Summary
Objective
In this review, we have highlighted the advances over the past year in genetics, genomics and epigenetics in the field of osteoarthritis (OA).
Methods
A literature search of PubMed was performed using the criteria: “osteoarthritis” and one of the following terms “genetic(s), genomic(s), epigenetic(s), polymorphism, noncoding ribonucleic acid (RNA), microRNA, long noncoding RNA, lncRNA, circular RNA, RNA sequencing (RNA-seq), single cell sequencing, transcriptomics, or deoxyribonucleic acid (DNA) methylation between April 01, 2020 and April 30, 2021.
Results
In total we identified 765 unique publications, which eventually reduced to 380 of relevance to the field as judged by two assessors. Many of these studies included multiple search terms. We summarised advances relating to genetics, functional genetics, genomics and epigenetics, focusing on our personal key papers during the year.
Conclusions
This year few studies have identified new genetic variants contributing to OA susceptibility, but a focus has been on refining risk loci or their functional validation. The use of new technologies together with investigating the cross-talk between multiple tissue types, greater sample sizes and/or better patient classification (OA subtypes) will continue to increase our knowledge of disease mechanisms and progress towards understanding and treating OA.
Keywords: Osteoarthritis, Genetics, Genomics, Epigenetics, microRNA, circRNA, lncRNA
Introduction
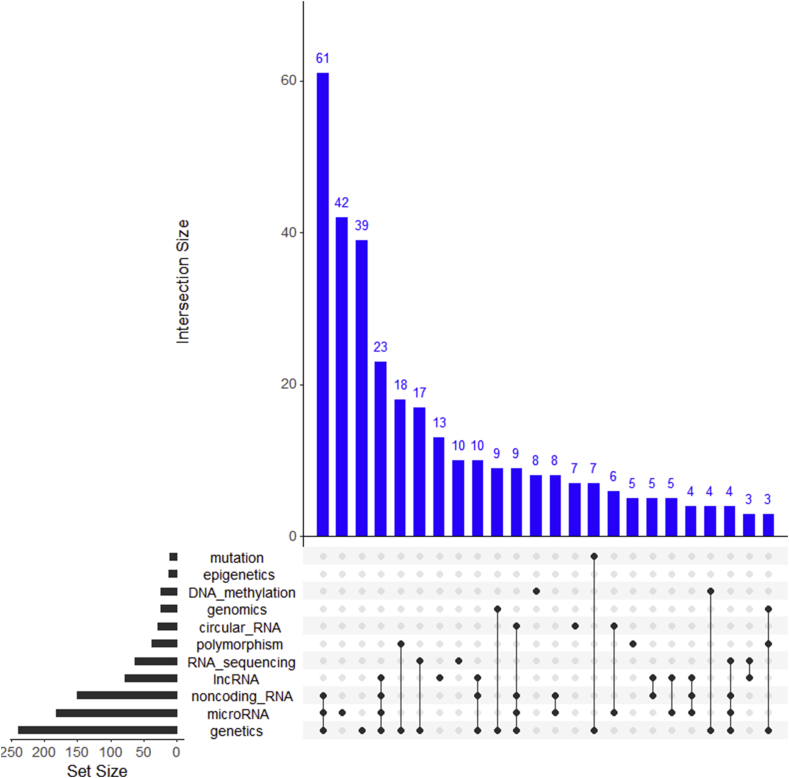
Osteoarthritis (OA) is the most prevalent musculoskeletal disease. The goal of this narrative review is to highlight key research studies published in OA genetics, genomics and epigenetics between April 1st 2020 and April 30th 2021. Using the defined search criteria we identified 765 articles, which when manually curated decreased to 380 deemed of relevance to the OA field (Supplementary File 1), many of which were identified by multiple search terms (Fig. 1). We also removed articles which contained potential irregularities identified in PubPeer (https://pubpeer.com/). Articles were chosen for discussion based on those we deemed to be novel, impactful to the field and fitted within the themes mentioned in this review.
Fig. 1.
Number of new publications indexed in PubMed with the defined search criteria after curation. Search was undertaken between April 1st 2020 and April 30th 2021. Set Size defines the total number of publications with the matching search term + ‘osteoarthritis’ after curation. Main Upset plot shows number of publications (≥3) with overlapping search criteria. Plotted using UpSetR version 1.4.073.
Genetics
This year saw a dearth of large genome-wide association studies (GWAS) for the major joints affected by OA, instead the focus was generally upon assessing previously identified genetic associations in differing ethnic populations (Table I). The growth differentiation factor 5 (GDF5) polymorphism rs143383 is reproducibly associated with OA, and this year was associated with OA-risk in the Kurdish population1. Oddly, in two meta-analyses examining the association of rs143383 with knee OA (KOA) in Asian individuals contradictory findings were observed2,3, while a new study in a Chinese Han population did find a link with KOA4. rs143383 is located in the 5′UTR of GDF5 and is associated with differential allelic expression of the gene, via altered DNA-binding of several transcriptional repressors5. However, recent compelling data, including a study discussed in last year's review6, suggests that the OA-risk mediated by the GDF locus could be via the control of the growth factor expression by highly conserved distal regulatory elements, with comprehensive work characterizing the effects of the rs6060369 polymorphism in mouse knee cartilage and OA-development7.
Table I.
Studies from April 12,020 to April 30 2021 examining genetic variants for OA risk are summarized above.
| Reference (PubMed ID) | OA joint site | Variant studied; nearest gene | OA risk | Sample size | Population | Study type |
|---|---|---|---|---|---|---|
| 31664512 | OA | rs2275913; IL17A rs763780; IL17F |
A (minor) rs2275913 and C (minor) rs763780 detrimental for KOA not HOA | 2,214 cases 2,474 Cx | various | Meta-analysis |
| 31713648 | KOA | rs2228611, rs2228612; DNMT1 rs2424913; DNMT3B |
C (minor) rs2228611 and C (minor) rs2228612 protective: C (minor) rs2424913 detrimental | 244 cases 244 Cx | Mexican mestizo | Candidate gene |
| 32103374 | KOA | rs4867568; LSPIP rs143383; GDF5; rs1558902; FTO |
T (minor) rs4867568 protective: C (minor) rs143383 protective: rs1558902; FTO - no association | 532 cases 927 Cx | Chinese (Han) | Candidate gene |
| 32144005 | KOA | rs2234693; ESR1 | T (major) detrimental | 1,033 cases 920 Cx | Chinese (Han) | Candidate gene |
| 32250658 | KOA | rs3740199, rs1871054; ADAM12 rs2073508; TGFB1 |
No association | 132 cases 164 Cx | Mexican | Candidate gene |
| 32364812 | KOA | rs217727, rs3741219; H19 rs7158663; MEG3 |
A (minor) rs217727 & A (minor) rs7158663 detrimental | 230 cases 230 Cx | Chinese (Han) | Candidate gene |
| 32452514 | HOA | rs10896015; LTBP3 | A (minor) protective | 884 cases 1896 Cx | Chinese (Han) | Candidate gene |
| 32460535 | KOA | rs10817595; AKNA | A (minor) detrimental | 824 cases 1,676 Cx | Chinese (Han) | Candidate gene |
| 32537881 | HOA | rs763780; IL17F | C (minor) detrimental | 796 cases 1854 Cx | Chinese (Han) | Candidate gene |
| 32557253 | KOA | rs4884, CKM | G (minor) protective | 87 cases 107 Cx | Mexican | Candidate gene |
| 32596320 | OA | rs1799750; MMP1/WTAPP1 | No general OA role, but association with temporomandibular joint | 1,245 cases 1,230 Cx | various | Meta-analysis |
| 32681364 | KOA | rs10116772, rs7045410, rs7032713; GLIS3 | Minor alleles (A, G, G) protective | 810 Cases 900 Cx | Chinese | Candidate gene |
| 32793194 | KOA | rs7097780; IL15RA | G (minor) symptomatic KOA | Symptomatic 403 vs asymptomatic 148 | UK | Candidate gene |
| 32928309 | KOA | rs143383; GDF5 | C (minor) protective - Caucasian only | 7997 KOA 12684 Cx | Caucasian/Asian | Meta-analysis |
| 33036557 | KOA | rs12700386; IL6 | C (minor) detrimental. C allele increases peripheral blood IL-6 expression | 352 cases 411 Cx | Chinese (Han) | Candidate gene |
| 33055079 | Hand OA | rs10916199; SNAP47 | A (major) detrimental – predicted causal gene WNT9A | 8,691 discovery participants. 1,203 replication | Dutch discovery US replication |
GWAS |
| 33112260 | OA | rs4946936; FOXO3 rs11769597; IGFBP3 |
rs4946936 minor allele (T) - protective male HOA: rs11769597 - C - detrimental female KOA | 337 cases 456 Cx | Dutch | Candidate gene |
| 33181041 | KOA | rs2862851; TGFA | T (minor) detrimental | 392 cases 808 Cx | Chinese (Han) | Candidate gene |
| 33590413 | KOA | rs2910164 and rs57095329; MIR146A | No association | 310 cases 379 Cx | Mexican (mestizo) | Candidate gene |
| 33861510 | hand OA and KOA | rs143383; GDF5 D-repeat; ASPN |
C (minor) rs143383 protective; ASPN D14 allele protective | 100 cases 100 Cx | Kurdish | Candidate gene |
| 33981765 | OA | rs28929474; SERPINA1 Z allele | Z allele protective | ∼16,000 heterozygous vs ∼300,000 Cx | Phe-WAS | |
| 34137553 | OA | rs12901499; SMAD3 | mild association | 2,403 cases 3,209 Cx | various | Meta-analysis |
| 34145804 | KOA | rs3819089; MMP13 rs3740199 and rs1871054; ADAM12 rs4747096; ADAMTS14 |
No association | 150 K-L ≥ III Cases; 150 K-L ≤ II Cx | Turkish | Candidate gene |
| 33640581 | OA | Various; Cu; Zn | High Cu/Zn detrimental | N/A | Mendelian randomisation |
∗HOA = hip OA, KOA= Knee OA, Cx = Control, K-L= Kellgren–Lawrence Grade. GWAS = genome-wide association study, Phe-WAS= Phenome-wide association study.
In a Phenome-wide association study using data from the UK Biobank heterozygosity the Z allele (rs28929474) of SERPINA1, homozygosity for which causes the rare condition α1-antitrypsin (AAT) deficiency, was associated with several musculoskeletal phenotypes, including a reduced risk of OA, but an increased risk of osteoporosis and lower bone mineral density (BMD)8. AAT can inhibit RANKL-induced osteoclast formation and bone resorption. Since heterozygosity for the Z allele reduces AAT levels, this could possibly explain both the lower BMD and associated OA risk. Recently, neutrophil elastase, for which AAT is an inhibitor, has been shown to fully activate MMP13, the major collagenase that promotes the irreversible destruction of cartilage collagen in OA9. Active MMP13 can proceed to inactivate AAT, thus AAT may play a complex role in multiple musculoskeletal tissues in OA.
GLIS3 is a transcriptional regulator which modulates hedgehog signalling, a pathway involved in limb development and OA severity10, but the factor itself is better associated with neonatal diabetes and congenital hypothyroidism, though single nucleotide polymorphisms (SNPs) around GLIS3 are associated with OA susceptibly11. This year Zhang et al. confirmed a similar OA association (rs10116772, rs7045410, rs7032713) in a Chinese population12. By correlating genotype with cartilage gene expression from 200 KOA patients the minor alleles of the same SNPs were associated with significantly lower GLIS3 expression, but higher expression of a non-coding GLIS3 nested transcript, GLIS3-AS1.
The IL-17 cytokine family comprises of six protein members (A-F). Studies are contradictory as to whether genetic polymorphisms in IL17A and IL17F are associated with OA. To resolve this Lee and Song conducted a meta-analysis of eight studies and concluded that rs2275913 in IL17A and rs763780 in IL17F are associated with general OA, but the latter not with hip OA (HOA)13. However in a more recent study, rs763780 was associated with HOA in a Han Chinese population14, indicating that perhaps ethnicity is partially responsible for the contradictory associations of IL17 genes with OA.
The main comprehensive GWAS within this year concentrated on hand OA and identified a novel risk locus centred upon the SNP rs1091619915. This study is particularly compelling as the authors stratified their cohort into three hand OA endophenotypes, based upon hierarchical clustering of radiograph-defined Kellgren and Lawrence grade across all 32 joints of both hands. This generated three hand OA phenotypes for analysis, essentially hand, finger and thumb. They then conducted their GWAS on each of the three hand OA phenotypes in the same cohort (∼8,700 individuals) with follow up in a replication cohort (∼1,200 individuals). This identified four OA associated loci, though only three were also significantly associated in the replication cohort. Two of these were novel (rs10916199 and rs2070852) and both associated with the thumb endophenotype. Of these rs10916199 was the most significant locus and is located on chromosome 1 near the ZNF678, WNT3A and WNT9A genes. To prioritise the causal gene the authors used human OA cartilage expression and methylation quantitative trait loci (eQTL and meQTL) datasets, and intersecting GWAS signals with accessible chromatin regions (ATAC-seq peaks) of human fetal cartilage and 3D chromatin conformation data from human mesenchymal stem cells (MSCs). These data, together with differential expression analysis between OA lesioned and preserved cartilage, identified WNT9A as the likely gene linked to rs10916199. Taken altogether, this paper demonstrates the power of assessment of stratified phenotypes in OA and that using such an approach could improve GWAS statistical power and provide novel insights into OA biology.
Although testing OA risk variants using candidate gene approaches, particularly with regards different ethnicities, can be useful these studies frequently use relatively few patients and controls. Therefore, the inclusion of diverse, non-European lineage, populations in GWAS OA studies is likely to be a productive way to identify novel OA risk variants and identify common pathogenic mechanisms across the globe16.
For a comprehensive summary of the current knowledge from GWAS of 124 OA-associated SNPs, encompassing 95 independent loci, we point the reader to a review article published during this year17.
Functional genetic analysis
This year several studies performed detailed functional studies of OA risk variants (Table II). As mentioned, an article published this year, but discussed in last year's review, elegantly described the functional characterization of the polymorphism rs6060369 which resides in an intron of UQCC1 yet is an enhancer variant controlling GDF5 expression, required for synovial joint formation and maintenance. The articles hypothesis is that during human evolution variants arose as we adapted to the biomechanical demands of bipedalism. However, although these variants had a positive role in limb formation, because of antagonistic pleiotropy, some, such as those in GDF5, are also deleterious to the long-term health of the joint, resulting in OA7.
Table II.
Studies from April 12,020 to April 30 2021 performing functional analysis of genetic variants for OA risk are summarized above
| OA Association SNP | Nearest ORF | Variant location | Identified Causal/regulated gene | Reference (PubMed ID) |
|---|---|---|---|---|
| rs11583641 | COLGALT2 | 3′UTR | COLGALT2 | 33760386 |
| rs10916199 | ZNF678 | Intronic | WNT9A | 33055079 |
| rs11780978 | PLEC | Intronic | PLEC | 32580029 |
| rs75621460 | TGFB1 | Intergenic | TGFB1 | 33760378 |
| rs6516886 | RWDD2B | Intergenic | RWDD2B | 32755071 |
| rs77245812 | MATN3 | ORF T303M (mouse T298M) | Matn3 | 33227438 |
| rs6060369 | UQCC1 | Intronic | GDF5/Gdf5 | 32220312 |
Mutation T303M of matrillin-3 was originally linked to hand OA in an Icelandic cohort18, though in other ethnicities these findings are only partially confirmed, though additional linkage to spinal disc generation is observed19. Seifer et al. generated a mouse model carrying the equivalent mutation (mouse T298M) in matrillin-3 and extensively characterized the animals. Developmentally the mice were essentially normal and though the human mutation is associated with hand OA no similar consequence was observed in the forepaw, perhaps due to species specific differential usage and mechanical forces. The cartilage of the mutant mice contained collagen fibrils with increased diameter compared to wild type (WT), increasing the compressive stiffness of the tissue. Despite this altered cartilage property, the mutant mice showed no increase in spontaneous OA severity with age but did for surgically-induced (meniscectomy) post-traumatic OA, suggesting the matrillin-3 T298M knock-in mouse could represent a model for investigating the pathogenic mechanisms involved in OA development20.
A T > A SNP at rs6516886, located in a region containing multiple genes, is associated with both KOA and HOA21. Parker et al. demonstrated that the OA-associated T allele of rs6516886 correlated with increased DNA methylation of neighbouring CpGs22. Differential allelic expression was identified for 3 genes in proximity to the SNP, but most notably for the gene RWDD2B, an RWD domain–containing protein coding gene of unknown function, across multiple joint tissues, with additional evidence of correlation with local CpG methylation. Finally, the authors were able to establish that experimental reduction of this CpG methylation by overexpression of a deactivated-(d)Cas9-Ten-eleven translocation methylcytosine dioxygenase 1-(TET1) fusion protein resulted in increased expression of RWDD2B.
Reported as an OA risk signal, the G>A polymorphism rs75621460 is positioned between CCDC97 and TGFB1 on chromosome 1923. TGFB1 of course encodes a ligand of the TGF-beta superfamily, with the pathway well described to regulate cartilage development and tissue homeostasis24. Rice et al. demonstrated differential enhancer activity and protein binding per allele25. Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9-mediated deletion of the locus implied regulation of only TGFB1, and correlation between DNA methylation of CpGs adjacent to the rs75621460 and TGFB1 expression were found, especially in cartilage tissue. Targeting methylation to the locus by a DNA methyl transferase 3a (DNMT3a)-dCas9 fusion reduced enhancer activity and TGFB1 expression, but in an allele-independent manner, implying downstream regulation potentially in response to allele-specific protein complex binding.
Similarly, Kehayova et al. examined HOA risk SNP rs11583641, located in the 3′UTR of COLGALT2, which encodes a β-galactosyltransferase that transfers β-galactose to hydroxylysine residues of collagen, identifying correlation between allelic expression and methylation of an upstream enhancer23,26. Experimental loss- and gain-of methylation again directed by dCas9 fusion transcriptional regulators modulated COLGALT2 expression, however there was variable allele-related expression and it remains unclear what impact the variation in the 3′UTR sequence might have directly on post-transcriptional regulation.
With the intention of improving functional gene discovery in OA, Butterfield et al. developed a three-mode imaging pipeline to comprehensively phenotype the mouse knee joint and its disease-related changes27. Iodine-contrast-enhancer (ICE) μCT determines several cartilage and bone volume, thickness and density characteristics, joint surface replication (JSR) quantifies cartilage surface damage, and subchondral bone X-ray microradiography (scXRM) determines mineral density. 100 WT mice were used to generate a baseline reference dataset for a phenotypically normal joint, which was then validated and contrasted against joints following OA induction by destablisation of the medial meniscus (DMM) surgery.
To establish the utility of the pipeline and identify genes associated with OA, the authors characterised 50 randomly selected mutant mouse lines generated by the international mouse phenotyping consortium. Surprisingly, 50% of the lines exhibited at least one phenotypic abnormality identified by the multi-modal imaging pipeline, suggesting many genes can impact the development and homeostasis of the joint. A thorough data integration strategy was taken to prioritise these identified genes, including data from knockout phenotypes, joint tissue expression, association with human disease and previous literature, which in this case identified Pitx1, Bhlhe40, Sh3bp4 and Unk. Having demonstrated the power of their joint phenotyping strategy three additional applications of the imaging pipeline were presented. Firstly, eight knockout mouse lines for genes differentially expressed in human OA were interrogated, of which joint phenotypes in 6 (Unk, Josd1, Gsdme, Arhgap30, Ccdc6, Col4a2) were found. Secondly, a comparison between 4- and 12-month-old mice identified age-related OA joint changes. Finally, a mutant Dio2 mouse, generated to reflect an OA-associated human polymorphism (rs225014), was examined and joint differences identified compared with the WT allele. The authors propose uptake of this novel joint phenotyping strategy by the OA research community, with its advantages over the traditional Osteoarthritis Research Society International (OARSI) method of damage scoring including increased sensitivity, and reduced cost and time, albeit requiring a large investment for the imaging facilities. Large scale screening of mutant lines for joint phenotypes (i.e., human OA GWAS hits) could provide data with reduced bias than researcher selected gene studies and provide valuable negative data for use in computational expansion of existing knowledge of joint damage associated genes28.
Transcriptomics and proteomics
Much of the existing OA genomics data is from cartilage tissues. This year saw several genomics studies examining other tissues of the joint and investigating the relationships between joint tissues. Tuerlings et al. examined subchondral bone using RNA sequencing (RNA-seq) to highlight the differentially expressed genes between paired preserved and lesional regions in 18 KOA and 6 HOA patients29. Comparison to articular cartilage revealed shared altered genes between joint sites, including the OA susceptibility genes IL11 and CHADL, which are candidates for future studies in in vivo OA models. However, genetic risk genes themselves may not represent the most drug tractable targets within an implicated signalling pathway. A future challenge from data such as this is to use this knowledge of the processes altered to gain a more mechanistic insight into what is driving them30.
In a multi-tissue omics study, Steinberg et al. performed both transcriptomics and proteomics of cartilage and synovium tissues as well as exome genotyping from peripheral blood in 115 OA patients31. The authors identified cis eQTLs for 1891 genes and 38 cis protein-eQTLS. The authors demonstrate the utility of this molecular map in determining the effector genes for non-coding GWAS lead variants. Statistical co-localisation of five OA GWAS signals were found with the molecular QTLs (ALDH1A2, NPC1, SMAD3, FAM53A and SLC44A2), two of which are not the nearest genes to the lead variant. Comparison of the high-grade and low-grade OA cartilage using transcriptomics and proteomics revealed 2,557 and 2,233 differentially abundant genes and proteins, respectively, of which 409 were dysregulated at both regulatory levels. The authors computationally screened for drug repurposing opportunities by testing for enrichment of the 148 upregulated proteins in damaged cartilage in a library of gene expression changes from in vitro drug treatments. The top enriched drugs such as IB-MECA (CF 101, Piclidenoson), VEGF-receptor-2-kinase-inhibitor-IV and nornicotine could potentially reduce the expression of the upregulated OA damaged cartilage signature. If these upregulated genes are detrimental in OA these drugs would be predicted to improve the cartilage phenotype and could potentially be repurposed to treat OA.
An important consideration for any future drug treatment in OA is the heterogeneity of the disease. Building on previous attempts to stratify OA patients with molecular data32 Steinberg and colleagues also assessed gene expression heterogeneity across a cohort of 113 KOA patient cartilage and synovium samples33. Patient subgroups were identified in both low-grade (intact) cartilage and synovium with the predominant discriminator being inflammation, although the patient subgroups in each tissue were independent of each other. A seven gene classifier was identified that could distinguish the subgroups and the corresponding main axis of molecular heterogeneity. Importantly, the classifier was verified in an independent validation cohort. An important future direction is translation of these tissue-based classifiers to more clinically assayable synovial fluid or serum-based biomarkers to enable longitudinal studies or examine differential response to putative therapeutics between the identified molecular subgroups.
Synovial fluid acts as a medium for joint tissue crosstalk with proteins such as growth factors, chemokines and cytokines secreted from the cartilage and synovium that may contribute to joint damage. The relative role of the synovium in driving OA remains unclear. Chou et al. applied single-cell sequencing to examine the crosstalk between matched synovial and damaged and intact cartilage tissue34. Using unsupervised clustering they identified 12 clusters of cells types in the synovium and 7 chondrocyte cell states. Detectable expression (>1% cells) of the commonly studied cytokines in OA such as TNF, IL6 and IL1B was restricted to synoviocytes, supporting previous cartilage RNA-seq based evidence that chondrocytes do not express these mediators32. In contrast, growth factors were expressed by both cell types and were more abundant in the synovial fluid than the cytokines. Future comparison of OA samples to non-OA controls to establish the healthy range and expression of mediators in the synovial fluid and tissues would assist in prioritising those proteins likely to be pathogenic. This work offers the potential for an assay or target against a particular cell population for monitoring progression or outcome of treatment. For instance, a subset of chondrocytes, enriched in damaged cartilage, were responsible for much of the cartilage protease expression.
The identification of the upstream regulators of the differentially expressed genes in each cell type in the Chou et al. study provides an insight into the communication between the synovial expressed cytokines and the chondrocytes34. Computational prediction of upstream drivers of the observed differential expression is challenging. There is risk of bias in their prediction due to the popularity of the regulators in the literature35 and these prediction approaches, as in the in silico drug screen described31, use data derived primarily from non-joint cell types so only conserved regulatory mechanisms and drug responses are likely to be identified. A study of the transcriptional responses to the cytokines and growth factors in joint tissues would provide useful information for computational inference of upstream regulators in the joint. For example, Mimpen et al. examined the transcriptional response of synovial fibroblasts and chondrocytes to members of the IL17 family, previously shown to be detectable in the synovial fluid of a subset of OA patients and, as described, genetically linked to the disease36. IL17A induced differential expression of genes, such as IL6, NFKBIZ, SOD2 and ZC3H12A which are both differentially expressed in human OA and alter joint phenotype in OA animal models37, 38, 39, 40. Data such as these could potentially allow more accurate inference of the main synovial fluid-based drivers of differential expression in affected tissues.
Transcriptomics of articular cartilage from mouse joints is technically challenging due to both contamination of dissected tissue with other cell types and the potential requirement for a long enzymatic digestion due to the extracellular matrix (ECM) rich nature of the cartilage. Sunkara et al. present a joint tissue cutting strategy where the distal section of the femur was segmented into three parts with known tissues and cell populations41. Importantly, treatment of the cells with the transcriptional inhibitor actinomycin-D during the ECM digestion revealed alterations in chondrocyte marker genes such as Col2a1, Sox9, and Comp, suggesting digestion itself can alter the measured chondrogenic phenotype of the studied cells. Single cell RNA-seq of the articular cartilage containing segment allowed the identification of a pure chondrocyte transcriptome from a small number of cells which when combined with bulk RNA-seq of the segments generated a high confidence chondrocyte gene signature. This chondrocyte signature could be used in computational deconvolution of both existing and future impure dissected articular cartilage bulk-RNA-seq into mixtures of cell types, to study chondrocyte-specific transcription responses42.
Epigenetics
DNA methylation
This year there were few new studies of global DNA methylation in OA, with an exception being in Kashin–Beck disease43. As discussed, several studies focused on OA genetic association loci correlating with different patterns of DNA methylation, meQTLs. Smeriglio et al.44 built upon the group's previous implied role of 5-hydroxymethylcytosine (5hmC) in OA, where levels increased at OA-associated genes in cartilage45, by showing an accumulation of 5hmC during DMM-induced OA in mice which corresponded with gene expression changes. 5-methylcytosine (5mC) oxidation to 5hmC is the first step in DNA demethylation, which can be mediated by the methylcytosine dioxygenase TET1. TET1 knockout (KO) mice following DMM surgery had reduced cartilage damage and osteophytes. Importantly, the therapeutic potential of targeting TET1 was investigated with frequent injections of TET inhibitor 2-HG post-DMM surgery in WT mice, the outcome being to phenocopy the effect of genetic ablation of TET1. The authors state that TET1-specific inhibitory peptides have recently been developed, opening the possibility for improved preclinical studies to establish the potential of this approach to protect against OA progression. However, TET1 acts as a tumor suppressor gene and TET1 inhibition is linked to cancer suggesting systemic delivery would be undesirable46.
Non-coding RNA
This year saw a huge number of studies evaluating non-coding RNA, particularly microRNAs in OA (Fig. 1, Fig. 2). There was also increasing interest in the concept of competing endogenous RNAs (ceRNAs) which includes circular RNAs (circRNAs) and long non-coding RNAs (lncRNAs), which act as microRNA sponges, though the physiological relevance of ceRNAs is questionable47. CircRNAs are highly stable transcripts produced as a back splicing event often from open reading frame transcript exons, while lncRNAs are defined as transcripts exceeding 200 nucleotides in length that are not translated into protein. For the first time, SnoRNAs altered in OA were profiled and several were studied through loss and gain of function experiments48. This year also saw the first study of tRNA-derived fragments (tRFs) in chondrocytes which have been ascribed microRNA-like functions leading to target mRNA repression49.
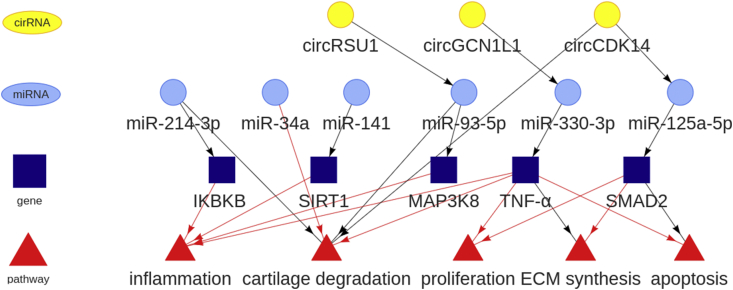
Fig. 2.
Targets and pathways of the selected non-coding RNAs studied in OA this year. The reported downstream signalling and interactions were extracted from the non-coding RNA papers reviewed within the main text52,55,60, 61, 62,74. Pathways are illustrated as red triangles, circRNA are yellow circles, microRNAs are light blue circles and gene/protein expression changes are denoted in dark blue squares. Black and red arrows indicate negative and positive regulation, respectively. Plotted using VisNetwork (v 2.0.9).
Many of the non-coding RNA studies were performed in in vitro chondrocyte cell lines under inflammatory (lipopolysaccharide (LPS) or IL1 treatment) conditions and generally present a linear cascade (axis) of signaling downstream of the studied RNA to inflammation or ECM degradation via a single target (Fig. 2). Robust high-throughput identification, validation and understanding of the interplay of the likely hundreds of targets of each non-coding RNAs remains challenging and is further exacerbated by complexities such as isomiRs, variants of microRNAs which can alter their target repertoire50.
MicroRNA
The difficulties of accessing joint tissues has attracted research into the possibility of delivering therapeutics into cartilage. Liang et al. sought a solution to this ‘grand challenge’ with the use of exosomes to deliver microRNAs into chondrocytes51. Utilising a previously identified chondrocyte-affinity peptide (CAP) fused to a lysosome membrane protein the authors were able to derive exosomes able to selectively deliver miR-140 into human chondrocytes, but not MSCs, and cause suppression of miR-140 targets ADAMTS5 and MMP13. Articular injection of fluorescently labelled miR-140-containing exosomes into rats indicated that the CAP-exosomes could penetrate cartilage in vivo and offer protection against cartilage damage in the DMM model of OA.
Ji et al. in a highly comprehensive study described the functional assessment of miR-141/200c cluster in OA52. These two microRNAs are co-expressed from the MIR200CHG gene on chromosome 12 (Gm15884 on chr6 in mouse). In a screen of differentially expressed microRNAs in end-stage OA cartilage, both microRNAs were upregulated correlating with hypomethylation of MIR200CHG gene promoter. Previously the same group, using gain- and loss-of-function studies in mouse models, showed that miR-141 was a key regulator of intervertebral disc degeneration, in part via regulating the deacetylase SIRT1 levels53. In the current work the group described a similar function of miR-141 in growth plate and articular cartilage chondrocytes. Adult miR-141/200c conditional knock-out (cKO) mice, either aged or subjected to OA-inducing DMM surgery, had reduced cartilage damage, indicating that the microRNAs are catabolic regulators of the process driving OA. The authors then performed detailed further work to deliver miR141/200c mimics and inhibitors to cartilage. Firstly, the cell-SELEX (systematic evolution of ligands by exponential enrichment) method54 was used to select a DNA aptamer, termed tgg2, which selectively bound to chondrocytes, potentially via Fibroblast Growth Factor Receptor 1 (FGFR1). This was conjugated with a dendritic polymer to create a stable nanoparticle capable of penetrating human OA cartilage explants ex vivo. Finally, delivery of miR-141/200c mimics or inhibitors using the nanoparticles via repeated intra-articular injection exacerbated or reduced DMM-induced OA damage, respectively. The proposed mechanism of how this occurs is via miR-141/200c regulation of IL-6 mediated inflammatory signalling, through the direct deacetylation of IL-6 by the miR-141 target SIRT1 (Fig. 2).
Endisha et al. investigated the role of miR-34a-5p in OA having previously identified its upregulation in late-stage OA synovial fluid55. After confirming the increase in miR-34a-5p in both human and experimental KOA, treatment of both OA chondrocytes and synoviocytes with miR-34a-5p mimic and inhibitor indicated the reciprocal regulation of OA anabolic and catabolic marker genes such as COL2A1 and MMP13 (Fig. 2). Importantly, these functional experiments were replicated in vivo where three fortnightly injections of a miR-34a-5p inhibitor reduced cartilage damage following DMM surgery in mice, while injection of miR-34a-5p mimic promoted spontaneous OA development. The authors also demonstrated that injection of miR-34a-5p inhibitors could suppress OA in a high fat diet-accelerated DMM model. MiR-34a-5p genetic ablation confirmed protection against DMM-induced cartilage damage while RNA-seq of the cartilage from the animals identified potential disease-relevant targets.
From the same group, Ali et al. characterised circulating microRNAs from the plasma of patients with early and late radiographic KOA, identifying a panel of microRNAs which differentiated between the two groups56. This adds to a small number of studies which have assessed circulating microRNAs in OA, but there currently appears little consensus in the microRNAs identified, potentially owing to differences in patient cohorts, study design and technological methodology utilized57.
Finally, Cao et al. identified reduced levels of miR-214-3p in human and mouse KOA cartilage. miR-214-3p inhibited the IL-1-mediated activation of the catabolic gene expression program in chondrocytes, potentially by directly targeting the NF-κB pathway kinase IKBKB to suppress NF-kB signalling. Intra-articular delivery of miR-214-3p mimic or inhibitor was able to respectively suppress or promote cartilage degradation in the DMM-induced OA mouse model, although only small numbers of mice were used and no evidence of alterations to miR-214-3p or IKBKB levels were presented (Fig. 2).
Long non-coding RNAs
To rationalize the lncRNAs most important in OA, van Hoolwerff et al. established lncRNA-mRNA co-expression networks using paired (lesion vs intact cartilage) RNA-seq data from almost 100 OA patients58. This identified 5,053 robustly expressed lncRNAs, of which 191 were significantly differentially expressed. These lncRNAs, which are predominantly intergenic or antisense transcripts, may regulate differentially expressed mRNAs in cis in cartilage.
Using RNA-seq of synovial fibroblasts Nanus et al. sought to profile the lncRNA changes in obesity-associated OA59. Nineteen lncRNAs were differentially expressed in fibroblasts of obese compared to normal-weight HOA patients, including the lncRNA MALAT1. Synovial fibroblasts from the obese group exhibited an inflammatory phenotype, which may, the authors conclude, be partially mediated by MALAT1.
CircRNAs
Yang et al., having demonstrated the increase in oxidative stress with age in OA, identified expression changes in 550 circRNAs following 5 days of H2O2 treatment of chondrocytes to induce oxidative stress60. The most upregulated was circRSU1, and experimental manipulation of its levels altered the balance of chondrocyte gene expression towards a catabolic profile with increased metalloproteinase expression and reduced matrix gene expression. The authors demonstrated that circRSU1 exhibited ceRNA activity against several miRNAs but most notably miR-93-5p. Remarkably, circRSU1 intra-articular delivery induced the same severity of OA in mice as DMM surgery, while mutation of the miR-93-5p binding site in the circRNA completely abrogated this effect. Finally they established a mechanism where miR-93-5p targets MAP3K8 thereby suppressing extracellular signal-regulated kinase (ERK) and NF-κB signalling, a scenario that would be reversed by the experimental or age-related upregulation of circRSU1 and the concomitant reduction in available miR-93-5p.
Further notable circRNA studies in the OA field briefly include, circGCN1L1 in the progression of OA in rats by acting as a sponge for miR-330-3p which targets TNF-α, thereby promoting synoviocyte proliferation and chondrocyte apoptosis61, and CircCDK14, which interacts with miR-125a-5p interfering with its repression of SMAD2, protecting against OA in rabbits by regulation of ECM and chondrocyte viability62 (Fig. 2).
Future approaches and summary
Driven by the decreasing costs of single cell technology and improvements in cell isolation techniques we expect to see growth in multimodal single cell technology applied to OA research. For example, ATAC-seq and RNA-seq could be performed simultaneously to gain greater insight into the transcriptional regulation in cell states in OA. Pooled CRISPR screening approaches with single cell read-out offer the opportunity characterize the function of hundreds of potential OA effector genes in relevant cell types simultaneously63. Another possible future sequencing approach is spatial RNA-seq which allows transcriptomics from intact histological sections while retaining valuable spatial tissue information64,65. Probabilistic methods could be used to integrate existing single cell data with spatial RNA-seq data to explore the spatial relationships between previously identified cell types and states66. This would allow investigation of the spatial distribution of single-cell seq identified cell types and give insights into the relative accessibility of these cells to synovial fluid mediators or any planned intra-articular interventions.
OA is increasingly seen as a complex heterogenous syndrome affecting the whole joint structure, not just cartilage. There is also limited synovial inflammation but to varying degrees for each patient. The heterogenous nature of the disease may partly explain the number and impact of GWAS-identified loci, which only explain a small proportion of the phenotypic variance23. Current GWAS have included thousands of patients with OA21,23,67, but a full meta-analysis of multiple populations will likely lead to the identification of further risk variants. This year Boer et al., elegantly demonstrated how analysis of well-characterized patient cohorts, where patients were segregated into distinct disease sub-types (in this case of hand OA), could increase the discovery power of a relatively modestly sized GWAS15. An expectation in the coming year(s) would be that further patient or disease stratification could improve GWAS-derived knowledge of disease-associated loci for OA of the other joint sites. Clinical adoption of stratification approaches using other molecular data such as global DNA methylation patterns68, 69, 70 and tissue RNA-seq as described this year33 and attempted previously32 are impeded by joint tissue inaccessibility. An integrated approach of molecular tissue characterization combined with assayable biomarker measurement such as from synovial fluid71 or circulating microRNA56, may help translation of these approaches and clarify if such genomically stratified patient groups are of clinical utility. Several new disease modifying therapies for OA are still being investigated in ongoing clinical trials and it will be of interest to speculate if these treatments could be improved if targeted to a more specific disease subtype72.
Contributions
All authors were involved in the design of the review, and analysis and interpretation of studies included. JS and DAY assessed the manuscripts to include. All authors drafted the article, and all approved the final version to be submitted.
Conflict of interest
All authors declare no conflicts of interest.
Acknowledgements
This work was supported by Versus Arthritis (22043); the Medical Research Council and Versus Arthritis as part of the MRC-Arthritis Research UK Centre for Integrated Research into Musculoskeletal Ageing (CIMA) (JXR 10641, MR/P020941/1); the JGW Patterson Foundation; and The Dunhill Medical Trust (R476/0516).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.joca.2021.11.004.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Moghimi N., Nasseri S., Ghafouri F., Jalili A. Frequency of Growth Differentiation Factor 5 rs143383 and asporin D-repeat polymorphisms in patients with hand and knee osteoarthritis in Kurdistan province, Iran. Int J Rheum Dis. 2021;24(5):694–700. doi: 10.1111/1756-185X.14097. [DOI] [PubMed] [Google Scholar]
- 2.Jia B., Jiang Y., Xu Y., Wang Y., Li T. Correlation between growth differentiation factor 5 (rs143383) gene polymorphism and knee osteoarthritis: an updated systematic review and meta-analysis. J Orthop Surg Res. 2021;16(1):146. doi: 10.1186/s13018-021-02269-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng L., Jin S., Lu J., Ouyang C., Guo J., Xie Z., et al. Association between growth differentiation factor 5 rs143383 genetic polymorphism and the risk of knee osteoarthritis among Caucasian but not Asian: a meta-analysis. Arthritis Res Ther. 2020;22(1):215. doi: 10.1186/s13075-020-02306-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y., Liu F., Xu X., Zhang H., Lu M., Gao W., et al. A novel variant near LSP1P3 is associated with knee osteoarthritis in the Chinese population. Clin Rheumatol. 2020;39(8):2393–2398. doi: 10.1007/s10067-020-04995-8. [DOI] [PubMed] [Google Scholar]
- 5.Syddall C.M., Reynard L.N., Young D.A., Loughlin J. The identification of trans-acting factors that regulate the expression of GDF5 via the osteoarthritis susceptibility SNP rs143383. PLoS Genet. 2013;9(6) doi: 10.1371/journal.pgen.1003557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratneswaran A., Kapoor M. Osteoarthritis year in review: genetics, genomics, epigenetics. Osteoarthritis Cartilage. 2021;29(2):151–160. doi: 10.1016/j.joca.2020.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Richard D., Liu Z., Cao J., Kiapour A.M., Willen J., Yarlagadda S., et al. Evolutionary selection and constraint on human knee chondrocyte regulation impacts osteoarthritis risk. Cell. 2020;181(2):362–381 e28. doi: 10.1016/j.cell.2020.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fawcett K.A., Song K., Qian G., Farmaki A.E., Packer R., John C., et al. Pleiotropic associations of heterozygosity for the SERPINA1 Z allele in the UK Biobank. ERJ Open Res. 2021;7(2) doi: 10.1183/23120541.00049-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkinson D.J., Falconer A.M.D., Wright H.L., Lin H., Yamamoto K., Cheung K., et al. Matrix metalloproteinase-13 is fully activated by neutrophil elastase and inactivates its serpin inhibitor, alpha-1 antitrypsin: implications for osteoarthritis. FEBS J. 2021 doi: 10.1111/febs.16127. [DOI] [PubMed] [Google Scholar]
- 10.Alman B.A. The role of hedgehog signalling in skeletal health and disease. Nat Rev Rheumatol. 2015;11(9):552–560. doi: 10.1038/nrrheum.2015.84. [DOI] [PubMed] [Google Scholar]
- 11.Casalone E., Tachmazidou I., Zengini E., Hatzikotoulas K., Hackinger S., Suveges D., et al. A novel variant in GLIS3 is associated with osteoarthritis. Ann Rheum Dis. 2018;77(4):620–623. doi: 10.1136/annrheumdis-2017-211848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J.L., Zhang C.L., Zhou B.G., Lei B.Y., Zhang B., Yang H.T. Association study of the functional variants of the GLIS3 gene with risk of knee osteoarthritis. Clin Rheumatol. 2021;40(3):1039–1046. doi: 10.1007/s10067-019-04871-0. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y.H., Song G.G. Association between IL-17 gene polymorphisms and circulating IL-17 levels in osteoarthritis: a meta-analysis. Z Rheumatol. 2020;79(5):482–490. doi: 10.1007/s00393-019-00720-2. [DOI] [PubMed] [Google Scholar]
- 14.Zhao T., Ma C., Wang W., Zhao B., Xie B., Liu J. Common variants in IL17F gene contributed to the risk of hip osteoarthritis susceptibility in Han Chinese population. Int J Rheum Dis. 2020;23(8):1050–1056. doi: 10.1111/1756-185X.13880. [DOI] [PubMed] [Google Scholar]
- 15.Boer C.G., Yau M.S., Rice S.J., Coutinho de Almeida R., Cheung K., Styrkarsdottir U., et al. Genome-wide association of phenotypes based on clustering patterns of hand osteoarthritis identify WNT9A as novel osteoarthritis gene. Ann Rheum Dis. 2020 doi: 10.1136/annrheumdis-2020-217834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boer C.G., Hatzikotoulas K., Southam L., Stefansdottir L., Zhang Y., Coutinho de Almeida R., et al. Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations. Cell. 2021;184(18):4784–4818 e17. doi: 10.1016/j.cell.2021.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aubourg G., Rice S.J., Bruce-Wootton P., Loughlin J. Osteoarthritis Cartilage; 2021. Genetics of Osteoarthritis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefansson S.E., Jonsson H., Ingvarsson T., Manolescu I., Jonsson H.H., Olafsdottir G., et al. Genomewide scan for hand osteoarthritis: a novel mutation in matrilin-3. Am J Hum Genet. 2003;72(6):1448–1459. doi: 10.1086/375556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Min J.L., Meulenbelt I., Riyazi N., Kloppenburg M., Houwing-Duistermaat J.J., Seymour A.B., et al. Association of matrilin-3 polymorphisms with spinal disc degeneration and osteoarthritis of the first carpometacarpal joint of the hand. Ann Rheum Dis. 2006;65(8):1060–1066. doi: 10.1136/ard.2005.045153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seifer P., Hay E., Fleischhauer L., Heilig J., Bloch W., Sonntag S., et al. The Matrilin-3 T298M mutation predisposes for post-traumatic osteoarthritis in a knock-in mouse model. Osteoarthritis Cartilage. 2021;29(1):78–88. doi: 10.1016/j.joca.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Zengini E., Hatzikotoulas K., Tachmazidou I., Steinberg J., Hartwig F.P., Southam L., et al. Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nat Genet. 2018;50(4):549–558. doi: 10.1038/s41588-018-0079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker E., Hofer I.M.J., Rice S.J., Earl L., Anjum S.A., Deehan D.J., et al. Multi-tissue epigenetic and gene expression analysis combined with epigenome modulation identifies RWDD2B as a target of osteoarthritis susceptibility. Arthritis Rheum. 2021;73(1):100–109. doi: 10.1002/art.41473. [DOI] [PubMed] [Google Scholar]
- 23.Tachmazidou I., Hatzikotoulas K., Southam L., Esparza-Gordillo J., Haberland V., Zheng J., et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat Genet. 2019;51(2):230–236. doi: 10.1038/s41588-018-0327-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Kraan P.M. The changing role of TGFbeta in healthy, ageing and osteoarthritic joints. Nat Rev Rheumatol. 2017;13(3):155–163. doi: 10.1038/nrrheum.2016.219. [DOI] [PubMed] [Google Scholar]
- 25.Rice S.J., Roberts J.B., Tselepi M., Brumwell A., Falk J., Steven C., et al. Genetic and epigenetic fine-tuning of TGFB1 expression within the human osteoarthritic joint. Arthritis Rheum. 2021;73(10):1866–1877. doi: 10.1002/art.41736. [DOI] [PubMed] [Google Scholar]
- 26.Kehayova Y.S., Watson E., Wilkinson J.M., Loughlin J., Rice S.J. Genetic and epigenetic interplay within a COLGALT2 enhancer associated with osteoarthritis. Arthritis Rheum. 2021;73(10):1856–1865. doi: 10.1002/art.41738. [DOI] [PubMed] [Google Scholar]
- 27.Butterfield N.C., Curry K.F., Steinberg J., Dewhurst H., Komla-Ebri D., Mannan N.S., et al. Accelerating functional gene discovery in osteoarthritis. Nat Commun. 2021;12(1):467. doi: 10.1038/s41467-020-20761-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soul J., Barter M.J., Little C.B., Young D.A. OATargets: a knowledge base of genes associated with osteoarthritis joint damage in animals. Ann Rheum Dis. 2020 doi: 10.1136/annrheumdis-2020-218344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuerlings M., van Hoolwerff M., Houtman E., Suchiman E., Lakenberg N., Mei H., et al. RNA sequencing reveals interacting key determinants of osteoarthritis acting in subchondral bone and articular cartilage: identification of IL11 and CHADL as attractive treatment targets. Arthritis Rheum. 2021;73(5):789–799. doi: 10.1002/art.41600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soul J., Hardingham T.E., Boot-Handford R.P., Schwartz J.M. SkeletalVis: an exploration and meta-analysis data portal of cross-species skeletal transcriptomics data. Bioinformatics. 2019;35(13):2283–2290. doi: 10.1093/bioinformatics/bty947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinberg J., Southam L., Roumeliotis T.I., Clark M.J., Jayasuriya R.L., Swift D., et al. A molecular quantitative trait locus map for osteoarthritis. Nat Commun. 2021;12(1):1309. doi: 10.1038/s41467-021-21593-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soul J., Dunn S.L., Anand S., Serracino-Inglott F., Schwartz J.M., Boot-Handford R.P., et al. Stratification of knee osteoarthritis: two major patient subgroups identified by genome-wide expression analysis of articular cartilage. Ann Rheum Dis. 2018;77(3):423. doi: 10.1136/annrheumdis-2017-212603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinberg J., Southam L., Fontalis A., Clark M.J., Jayasuriya R.L., Swift D., et al. Linking chondrocyte and synovial transcriptional profile to clinical phenotype in osteoarthritis. Ann Rheum Dis. 2021 doi: 10.1136/annrheumdis-2020-219760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou C.H., Jain V., Gibson J., Attarian D.E., Haraden C.A., Yohn C.B., et al. Synovial cell cross-talk with cartilage plays a major role in the pathogenesis of osteoarthritis. Sci Rep. 2020;10(1):10868. doi: 10.1038/s41598-020-67730-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Browaeys R., Saelens W., Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2020;17(2):159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 36.Snelling S.J., Bas S., Puskas G.J., Dakin S.G., Suva D., Finckh A., et al. Presence of IL-17 in synovial fluid identifies a potential inflammatory osteoarthritic phenotype. PLoS One. 2017;12(4) doi: 10.1371/journal.pone.0175109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Hooge A.S., van de Loo F.A., Bennink M.B., Arntz O.J., de Hooge P., van den Berg W.B. Male IL-6 gene knock out mice developed more advanced osteoarthritis upon aging. Osteoarthritis Cartilage. 2005;13(1):66–73. doi: 10.1016/j.joca.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 38.Koike M., Nojiri H., Ozawa Y., Watanabe K., Muramatsu Y., Kaneko H., et al. Mechanical overloading causes mitochondrial superoxide and SOD2 imbalance in chondrocytes resulting in cartilage degeneration. Sci Rep. 2015;5:11722. doi: 10.1038/srep11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi M.C., MaruYama T., Chun C.H., Park Y. Alleviation of murine osteoarthritis by cartilage-specific deletion of IkappaBzeta. Arthritis Rheum. 2018;70(9):1440–1449. doi: 10.1002/art.40514. [DOI] [PubMed] [Google Scholar]
- 40.Yang J.I., Chun J.S. Upregulated endonuclease Regnase-1 suppresses osteoarthritis by forming a negative feedback loop of catabolic signaling in chondrocytes. Arthritis Res Ther. 2021;23(1):114. doi: 10.1186/s13075-021-02485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sunkara V., Heinz G.A., Heinrich F.F., Durek P., Mobasheri A., Mashreghi M.F., et al. Combining segmental bulk- and single-cell RNA-sequencing to define the chondrocyte gene expression signature in the murine knee joint. Osteoarthritis Cartilage. 2021;29(6):905–914. doi: 10.1016/j.joca.2021.03.007. [DOI] [PubMed] [Google Scholar]
- 42.Dong M., Thennavan A., Urrutia E., Li Y., Perou C.M., Zou F., et al. SCDC: bulk gene expression deconvolution by multiple single-cell RNA sequencing references. Briefings Bioinf. 2021;22(1):416–427. doi: 10.1093/bib/bbz166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan Y., Gao D., Zhang Y., Zhu J., Zhang F., Wang L., et al. Genome-wide differentially methylated region analysis to reveal epigenetic differences of articular cartilage in kashin-beck disease and osteoarthritis. Front Cell Dev Biol. 2021;9:636291. doi: 10.3389/fcell.2021.636291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smeriglio P., Grandi F.C., Davala S., Masarapu V., Indelli P.F., Goodman S.B., et al. Inhibition of TET1 prevents the development of osteoarthritis and reveals the 5hmC landscape that orchestrates pathogenesis. Sci Transl Med. 2020;12(539) doi: 10.1126/scitranslmed.aax2332. [DOI] [PubMed] [Google Scholar]
- 45.Taylor S.E., Li Y.H., Wong W.H., Bhutani N. Genome-wide mapping of DNA hydroxymethylation in osteoarthritic chondrocytes. Arthritis Rheum. 2015;67(8):2129–2140. doi: 10.1002/art.39179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cimmino L., Dawlaty M.M., Ndiaye-Lobry D., Yap Y.S., Bakogianni S., Yu Y., et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol. 2015;16(6):653–662. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 48.Peffers M.J., Chabronova A., Balaskas P., Fang Y., Dyer P., Cremers A., et al. SnoRNA signatures in cartilage ageing and osteoarthritis. Sci Rep. 2020;10(1):10641. doi: 10.1038/s41598-020-67446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Green J.A., Ansari M.Y., Ball H.C., Haqqi T.M. tRNA-derived fragments (tRFs) regulate post-transcriptional gene expression via AGO-dependent mechanism in IL-1beta stimulated chondrocytes. Osteoarthritis Cartilage. 2020;28(8):1102–1110. doi: 10.1016/j.joca.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woods S., Charlton S., Cheung K., Hao Y., Soul J., Reynard L.N., et al. microRNA-seq of cartilage reveals an overabundance of miR-140-3p which contains functional isomiRs. RNA. 2020;26(11):1575–1588. doi: 10.1261/rna.075176.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang Y., Xu X., Li X., Xiong J., Li B., Duan L., et al. Chondrocyte-targeted MicroRNA delivery by engineered exosomes toward a cell-free osteoarthritis therapy. ACS Appl Mater Interfaces. 2020;12(33):36938–36947. doi: 10.1021/acsami.0c10458. [DOI] [PubMed] [Google Scholar]
- 52.Ji M.L., Jiang H., Wu F., Geng R., Ya L.K., Lin Y.C., et al. Precise targeting of miR-141/200c cluster in chondrocytes attenuates osteoarthritis development. Ann Rheum Dis. 2020 doi: 10.1136/annrheumdis-2020-218469. [DOI] [PubMed] [Google Scholar]
- 53.Ji M.L., Jiang H., Zhang X.J., Shi P.L., Li C., Wu H., et al. Preclinical development of a microRNA-based therapy for intervertebral disc degeneration. Nat Commun. 2018;9(1):5051. doi: 10.1038/s41467-018-07360-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sefah K., Shangguan D., Xiong X., O'Donoghue M.B., Tan W. Development of DNA aptamers using Cell-SELEX. Nat Protoc. 2010;5(6):1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 55.Endisha H., Datta P., Sharma A., Nakamura S., Rossomacha E., Younan C., et al. MicroRNA-34a-5p promotes joint destruction during osteoarthritis. Arthritis Rheum. 2021;73(3):426–439. doi: 10.1002/art.41552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ali S.A., Gandhi R., Potla P., Keshavarzi S., Espin-Garcia O., Shestopaloff K., et al. Sequencing identifies a distinct signature of circulating microRNAs in early radiographic knee osteoarthritis. Osteoarthritis Cartilage. 2020;28(11):1471–1481. doi: 10.1016/j.joca.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 57.Jones T.L., Esa M.S., Li K.H.C., Krishnan S.R.G., Elgallab G.M., Pearce M.S., et al. Osteoporosis, fracture, osteoarthritis & sarcopenia: a systematic review of circulating microRNA association. Bone. 2021;152:116068. doi: 10.1016/j.bone.2021.116068. [DOI] [PubMed] [Google Scholar]
- 58.van Hoolwerff M., Metselaar P.I., Tuerlings M., Suchiman H.E.D., Lakenberg N., Ramos Y.F.M., et al. Elucidating epigenetic regulation by identifying functional cis-acting long noncoding RNAs and their targets in osteoarthritic articular cartilage. Arthritis Rheum. 2020;72(11):1845–1854. doi: 10.1002/art.41396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nanus D.E., Wijesinghe S.N., Pearson M.J., Hadjicharalambous M.R., Rosser A., Davis E.T., et al. Regulation of the inflammatory synovial fibroblast phenotype by metastasis-associated lung adenocarcinoma transcript 1 long noncoding RNA in obese patients with osteoarthritis. Arthritis Rheum. 2020;72(4):609–619. doi: 10.1002/art.41158. [DOI] [PubMed] [Google Scholar]
- 60.Yang Y., Shen P., Yao T., Ma J., Chen Z., Zhu J., et al. Novel role of circRSU1 in the progression of osteoarthritis by adjusting oxidative stress. Theranostics. 2021;11(4):1877–1900. doi: 10.7150/thno.53307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu H., Hu Y., Wang C., Zhang X., He D. CircGCN1L1 promotes synoviocyte proliferation and chondrocyte apoptosis by targeting miR-330-3p and TNF-alpha in TMJ osteoarthritis. Cell Death Dis. 2020;11(4):284. doi: 10.1038/s41419-020-2447-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen P., Yang Y., Liu G., Chen W., Chen J., Wang Q., et al. CircCDK14 protects against Osteoarthritis by sponging miR-125a-5p and promoting the expression of Smad2. Theranostics. 2020;10(20):9113–9131. doi: 10.7150/thno.45993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dixit A., Parnas O., Li B., Chen J., Fulco C.P., Jerby-Arnon L., et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167(7):1853–1866 e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stahl P.L., Salmen F., Vickovic S., Lundmark A., Navarro J.F., Magnusson J., et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353(6294):78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 65.Rodriques S.G., Stickels R.R., Goeva A., Martin C.A., Murray E., Vanderburg C.R., et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363(6434):1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andersson A., Bergenstrahle J., Asp M., Bergenstrahle L., Jurek A., Fernandez Navarro J., et al. Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography. Commun Biol. 2020;3(1):565. doi: 10.1038/s42003-020-01247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Styrkarsdottir U., Lund S.H., Thorleifsson G., Zink F., Stefansson O.A., Sigurdsson J.K., et al. Meta-analysis of Icelandic and UK data sets identifies missense variants in SMO, IL11, COL11A1 and 13 more new loci associated with osteoarthritis. Nat Genet. 2018;50(12):1681–1687. doi: 10.1038/s41588-018-0247-0. [DOI] [PubMed] [Google Scholar]
- 68.Fernandez-Tajes J., Soto-Hermida A., Vazquez-Mosquera M.E., Cortes-Pereira E., Mosquera A., Fernandez-Moreno M., et al. Genome-wide DNA methylation analysis of articular chondrocytes reveals a cluster of osteoarthritic patients. Ann Rheum Dis. 2014;73(4):668–677. doi: 10.1136/annrheumdis-2012-202783. [DOI] [PubMed] [Google Scholar]
- 69.Rushton M.D., Reynard L.N., Barter M.J., Refaie R., Rankin K.S., Young D.A., et al. Characterization of the cartilage DNA methylome in knee and hip osteoarthritis. Arthritis Rheum. 2014;66(9):2450–2460. doi: 10.1002/art.38713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.den Hollander W., Ramos Y.F., Bos S.D., Bomer N., van der Breggen R., Lakenberg N., et al. Knee and hip articular cartilage have distinct epigenomic landscapes: implications for future cartilage regeneration approaches. Ann Rheum Dis. 2014;73(12):2208–2212. doi: 10.1136/annrheumdis-2014-205980. [DOI] [PubMed] [Google Scholar]
- 71.van Spil W.E., Szilagyi I.A. Osteoarthritis year in review 2019: biomarkers (biochemical markers) Osteoarthritis Cartilage. 2020;28(3):296–315. doi: 10.1016/j.joca.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 72.Oo W.M., Little C., Duong V., Hunter D.J. The development of disease-modifying therapies for osteoarthritis (DMOADs): the evidence to date. Drug Des Dev Ther. 2021;15:2921–2945. doi: 10.2147/DDDT.S295224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Conway J.R., Lex A., Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33(18):2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cao Y., Tang S., Nie X., Zhou Z., Ruan G., Han W., et al. Decreased miR-214-3p activates NF-kappaB pathway and aggravates osteoarthritis progression. EBioMedicine. 2021;65:103283. doi: 10.1016/j.ebiom.2021.103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.