Abstract
Background: Accumulating evidence has suggested that there is a positive association between asthma and cardiovascular diseases (CVDs), implying a common architecture between them. However, the shared genetic architecture and causality of asthma and CVDs remain unclear.
Methods: Based on the genome-wide association study (GWAS) summary statistics of recently published studies, our study examined the genetic correlation, shared genetic variants, and causal relationship between asthma (N = 127,669) and CVDs (N = 86,995–521,612). Statistical methods included high-definition likelihood (HDL), cross-trait meta-analyses of large-scale GWAS, transcriptome-wide association studies (TWAS), and Mendelian randomization (MR).
Results: First, we observed a significant genetic correlation between asthma and heart failure (HF) (Rg = 0.278, P = 5 × 10−4). Through cross-trait analyses, we identified a total of 145 shared loci between asthma and HF. Fifteen novel loci were not previously reported for association with either asthma or HF. Second, we mapped these 145 loci to a total of 99 genes whose expressions are enriched in a broad spectrum of tissues, including the seminal vesicle, tonsil, appendix, spleen, skin, lymph nodes, breast, cervix and uterus, skeletal muscle, small intestine, lung, prostate, cardiac muscle, and liver. TWAS analysis identified five significant genes shared between asthma and HF in tissues from the hemic and immune system, digestive system, integumentary system, and nervous system. GSDMA, GSDMB, and ORMDL3 are statistically independent genetic effects from all shared TWAS genes between asthma and HF. Third, through MR analysis, genetic liability to asthma was significantly associated with heart failure at the Bonferroni-corrected significance level. The odds ratio (OR) is 1.07 [95% confidence interval (CI): 1.03–1.12; p = 1.31 × 10−3] per one-unit increase in loge odds of asthma.
Conclusion: These findings provide strong evidence of genetic correlations and causal relationship between asthma and HF, suggesting a shared genetic architecture for these two diseases.
Keywords: asthma, heart failure, shared genetics, genetic correlation, Mendelian randomization
Introduction
Asthma is a heterogeneous disease with a strong genetic basis and is a major global public health problem worldwide due to its high prevalence and inadequate disease control (Barnes, 2011; Yang et al., 2017). Cardiovascular diseases (CVDs) including heart failure (HF) are a rapidly growing public health threat globally (Roth et al., 2017) and confer a substantial burden to the healthcare system (Ziaeian and Fonarow, 2016). Globally, both asthma and CVDs have increased over the past several decades, measured by disease incidence rate (for asthma) or prevalence (for CVDs) (Gottdiener, 2017).
Previous studies have reported overall convincing positive associations between asthma and CVDs (Tattersall et al., 2016; Sun et al., 2017). Observational studies have found that a history of asthma was associated with higher adulthood left ventricular mass index (LVMI) and hypertension (Sun et al., 2017). A large-cohort study of adults found late-onset asthmatics had an increased risk of incident CVDs that persisted after adjustment for age, sex, and CVD risk factors (Tattersall et al., 2016). In previous large multi-ethnic cohort studies, persistent asthmatics had a higher CVD event rate than non-asthmatics (Tattersall et al., 2015; Tattersall et al., 2020; Wee et al., 2021) and adult patients with asthma had increased risks of HF incident (Iribarren et al., 2012). Patients with asthma–chronic obstructive pulmonary disease (COPD) overlap syndrome (ACOS) were associated with a higher risk of HF (Yeh et al., 2017). It has been well known that observational studies are susceptible to reverse causality and confounding; therefore, whether asthma plays a causal role in the development of CVDs or vice versa remains largely unknown (Gottdiener, 2017). In particular, inflammation is involved in the pathophysiology of both diseases and therefore becomes a key confounder to study the causal effect between these two traits (Gottdiener et al., 2000; Fahy, 2015). Genetic studies showed that genetic markers of inflammation play important roles in the development of both CVDs and asthma (Raman et al., 2013), implying a common genetic architecture between asthma and CVDs. Besides the investigation of inflammation and potentially a few other similar focused studies, to our knowledge, no large-scale, genome-wide study has systematically reported the shared genetic loci between asthma and CVDs.
In this study, we conducted genome-wide, large-scale, cross-trait analyses to examine the genetic correlation and identify shared variants between asthma and CVDs. These included genome-wide genetic correlation analyses, Mendelian randomization (MR)-based causal inference, and functional enrichments analyses.
Methods
Genome-Wide Cross-Trait Analysis Study Design
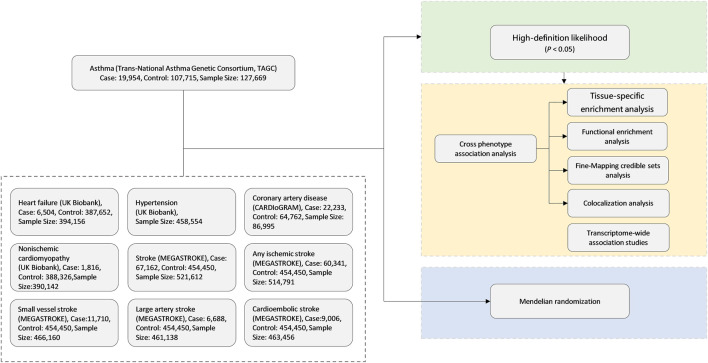
The overall study design is shown in Figure 1. We conducted a large-scale, genome-wide, cross-trait meta-analysis to identify the shared genetic architecture between asthma and CVDs. Our study contains three parts. Firstly, we adopted high-definition likelihood (HDL), a recently developed method, for calculation of genetic correlation between asthma and CVDs to find the significant trait pairs. Secondly, for those significant trait pairs in the first step, we run cross-trait association and meta-analyses to identify the shared loci of significant trait pairs. Finally, we applied MR analyses to evaluate whether asthma is causally associated with CVDs, especially the ones with significant genetic correlations in the first step.
FIGURE 1.
Genome-wide cross-trait analysis study design. CARDIoGRAM, Coronary ARtery Disease Genome-Wide Replication And Meta-Analysis; MEGASTROKE, multi-ancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes.
Through literature review and based on the availability of full genome-wide association study (GWAS) data, we chose the following published GWAS for our analyses: 1) asthma (N = 127,669), from the Trans-National Asthma Genetic Consortium (TAGC) (Demenais et al., 2018); 2) HF (N = 394,156) and non-ischemic cardiomyopathy (NICM) (N = 390,142), from the study of Aragam et al. (2018); 3) coronary artery disease (CAD) (N = 22,233), from the Coronary ARtery Disease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) (Schunkert et al., 2011); and 4) all-cause stroke and subtype-specific stroke [any ischemic stroke (AIS), large-artery stroke (LAS), cardioembolic stroke (CES), and small-vessel stroke (SVS)] (N = 6,688–67,162), from MEGASTROKE (Malik et al., 2018). The full GWAS summary statistics for all these traits are publicly available for downloading (Supplementary Table S1). All of the datasets were from European populations.
Genetic Correlation Analysis
HDL was used to estimate the genetic correlation between asthma and CVDs in our genome-wide, cross-trait genetic correlation analysis. HDL used summary data from GWAS to compute the genetic correlation (ranging from −1 to 1), which is similar to linkage disequilibrium score regression (LDSC). However, HDL was reported to reduce the variance of genetic correlation estimates and is therefore more powerful to reveal the potential connections between human complex traits (Ning et al., 2020).
Cross-Phenotype Association Analyses
GWAS has identified genetic variation for many complex traits, but studies have shown that many detected loci can be associated with multiple traits at the same time. This phenomenon is called cross-phenotype (CP) association, or simply put, pleiotropy (Solovieff et al., 2013). Detection of shared loci for cross-phenotype associations can improve statistical testing and uncover more genetic information between different traits. We used cross-phenotype association analyses (CPASSOC) to identify shared loci between asthma and CVDs in genome-wide scale. We chose CPASSOC over other similar methods/tools because it allows heterogeneity test between traits, controls for population structure and cryptic relatedness, and effectively accounts for sample overlapping. Considering the heterogeneity of different traits in this study, we chose S Het as the main statistic. We used the threshold of P SHet <5e−8 and p < 0.05 to identify CP-associated single-nucleotide polymorphisms (SNPs) that have effects on asthma and each of the CVDs that are included in this study (Zhu et al., 2015). CP-associated SNPs were compared with reported gene and SNPs of asthma and HF with the NHGRI-EBI GWAS catalog (July 2021) in order to test independent replication and find novel loci. Independent loci were extracted after performing SNP prioritization based on linkage disequilibrium for CP-associated SNPs.
Tissue-Specific Enrichment Analysis
The analyses mentioned above only studied genetics at the DNA level. We next went downstream the chain of the central dogma of molecular biology, to examine tissue-specific gene expression. We first run tissue-specific enrichment analysis (TSEA) to identify gene expressions that are highly enriched or expressed in certain given tissues (Bindea et al., 2009). In our study, we used the gene sets obtained in cross-phenotypic association analysis to calculate tissue-specific gene enrichment and identify tissues that share enriched gene expression between asthma and CVDs. We calculated tissue-specific gene enrichment statistic using the hypergeometric test in the “TissueEnrich” R package (Jain and Tuteja, 2019). The RNA-seq data used in this R package was from the GTEx project (GTEx Consortium, 2015) and Human Protein Atlas (HPA) (Uhlen et al., 2015). In brief, GTEx analyzed RNA sequencing data from a total of 1,641 samples from 43 tissues of 145 individuals to compile a catalog of thousands of tissue-specific and shared regulatory expression quantitative trait loci (eQTL) variants (GTEx Consortium, 2015).
Functional Enrichment Analysis
Functional enrichment analysis is a method to find a series of over-represented genes or proteins. We used the ClueGO software to assess the enrichment of gene sets in Gene Ontology (GO) biologic process, reactome pathways, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Bindea et al., 2009). GO annotates genes to biological process, molecular function, and cellular component in a hierarchically structured way (Ashburner et al., 2000). KEGG assigns genes to functional pathways (Kanehisa et al., 2002). The reactome pathway (https://reactome.org) provides molecular details of signal transduction, transport, DNA replication, metabolism, and other cellular processes (Jassal et al., 2020). In this study, we used the gene sets corresponding to significant CP-associated loci obtained in CPASSOC to explore the enrichment and relationship between shared gene sets and GO terms, reactome, and KEGG pathways (Ashburner et al., 2000; Kanehisa et al., 2002; The Gene Ontology Consortium, 2019; Jassal et al., 2020).
Transcriptome-Wide Association Studies
TWAS enables to evaluate tissue-specific expression–trait associations and the shared expression–trait associations between asthma and CVDs by integrating GWAS summary data and gene expression datasets (Gusev et al., 2016). In our study, we conducted TWAS based on 48 GTEx (version 7) tissue expression weights of the FUSION software package and compared the results with those obtained by TSEA (GTEx Consortium, 2015). We applied Bonferroni correction on TWAS p-values, and P Bonferroni < 0.05 was considered significant.
Fine-Mapping Credible Set Analysis
A credible variant set most likely to be causally related in each risk locus can be obtained from fine mapping credible set analysis, replacing the original independent significant loci for subsequent functional studies, with a higher probability to obtain the true causal loci (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014; Fachal et al., 2020). In our study, we extracted variants within 500 kb in every locus that were highly linked (R 2 > 0.4) with CP-associated loci of asthma and CVDs and then used Bayesian likelihood fine-mapping algorithm to determine a 99% plausible set of causal loci, making the identified regions more precise (Wellcome Trust Case Control Consortium et al., 2012).
Colocalization Analysis
The colocalization analysis is used to assess the colocalization of association loci in two different GWAS by fully Bayesian colocalization analyses, which can help to gain insight into the molecular pathways of complex diseases (Giambartolomei et al., 2014; Hukku et al., 2021). In our study, we used the “coloc.abf” function from the “coloc” R package to conduct genetic colocalization analysis of asthma and CVDs, calculating the probability that each CP-associated locus is a shared genetic causal variant (H4). Loci with posterior probabilities greater than 0.5 were considered colocalized.
Mendelian Randomization
Mendelian randomization takes advantage of the innate stability and ease of measurement of genetic variation, using it as an instrumental variable (IV) for analysis and thereby obtaining a causal relationship between exposure and outcome (Emdin et al., 2017). Observational studies have found that lung disease is independently associated with CVDs, especially ischemic heart disease and HF (Carter et al., 2019). However, the actual relationship between asthma and CVDs remains unclear. Inverse variance weighting (IVW) was used as the main method to assess causality. Two median-based methods (simple and weighted), MR-Egger and MR-Pleiotropy RESidual Sum and Outlier (MR-PRESSO) methods, were used as sensitivity analyses.
Results
Genetic Correlation
Among nine CVDs, we found a significant genetic correlation between asthma and HF (Rg = 0.278, p = 0.0005). HDL analyses did not support a significant genetic correlation between asthma and CAD, hypertension (Hp), NICM, stroke, SVS, AIS, CES, and LAS. Estimates of genetic correlation using GWAS summary statistics data are shown in Table 1 (SNP-based heritability estimated by HDL is shown in Supplementary Table S2).
TABLE 1.
Genetic correlation between asthma and cardiovascular diseases.
| Correlation | Rg | Rg SE | p |
|---|---|---|---|
| Heart failure | 0.278 | 0.080 | 0.0005 |
| Coronary artery disease | 0.003 | 0.191 | 0.9894 |
| Hypertension | 0.091 | 0.059 | 0.1236 |
| Non-ischemic cardiomyopathy | 0.155 | 0.137 | 0.2571 |
| Stroke | 0.133 | 0.076 | 0.0796 |
| Small-vessel stroke | 0.156 | 0.162 | 0.3347 |
| Any ischemic stroke | 0.128 | 0.076 | 0.0927 |
| Cardioembolic stroke | 0.145 | 0.086 | 0.0923 |
| Large-artery stroke | 0.253 | 0.154 | 0.1008 |
Rg, genetic correlation estimate; SE, standard error.
Cross-Phenotype Association Analysis
CPASSOC analyses identified a total of 145 independent loci (P SHet <5 × 10−8 and trait-specific p < 0.05), which maps to 99 genes (Supplementary Table S3). We compared the reported gene and SNPs of asthma and HF with the NHGRI-EBI GWAS catalog (July 2021) and found that 15 of 145 SNPs were novel associations not previously reported for either asthma or HF (Supplementary Table S4). Independent loci were extracted after performing SNP prioritization based on linkage disequilibrium (LD) for CP-associated SNPs and are shown in Table 2. The threshold P of LD is 5e−5, with clump results using a distance of 500 kb and using an R 2 of 0.4. The strongest association signal was observed near the GSDMB gene on chromosome 17 (P SHet = 7.13 × 10−44). We also observed the association signal near ORMLD3 (P SHet = 1.82 × 10−43) as well as IKZF3 (P SHet = 1.07 × 10−41) on chromosome 17. Previous studies reported that GSDMB and ORMDL3 are associated with asthma (Zhao et al., 2015), but their association with HF was not reported. IKZF3 is associated with the IL-2 signaling pathway and NF-κB signaling pathway, which are involved in regulating BCL2 expression and controlling apoptosis (Kioussis, 2007). Our HDL findings suggest that the overall strong positive genetic correlation between asthma and HF is driven by these identified shared genetic loci.
TABLE 2.
Summary of independent loci associated with asthma and heart failure in cross-phenotype association analysis.
| SNP | CHR | POS | Alt. allele | Ref. allele | Asthma P | HF P | Meta-analysis P | DIST | Nearest gene to the loci |
|---|---|---|---|---|---|---|---|---|---|
| rs2305479 | 17 | 38062217 | C | T | 1.00E−42 | 3.76E−04 | 7.13E−44 | 0 | GSDMB |
| rs4795408 | 17 | 38107627 | G | A | 2.12E−36 | 2.18E−04 | 3.12E−37 | 6.64 | LRRC3C |
| rs7207600 | 17 | 38091660 | G | A | 4.72E−29 | 8.34E−03 | 1.58E−29 | 6.066 | LRRC3C |
| rs2952156 | 17 | 37876835 | A | G | 7.55E−29 | 2.14E−02 | 2.60E−29 | 0 | ERBB2 |
| rs3859192 | 17 | 38128648 | C | T | 5.38E−28 | 1.92E−02 | 2.03E−28 | 0 | GSDMA |
| rs11078930 | 17 | 38141955 | C | T | 1.49E−20 | 3.67E−02 | 1.24E−20 | 0 | PSMD3 |
| rs876493 | 17 | 37824545 | G | A | 1.92E−16 | 1.07E−02 | 2.42E−16 | 0 | PNMT |
| rs8069451 | 17 | 37504933 | T | C | 1.50E−08 | 4.67E−02 | 3.70E−08 | 0 | FBXL20 |
| rs2158177 | 5 | 131984058 | A | G | 6.18E−11 | 2.32E−02 | 1.28E−10 | 3.745 | RAD50 |
| rs2248116 | 5 | 131804347 | C | A | 6.67E−11 | 3.29E−02 | 1.38E−10 | 0 | C5orf56 |
| rs12521097 | 5 | 131575338 | G | A | 1.70E−08 | 1.91E−02 | 4.22E−08 | 11.782 | P4HA2 |
| rs10124250 | 9 | 6161686 | C | T | 2.40E−12 | 3.70E−02 | 4.41E−12 | 54.099 | IL33 |
| rs1333042 | 9 | 22103813 | A | G | 2.97E−04 | 3.23E−07 | 2.53E−09 | 0 | CDKN2B-AS1 |
| rs2383206 | 9 | 22115026 | A | G | 4.05E−04 | 4.95E−07 | 4.95E−09 | 0 | CDKN2B-AS1 |
| rs10811650 | 9 | 22067593 | A | G | 1.15E−03 | 1.68E−06 | 3.74E−08 | 0 | CDKN2B-AS1 |
SNP, single-nucleotide polymorphism; CHR, chromosome; POS, position of the clumping region on the chromosome; Alt. allele, alternative allele; Ref. allele, reference allele; HF, heart failure; DIST: the smallest (in absolute value) signed distance of the nearest gene to the loci. The threshold P of linkage disequilibrium is 5e−5. Clump results use a distance of 500 kb and R 2 of 0.4. SNP positions are reported according to Build GRch37.
Tissue-Specific Enrichment Analysis
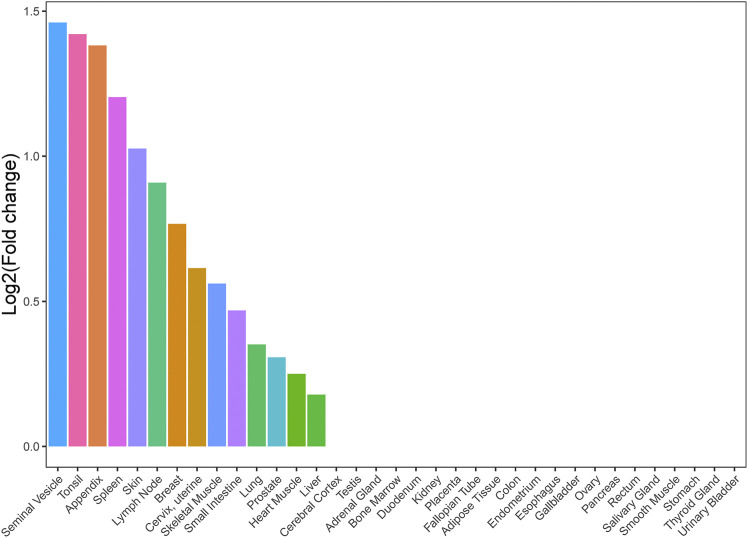
We conducted a TSEA for asthma and HF with significant genetic correlation using GTEx pilot data to assess whether shared genes between the two traits are enriched for expression in tissues. We chose all tissues in GTEx in order to provide complete analysis results and explore some potential tissues associated with shared genes between asthma and HF. We found that shared genes between asthma and HF were significantly enriched in 14 tissues, including the seminal vesicle, tonsil, appendix, spleen, skin, lymph nodes, breast, cervix and uterus, skeletal muscle, small intestine, lung, prostate, cardiac muscle, and liver (Figure 2). These results support that there is a suite of complex biological process underlying the shared genetic architecture between asthma and HF. It is not driven by a single tissue type or a single biological process such as inflammation.
FIGURE 2.
Tissue enrichment analysis result of shared genes between asthma and HF. Vertical axis: the logarithm of tissue expression enrichment fold change based on two; horizontal axis: 35 independent tissues. HF, heart failure.
Functional Enrichment Analysis
In order to understand the biological processes between asthma and HF, we conducted an analysis of GO biologic process, KEGG pathways, and reactome pathways (false discovery rate <0.05). The GO analysis indicated that shared genes between asthma and HF were significantly enriched in nuclear receptor activity, mating behavior, cyclin-dependent protein serine/threonine kinase regulator activity, granulocyte differentiation, regulation of B cell proliferation, negative regulation of lymphocyte differentiation, regulation of transcription initiation from RNA polymerase II promoter, and regulation of leukocyte apoptotic process (Supplementary Figure S1; Supplementary Table S5). To further know the functional pathways, KEGG pathway analyses indicated that shared genes were significantly enriched in hematopoietic cell lineage, Fc epsilon RI signaling pathway, and acute myeloid leukemia (Supplementary Table S6). In additional analyses of reactome pathways, we found that shared association signals for asthma and HF were significantly enriched in the nuclear receptor transcription pathway and interleukin receptor SHC signaling (Supplementary Table S7).
Transcriptome-Wide Association Studies
TWAS analyses were used to evaluate tissue-specific expression–trait associations and the shared expression–trait associations between asthma and HF, to identify shared genes revealing the shared genetic architecture or mechanisms. We compared the asthma and HF reported gene with the NHGRI-EBI GWAS catalog in July 2021, respectively, and found that 829 of 1,619 asthma genes were novel to asthma and 199 of 254 HF genes were novel to HF (significant genes were identified by P Bonferroni <0.05). TWAS results of asthma and HF are shown in Supplementary Tables S8 and S9.
A large number of TWAS-significant associations for asthma were identified in tissues that are part of the nervous system, digestive system, exo-/endocrine system, as well as cardiovascular system. The strongest expression–trait associations for asthma were observed at PNMT (phenylethanolamine N-methyltransferase) on chromosome 17 (the strongest association at the adipose visceral omentum, P TWAS = 6.58 × 10−46), which catalyzes the synthesis of epinephrine from norepinephrine (Mahmoodi et al., 2020). In addition, GSDMB (the strongest association at the small intestine terminal ileum, P TWAS = 1.37 × 10−44), RP11-387H17.4 (the strongest association at the coronary artery, P TWAS = 1.43 × 10−43), GSDMB (the strongest association at transverse colon, P TWAS = 1.00 × 10−42), and ORMDL3 (the strongest association at cell-transformed fibroblasts, P TWAS = 7.81 × 10−43) were also TWAS significant (Supplementary Figure S2). These results were consistent with those reported by Demenais et al. (2018), which studied asthma risk loci that colocalize with immune cell enhancer marks.
A large number of TWAS-significant associations for HF were identified in tissues that are part of the digestive system, nervous system, exo-/endocrine system, and cardiovascular system, similar with asthma TWAS. The strongest expression–trait associations for asthma were observed at RP11-378J18.8 on chromosome 1 (the strongest association at the uterus, P TWAS = 1.43 × 10−7). In addition, MAP3K7CL (the strongest association at cell-transformed fibroblasts, P TWAS = 4.86 × 10−7), LINC00189 (the strongest association at the spleen, P TWAS = 1.25 × 10−6), and MRPL35 (strongest association at coronary artery, P TWAS = 1.66 × 10−6; the strongest association at the sigmoid colon, P TWAS = 1.51 × 10−6) were also TWAS significant (Supplementary Figure S3).
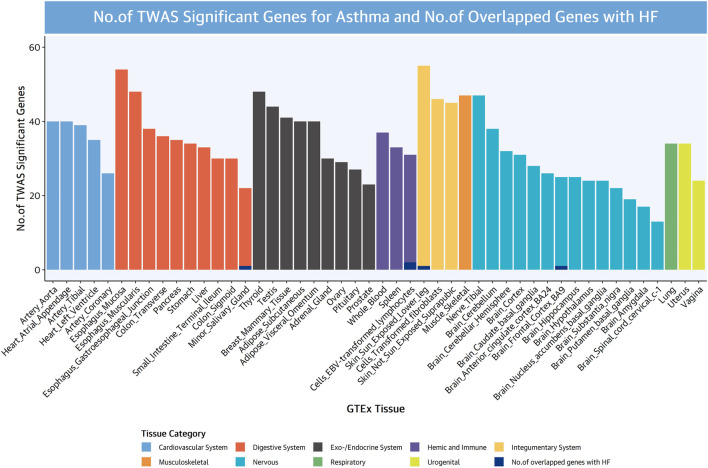
We examined TWAS genes shared between asthma and HF and identified five TWAS-significant genes shared between asthma and HF, mainly from the hemic and immune systems (two TWAS-significant genes at EBV-transformed lymphocytes), digestive system (one TWAS-significant gene at the minor salivary gland), integumentary system (one TWAS-significant at the skin), and nervous system (one TWAS-significant at the brain frontal cortex) (Figure 3).
FIGURE 3.
Numbers of significant genes for asthma and numbers of overlapped genes with HF. Significant genes were identified by P Bonferroni <0.05.
We identified three genes (GSDMA in chr17, GSDMB in chr17, and ORMDL3 in chr17) with statistically independent genetic effects from all of the shared TWAS genes between asthma and HF (Table 3). GSDMA and GSDMB induce inflammatory response and cell pyroptosis (Ding et al., 2016). ORMLD3 could facilitate the survival of splenic B cells via an ATF6α-endoplasmic reticulum stress-beclin1 autophagy regulatory pathway (Dang et al., 2017).
TABLE 3.
TWAS significant genes shared between asthma and HF.
| Gene | HF | Asthma | ||||||
|---|---|---|---|---|---|---|---|---|
| BEST. GWAS.ID | TWAS.Z | P Bonferroni | Tissue | BEST. GWAS.ID | TWAS.Z | P Bonferroni | Tissue | |
| GSDMA | rs11557467 | −4.0641 | 0.0283 | Sun-exposed lower leg skin | rs2305479 | −12.9856 to 10.9691 | 1.12E−35 to 2.60E−19 | Adipose subcutaneous, adipose visceral omentum, adrenal gland, aortic artery, coronary artery, tibial artery, brain substantia nigra, breast mammary tissue, cell-transformed fibroblasts, sigmoid colon, transverse colon, gastroesophageal junction of the esophagus, esophageal mucosa, esophagus muscularis, heart atrial appendage, heart left ventricle, lung, minor salivary gland, skeletal muscle, tibial nerve, pancreas, prostate, sun-exposed lower leg skin, small intestine terminal ileum, spleen, stomach, testis, and thyroid |
| GSDMB | rs11557467 | 3.7006 to 3.7704 | 0.0209 to 0.0441 | Minor salivary gland, cells EBV-transformed lymphocytes | rs2305479 | −8 to 14.0094 | 3.96E−42 to 9.58E−09 | Adipose subcutaneous, adrenal gland, tibial artery, brain cortex, brain nucleus accumbens basal ganglia, cells EBV-transformed lymphocytes, cell-transformed fibroblasts, transverse colon, esophageal mucosa, esophagus muscularis, lung, minor salivary gland, tibial nerve, ovary, pancreas, sun-exposed lower leg skin, small intestine terminal ileum, spleen, stomach, and whole blood |
| ORMDL3 | rs11557467 | 3.7006 to 3.8588 | 0.0247 to 0.0441 | Brain frontal cortex BA9, cells EBV-transformed lymphocytes | rs2305479 | −12.7 to 13.7191 | 5.08E−40 to 9.07E−06 | Adipose subcutaneous, adrenal gland, aortic artery, brain frontal cortex BA9, breast mammary tissue, cells EBV-transformed lymphocytes, cell-transformed fibroblasts, sigmoid colon, esophageal mucosa, esophagus muscularis, lung, ovary, suprapubic skin not sun exposed, sun-exposed lower leg skin, spleen, stomach, thyroid, and whole blood |
CHR, chromosome; HF, heart failure; TWAS, transcriptome-wide association scan.
Fine-Mapping and Colocalization Analyses
To more accurately assess whether asthma and HF share common genetic variants within a given region, we conducted a fine-mapping analysis. The credible set of SNPs for each shared locus is shown in the Supplementary Material (Supplementary Table S10). The colocalization analysis showed that 22 out of 145 loci (H4 > 0.5) share causal variants between asthma and HF (Supplementary Table S11).
Mendelian Randomization
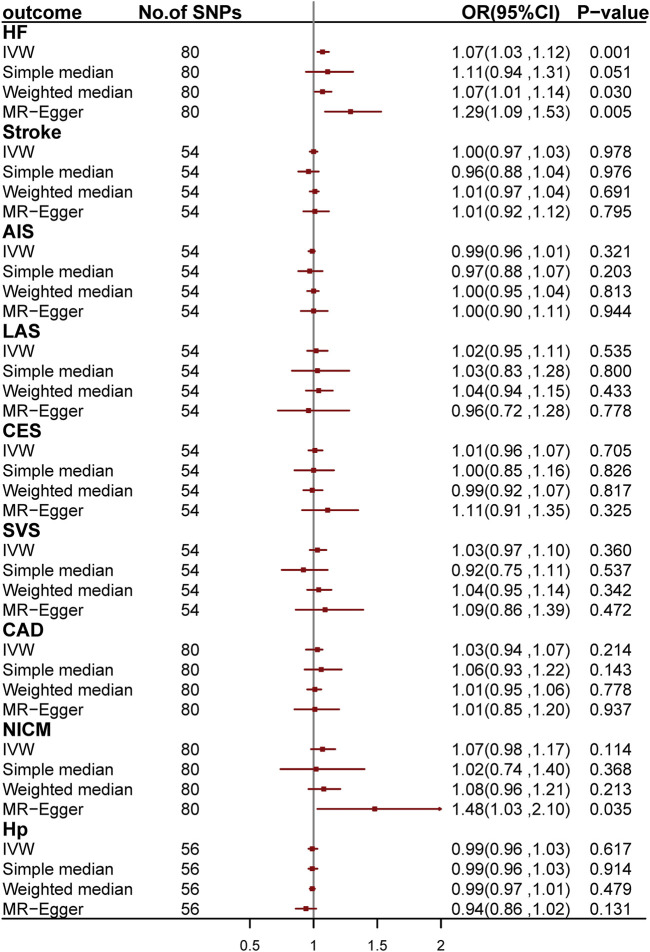
We conducted two-sample MR analyses to investigate the causal relationship between asthma and CVDs by using 54, 80, 80, 80, and 56 genetic variants as IV for stroke and its subgroups, CAD, HF, NICM, and Hp, respectively (Figure 4; Supplementary Tables S11–S21). After Bonferroni correction, we found a causal relationship between asthma and HF. The odds ratio (OR) is 1.07 [95% confidence interval (CI): 1.03–1.12; p = 1.31 × 10−3] per one-unit increase in loge odds of asthma. We further validated the causal effect by applying MR-PRESSO regression, which is robust to bias due to horizontal pleiotropy (Bowden et al., 2015; Burgess and Thompson, 2017).
FIGURE 4.
Odds ratio for the association among genetically predicted asthma and cardiovascular-related traits. OR, odds ratio; 95%CI, 95% confidence interval; AIS, any ischemic stroke; CES, cardioembolic stroke; LAS, large-artery stroke; SVS, small-vessel stroke; CAD, coronary artery disease; HF, heart failure; NICM, non-ischemic cardiomyopathy; Hp, hypertension.
Discussion
We used large-scale GWAS summary statistics to analyze the shared genetic architecture and causal associations between asthma and CVDs. First, we found a strong positive genetic correlation and further identified 145 shared significant loci between asthma and HF, of which 15 were novel. Second, we found that these shared loci were significantly enriched in 14 tissues and B-cell proliferation, Fc epsilon RI signaling, and nuclear receptor transcription pathway using functional analysis. Third, we found that shared genes between asthma and HF are focused on the hemic and immune systems, digestive system, integumentary system, and nervous system using TWAS analysis. Finally, we found a causal relationship between asthma and HF using MR analysis.
A previous study showed that the risks of HF were increased in asthma, COPD, and ACOS (Ingebrigtsen et al., 2020). In the present study, we have identified 145 shared loci between asthma and HF through large-scale, genome-wide, cross-trait meta-analysis, of which 15 were novel associations, and all the 145 shared loci did not achieve genome-wide trait-specific significance for both asthma and HF GWAS summary statistics, which demonstrated the great statistical power of our cross-trait meta-analysis.
The top locus common to the asthma and HF was rs2305479 near GSDMB. GSDMB is an important member of the GSDMS family, like GSDMA confirmed in TWAS, whose most important function is to induce inflammatory response and cell pyroptosis (Ding et al., 2016). Therefore, GSDMB may affect myocardial cell survival through its involvement in the process of pyroptosis. In addition, we found that asthma was causally related with HF (per one-unit increase in log OR of asthma). The result was consistent with a large cohort study (n = 203,595), which found that asthma was associated with a 2.14-fold (95% CI: 2.06, 2.22) hazard of HF (Iribarren et al., 2012). Previous studies have demonstrated that inflammatory processes are likely involved in the pathophysiology of asthma, atherosclerosis, and endothelial dysfunction (Fenta et al., 2010; Yun et al., 2012). A previous study indicates that cytokines that are up-regulated in asthma may promote the migration and activation of inflammatory cells implicated in atherogenesis (Halasz et al., 2002; Zietkowski et al., 2007; Zietkowski et al., 2010), and cytokines are also important mediators of bronchoconstriction in asthma (Hiemstra et al., 2015). Leukotrienes as a kind of cytokines found in asthmatic bronchioles also exhibits strong proinflammatory activities in cardiovascular tissues (Spanbroek et al., 2003; Gurgone et al., 2020). Previous studies have shown that pyroptosis not only plays an important role in infectious diseases but is also associated with CVDs, central nervous system diseases, and tumors (Gong et al., 2020; Zhou et al., 2020). TSEA showed that shared genes were enriched in 14 tissues, including the seminal vesicle, tonsil, appendix, spleen, skin, lymph nodes, breast, cervix and uterus, skeletal muscle, small intestine, lung, prostate, cardiac muscle, and liver. Prostaglandins related to the seminal vesicle are suggested to regulate allergic diseases (Peebles, 2019). Prostaglandins and leukotrienes are involved in numerous homeostatic biological functions and inflammation (Funk, 2001). Our TWAS reported tissue enrichment from the hemic and immune systems, digestive system, integumentary system, and nervous system. Another top association for asthma and HF was ORMDL3, which could facilitate the survival of splenic B cells via an ATF6α-endoplasmic reticulum stress-beclin1 autophagy regulatory pathway (Dang et al., 2017). GSDMB and ORMDL3 were also confirmed to be shared between asthma and HF using TWAS in our study. In addition, IKZF3 was confirmed in CPASSOC to be associated with the IL-2 signaling pathway and NF-κB signaling pathway, which are involved in regulating BCL2 expression and controlling apoptosis (Kioussis, 2007).
The functional analysis provided biologic insights into the shared genes between asthma and HF, and nuclear receptor-related pathway was found significant in GO and reactome pathway. The nuclear receptor superfamily includes retinoic acid receptors (RAR) and retinoid X receptors (RXR), whose effects on cell growth and survival can be modulated therapeutically by small-molecule ligands (de Lera et al., 2007). All-trans retinoic acid (ATRA) is an active metabolite of vitamin A, which is associated with increased ORMDL3 and shows protective effects on asthma, including maintaining airway epithelial integrity, inhibiting asthma effector cell differentiation, modulating immune response, etc. (Day et al., 2006; Zhuang et al., 2013; Garcia-Sanchez et al., 2016). Some studies have reported that retinoic acid (RA) is important for the development of the heart (Pan and Baker, 2007; Olsen and Blomhoff, 2020). Experimental studies showed that RA treatment could improve cardiac injury of gestational diabetes mellitus mice (Liu et al., 2020).
This study has several strengths. This is the first analysis to identify the shared genetic architecture of asthma and HF using the large GWAS data. HDL that was used to explore genetic correlations in this study and was able to reduce the genetic correlation variance by 60%, equivalent to a 2.5-fold increase in sample size (Ning et al., 2020). We used CPASSOC to better understand the shared genetic architecture between asthma and HF. CPASSOC allows for heterogeneity between traits, controls population structure and invisible relatedness, and allows for sample overlap. We found the seminal vesicle to be one of the major tissues and the hemic and immune systems to be one of the major systems that were significant between asthma and HF. These results contribute to further understanding of the association of asthma and HF and provide additional suggestions for possible shared mechanisms.
This study has some limitations. First, the data used in the present study was limited by diseases diagnosis, medication administration, and so on. However, GWAS summary statistics conducted study-specific quality control to ensure the quality. Second, the summary statistics limit us to segregate asthma patients by disease onset (childhood vs. adulthood), and lung function test data that reflect asthma severity are missing. Further studies are required to explore the genetic association of different asthma types with CVD risk. Third, our study is only limited to assessing the shared genetic architecture between asthma and CVDs, but environmental factors are different for specific traits. Therefore, a gene–environment interaction study between asthma and CVDs is encouraged. Finally, our study was limited to European ancestry, and shared genetic architecture in other ethnic groups are uncertain.
Conclusion
In summary, our findings provide strong evidence of genetic correlations and causal association between asthma and HF, which support that early preventive measures of CVDs can be taken in asthmatic patients. These results provide a novel insight of the shared genetic basis between asthma and HF from molecular and functional levels and contribute to further laboratory investigation and clinical studies.
Acknowledgments
The authors thank all the participants and researchers who contributed and collected data. The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgments.html. Data on coronary artery disease have been contributed by CARDIoGRAM investigators and have been downloaded from www.CARDIOGRAMPLUSC4D.ORG.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
YZ and JH designed the study. YZ and Z-SL collected the data and performed the analyses. YZ, TH, and JH wrote the manuscript. YZ and Z-SL provided technical support for software installation and statistical method selection. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (2021YFC0863400).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.775591/full#supplementary-material
References
- Aragam K. G., Chaffin M., Levinson R. T., Mcdermott G., Choi S. H., Shoemaker M. B., et al. (2018). Phenotypic Refinement of Heart Failure in a National Biobank Facilitates Genetic Discovery. Circulation [Epub ahead of print]. 10.1161/CIRCULATIONAHA.118.035774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., et al. (2000). Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes K. C. (2011). Genetic Studies of the Etiology of Asthma. Proc. Am. Thorac. Soc. 8, 143–148. 10.1513/pats.201103-030ms [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., et al. (2009). ClueGO: a Cytoscape Plug-In to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 25, 1091–1093. 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J., Davey Smith G., Burgess S. (2015). Mendelian Randomization with Invalid Instruments: Effect Estimation and Bias Detection through Egger Regression. Int. J. Epidemiol. 44, 512–525. 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S., Thompson S. G. (2017). Interpreting Findings from Mendelian Randomization Using the MR-Egger Method. Eur. J. Epidemiol. 32, 377–389. 10.1007/s10654-017-0255-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P., Lagan J., Fortune C., Bhatt D. L., Vestbo J., Niven R., et al. (2019). Association of Cardiovascular Disease with Respiratory Disease. J. Am. Coll. Cardiol. 73, 2166–2177. 10.1016/j.jacc.2018.11.063 [DOI] [PubMed] [Google Scholar]
- Dang J., Bian X., Ma X., Li J., Long F., Shan S., et al. (2017). ORMDL3 Facilitates the Survival of Splenic B Cells via an ATF6alpha-Endoplasmic Reticulum Stress-Beclin1 Autophagy Regulatory Pathway. J. Immunol. 199, 1647–1659. 10.4049/jimmunol.1602124 [DOI] [PubMed] [Google Scholar]
- Day R. M., Lee Y. H., Park A. M., Suzuki Y. J. (2006). Retinoic Acid Inhibits Airway Smooth Muscle Cell Migration. Am. J. Respir. Cel. Mol. Biol. 34, 695–703. 10.1165/rcmb.2005-0306oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lera A. R., Bourguet W., Altucci L., Gronemeyer H. (2007). Design of Selective Nuclear Receptor Modulators: RAR and RXR as a Case Study. Nat. Rev. Drug Discov. 6, 811–820. 10.1038/nrd2398 [DOI] [PubMed] [Google Scholar]
- Demenais F., Margaritte-Jeannin P., Barnes K. C., Cookson W. O. C., Altmuller J., Ang W., et al. (2018). Multiancestry Association Study Identifies New Asthma Risk Loci that Colocalize with Immune-Cell Enhancer marks. Nat. Genet. 50, 42–53. 10.1038/s41588-017-0014-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Wang K., Liu W., She Y., Sun Q., Shi J., et al. (2016). Pore-forming Activity and Structural Autoinhibition of the Gasdermin Family. Nature 535, 111–116. 10.1038/nature18590 [DOI] [PubMed] [Google Scholar]
- Emdin C. A., Khera A. V., Kathiresan S. (2017). Mendelian Randomization. JAMA 318, 1925–1926. 10.1001/jama.2017.17219 [DOI] [PubMed] [Google Scholar]
- Fachal L., Aschard H., Beesley J., Barnes D. R., Allen J., Kar S., et al. (2020). Fine-mapping of 150 Breast Cancer Risk Regions Identifies 191 Likely Target Genes. Nat. Genet. 52, 56–73. 10.1038/s41588-019-0537-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy J. V. (2015). Type 2 Inflammation in Asthma-Present in Most, Absent in many. Nat. Rev. Immunol. 15, 57–65. 10.1038/nri3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenta Y. A., Tello N., Jung J. A., Urm S. H., Loftus E. V., Jr., Yawn B. P., et al. (2010). Inflammatory Bowel Disease and Asthma: a Population-Based, Case-Control Study. Inflamm. Bowel Dis. 16, 1957–1962. 10.1002/ibd.21277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk C. D. (2001). Prostaglandins and Leukotrienes: Advances in Eicosanoid Biology. Science 294, 1871–1875. 10.1126/science.294.5548.1871 [DOI] [PubMed] [Google Scholar]
- Garcia-Sanchez A., Marcos-Vadillo E., Sanz C., Hernandez-Hernandez L., Cerutti-Muller G., Marques-Garcia F., et al. (2016). Retinoic Acid Modulates PTGDR Promoter Activity. J. Investig. Allergol. Clin. Immunol. 26, 249–255. 10.18176/jiaci.0042 [DOI] [PubMed] [Google Scholar]
- Giambartolomei C., Vukcevic D., Schadt E. E., Franke L., Hingorani A. D., Wallace C., et al. (2014). Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. Plos Genet. 10, e1004383. 10.1371/journal.pgen.1004383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W., Shi Y., Ren J. (2020). Research Progresses of Molecular Mechanism of Pyroptosis and its Related Diseases. Immunobiology 225, 151884. 10.1016/j.imbio.2019.11.019 [DOI] [PubMed] [Google Scholar]
- Gottdiener J. S., Arnold A. M., Aurigemma G. P., Polak J. F., Tracy R. P., Kitzman D. W., et al. (2000). Predictors of Congestive Heart Failure in the Elderly: the Cardiovascular Health Study. J. Am. Coll. Cardiol. 35, 1628–1637. 10.1016/s0735-1097(00)00582-9 [DOI] [PubMed] [Google Scholar]
- Gottdiener J. S. (2017). Intersection of 2 Epidemics: Asthma and Cardiovascular Disease. JACC Heart Fail. 5, 505–506. 10.1016/j.jchf.2017.05.003 [DOI] [PubMed] [Google Scholar]
- GTEx Consortium (2015). Human Genomics. The Genotype-Tissue Expression (GTEx) Pilot Analysis: Multitissue Gene Regulation in Humans. Science 348, 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurgone D., Mcshane L., Mcsharry C., Guzik T. J., Maffia P. (2020). Cytokines at the Interplay Between Asthma and Atherosclerosis? Front. Pharmacol. 11, 166. 10.3389/fphar.2020.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B. W., et al. (2016). Integrative Approaches for Large-Scale Transcriptome-wide Association Studies. Nat. Genet. 48, 245–252. 10.1038/ng.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halasz A., Cserhati E., Magyar R., Kovacs M., Cseh K. (2002). Role of TNF-Alpha and its 55 and 75 kDa Receptors in Bronchial Hyperreactivity. Respir. Med. 96, 262–267. 10.1053/rmed.2001.1256 [DOI] [PubMed] [Google Scholar]
- Hiemstra P. S., Mccray P. B., Jr., Bals R. (2015). The Innate Immune Function of Airway Epithelial Cells in Inflammatory Lung Disease. Eur. Respir. J. 45, 1150–1162. 10.1183/09031936.00141514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukku A., Pividori M., Luca F., Pique-Regi R., Im H. K., Wen X. (2021). Probabilistic Colocalization of Genetic Variants from Complex and Molecular Traits: Promise and Limitations. Am. J. Hum. Genet. 108, 25–35. 10.1016/j.ajhg.2020.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingebrigtsen T. S., Marott J. L., Vestbo J., Nordestgaard B. G., Lange P. (2020). Coronary Heart Disease and Heart Failure in Asthma, COPD and Asthma-COPD Overlap. BMJ Open Respir. Res. 7, e000470. 10.1136/bmjresp-2019-000470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iribarren C., Tolstykh I. V., Miller M. K., Sobel E., Eisner M. D. (2012). Adult Asthma and Risk of Coronary Heart Disease, Cerebrovascular Disease, and Heart Failure: a Prospective Study of 2 Matched Cohorts. Am. J. Epidemiol. 176, 1014–1024. 10.1093/aje/kws181 [DOI] [PubMed] [Google Scholar]
- Jain A., Tuteja G. (2019). TissueEnrich: Tissue-specific Gene Enrichment Analysis. Bioinformatics 35, 1966–1967. 10.1093/bioinformatics/bty890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., et al. (2020). The Reactome Pathway Knowledgebase. Nucleic Acids Res. 48, D498–D503. 10.1093/nar/gkz1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Nakaya A. (2002). The KEGG Databases at GenomeNet. Nucleic Acids Res. 30, 42–46. 10.1093/nar/30.1.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioussis D. (2007). Aiolos: an Ungrateful Member of the Ikaros Family. Immunity 26, 275–277. 10.1016/j.immuni.2007.03.003 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhao J., Lu M., Wang H., Tang F. (2020). Retinoic Acid Attenuates Cardiac Injury Induced by Hyperglycemia in Pre- and post-delivery Mice. Can. J. Physiol. Pharmacol. 98, 6–14. 10.1139/cjpp-2019-0009 [DOI] [PubMed] [Google Scholar]
- Mahmoodi N., Harijan R. K., Schramm V. L. (2020). Transition-State Analogues of Phenylethanolamine N-Methyltransferase. J. Am. Chem. Soc. 142, 14222–14233. 10.1021/jacs.0c05446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik R., Chauhan G., Traylor M., Sargurupremraj M., Okada Y., Mishra A., et al. (2018). Multiancestry Genome-wide Association Study of 520,000 Subjects Identifies 32 Loci Associated with Stroke and Stroke Subtypes. Nat. Genet. 50, 524–537. 10.1038/s41588-018-0058-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z., Pawitan Y., Shen X. (2020). High-definition Likelihood Inference of Genetic Correlations across Human Complex Traits. Nat. Genet. 52, 859–864. 10.1038/s41588-020-0653-y [DOI] [PubMed] [Google Scholar]
- Olsen T., Blomhoff R. (2020). Retinol, Retinoic Acid, and Retinol-Binding Protein 4 Are Differentially Associated with Cardiovascular Disease, Type 2 Diabetes, and Obesity: An Overview of Human Studies. Adv. Nutr. 11, 644–666. 10.1093/advances/nmz131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Baker K. M. (2007). Retinoic Acid and the Heart. Vitam Horm. 75, 257–283. 10.1016/s0083-6729(06)75010-5 [DOI] [PubMed] [Google Scholar]
- Peebles R. S., Jr. (2019). Prostaglandins in Asthma and Allergic Diseases. Pharmacol. Ther. 193, 1–19. 10.1016/j.pharmthera.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman K., Chong M., Akhtar-Danesh G. G., D'mello M., Hasso R., Ross S., et al. (2013). Genetic Markers of Inflammation and Their Role in Cardiovascular Disease. Can. J. Cardiol. 29, 67–74. 10.1016/j.cjca.2012.06.025 [DOI] [PubMed] [Google Scholar]
- Roth G. A., Johnson C., Abajobir A., Abd-Allah F., Abera S. F., Abyu G., et al. (2017). Global, Regional, and National Burden of Cardiovascular Diseases for 10 Causes, 1990 to 2015. J. Am. Coll. Cardiol. 70, 1–25. 10.1016/j.jacc.2017.04.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological Insights from 108 Schizophrenia-Associated Genetic Loci. Nature 511, 421–427. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunkert H., Konig I. R., Kathiresan S., Reilly M. P., Assimes T. L., Holm H., et al. (2011). Large-Scale Association Analysis Identifies 13 New Susceptibility Loci for Coronary Artery Disease. Nat. Genet. 43, 333–338. 10.1038/ng.784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovieff N., Cotsapas C., Lee P. H., Purcell S. M., Smoller J. W. (2013). Pleiotropy in Complex Traits: Challenges and Strategies. Nat. Rev. Genet. 14, 483–495. 10.1038/nrg3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanbroek R., Grabner R., Lotzer K., Hildner M., Urbach A., Ruhling K., et al. (2003). Expanding Expression of the 5-lipoxygenase Pathway within the Arterial wall during Human Atherogenesis. Proc. Natl. Acad. Sci. U S A. 100, 1238–1243. 10.1073/pnas.242716099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Wang T., Heianza Y., Lv J., Han L., Rabito F., et al. (2017). A History of Asthma from Childhood and Left Ventricular Mass in Asymptomatic Young Adults: The Bogalusa Heart Study. JACC Heart Fail. 5, 497–504. 10.1016/j.jchf.2017.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall M. C., Guo M., Korcarz C. E., Gepner A. D., Kaufman J. D., Liu K. J., et al. (2015). Asthma Predicts Cardiovascular Disease Events: the Multi-Ethnic Study of Atherosclerosis. Arterioscler Thromb. Vasc. Biol. 35, 1520–1525. 10.1161/atvbaha.115.305452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall M. C., Barnet J. H., Korcarz C. E., Hagen E. W., Peppard P. E., Stein J. H. (2016). Late-Onset Asthma Predicts Cardiovascular Disease Events: The Wisconsin Sleep Cohort. J. Am. Heart Assoc. 5, e003448. 10.1161/JAHA.116.003448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall M. C., Dasiewicz A. S., Mcclelland R. L., Gepner A. D., Kalscheur M. M., Field M. E., et al. (2020). Persistent Asthma Is Associated with Increased Risk for Incident Atrial Fibrillation in the MESA. Circ. Arrhythm Electrophysiol. 13, e007685. 10.1161/CIRCEP.119.007685 [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2019). The Gene Ontology Resource: 20 Years and Still GOing strong. Nucleic Acids Res. 47, D330–D338. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M., Fagerberg L., Hallstrom B. M., Lindskog C., Oksvold P., Mardinoglu A., et al. (2015). Proteomics. Tissue-Based Map of the Human Proteome. Science 347, 1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Wee J. H., Park M. W., Min C., Byun S. H., Park B., Choi H. G. (2021). Association between Asthma and Cardiovascular Disease. Eur. J. Clin. Invest. 51, e13396. 10.1111/eci.13396 [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Maller J. B., Mcvean G., Byrnes J., Vukcevic D., Palin K., et al. (2012). Bayesian Refinement of Association Signals for 14 Loci in 3 Common Diseases. Nat. Genet. 44, 1294–1301. 10.1038/ng.2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I. V., Lozupone C. A., Schwartz D. A. (2017). The Environment, Epigenome, and Asthma. J. Allergy Clin. Immunol. 140, 14–23. 10.1016/j.jaci.2017.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh J. J., Wei Y. F., Lin C. L., Hsu W. H. (2017). Association of Asthma-Chronic Obstructive Pulmonary Disease Overlap Syndrome with Coronary Artery Disease, Cardiac Dysrhythmia and Heart Failure: a Population-Based Retrospective Cohort Study. BMJ Open 7, e017657. 10.1136/bmjopen-2017-017657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun H. D., Knoebel E., Fenta Y., Gabriel S. E., Leibson C. L., Loftus E. V., Jr., et al. (2012). Asthma and Proinflammatory Conditions: a Population-Based Retrospective Matched Cohort Study. Mayo Clin. Proc. 87, 953–960. 10.1016/j.mayocp.2012.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C. N., Fan Y., Huang J. J., Zhang H. X., Gao T., Wang C., et al. (2015). The Association of GSDMB and ORMDL3 Gene Polymorphisms with Asthma: A Meta-Analysis. Allergy Asthma Immunol. Res. 7, 175–185. 10.4168/aair.2015.7.2.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., He H., Wang K., Shi X., Wang Y., Su Y., et al. (2020). Granzyme A from Cytotoxic Lymphocytes Cleaves GSDMB to Trigger Pyroptosis in Target Cells. Science 368, eaaz7548. 10.1126/science.aaz7548 [DOI] [PubMed] [Google Scholar]
- Zhu X., Feng T., Tayo B. O., Liang J., Young J. H., Franceschini N., et al. (2015). Meta-analysis of Correlated Traits via Summary Statistics from GWASs with an Application in Hypertension. Am. J. Hum. Genet. 96, 21–36. 10.1016/j.ajhg.2014.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang L. L., Huang B. X., Feng J., Zhu L. H., Jin R., Qiu L. Z., et al. (2013). All-trans Retinoic Acid Modulates ORMDL3 Expression via Transcriptional Regulation. PLoS One 8, e77304. 10.1371/journal.pone.0077304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziaeian B., Fonarow G. C. (2016). Epidemiology and Aetiology of Heart Failure. Nat. Rev. Cardiol. 13, 368–378. 10.1038/nrcardio.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietkowski Z., Skiepko R., Tomasiak M. M., Bodzenta-Lukaszyk A. (2007). Endothelin-1 in Exhaled Breath Condensate of Allergic Asthma Patients with Exercise-Induced Bronchoconstriction. Respir. Res. 8, 76. 10.1186/1465-9921-8-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietkowski Z., Skiepko R., Tomasiak-Lozowska M. M., Mroczko B., Szmitkowski M., Bodzenta-Lukaszyk A. (2010). RANTES in Exhaled Breath Condensate of Allergic Asthma Patients with Exercise-Induced Bronchoconstriction. Respiration 80, 463–471. 10.1159/000264923 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.