Abstract
The pandemic of novel Coronavirus Disease 2019 (COVID-19) is widespread all over the world causing serious health problems as well as serious impact on the global economy. Reliable and fast testing of the COVID-19 has been a challenge for researchers and healthcare practitioners. In this work, we present a novel machine learning (ML) integrated X-ray device in Healthcare Cyber-Physical System (H-CPS) or smart healthcare framework (called “CoviLearn”) to allow healthcare practitioners to perform automatic initial screening of COVID-19 patients. We propose convolutional neural network (CNN) models of X-ray images integrated into an X-ray device for automatic COVID-19 detection. The proposed CoviLearn device will be useful in detecting if a person is COVID-19 positive or negative by considering the chest X-ray image of individuals. CoviLearn will be useful tool doctors to detect potential COVID-19 infections instantaneously without taking more intrusive healthcare data samples, such as saliva and blood. COVID-19 attacks the endothelium tissues that support respiratory tract, and X-rays images can be used to analyze the health of a patient’s lungs. As all healthcare centers have X-ray machines, it could be possible to use proposed CoviLearn X-rays to test for COVID-19 without the especial test kits. Our proposed automated analysis system CoviLearn which has 98.98% accuracy will be able to save valuable time of medical professionals as the X-ray machines come with a drawback as it needed a radiology expert.
Keywords: Smart healthcare, Healthcare-Cyber-Physical System (H-CPS), Machine learning, Deep neural network (DNN), COVID-19, X-ray
Introduction
Coronavirus disease (COVID-19) is a respiratory tract infectious disease that has spread across the world [1]. It belongs to a family of viruses whose infection can cause complications that vary from typical cold to shortness of breath [2]. Patients also develop pneumonia termed, Novel Coronavirus Pneumonia (NCP), that results in acute respiratory failure with a very poor prognosis and high mortality [3, 4]. Subsequently, the pandemic nature of the coronavirus and the absence of reliable vaccines make COVID-19 diagnosis an urgent medical crisis.
At present, the standard testing method for COVID-19 diagnosis is the real-time Reverse Transcription Polymerase Chain Reaction (rRT-PCR) test. In this test, nasal swab is collected from the patient and kept in a special medium called the “virus transport medium”, to protect the RNA. Upon reaching the lab, the swab is further processed to determine whether or not the patient is positive for the coronavirus [5]. The entire process takes several hours and the results generally arrive after a day or two depending on the time taken from the swab to reach the lab.
The spread of the COVID-19 virus at this point advocates the requirement of its quick diagnosis and treatment. Studies such as [6, 7] have proved that the COVID-19 virus infects the lungs and creates smooth and thick mucus in the patient’s affected lungs that is visible when chest X-rays and CT scans are performed. However, the analysis of X-ray images is a tedious task and require expert radiologists. In this endeavor, several computer algorithms and diagnosis tools such as [8, 9] have been proposed to get detailed insights from the X-ray images. Although these studies have performed efficiently, they lack in terms of higher accuracy, generalization, computational time, and error rate. To mitigate the shortcomings, recent studies such as [10–13] have incorporated machine learning (ML) and deep learning (DL) tools to investigate the chest X-ray images. The selection of proper DL-based automated analyzer and predictor for coronavirus patients will be very beneficial and helpful for the medical department and society. Additionally, ML-DL approaches can provide test results faster and more economically as compared to the laboratory-based tests.
Furthermore, as COVID-19 is spreading rapidly through person-to-person contact, hospitals and healthcare professionals are becoming increasing overburdened, sometimes to the point of complete breakdown. Clearly, an alternative, remote-based, online diagnostic and testing solution is required to fill this urgent and unmet need. The Internet of Medical Things (IoMT) could be extended to achieve this healthcare-specific solution. With this motivation, the present work proposes an AI-based Healthcare Cyber-Physical System (H-CPS) that incorporates convolutional neural networks (CNNs) (see Fig. 1). The model allows healthcare practitioners to promptly and automatically screen positive and negative COVID-19 patients by considering their chest X-ray images.
Fig. 1.
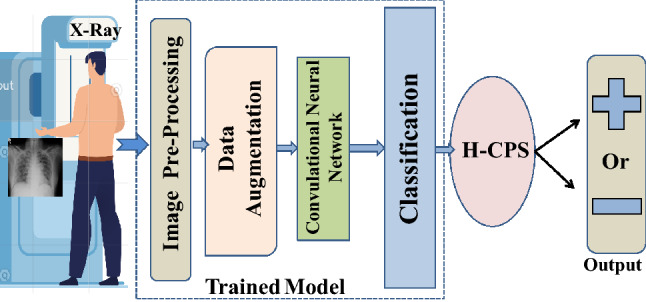
Process flow of proposed COVID-19 classification
The organization of the paper is as follows: “Related prior research works” discusses the working of existing COVID-19 detection models, their shortcomings, and our contributions in the H-CPS framework. “Proposed CoviLearn model for automatic initial screening of COVID-19” explains the proposed solution and its functioning, followed by “Performance evaluation” that validates the model using real-life data. Finally, “Conclusions and future scope” gives a compact conclusion and mentions the area of future study.
Related Prior Research Works
How Existing Research Models Function
Over the course of 2 years, many techniques have been proposed for effective COVID-19 detection [14]. However, from the exhaustive list of works, we have selected some of the state-of-the-art methods focusing only on the deep learning based COVID-19 detection. A CNN called COVIDNet was trained in [15] using more than 15000 chest radiography images of COVID-19 positive and negative cases. The deep neural network (DNN) reported accuracy of 92.4 and sensitivity of 80. A three-dimensional convolutional ResNet-50 network, termed COVNet, was proposed in [16] that utilized volumetric chest CT images consisting of community acquired pneumonia (CAP) and other non-pneumonia cases. The reported AUC metric by the model was 0.96. A similar ResNet-50 model proposed by [17] reported an AUC of 0.996 although tested to a much lesser dataset. In [18], a location-attention network using ResNet-18 was proposed using disparate CT samples from COVID-19 patients, influenza-A infected, and healthy individuals to classify COVID-19 cases, that reported an accuracy of 86.7. Samples from 4 classes: healthy, bacterial pneumonia, non-COVID-19 pneumonia, and COVID-19 were used in [19] to train drop-weight-based Bayesian CNNs that reported an accuracy of 89.92.
In [20], a modified inception transfer-learning model that reported an accuracy of 79.3, specificity of 0.83, and sensitivity of 0.67 was proposed. In [21], a multilayer perceptron combined with an LSTM neural network was implemented, that was trained using clinical data collected from 133 patients out of which 54 belonged to the critical care domain. The authors in [22] implemented a two-dimensional deep CNN architecture, while the authors in [23] combined three-dimensional UNet and ResNet-50 architectures. Both were trained using volumetric CT scanned data of patients categorized as COVID-19 positive and negative. The method in [24] used a pre-trained ResNet-50 network using chest X-ray images from 50 COVID-19 positive and 50 COVID-19 negative patients and reported an accuracy of 98. In [25] four state-of-the-art DNNs: AlexNet, Resnet-18, DenseNet-201, and SqueezeNet were ensembled. The model also used chest X-ray images of normal, viral pneumonia, and COVID-19 cases. A novel CNN augmented with a pre-trained AlexNet using transfer learning was proposed in [26]. The model was tested on both X-ray and CT scanned images with reported accuracies of 98 and 94.1, respectively.
Shortcomings in the Existing Research Works
Although the domain is very new and many studies pertaining to the deep learning-based methodology have been proposed, most of them suffer from shortcomings such as lower accuracy, model generalization, computational cost, and error rate. Even when certain research works achieve higher accuracy, they either suffer from lower sensitivity, specificity or have a small test dataset. Moreover, the prospect of augmenting IoMT frameworks with COVID-19 diagnosis is new and its incorporation can further assist the existing healthcare system to cope in this difficult times. Also, the training dataset for certain methods is limited because of class imbalance, that is, less number of coronavirus images as compared to normal lung images. This problem of dataset imbalance results in lesser model accuracy and less efficient. Table 1 provides a comprehensive comparison of the existing research works.
Table 1.
Comparative perspective with related AI works for COVID-19 detection
| Model | Dataset used | Method | Results | Shortcoming (s) |
|---|---|---|---|---|
| Wang et al. [15] | 15000 chest radiography images of confirmed COVID-19 positive and negative cases | Deep convolutional neural network called COVIDNet | Accuracy-92.4, Sensitivity-80 | Comparatively lower accuracy and sensitivity |
| Li et al. [16] | 4356 Volumetric chest CT images that included community acquired pneumonia (CAP) and other non-pneumonia cases | 3-Dimensional Convolutional ResNet-50 network, termed COVNetDeep | AUC-0.96 | High computational cost and requirement of professionals to analyze the results |
| Gozes et al. [17] | CT images from 157 COVID affected patients | ResNet-50 | AUC-0.996 | Relatively small testing dataset |
| Xu et al. [18] | 618 CT samples from COVID-19 patients (219), influenza-A infected (224), and healthy individuals (175) | Location attention network using ResNet-18 | Accuracy-86.7 | Lower accuracy |
| Ghoshal et al. [19] | 5941 Chest radiography images samples from 4 classes: healthy, bacterial pneumonia, non-COVID-19 pneumonia | Drop-weights based Bayesian CNNs | Accuracy-89.92 | Lower accuracy |
| Wang et al. [20] | 1065 CT images (325 COVID, 740 Viral Pneumonia) | Modified inception transfer-learning model | Accuracy-79.3, Specificity-83, Sensitivity-67 | Lower accuracy and imbalanced dataset |
| Fang et al. [21] | 133 CT images of COVID-19 patients | Multilayer perceptron combined with an LSTM | AUC-0.954 | Relatively smaller dataset size and lower accuracy |
| Jin Feng et al. [22] | 970 CT images of COVID-19 positive and 1385 COVID-19 negative patients | 2-Dimensional CNN | Accuracy-94.98, AUC-0.979 | Lower accuracy and lack of generalization |
| Jin et al. [23] | 1136 CT images (723 COVID-19 positive) | 3-Dimensional UNet and ResNet-50 | Specificity-92.2, Sensitivity-97.4 | Lower accuracy |
| Narin et al. [24] | Chest X-ray images from 50 COVID-19 positive and 50 COVID-19 negative patients | ResNet-50 | Accuracy-98 | Relatively small testing dataset |
| Chowdhury et al. [25] | 1341 Normal, 1345 Viral Pneumonia and 190 COVID-19 chest X-ray images | Combination of AlexNet, ResNet-18, DenseNet-201, and SqueezeNet | Accuracy-98.3 | High computational cost, large number of training hyperparameters, and class imbalance |
| Maghdid et al. [26] | 170 X-ray and 361 CT images | CNN augmented with a pre-trained AlexNet using transfer learning | Accuracy-98 for X-ray images, Accuracy-94.1 for CT images | High computational cost and lack of implementation in smart healthcare |
Our Vision of CoviLearn in the H-CPS Framework
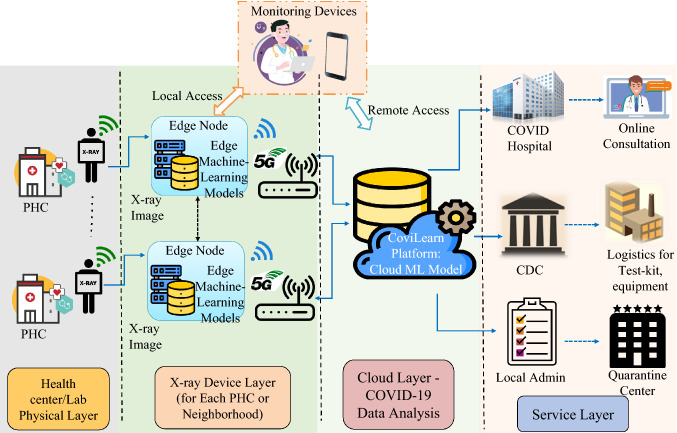
We propose an AI-based H-CPS framework termed “CoviLearn” to provide healthcare professionals the leverage to perform automatic screening of COVID-19 patients using their chest X-ray images. With a deep neural network (DNN) in its core, the CoviLearn model is implemented on the server for ubiquitous deployment. The hyperparameters of the DNN have been adjusted to make its functioning reliable, accurate, and specific. By just uploading the X-ray images, the model automatically identifies the symptoms and reports unbiased results. CoviLearn augmented with H-CPS brings patients, doctors, and test lab in a single smart healthcare platform, as illustrated in Fig. 2. The reported results can be uploaded to the IoMT platform from where it may be transferred to nearby COVID-care hospitals, the Center for Disease Control (CDC), and state and local health bureaus. Hospitals could subsequently offer online health consultations based on the patient’s condition and monitor vital equipments and quarantine requirements. Therefore, the proposed H-CPS provides people the leverage to dynamically monitor their disease status, receive proper medical needs, and eventually curb the spread of the virus.
Fig. 2.
Schematic representation of the Healthcare Cyber-Physical System (H-CPS) ecosystem concept for combating COVID-19
Novel Contributions of CoviLearn
The major contributions of the work are:
An architecture of H-CPS framework augmented with a next generation smart X-ray machine architecture at the interface is proposed to combat the spread of COVID-19.
An efficient heuristic search technique is incorporated which automatically finds an optimal feature subset present in the input chest X-ray images.
An end-to-end automatic functioning DNN model that extracts the features from X-ray images is incorporated.
The CNN blocks are reliable, accurate, and very specific that makes the overall model very effective. Furthermore, the model can be easily integrated into embedded and mobile devices, thereby assisting health practitioners to effectively diagnose COVID-19.
Proposed CoviLearn Model for Automatic Initial Screening of COVID-19
The CoviLearn Device for Next-Generation X-ray Screening
As discussed in the earlier sections, COVID-19 and other related pneumonia diseases can be screened and diagnosed by analyzing chest X-ray images. However, the existing X-ray diagnosis suffers from limited access and lack of experienced personnel. To address this issue, we propose a next-generation X-ray system in the H-CPS perspective. The H-CPS and IoMT together bring all the necessary agents of smart healthcare in a universal communication and connectivity platform. This linking of technologies extends the efficiency services such as telemedicine, teleconsultation, and endorse smart-medical care.
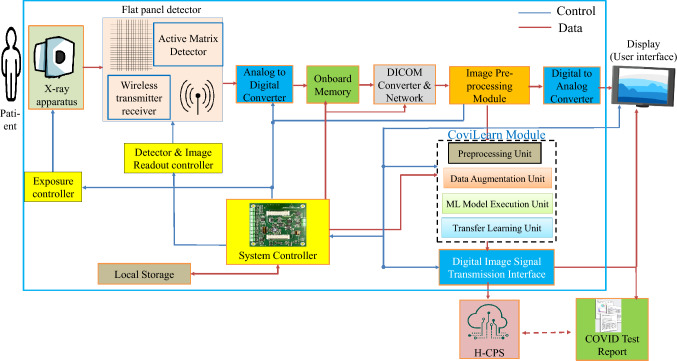
Figure 3 shows the system-level block diagram of the next-generation X-ray machine integrated with CoviLearn for automatic screening of infectious diseases. It identifies most of its components, such as X-ray apparatus (tube), flat panel detector, onboard memory, DICOM protocol converter, Image processing, CoviLearn diagnosis, wired/wireless data communication, display, or user interface, along with system controller. In the proposed X-ray machine, X-ray image is captured by an array of sensor in the digital and radiography flat panel detector. The flat panel also includes the devices of communication to next stages. The image is then saved and converted to DICOM X-ray image. Subsequently, the image is processed and based on the quality and requirement the exposure of the X-ray tube is adjusted. The captured image is stored temporarily in the local memory, after which it is displayed on monitor screen with the help of the controller. After acquiring the quality assured image, it is then transferred to the CoviLearn model which automatically classifies the image either as normal or COVID-19 affected. The image classification is performed either locally in the presence of sufficient resources or on cloud by transmitting the images over network. The test results automatically synchronize with the H-CPS platform for necessary medical and administrative actions. The controller unit is responsible for controlling the entire sequence of events.
Fig. 3.
The proposed next-generation X-ray device of CoviLearn integrated with machine learning models
Dataset Used for Validating the Proposed CoviLearn System
To overcome the problem of class imbalance, we have manually collected chest X-ray of patients having coronavirus. These images are from various resources such as pyimagesearch, radiopedia, sirm, and eurorad. For the normal chest X-ray, we have used the chest X-ray dataset from the National Institute of Health (NIH), USA [27]. The count of images from both the sources was 250. Subsequently, the dataset has been divided into two classes: patient’s diagnosed as COVID-19 positive and negative. For training 80 of the dataset ( 200 images) is used from which 30 is used for validation ( 60 images). The testing of the model is performed on 20 ( 50 images) of the dataset. Based on this validation dataset, the loss and validation graphs have been plotted. All the images are processed and mixed to prevent undue biasing as discussed in the following subsections.
Data Pre-processing
All the captured images have different sizes, and therefore, data pre-processing was essential before doing further analysis. The pre-processing is performed in three stages: first, the individual data are normalized by subtracting the mean RGB values; second, all the pixels in the input image data are scaled within the range of 0 to 1. Finally, the tensor is reshaped appropriately, so that it fits the model (in this case, the tensor is reshaped into pixels).
Data Augmentation
Deep learning models are ravenous for data and since our model only has around 250 images for each class; hence, the volume of our data needs to be increased and this can be achieved through data augmentation. Therefore, similar to the process mentioned in [28], the input images are augmented by random crop, adjust contrast, flip, rotation, adjust brightness, horizontal–vertical shift, aspect ratio, random shear, zoom, and pixel jitter. As a result of this augmentation, the proposed CoviLearn system became more efficient.
The Proposed Transfer Learning for Deep Neural Network in CoviLearn
CoviLearn uses transfer learning to predict the classification results. Transfer learning substitutes for the requirement of large dataset and has been used in different applications, such as healthcare, manufacturing, etc. It uses the knowledge learned in training a large dataset and transfers that same knowledge in some different and smaller dataset. In the present work, four different DNNs: ResNet-50, ResNet-101, DenseNet-121, and DenseNet-169, along with different blocks to train the individual networks. The hyperparameters have been adjusted to report the highest accuracy. Detailed structural organizational of network layers is as illustrated in Fig. 4 where each network is divided into phases, starting from getting an image input, followed by training the model by sequentially passing the set of images into convolutional networks, to finally predicting the results using a classification layer. Following subsection discusses the base classifiers and the difference between them.
Fig. 4.
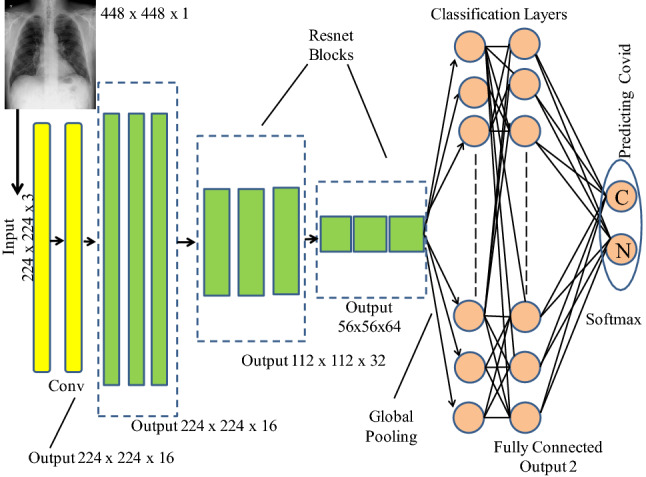
Organization of the DNN with classification layers
Deep Neural Base Classifiers
The CoviLearn model uses four deep neural networks as the base classifiers. Two of these belong to the ResNet family [29] (ResNet-50 and ResNet-101) and remaining two belong to the DenseNet family [30] (DenseNet-121 and DenseNet-169). As the convolutional neural networks become deeper, the back propagated error from any layer is required to traverse the entire depth where repeated weight multiplications occur. As a result of these multiplications, the original error significantly diminishes and the neural network’s performance is satisfactorily affected. To combat this, researchers have proposed many architectures, out of which the current state-of-the-art includes the DenseNet and the ResNet models.
DenseNet or Dense Convolutional Network solves the problem using shorter connections between the layers. In other words, inside the DenseNet network, the each layer is connected to all its higher layers. Equation (1) represents the learning equation for a traditional CNN
| 1 |
where P represents the lth layer of the network, and T denotes the feature learned in the previous layer. For a DenseNet, the equation changes to (2)
| 2 |
This arrangement allows feature reusing without having to travel the entire depth or entire depth of the network. In comparison to a traditional CNN, DenseNet requires fewer parameters, because features learned in one layer are sent to the higher layers, thereby eliminating redundancy. A typical DenseNet architecture involves a convolution layer, followed by a pooling layer. These are followed by 4 dense blocks and 3 transition blocks placed one after the other. Inside the dense block, there are two convolutional layers with filters of different sizes, while the transition layer involves an average pooling layer. The dissimilarity between the DenseNet-121 and DenseNet-169 networks is with respect to the number of hidden layers. For the former, the total number of convolution layers in the four Dense Blocks is 121, while for the latter that is 169. Increasing the layers does not necessarily improve the accuracy and depends upon the particular situation.
Residual Networks or ResNet solves the problem of vanishing gradient decent by utilizing a skip connection between the original input and the final convolution layers. By overlooking the in between layers and attaching the given input directly to the output allows the presence of an additional path for the back propagated error to flow and therefore solving the problem of vanishing gradient descent. For a DenseNet, the equation changes to (3)
| 3 |
A typical ResNet architecture involves four stages. The first stage is responsible for performing zero-padding operation on the input data. The second stage is made up of convolutional blocks that performs convolution along with batch-normalization and max-pooling. The penultimate layer consists of identity blocks augmented with filters, followed by the final stage that comprises a GAP layer, a fully connected dense layer, and classifier function. All convolution layers use ReLU as the activation function. Similar to DenseNet, the two types of ResNets that is ResNet-50 and ResNet-101 differ in the depth of the network. It has been observed that certain variations of ResNet have redundant layers that barely contribute. The presence of them results in ResNet handling larger parameters and weights. On the other hand, DenseNet are relatively narrow (fewer number of filters) and simply add the new feature maps. Another difference between the DenseNet and ResNet models is that the former does not sum the output feature maps of the preceding layers but rather concatenates them, unlike the latter where summation happens. This is evident from Eqs. (2) and (3).
Training and Testing of the Proposed Model
The CoviLearn model takes the input image, swaps the color channels, and resizes it to 224 224 pixels. Afterwards, the data and label list are converted into an array, while the pixel intensities are normalized between 0 and 1, by dividing the entire input image by 255. Subsequently, one-hot encoding is performed on the labels, following which various models are loaded one at a time by freezing few upper layers and a base layer is created with dropout. Finally, the input tensor of size 224 224 is loaded onto the model and compiled using Adam optimizer and binary cross entropy loss.
Performance Evaluation
Experimental Setup
To compare the performance of different models, three evaluation parameters: accuracy, sensitivity, and specificity have been considered. As the test images are converted into 224 224 tensor, the model predicts the above-mentioned three metrics. Table 2 illustrates the comparison of results between the four models: DNN I (ResNet-50), DNN II (ResNet-101), DNN III (DenseNet-121), and DNN IV (DenseNet-169). A confusion matrix (see Fig. 5 compares the True Positive, True Negative, False Positive, and False Negative values. Moreover, loss-accuracy versus epoch graph is also provided to project how the training loss, validation loss, training accuracy, and validation accuracy vary with each epoch.
Table 2.
Performance metrics for different deep learning techniques
| Models explored | Accuracy | Sensitivity | Specificity | Total parameter | AUC area |
|---|---|---|---|---|---|
| DNN I | 0.9592 | 0.9583 | 0.9600 | 23,696,066 | 0.959 |
| DNN II | 0.9694 | 0.9792 | 0.9600 | 42,757,826 | 0.970 |
| DNN III | 0.9898 | 1.0000 | 0.9800 | 7,103,234 | 0.990 |
| DNN IV | 0.9796 | 1.0000 | 0.9600 | 12,749,570 | 0.980 |
Fig. 5.
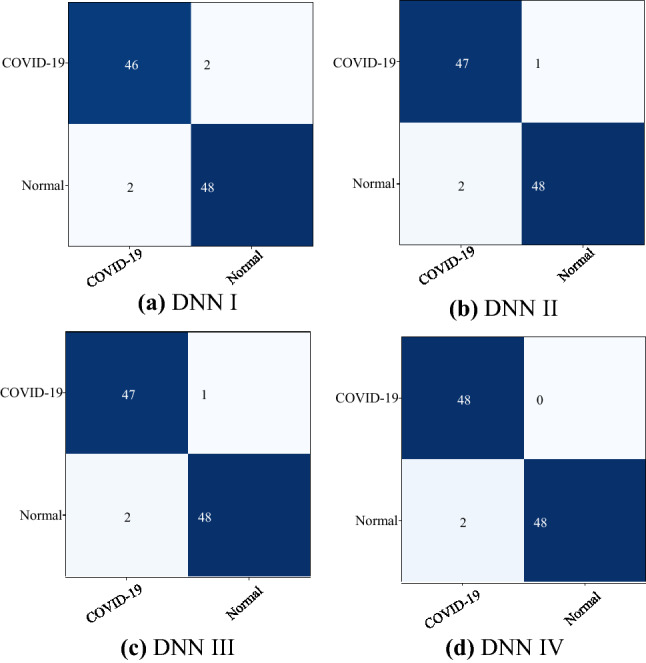
Confusion matrix for a DNN I, b DNN II, c DNN III, and d DNN IV
Result Analysis
In context of coronavirus detection, True Positive (TP) is when the patient has coronavirus and the model detects coronavirus, True Negative is when the patient does not have coronavirus and the model also predicts the same. False Positive (FP) is when the the patient is not infected with the coronavirus, but the model predicts the opposite, while False Negative (FN) is when the patient has coronavirus, but the model says otherwise. Accuracy specifies the correct number of predictions made by the CoviLearn model with respect to the total number of patients and is represented by Equation (4). Additional metrics such as sensitivity—the ability to identify coronavirus patients correctly—and selectivity—the ability to identify non-coronavirus patients correctly—are as defined by Eqs. (5) and (6), respectively
| 4 |
| 5 |
| 6 |
Table 2 summarizes the performance matrix for different deep learning model tested for the different classification schemes. DNN III, which has DenseNet-121 architecture, performs best over other models in classification yielding an accuracy of 98.98, sensitivity of 100, and specificity value of 98. Whereas, DNN I has the lowest performance value with an accuracy of 95.92, sensitivity of 95.83, and specificity value of 96.
Figure 5 shows the confusion matrices of COVID-19 and normal test results of the different pre-trained models. The graphs show a well-defined pattern of the training–validation accuracy that increases, and the training–validation loss that decreases, with increasing epochs. Because of the limited computational resources, the comparison between different parameters is done for 25 epochs only. Besides the confusion matrix, receiver-operating characteristic (ROC) curve plots and areas for each model are given in Fig. 6. DNNs which are trained with DenseNet pre-trained blocks appear to be very higher than DNN trained with ResNet blocks, with DNN III having the highest AUC of 99. One of the interesting findings is the DNN which when used with the ability of the DenseNet model achieves higher sensitivity and specificity. This ensures the reduction of false positives for both the COVID-19 and the healthy classes. As is evident from the relationship between accuracy and epoch, DNN-III shows the highest accuracy followed by DNN-IV, DNN-II, and DNN-I. The accuracy increases with each subsequent epoch except at few as illustrated in Fig. 7. A similar trend is shown in loss graphs where the loss decreases with each subsequent epochs and a similar trend is followed, that is, DNN-III shows the lowest loss followed by DNN-IV, DNN-II, and DNN-I (see Fig. 8). The results as reported by the proposed CoviLearn model are compared with the existing research works and tabulated in Table 3. In [18], detects COVID-19 using classification of CT samples by CNN models with an accuracy of 86.7, sensitivity of 98.2, and specificity of 92.2. CovidNet in [15] reported an accuracy of 93.3. The CNN-based DarkCovidNet model [31] to detect COVID-19 from chest X-ray also has an accuracy of 98.08. In comparison, the proposed model has an accuracy of 98.98, sensitivity of 0.984, and specificity of 0965. CoviLearn has significantly outperformed existing deep learning-based COVID-19 detection techniques such as [15, 18–20, 23]. Also, the sensitivity of the proposed model has outperformed existing models such as [15, 20, 23] both in terms of sensitivity and specificity. [17, 21, 24] achieved similar accuracy; however, their test dataset size is relatively smaller than the one used in the current work. The deep neural architectures proposed in [25, 26] involved many hyperparameters, estimation of which increased the overall computation cost and resulted in ubiquitous deployment. On the other hand, CoviLearn because of its transfer-learning ability and selected deep neural networks has the advantage of rejecting redundant parameters and thereby reducing the overall computational cost. Finally, all these models lacked a smart healthcare framework, which has been proposed and implemented in CoviLearn in the form of H-CPS. The comparison of existing research works is compactly summarized in Tables 3 and 4 .
Fig. 6.
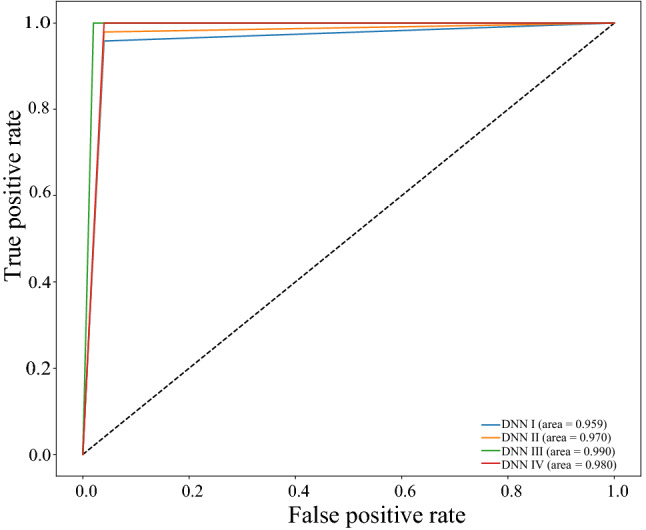
Comparison of the receiver-operating characteristics (ROC)
Fig. 7.
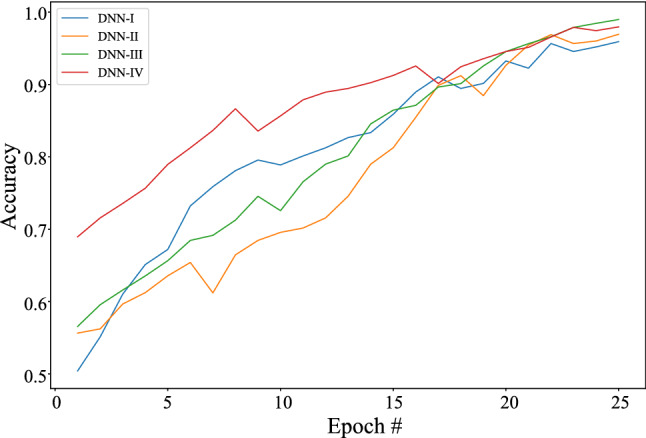
Classification accuracy in the deep learning system validation
Fig. 8.
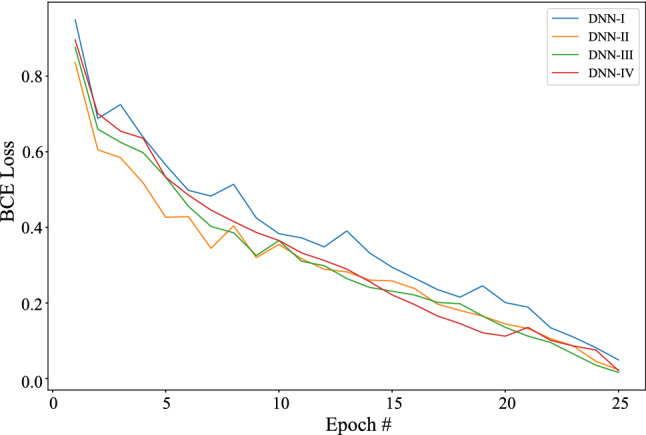
Binary cross entropy loss in the deep learning system validation
Table 3.
Comparison of results with existing recent similar works
Table 4.
Comparison with existing deep learning-based COVID-19 detection model
Effectiveness of the Transfer-Learning Concept
The initial neural network when trained reported accuracy, sensitivity, and specificity values of 0.5981, 0.6041, and 0.5923, respectively. To improve these substantially, we used the concept of transfer learning. It is done by freezing the layers of the existing models and replacing with the penultimate layer (the layer responsible for performing classification) of state-of-the-art neural networks trained on larger datasets to perform final classification. This step improved the accuracy, sensitivity, and specificity metrics to 0.9225, 0.9319, and 0.9135, respectively. Following this step, fine-tuning is performed on the model’s hyperparameters to further improve the model’s performance by . Therefore, despite a small training dataset of 250 images, embedding the transfer learning helped improve the model’s classification performance significantly. Table 5 compares the metrics obtained in each of the stages.
Table 5.
Performance metrics at different stages of training
| Stage | Network | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| Initial training | DNN I | 0.5887 | 0.5882 | 0.5892 |
| DNN II | 0.5949 | 0.6009 | 0.5892 | |
| DNN III | 0.6075 | 0.6137 | 0.6015 | |
| DNN IV | 0.6012 | 0.6137 | 0.5892 | |
| Transfer learning | DNN I | 0.9081 | 0.9072 | 0.9088 |
| DNN II | 0.9177 | 0.9270 | 0.9088 | |
| DNN III | 0.9370 | 0.9467 | 0.9278 | |
| DNN IV | 0.9274 | 0.9467 | 0.9088 | |
| Fine tuning | DNN I | 0.9592 | 0.9583 | 0.9600 |
| DNN II | 0.9694 | 0.9792 | 0.9600 | |
| DNN III | 0.9898 | 1.0000 | 0.9800 | |
| DNN IV | 0.9796 | 1.0000 | 0.9600 |
Conclusions and Future Scope
The study presents CoviLearn, a DNN-based transfer-learning approach in Healthcare Cyber-Physical System framework to perform automatic initial screening of COVID-19 patients using their chest X-ray image data. An architecture of next-generation smart X-ray machine for automatic screening of COVID-19 is proposed at the interface of H-CPS. Four different DNNs: ResNet-50, ResNet-101, DenseNet-121, and DenseNet-169 are trained and tested for classification of the X-ray images from healthy and corona disease-infected patients. DenseNet-121 showed the highest accuracy close to 98.98 followed by DenseNet-169 , ResNet-50, and ResNet-101. Similarly, the sensitivity of DenseNet-121 and DenseNet-169 are 100, while that of ResNet-50 and ResNet-101 are close to 97. The highest specificity of DNN III is 98%. Therefore, all these results clearly indicate the ability to classify the deadly coronavirus correctly.
The present CoviLearn platform will be very useful tool doctors to diagnosis the coronavirus disease at a lower cost despite being economical and automatic. However, additional study and medical trial are required to full proof the extracted features extracted by machine learning as reliable bio-markers for COVID-19. Furthermore, these machine learning models can be extended to diagnose other chest-related diseases including tuberculosis and pneumonia. A limitation of the study is the use of a limited number of COVID-19 X-ray images. Therefore, in the future, a larger dataset and a cloud based system can be ventured to make the model ubiquitous and more robust. In fact, the results can be used to detect the highly prone corona positive patients in a timely application of quarantine measure, until the rRT-PCR test examinations results are obtained. The proposed CoviLearn can be added to our healthcare CPS framework CoviChain for reliable information sharing right from the source to destination end while accommodating various stake holders [34].
Acknowledgements
We would like to acknowledge IIIT-NR undergraduate students Chirag Samal, Deewanshu Ukey, and Gourav Chowdhary for their help on this paper.
Declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Debanjan Das, Email: debanjan@iiitnr.edu.in.
Sagnik Ghosal, Email: sagnikghosal1999@gmail.com.
Saraju P. Mohanty, Email: saraju.mohanty@unt.edu
References
- 1.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu T, Hu J, Kang M, Lin L, Zhong H, Xiao J, He G, Song T, Huang Q, Rong Z, Deng A, Zeng W, Tan X, Zeng S, Zhu Z, Li J, Wan D, Lu J, Deng H, He J, Ma W. Transmission dynamics of 2019 novel coronavirus (2019-ncov). 2020.
- 3.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Challen R, Brooks-Pollock E, Read JM, Dyson L, Tsaneva-Atanasova K, Danon L. Risk of mortality in patients infected with sars-cov-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372–81. [DOI] [PMC free article] [PubMed]
- 5.Jiang F, Deng L, Zhang L, Cai Y, Cheung CW, Xia Z. Review of the clinical characteristics of coronavirus disease 2019 (COVID-19). J Gen Int Med. 2020;80(6):656–65. [DOI] [PMC free article] [PubMed]
- 6.Sethy PK, Behera SK. Detection of coronavirus disease (COVID-19) based on deep features. Preprints. 2020;2020030300:2020. [Google Scholar]
- 7.Simpson S, Kay FU, Abbara S, Bhalla S, Chung JH, Chung M, Henry TS, Kanne JP, Kligerman S, Ko JP, Litt H. Radiological society of North America expert consensus statement on reporting chest CT findings related to COVID-19. Endorsed by the society of thoracic radiology, the American college of radiology, and RSNA. Radiol Cardiothorac Imaging. 2020;2(2):e200152. doi: 10.1148/ryct.2020200152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pattrapisetwong P, Chiracharit W. Automatic lung segmentation in chest radiographs using shadow filter and multilevel thresholding. In: Proceedings of ICSEC. 2016;pp. 1–6.
- 9.Qin C, Yao D, Shi Y, Song Z. Computer-aided detection in chest radiography based on artificial intelligence: a survey. Biomed Eng Online. 2018;17(1):113. doi: 10.1186/s12938-018-0544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasa F, Golkov V, Pfeiffer F, Cremers D, Pfeiffer D. Efficient deep network architectures for fast chest X-ray tuberculosis screening and visualization. Sci Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-42557-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ausawalaithong W, Thirach A, Marukatat S, Wilaiprasitporn T. Automatic lung cancer prediction from chest X-ray images using the deep learning approach. In: Proceedings of the international conference on biomedical engineering, 2018;pp. 1–5.
- 12.Drozdov I, Forbes D, Szubert B, Hall M, Carlin C, Lowe DJ. Supervised and unsupervised language modelling in chest X-ray radiological reports. PLoS ONE. 2020;15(3):e0229963. doi: 10.1371/journal.pone.0229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan F, Gao Y, Wang J, Shi W, Shi N, Han M, Xue Z, Shi Y. Lung infection quantification of COVID-19 in CT images with deep learning. arXiv preprint arXiv:2003.04655. 2020.
- 14.Thapliyal H, Michael K, Mohanty SP, Srinivas M, Ganapathiraju MK. Consumer technology-based solutions for COVID-19. IEEE Consumer Electron Mag. 2021;10(2):64–65. doi: 10.1109/MCE.2020.3040513. [DOI] [Google Scholar]
- 15.Wang L, Lin ZQ, Wong A. Covid-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest x-ray images. Sci Rep. 2020;10(1):1–12. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q, Cao K, Liu D, Wang G, Xu Q, Fang X, Zhang S, Xia J, Xia J. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest ct. Radiology. 2020;296;E65–71. [DOI] [PMC free article] [PubMed]
- 17.Gozes O, Frid-Adar M, Greenspan H, Browning P. D, Zhang H, Ji W, Bernheim A, Siegel E. Rapid ai development cycle for the coronavirus (COVID-19) pandemic: Initial results for automated detection & patient monitoring using deep learning CT image analysis. arXiv preprint arXiv:2003.05037, 2020.
- 18.Xu X, Jiang X, Ma C, Du P, Li X, Lv S, Yu L, Ni Q, Chen Y, Su J, Lang G, Li Y, Zhao H, Liu J, Xu K, Ruan L, Sheng J, Qiu Y, Wu W, Liang T, Li L. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghoshal B, Tucker A. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv preprint arXiv:2003.10769, 2020.
- 20.Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, Yang J, Li Y, Meng X, Xu B. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). Eur Radiol 2021;31:6096–104. [DOI] [PMC free article] [PubMed]
- 21.Fang C, Bai S, Chen Q, Zhou Y, Xia L, Qin L, Gong S, Xie X, Zhou C, Tu D, Zhang C, Liu X, Chen W, Bai X, Torr PH. Deep learning for predicting COVID-19 malignant progression. Med Image Anal. 2021;72:102096. doi: 10.1016/j.media.2021.102096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin C, Chen W, Cao Y, Xu Z, Tan Z, Zhang X, Deng L, Zheng C, Zhou J, Shi H, Feng J. Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat Commun. 2020;11:5088. [DOI] [PMC free article] [PubMed]
- 23.Jin S, Wang B, Xu H, Luo C, Wei L, Zhao W, Hou X, Ma W, Xu Z, Zheng Z, Sun W, Lan L, Zhang W, Mu X, Shi C, Wang Z, Lee J, Jin Z, Lin M, Jin H, Zhang L, Guo J, Zhao B, Ren Z, Wang S, You Z, Dong J,. Wang X, Wang J, Xu W. Ai-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical ai system in four weeks. Appl. Soft Comput. 2021;98:106897. [DOI] [PMC free article] [PubMed]
- 24.Narin A, Kaya C, Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using x-ray images and deep convolutional neural networks. Pattern Anal Appl. 2021;24:1207–20. [DOI] [PMC free article] [PubMed]
- 25.Chowdhury MEH, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Emadi NA, Reaz MBI, Islam MT. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- 26.Maghdid HS, Asaad AT, Ghafoor KZ, Sadiq AS, Mirjalili S, Khan MK. Diagnosing COVID-19 pneumonia from x-ray and CT images using deep learning and transfer learning algorithms. In: Multimodal Image Exploitation and Learning 2021. vol. 11734. International Society for Optics and Photonics, Bellingham. 2021;p. 117340E.
- 27.NIH. Nih clinical center—cxr8. 2020. https://nihcc.app.box.com/v/ChestXray-NIHCC. Accessed 2 Sept 2017.
- 28.Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J. Big Data. 2019;6(1):60. doi: 10.1186/s40537-019-0197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of IEEE conference on computer vision pattern recognition. 2016; pp. 770–8.
- 30.Huang G, Liu Z, Van Der Maaten L, Weinberger K. Q. Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2017; pp. 4700–4708.
- 31.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. [DOI] [PMC free article] [PubMed]
- 32.Khatri A, Jain R, Vashista H, Mittal N, Ranjan P, Janardhanan R. Pneumonia identification in chest X-Ray images using EMD. Trends Commun Cloud Big Data. 2020;99:87–98.
- 33.Toğaçar M, Ergen B, Cömert Z, Özyurt F. A deep feature learning model for pneumonia detection applying a combination of mrmr feature selection and machine learning models. Irbm. 2020;41:212–222. doi: 10.1016/j.irbm.2019.10.006. [DOI] [Google Scholar]
- 34.Vangipuram SLT, Mohanty SP, Kougianos E. CoviChain: a blockchain based framework for nonrepudiable contact tracing in healthcare cyber-physical systems during pandemic outbreaks. SN Comput Sci. 2021;2(5):346. doi: 10.1007/s42979-021-00746-x. [DOI] [PMC free article] [PubMed] [Google Scholar]