bacterial isolates.
A total of 499 isolates previously identified as MRSA by the collaborating centers—28 from Mexico, 89 from Argentina, 102 from Uruguay, 118 from Chile, and 162 from Brazil—collected during a 3-year period (1996 to 1998) were analyzed in this study. A common specimen sheet with information concerning the origin of each isolate was provided by the collecting centers. The information included not only demographic data, such as patient name, gender, date of birth, immunodeficiency, origin (nosocomial or community), clinical service, and hospitalization in intensive care unit (ICU) but also specimen data such as the date of collection and the source. The majority of the isolates (81%) were responsible for infection, and all were consecutively recovered from single patients from various sources: 35% from respiratory sources, 25% from wound, 25% from blood, 8% from abscess, 2% from urine, and 5% from other nonspecified sources. The Mexican isolates were mainly from blood (57%), while the Chilean isolates were predominantly from wounds (52%). All isolates were identified at the collaborating institutions and sent to the central laboratory, i.e., the Laboratory of Molecular Genetics located at Instituto de Technologia Química e Biológica da Universidade Nova de Lisboa (ITQB/UNL), Oeiras, Portugal, for pheno- and genotypic characterization.
Demographic information.
Globally, 51% of the isolates were recovered from adults, 29% from elderly individuals (over 65 years old), and 20% from children (under 12 years old). However, hospitals from Mexico and Argentina provided predominantly isolates from children (100 and 44%, respectively), in contrast to Uruguay and Brazil, where only 3 and 7% of the isolates were from patients under 12 years old. The great majority of the isolates (85%) were recovered from inpatients hospitalized for more than 48 h, out of which 44% were in ICUs. The patients were hospitalized or treated in different clinical services: 48% in medicine, 25% in surgery, 17% in pediatrics, 1% in gynecology, and 9% in other services. Eleven percent of the isolates were from immunocompromized patients.
Control strains.
S. aureus ATCC 25923 and Escherichia coli ATCC 25922 were included in the assays for quality control of susceptibility testing. The partially dalfopristin- and quinupristin-resistant S. aureus clinical isolates Syn1 and Syn25 (provided by Rhône-Poulenc Rorer, S.A.) were included as quality control strains in the determination of dalfopristin-quinupristin susceptibility testing. S. aureus ATCC 29213 and S. aureus PC3 (34) were used as controls in the vancomycin resistance screen assay. Reference strains for ClaI-mecA and Tn554 patterns and representative strains used to compare the pulsed-field gel electrophoresis (PFGE) profiles were from the MRSA strain collection of the Laboratory of Molecular Genetics at ITQB/UNL. Control strains for ClaI-mecA patterns were HPV107 (pattern I) (33), BK71 (II), RN7164 (III), RN6322 (V) (17), PER168 (IX) (12), HU25 (XI) (2, 35), and PL48 (XX) (18). Control strains for ClaI-Tn554 patterns were RN7178 (pattern A), BK798 (AA), RN7174 (B), RN7538 (M) (17), HPV107 (E) (33), POL2 (κ) (18), and ITA33 (ξ) (19). Control strains for comparison of PFGE patterns were representatives of the Iberian clone HPV107 (33), the Brazilian clone HU25 (35), the Pediatric clone HDE288 (29), COB5 (15), the Archaic clones E2125 and E2213 (7), and the New York-Tokyo clone JP1 (1), as well as representatives of clonal types found in other countries, including Poland (POL1 [18]), PLN49 (R. Alves et al., unpublished results), Argentina (ARG33 [4]), Turkey (TUR5 [S. Kocagoz et al., unpublished results), Hungary (HUR24 [22]), Greece (GRE5 [I. Spiliopoulou et al., unpublished results), and Italy (ITL262 and ITL370 [R. Mato et al., unpublished results).
Antimicrobial susceptibility testing.
Susceptibility to 12 antimicrobial agents (penicillin, oxacillin, trimethoprim-sulfamethoxazole, ciprofloxacin, chloramphenicol, clindamycin, erythromycin, gentamicin, rifampin, tetracycline, vancomycin, and teicoplanin; Oxoid, Basingstoke, Hampshire, England) was confirmed at the central laboratory by disk diffusion according to the National Committee for Clinical Laboratory Standards guidelines (21). Susceptibility to dalfopristin-quinupristin (provided by Rhône-Poulenc Rorer, S.A.) was evaluated by the agar dilution technique as previously described (32).
Expression of oxacillin resistance.
The mode of expression of oxacillin resistance was tested by population analysis profiles (PAPs) by using cultures grown overnight in tryptic soy broth (Difco, Oxoid, Hampshire, United Kingdom) (8, 9). Twenty isolates representative of the three major clonal types found in this study were tested. Strains were assigned to phenotypic expression classes on the basis of the oxacillin MIC that was arbitrarily defined as the lowest concentration of the antibiotic that prevented the appearance of 99.9% of bacterial colonies (39).
Vancomycin agar screen.
Detection of vancomycin-resistant S. aureus (VRSA) strains was done by vancomycin agar screen. A suspension of cells from a 20- to 24-h tryptic soy agar plate (Difco) was prepared in sterile water to a turbidity equivalent to that of 0.5 McFarland standard. Ten microliters of the inoculum were plated onto Mueller-Hinton agar (MH; Difco) containing 2 μg of vancomycin per ml. Growth was recorded following 24 and 48 h of incubation at 35°C. A reduction of the vancomycin concentration to 2 μg/ml (37) and an incubation time of 48 h decrease the likelihood of not detecting VRSA strains. The MIC was subsequently evaluated by E-test (AB Biodisk, Solne, Sweden) according to manufacturer's instructions for all strains that presented growth of at least one colony. The E-test was performed in MH with an incubation for 24 h at 35°C.
DNA probes, mecA-Tn554 polymorphisms, and PFGE.
Protocols for DNA probes (mecA, mecI, and Tn554), preparation and labeling for ClaI::mecA and ClaI::Tn554 patterns, as well as PFGE were performed as previously reported (3, 5, 8, 17). PFGE patterns were compared by visual inspection (36), followed by computer analysis using Whole Band Analyzer version 3.3 (BioImage, Ann Arbor, Mich.) software for UNIX SparcStation 4 running under SunOS version 5.5.1 operating systems (26).
RESULTS
MRSA clonal classification.
Strains were classified into clonal types on the basis of a combination of their particular mecA and Tn554 polymorphisms and PFGE patterns (mecA::Tn554::PFGE types). In addition, the sequences of mecA, mecl, and Tn554 were located in the bacterial chromosome (see below).
Analysis of the mecA vicinity and Tn554 insertion patterns and PFGE patterns distributed 494 MRSA isolates into nine different mecA polymorphs, 17 Tn554 patterns, and 11 PFGE patterns. However, mecA polymorph XI (12), its variants III and IX (12), Tn554 type B (17), or its relative B′(7), and PFGE pattern B were clearly predominant, representing 78, 62, and 79% of the isolates, respectively. Of the 499 isolates, 5 were found not to carry the mecA gene when analyzed at the central laboratory (see below). Figures 1 and 2A show the ClaI-Tn554 patterns identified among the Latin American strains and the PFGE patterns representing the three major clonal types found in this MRSA collection.
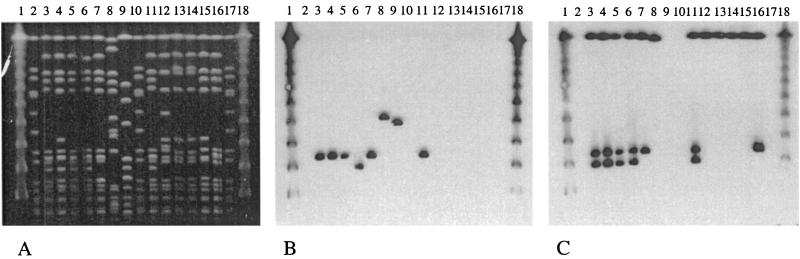
FIG. 1.
ClaI-Tn554 patterns identified among the Latin American strains. Patterns A, B, E, AA, M (17), κ (18), B′ (7), and ξ (19) were previously described. Patterns E′, η, MM, NN, OO, PP, and QQ correspond to novel patterns described in this study. Lanes 1 and 17, molecular weight markers, 1-kb ladder; lane 2, AGT55 (pattern E); lane 3, CHL5 (pattern E′); lane 4, CHL1 (pattern κ); lane 5, AGT115 (pattern A); lane 6, AGT84 (pattern M); lane 7, BRA131 (pattern AA); lane 8, URU105 (pattern B); lane 9, URU149 (pattern B′); lane 10, AGT27 (pattern PP); lane 11, AGT8 (pattern QQ); lane 12, BRA135 (pattern MM); lane 13, BRA145 (pattern NN); lane 14, BRA162 (pattern OO); lane 15, CHL18 (pattern ξ); lane 16, CHL113 (pattern η).
FIG. 2.
(A) SmaI PFGE patterns of the major MRSA clones found in five countries of Latin America (Argentina, Brazil, Uruguay, Chile, and Mexico). Lanes 1 and 18, lambda ladder; lanes 2, 10, and 17, NCTC 8325; lanes 3 to 7 and 11, representatives of the Brazilian clone: lanes 3 and 11 (HU25, XI::B::B, Brazil [35]), lane 4 (BRA10, XI::B::B, Brazil), lane 5 (AGT35, XI::B::B, Argentina), lane 6 (URU1, XI::B′::B, Uruguay), lane 7 (CHL1, XI::κ::B, Chile). Lane 8, representative of the Chilean clone (CHL93, II::E′::F, Chile). Lane 9, representative of the Mexican clone (MEX58, I::NH::M, Mexico). Lanes 12 to 16, MSSA strains from Argentina: ARG38, ARG102, ARG110, ARG111, and ARG119, respectively. (B) SmaI PFGE of Fig. 2A hybridized with a mecA probe. (C) SmaI PFGE of Fig. 2A hybridized with a Tn554 probe.
PFGE pattern B was predominant in all countries, except Mexico, showing between 13 (Chile) and 30 subtypes (Brazil). Although PFGE type B was generally associated with ClaI-mecA polymorph XI (or its variants III and IX), it was related to different ClaI-Tn554 patterns, and this feature was characteristic of isolates of a given country. In fact, PFGE type B was mainly associated with Tn554 patterns B in Brazil and B or B′ in Argentina and Uruguay and with pattern κ (18) in Chile. The PFGE analysis also identified two other important fingerprints: pattern F (11%) found in Chile associated with ClaI-mecA polymorph II (17) and Tn554 pattern E′ (corresponding to a small variation of pattern E [17]) and PFGE pattern M (6%) characteristic of all isolates collected in Mexico with no homology to Tn554 and associated with ClaI-mecA polymorph I (17).
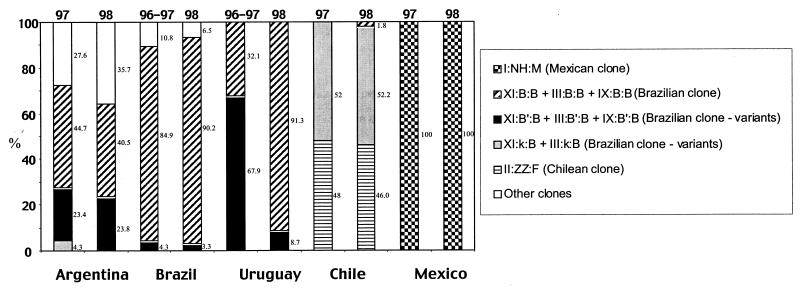
Although the 494 MRSA isolates were distributed into 39 clones, three major clonal families were noticeable (see Table 2). The first lineage was found with an indubitable preponderance (79%) and was represented by the Brazilian clone (XI::B::B) and its closely relatives having variable mecA or Tn554 patterns but similar PFGE profiles: XI::B′::B, III::B::B, III::B′::B, XI::κ::B, and III::κ::B. This lineage was clearly dominant in Uruguay (100%), Brazil (97%), and Argentina (86%) and represented approximately half of the isolates from Chile (53%). The remaining isolates of Chile (47%) belonged to the second family, the Chilean clone (II::E′::F). The third important and unique lineage was the Mexican clone (I::NH::M) that included all the isolates recovered at one pediatric hospital from Mexico.
TABLE 2.
Genotypic properties and antibiotypes of MRSA isolates belonging to the three major clones found in five countries of Latin America
| Clone; no. of isolates (%) | Characteristic antibiotypea | Source country; no. of isolates (%) | ClaI-mecA:Tn554: PFGE typeb; no. of isolates (%) | ClaI-mecA variants | ClaI-Tn554 variants | Exceptions to characteristic antibiotypec |
|---|---|---|---|---|---|---|
| Brazilian; 388 (79) | PR, OR, SxtR, CiR, CR, ClR, ER, GR, RR, TR, QDS, VS, TeS | Brazil; 151 (97) | XI:B:B; 122 (75) | |||
| III:B:B; 8 (5) | ||||||
| XI:B′:B; 5 (3) | ||||||
| III:B′:B; 3 (2) | ||||||
| Other variants; 13 (12) | XX, IX | AA, κ, MM, NN, OO | ||||
| Argentina; 73 (86) | XI:B:B; 36 (43) | |||||
| XI:B′:B; 22 (26) | CS, RS | |||||
| Other variants; 15 (17) | IX | PP, QQ, A, κ, M | ||||
| Uruguay; 102 (100) | XI:B′:B; 40 (39) | RS | ||||
| XI:B:B; 27 (26) | RS | |||||
| IX:B:B; 23 (23) | RS | |||||
| III:B:B; 10 (10) | RS | |||||
| III:B′:B; 2 (2) | GS, RS | |||||
| Chile; 62 (53) | XI:κ:B; 50 (42) | CS | ||||
| III:κ:B; 9 (8) | ||||||
| Other variants; 3 (3) | η | RS | ||||
| Chilean; 56 (11) | PR, OR, SxtS, CiR, CR, ClR, ER, GR, RS, TS, QDS, VS, TeS | Chile; 56 (47) | II:E′:F (41, 48) Variants; 8 (6) | V, IX′ | ξ | |
| Mexican; 28 (6) | PR, OR, SxtS, CiS, CS, ClS, ES, GR, RS, TS, QDS, VS, TeS | Mexico; 28 (100) | I:NH:M |
The characteristic antibiotype is indicated for the major clones: P, penicillin; O, oxacillin; Sxt, trimethoprim-sulfamethoxazole; Ci, ciprofloxacin; C, chloramphenicol; Cl, clindamycin; E, erythromycin; G, gentamicin; R, rifampin; T, tetracycline; QD, quinupristin-dalfopristin; V, vancomycin; T, teicoplanin. Superscripts: R, resistant; S, susceptible.
ClaI-mecA patterns I, II, III, V (17), IX, XI (12), and XX (18) were previously described. Pattern IX corresponds to a small variation of pattern IX. ClaI-Tn554 patterns A, B, AA, M (17), B′ (7), κ (18), and ξ (19) were previously described. NH, no homology. Patterns η, MM, NN, OO, PP, and QQ correspond to novel patterns. Pattern E′ corresponds to a small variation of pattern E (17).
An exception is considered when at least 50% of the isolates show a different susceptibility pattern relative to the characteristic antibiotype.
Characteristics of the major MRSA clones. (i) Resistance to antimicrobial agents.
The resistance profile of each major clonal type is shown in Table 2. The Brazilian clone (XI::B::B and its relatives) was resistant to penicillin (100%), oxacillin (99%), trimethoprim-sulfamethoxazole (99%), ciprofloxacin (98%), chloramphenicol (79%), clindamycin (97%), erythromycin (99%), gentamicin (92%), rifampin (53%), and tetracycline (98%). Interestingly, all strains from Uruguay related to this clonal type were susceptible to rifampin. Strains from Chile belonging to clone II::E′::F or its variants were resistant to penicillin (100%), oxacillin (100%), ciprofloxacin (100%), chloramphenicol (70%), clindamycin (98%), erythromycin (98%), and gentamicin (98%). Strains of the Mexican clone (I::NH::M) were only resistant to penicillin (100%), oxacillin (100%), and gentamicin (79%). All 494 isolates were susceptible to teicoplanin, vancomycin, and dalfopristin-quinupristin. Of the 499 strains screened for VRSA, 37 isolates exhibited growth at 2 μg of vancomycin per milliliter after 48 h (between 1 and ca. 100 colonies). However, the vancomycin MICs ranged from 1 to 2 μg/ml. The three major clonal families could be distinguished by the mode of oxacillin expression: the Brazilian clone showed high-level resistance (MIC = 400 μg/ml) and homogeneous expression class 4, the Chilean clone showed intermediate profile (MIC = 100 μg/ml) and heterogeneous expression class 3, and the Mexican clone showed low-level resistance (MIC = 12 μg/ml) and also heterogeneous phenotype class 2. Isolates belonging to the Mexican clone do not grow at concentrations of >25μg/ml, and isolates belonging to the Chilean clone do not grow at concentrations of >800 μg/ml (data not shown).
(ii) SmaI-hybridization bands.
Strains belonging to the Brazilian clone (XI::B::B) usually carry the mecA gene in a SmaI fragment of 119 kb. The variant XI::B′::B had the mecA gene located in a 95-kb fragment. The three copies of Tn554 were identified in SmaI fragments of approximately 650, 119, and 95 kb among isolates of clones XI::B::B and XI::B′::B. Strains belonging to the Chilean variant of the Brazilian clone (III::κ::B or XI::κ::B) carry only two copies of Tn554 (650 and 119 kb). Strains belonging to the Mexican clone (I::NH::M) carried the mecA gene in a fragment of approximately 190 kb, while strains of the Chilean clone (II::E′::F) carry the gene in a fragment of 195 kb. Strains of this last clone contain only one copy of the transposon in its primary attachment site (SmaI fragment of 650 kb), while strains belonging to the Mexican clone do not have homology with Tn554. Figures 2B and C represent the hybridization results of the mecA gene and transposon Tn554 in the chromosomal SmaI fragments. Hybridization with a mecI probe located this gene in the same fragment as mecA in strains of the Brazilian clone, whereas strains belonging to the Mexican and Chilean clones did not carry mecI.
Five S. aureus strains collected in hospital 2 in Buenos Aires, Argentina, initially classified by the collaborating institution as MRSA, did not hybridize with the mecA (Fig. 2B) nor the mecI probes (result not shown) and displayed susceptibility to oxacillin when the antibiogram and the molecular typing were performed at the central laboratory. However, these five strains had a multiresistant antibiotype: they showed resistance to penicillin, trimethoprim-sulfamethoxazole, ciprofloxacin, erythromycin, gentamicin, rifampin, tetracycline, clindamycin, and chloramphenicol (three isolates). Interestingly, these isolates have PFGE patterns similar to the PFGE pattern B characteristic of the Brazilian clone (Fig. 2A) and carried one (pattern E, four isolates) or two copies (pattern AA, one isolate) of Tn554 (Fig. 2C).
MRSA clonal evolution in Latin American hospitals during 1996 to 1998.
Figure 3 represents the major clonal types found in each country in two periods of time: 1996 to 1997 and 1998. In Argentina, Brazil, Chile, and Mexico there was no significant difference in the frequency of the major clonal types in both periods. In contrast, in Uruguay, during 1996 to 1997, clonal type XI::B′::B and its variant associated with Tn554 pattern B′ (III::B′::B) represented more than half of the isolates (68%), whereas clone XI::B::B and its variants associated with Tn554 pattern B (IX::B::B and III::B::B) represented only 32%. In 1998 this last clone represented almost all isolates (91%), whereas clone XI::B′::B was reduced to 9%.
FIG. 3.
Evolution of the major clonal types found in five countries of Latin America through two periods of time: 1996 to 1997 and 1998.
Geographic dissemination of clonal types.
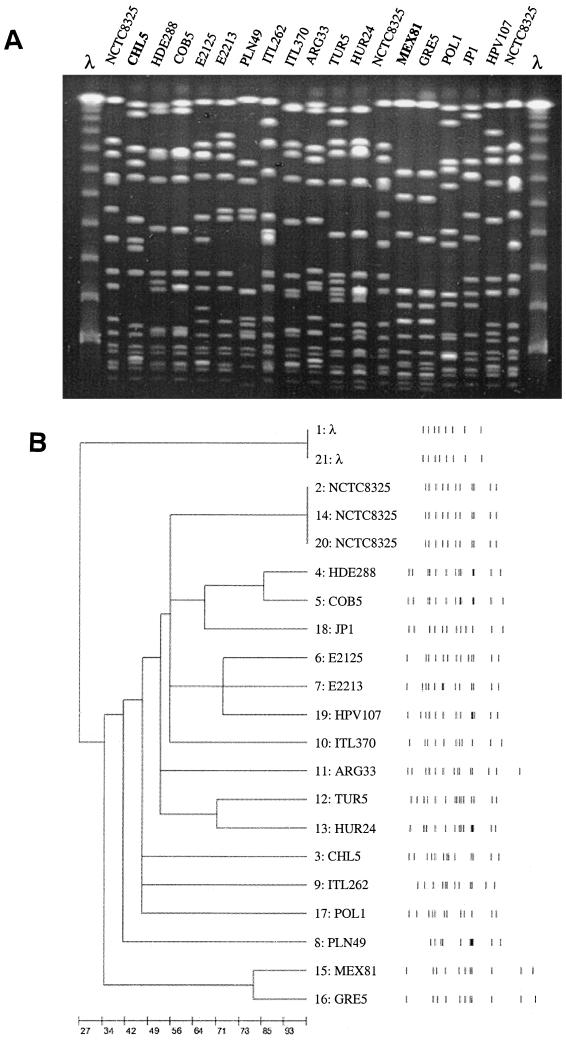
PFGE patterns B, F, and M, characteristic of, respectively, the Brazilian, the Chilean, and the Mexican MRSA clones were compared with representative PFGE patterns of other MRSA strains stored in the CEM/NET database. PFGE type B found in Brazil, Argentina, Uruguay, and Chile showed a high degree of similarity with the prototype of the MRSA Brazilian clone (35) (Fig. 2A). PFGE type M from Mexico was found to be closely related to a representative of the dominant clone in 1993 and 1998 of a hospital in Patras, Greece (I. Spiliopoulou et al., unpublished), while type F from Chile had no resemblance to any other profiles of MRSA clones found so far (Fig. 4).
FIG. 4.
(A) Comparison of PFGE patterns of the Mexican and Chilean clones with international PFGE patterns stored in the CEM/NET database. Representatives from different countries are shown. Lanes 1 and 21, lambda ladder. Lanes 2, 14, and 20, NCTC 8325. Lane 3, CHL5, II::E′::F (Chile); lane 4, HDE288, II::NH::D (Portugal) (29); lane 5, COB5, II::NH::D (Colombia) (15); lane 6, E2125, II::NH::A1 (Denmark) (7); lane 7, E2213, II::NH::A9 (Denmark) (7); lane 8, PLN49, II::NH::I (Poland); lane 9, ITL262, II::NH::C (Italy); lane 10, ITL370, II::E::E (Italy); lane 11, ARG33, II::E::C (Argentina) (4); lane 12, TUR5, II::ηη::A (Turkey); lane 13, HUR24, II::q::D (Hungary) (22); lane 15, MEX81, I::NH::M (Mexico); lane 16, GRE5, VII′::NH::A (Greece); lane 17, POL1, I::NH::A (Poland) (18); lane 18, JP1, I::A::A (Japan) (1); lane 19, HPV 107, I::E::A (Portugal) (33). (B) Computer-generated dendogram of the PFGE of Fig. 4. Results are from an analysis of similarity by Dice's coeficient; clustering was done by the minimum linkage method (26).
DISCUSSION
In this work, a collection of 494 MRSA isolates was analyzed by applying the same set of molecular typing methods used in other MRSA typing studies performed in our laboratory. Interestingly, only a few clonal types were found among these large collections of isolates recovered in 1996 to 1998 from five different countries of Latin America (Argentina, Brazil, Chile, Uruguay, and Mexico).
The Brazilian multiresistant clonal type (XI::B::B), together with its relatives, represented 79% of the 494 MRSA isolates. Previous studies documented the preponderance and extensive geographic spread in Brazil of a unique MRSA clone XI::B::B. This multiresistant and epidemic clone was reported as the most frequent among MRSA isolates recovered from several hospitals in seven cities located in the Northern as well as in the Southern parts of the country and separated by a distance of several thousand of kilometers from one another (13, 25, 28, 30, 31, 35). Our study reports the predominance (97%) and persistence of clone XI::B::B in Brazil among isolates from 1996 to 1998. This clone has been maintained in the country for a considerable period, at least since 1990 (28), which may explain the large variability associated with PFGE (30 subtypes).
Previous studies have reported the spread of the Brazilian clone to three different cities of Argentina (Buenos Aires, Posadas, and San Miguel de Tucumán) separated by a distance of over 1,000 km from one another (4, 6). Our study confirms the predominance (86%) of the Brazilian clone in 1996 to 1998 in three hospitals of Buenos Aires.
Impressively, the Brazilian clone was the unique clonal type present in Uruguay; clonal type XI::B::B and its variants (III::B::B, IX::B::B, III::B′::B, and XI::B′::B) represented all isolates recovered from 1996 to 1998 in this country, confirming the capacity of spread and high epidemicity of this MRSA clone.
A variant of the Brazilian clone (XI::κ::B), which contains a different Tn554 type (pattern κ), was detected with a frequency of 53% in Chile during 1997 to 1998. Recently, Gales et al. (14) reported an identical observation, among multidrug-resistant MRSA collected during 1997 to 1998 in Brazil, Chile, and Argentina which exhibited an identical ribotype and similar SmaI PFGE pattern to a representative of the Brazilian MRSA clone. Besides the ability to acquire new genes, including high-level mupirocin resistance (25), the Brazilian clone seems to spread alarmingly fast and have a great capacity for displacement of other very epidemic clones, as seen in Portugal (2, 23) and the Czech Republic (20).
Our study clearly documents the massive dissemination of the Brazilian clone in four countries of South America, Brazil, Argentina, Uruguay, and Chile, all sharing directly or indirectly national borders. The presence of this clone in Mexican hospitals cannot be excluded since we have analyzed MRSA isolates exclusively from one Mexican pediatric hospital.
In contrast to the recent report of emergence of a vancomycin and teicoplanin heterogeneously resistant subpopulation among MRSA isolates belonging to the Brazilian clone widely spread in a hospital in the Northeast region of Brazil (13), no VRSA or hetero-VRSA strains have been found among our collection of 499 isolates from Latin America. These strains may be rare, which means it is necessary to continue the surveillance for this phenotype.
Identification of five methicillin-susceptible S. aureus strains with a relatively high drug resistance profile, Tn554 insertions, and PFGE type similar to the PFGE pattern B suggests that they were probably derived from the Brazilian clone by deletion of mecA and mecI. Moreover, four of these strains lack the two Tn554 insertions located in the usual SmaI fragments of mecA (119 or 95 kb), suggesting the loss of these copies of the transposon together with the mecA gene. Recently, two cassette chromosome recombinase genes (ccrA and ccrB) were found to catalyze excision of the 52-kb staphylococcal cassette chromosome mec (16). However, we cannot exclude the hypothesis that these MSSA strains (or strains with a similar background) may be a progenitor to the Brazilian clone.
Clonal type II::E′::F was highly represented in Chile (47%) in coexistence with the Brazilian clone and designated as the Chilean MRSA clone for being exclusively identified in this country so far. The Chilean and the Brazilian clones were represented by similar percentages during 1997 and 1998 (47 versus 53%), which might signify that clone II::E′::F is as epidemic as the Brazilian clone. It would be interesting to monitor the evolution of these two clones in isolates from 1999 to 2000 not only in Chile but also in other countries of South America.
Comparative analysis of MRSA from different geographic origins revealed a high degree of similarity between the single clonal type found in Mexico, designated the Mexican clone (I::NH::M), and the predominant clone found among samples of 1993 and 1998 in a hospital in Patras, Greece (I. Spiliopoulou et al, unpublished). This is probably one more example of the long-distance capacity of spread of certain MRSA clones. Clonal type I::NH::M was found in Mexico among strains recovered from a single hospital and had a relatively limited drug resistance profile; strains showed resistance to penicillin, oxacillin, and gentamicin only and belonged to the phenotypic oxacillin expression class 2 (MIC = 12 μg/ml), which may be explained by its pediatric origin. Isolates belonging to the Pediatric MRSA clone were found to lack Tn554 transposon, belong to oxacillin expression class 1 or 2, and show lower-resistance phenotypes, which are probably related to the antibiotic policy practiced in pediatric settings (29).
Among strains belonging to the Brazilian, the Chilean, and the Mexican clones, only representatives of the Brazilian clone had homology with the mecI gene. Strains belonging to the Pediatric MRSA clone (29), as well as historically early MRSA isolates belonging to the Archaic clone (7), were found to lack sequences that hybridize with mecI. We might speculate that the Chilean and Mexican clones represent chronologically old MRSA clones that survived into the contemporary era.
There is no considerable difference between both periods (1996 to 97 and 1998) except for Uruguay, where clonal types associated with the Tn554 pattern B′ tended to decrease relative to clones associated with Tn554 pattern B. This fact was not observed in other countries, and the reason for this remains unclear. A great number of variants of the major clonal types were predominantly found in larger hospitals, elderly patients, patients not immunocompromized, and patients interned in the Medicine service, which may be related to a long period of hospitalization.
This study provides a perspective on the increasing uniformity of the MRSA clones in South America, which seems to be due to the continued geographic spread of a relative few very epidemic strains among hospitals, across national boundaries and across continents.
ACKNOWLEDGMENTS
This work was partially supported by 2/2.1/SAU/1295/95 and ESP/34872/99-00 from the Fundação para a Ciência e Tecnologia, Portugal, and by a grant from the Fundação Caloust Gulbenkian awarded to H. de Lencastre. The 1997 and 1998 strains were obtained with a grant from Rhône-Poulenc Rorer, S. A., to Alexander Tomasz and Hermínia de Lencastre. M. Aires de Sousa and S. Ávila were supported by grants BD13731/97 and 002/99/BTI/P, respectively, from PRAXIS XXI. I. Adamson, S. T. Casagrande, R. Palacio, and L. Dell'Acqua received CEM/NET fellowships from Fundação Calouste Gulbenkian. M. Miragaia was supported by a grant from the Fundação Calouste Gulbenkian.
We thank F. Valdetaro, F. Vitorino, and M. F. A. Moreira from Casa de Saude Santa Marcelina, São Paulo, Brazil, for technical support. We thank J. Liñares for providing protocols for testing VRSA and hetero-VRSA and A. Corso for critical reading of the manuscript.
REFERENCES
- 1.Aires de Sousa M, de Lencastre H, Santos Sanches I, Kikuchi K, Totsuka K, Tomasz A. Similarity of antibiotic resistance patterns and molecular typing properties of methicillin-resistant Staphylococcus aureus widely spread in hospitals in New York City and in a hospital in Tokyo, Japan. Microb Drug Resist. 2000;6:221–226. doi: 10.1089/mdr.2000.6.253. [DOI] [PubMed] [Google Scholar]
- 2.Aires de Sousa M, Sanches I S, Ferro M L, Vaz M J, Saraiva Z, Tendeiro T, Serra J, de Lencastre H. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus (MRSA) clone. J Clin Microbiol. 1998;36:2590–2596. doi: 10.1128/jcm.36.9.2590-2596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung M, de Lencastre H, Matthews P, Tomasz A Multilab Project Collaborators I. Adamsson, M. Aires de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sá-Leão, I. S Sanches, J. H. Song, P. T. Tassios, and P. Villari. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb Drug Resist. 2000;6:189–198. doi: 10.1089/mdr.2000.6.189. [DOI] [PubMed] [Google Scholar]
- 4.Corso A, Santos Sanches I, Aires de Sousa M, Rossi A, de Lencastre H. Spread of a dominant methicillin-resistant multiresistant Staphylococcus aureus (MRSA) clone in Argentina. Microb Drug Resist. 1998;4:277–288. doi: 10.1089/mdr.1998.4.277. [DOI] [PubMed] [Google Scholar]
- 5.Couto I, Santos Sanches I, Leão R S, de Lencastre H. Molecular characterization of Staphylococcus sciuri strains isolated from humans. J Clin Microbiol. 2000;38:1136–1143. doi: 10.1128/jcm.38.3.1136-1143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Da Silva-Coimbra M V, Teixeira L A, Ramos R L, Predari S C, Castello L, Famiglietti A, Vay C, Klan L, Figueiredo A M. Spread of the Brazilian epidemic clone of a multiresistant MRSA in two cities in Argentina. J Med Microbiol. 2000;49:187–192. doi: 10.1099/0022-1317-49-2-187. [DOI] [PubMed] [Google Scholar]
- 7.De Lencastre H, Chung M, Westh H. Archaic strains of methicillin-resistant Staphylococcus aureus: molecular and microbiological properties of isolates from the 1960s in Denmark. Microb Drug Resist. 2000;6:1–10. doi: 10.1089/mdr.2000.6.1. [DOI] [PubMed] [Google Scholar]
- 8.De Lencastre H, Couto I, Santos I, Melo-Cristino J, Torres-Pereira A, Tomasz A. Methicillin-resistant Staphylococcus aureus disease in a Portuguese hospital: Characterization of clonal types by a combination of DNA typing methods. Eur J Clin Microbiol Infect Dis. 1994;13:64–73. doi: 10.1007/BF02026129. [DOI] [PubMed] [Google Scholar]
- 9.De Lencastre H, Figueiredo A, Urban C, Rahal J, Tomasz A. Multiple mechanisms of methicillin resistance and improved methods for detection in clinical isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 1991;35:632–669. doi: 10.1128/aac.35.4.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Lencastre H, Santos Sanches I, Tomasz A. CEM/NET: clinical microbiology and molecular biology in alliance. In: Tomasz A, editor. Streptococcus pneumoniae: molecular biology and mechanisms of disease. Larchmont, N.Y: Mary Ann Liebert, Inc.; 2000. pp. 451–456. [Google Scholar]
- 11.Diekema D J, Pfaller M A, Jones R N, Doern G V, Kugler K C, Beach M L, Sader H S the SENTRY Participants Group. Trends in antimicrobial susceptibility of bacterial pathogens isolated from patients with bloodstream infections in the USA, Canada, and Latin America. Int J Antimicrob Agents. 2000;13:257–271. doi: 10.1016/s0924-8579(99)00131-4. [DOI] [PubMed] [Google Scholar]
- 12.Dominguez M A, de Lencastre H, Liñares J, Tomasz A. Spread and maintenance of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J Clin Microbiol. 1994;32:2081–2087. doi: 10.1128/jcm.32.9.2081-2087.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dos Santos Soares M J, da Silva-Carvalho M C, Ferreira-Carvalho B T, Figueiredo A M. Spread of methicillin-resistant Staphylococcus aureus belonging to the Brazilian epidemic clone in a general hospital and emergence of heterogenous resistance to glycopeptide antibiotics among these isolates. J Hosp Infect. 2000;44:301–308. [PubMed] [Google Scholar]
- 14.Gales A C, Jones R N, Pfaller M A, Gordon K A, Sader H S, Sampaio J, Zoccoli C, Casellas J M, Smayevsky J, Palavecino E, Prado V, Robledo J A, Sifuentes-Osornio J, Guzman-Blanco M, Bagnulo H. Two-year assessment of the pathogen frequency and antimicrobial resistance patterns among organisms isolated from skin and soft tissue infections in Latin American hospitals: results from the SENTRY antimicrobial surveillance program, 1997–98. Int J Infect Dis. 2000;4:75–84. doi: 10.1016/s1201-9712(00)90098-5. [DOI] [PubMed] [Google Scholar]
- 15.Gomes A R, Santos Sanches I, Aires de Sousa M, Castañeda E, de Lencastre H. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Colombian hospitals: dominance of a single unique multidrug-resistant clone. Microb Drug Resist. 2001;7:23–32. doi: 10.1089/107662901750152729. [DOI] [PubMed] [Google Scholar]
- 16.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreiswirth B, Kornblum J, Arbeit R D, Eisner W, Maslow J N, McGeer A, Low D E, Novick R P. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- 18.Leski T, Oliveira D, Trzcinski K, Sanches I S, de Sousa M A, Hryniewicz W, de Lencastre H. Clonal distribution of methicillin-resistant Staphylococcus aureus in Poland. J Clin Microbiol. 1998;36:3532–3539. doi: 10.1128/jcm.36.12.3532-3539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mato R, Santos Sanches I, Venditti M, Platt D J, Brown A, de Lencastre H. Spread of the multiresistant Iberian clone of methicillin-resistant Staphylococcus aureus (MRSA) to Italy and Scotland. Microb Drug Resist. 1998;4:107–112. doi: 10.1089/mdr.1998.4.107. [DOI] [PubMed] [Google Scholar]
- 20.Melter O, Santos Sanches I, Schindler J, Aires de Sousa M, Mato R, Kovarova V, Zemlickova H, de Lencastre H. Methicillin-resistant Staphylococcus aureus (MRSA) clonal types in the Czech Republic. Microb Drug Resist. 1999;37:2798–2803. doi: 10.1128/jcm.37.9.2798-2803.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. Vol. 15 1995. , no. 14. Approved standard M2–A7. NCCLS, Villanova, Pa. [Google Scholar]
- 22.Oliveira D C, Crisóstomo I, Santos-Sanches I, Major P, Alves C R, Aires de Sousa M, Konkoly-Thege M, de Lencastre H. Comparison of DNA sequencing of the protein A gene polymorphic region with other molecular typing techniques for typing two epidemiologically diverse collections of MRSA. J Clin Microbiol. 2001;39:574–580. doi: 10.1128/JCM.39.2.574-580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira D, Santos-Sanches I, Mato R, Tamayo M, Ribeiro G, Costa D, de Lencastre H. Virtually all methicillin-resistant Staphylococcus aureus (MRSA) infections in the largest Portuguese teaching hospital are caused by two internationally spread multiresistant strains: the “Iberian” and the “Brazilian” clones of MRSA. Clin Microbiol Infect. 1998;4:373–384. doi: 10.1111/j.1469-0691.1998.tb00081.x. [DOI] [PubMed] [Google Scholar]
- 24.Pfaller M A, Jones R N, Doern G V, Sader H S, Kugler K C, Beach M L. Survey of blood stream infections attributable to gram-positive cocci: frequency of occurence and antimicrobial susceptibility of isolates collected in 1997 in the United States, Canada, and Latin America from the SENTRY Antimicrobial Surveillance Program. SENTRY Participants Group. Diagn Microbiol Infect Dis. 1999;33:283–297. doi: 10.1016/s0732-8893(98)00149-7. [DOI] [PubMed] [Google Scholar]
- 25.Ramos R L, Teixeira L A, Ormonde L R, Siqueira P L, Santos M S, Marangoni D, Figueiredo A M. Emergence of mupirocin resistance in multiresistant Staphylococcus aureus clinical isolates belonging to Brazilian epidemic clone III::B::A. J Med Microbiol. 1999;48:303–307. doi: 10.1099/00222615-48-3-303. [DOI] [PubMed] [Google Scholar]
- 26.Roberts R B, de Lencastre A, Eisner W, Severina E P, Shopsin B, Kreiswirth B N, Tomasz A the MRSA Collaborative Study Group. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J Infect Dis. 1998;178:164–171. doi: 10.1086/515610. [DOI] [PubMed] [Google Scholar]
- 27.Sader H S, Jones R N, Gales A C, Winokur P, Kugler K C, Pfaller M A, Doern G V. Antimicrobial susceptibility patterns for pathogens isolated from patients in Latin American medical centers with a diagnosis of pneumonia: analysis of results from the SENTRY Antimicrobial Surveillance Program (1997), SENTRY Latin America Study Group. Diagn Microbiol Infect Dis. 1998;32:289–301. doi: 10.1016/s0732-8893(98)00124-2. [DOI] [PubMed] [Google Scholar]
- 28.Sader H S, Pignatari A C, Hollis R J, Jones R N. Evaluation of interhospital spread of methicillin-resistant Staphylococcus aureus in São Paulo, Brazil, using pulsed-field gel electrophoresis of chromosomal DNA. Rev Saude Publica. 1994;28:406–409. doi: 10.1086/646921. [DOI] [PubMed] [Google Scholar]
- 29.Sá-Leāo R, Santos Sanches I, Dora Dias D, Peres I, Barros R M, de Lencastre H. Detection of an Archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J Clin Microbiol. 1999;37:1913–1920. doi: 10.1128/jcm.37.6.1913-1920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos K R, Teixeira L M, Leal G S, Fonseca L S, Gontijo Filho P P. DNA typing of methicillin-resistant Staphylococcus aureus: isolates and factors associated with nosocomial acquisition in two Brazilian university hospitals. J Med Microbiol. 1999;48:17–23. doi: 10.1099/00222615-48-1-17. [DOI] [PubMed] [Google Scholar]
- 31.Santos-Filho L, Sader H S, Bortolotto V I, Gontijo Filho P P, Pignatari A C. Analysis of the clonal diversity of Staphylococcus aureus methicillin-resistant strains isolated at João Pessoa, state of Paraiba, Brazil, Mem. Inst Oswaldo Cruz. 1996;91:101–105. doi: 10.1590/s0074-02761996000100018. [DOI] [PubMed] [Google Scholar]
- 32.Santos Sanches I, Mato R, de Lencastre H, Tomasz A CEM/NET Collaborators S. Nunes, C. R. Alves, M. Miragaia, J. Carriço, I. Couto, I. Bonfim, M. A. de Sousa, D. Oliveira, A. Gomes, M. Vaz, S. Fernandes, S. C. Verde, A. Ávila, F. Antunes, R. Sá-Leão, J. Almeida, O. Melter, and M. Chung; International Collaborators M. C. Brandileone, E. Castañeda, C. Cocuzza, G. Echaniz-Aviles, I. Heitmann, M. Hortal, W. Hryniewicz, F. Jia, K. Kikuchi, M. Konkoly-Thege, K. G. Kristinsson, J. Liñares, A. Rossi, E. Z. Savov, J. Schindler, F. Solorzano-Santos, K. Totsuka, M. Venditti, P. Villari, H. Westh, J.-S. Wu, and R. C. Zanella. Patterns of multidrug resistance among methicillin-resistant hospital isolates of coagulase-positive and coagulase-negative staphylococci collected in the International Multicenter Study RESIST in 1997 and 1998. Microb Drug Resist. 2000;6:199–211. doi: 10.1089/mdr.2000.6.199. [DOI] [PubMed] [Google Scholar]
- 33.Santos Sanches L, Ramirez M, Troni H, Abecassis M, Padua M, Tomasz A, de Lencastre H. Evidence for the geographic spread of a methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J Clin Microbiol. 1995;33:1243–1246. doi: 10.1128/jcm.33.5.1243-1246.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sieradzki K, Roberts R, Haber S, Tomasz A. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. New Engl J Med. 1999;340:517–523. doi: 10.1056/NEJM199902183400704. [DOI] [PubMed] [Google Scholar]
- 35.Teixeira L, Resende C A, Ormonde L R, Rosenbaum R, Figueiredo A M S, de Lencastre H, Tomasz A. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J Clin Microbiol. 1995;33:2400–2404. doi: 10.1128/jcm.33.9.2400-2404.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tenover F C, Lancaster M V, Hill B C, Steward C D, Stocker S A, Hancock G A, O'Hara C M, Clark N C, Hiramatsu K. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J Clin Microbiol. 1998;36:1020–1027. doi: 10.1128/jcm.36.4.1020-1027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomasz A, de Lencastre H. Molecular microbiology and epidemiology: coexistence or alliance? In: Wenzel R P, editor. Prevention and control of nosocomial infections. Baltimore, Md: Williams & Wilkins; 1997. pp. 309–321. [Google Scholar]
- 39.Tomasz A, Nachman S, Leaf H. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother. 1991;35:124–129. doi: 10.1128/aac.35.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]