Abstract
We describe rapid PCR-biprobe identification of Campylobacter spp.. This is based on real-time PCR with product analysis in the same system. The assay identifies enteropathogenic campylobacters to the species level on the basis of their degree of hybridization to three 16S ribosomal DNA (rDNA) biprobes. First-round symmetric PCR is performed with genus-specific primers which selectively target and amplify a portion of the 16S rRNA gene common to all Campylobacter species. Second-round asymmetric PCR is performed in a LightCycler in the presence of one of three biprobes; the identity of an amplified DNA-biprobe duplex is established after determination of the species-specific melting peak temperature. The biprobe specificities were determined by testing 37 reference strains of Campylobacter, Helicobacter, and Arcobacter spp. and 59 Penner serotype reference strains of Campylobacter jejuni and C. coli. From the combination of melting peak profiles for each probe, an identification scheme was devised which accurately detected the five taxa pathogenic for humans (C. jejuni/C. coli, C. lari, C. upsaliensis, C. hyointestinalis, and C. fetus), as well as C. helveticus and C. lanienae. The assay was evaluated with 110 blind-tested field isolates; when the code was broken their previous phenotypic species identification was confirmed in every case. The PCR-biprobe assay also identified campylobacters directly from fecal DNA. PCR-biprobe testing of stools from 38 diarrheic subjects was 100% concordant with PCR–enzyme-linked immunosorbent assay identification (13, 20) and thus more sensitive than phenotypic identification following microaerobic culture.
Campylobacter is the most common bacterial cause of human gastrointestinal infection worldwide. Incidence rates are estimated at 1% of the population per year in both the United Kingdom and United States, representing a major public health and economic burden (2, 11, 19, 25). The species most frequently isolated from humans are Campylobacter jejuni and C. coli; C. upsaliensis, C. hyointestinalis, C. lari, and C. fetus have also been implicated, though rarely (13). Campylobacter sputorum subsp. sputorum and Campylobacter sputorum subsp. bubulus occasionally cause disease in humans, while five species (C. concisus, C. curvus, C. gracilis, C. rectus, and C. showae) are not gastrointestinal pathogens but are found in association with the oral cavity. C. mucosalis, C. helveticus, and Campylobacter sputorum subsp. faecalis are commensals or pathogens of animals (21, 24).
Rapid diagnosis of Campylobacter infections is complicated by their slow growth rate and by difficulties in phenotypic identification to species level due to the fact that few informative biochemical tests can be applied to them. Furthermore, the frequency of non-C. jejuni, non-C. coli campylobacter infections is probably underestimated, since the isolation methods used (incubation at 42°C on selective antibiotic-containing media) are inhibitory to these species. Genotypic methods may therefore offer an attractive alternative to culture for accurate identification of the range of Campylobacter spp. involved in human infection (13).
We have previously described a PCR–enzyme-linked immunosorbent assay (PCR-ELISA) where identification of a genus-specific PCR product was carried out by capture with species-specific oligonucleotides immobilized on a microtiter plate (20). The present study is based on the LightCycler, a temperature-controlled microvolume fluorimeter which provides rapid real-time PCR and product analysis in a single closed-tube system (35). This real-time PCR platform can support several different sequence-specific fluorescent probe detection systems such as hybridization probes (34), TaqMan probes (17), Molecular Beacons (26), and biprobes (3). The term “biprobe” was coined by the user community subsequent to issue of a patent for the system (Roderick Fuerst [Bio/Gene Ltd.], personal communication). In the present report a biprobe assay is described where the hybridization of a biprobe to a target sequence results in intercalation of a double-stranded DNA-specific fluorophore (SYBR Green I). Due to the close proximity of SYBR Green I to the biprobe (which is labeled with the red fluorophore Cy5), there is increased light emission from Cy5, a phenomenon termed fluorescent resonance energy transfer (FRET) (23). The Cy5-labeled probe is blocked with biotin at the 3′ end to prevent its acting as a primer. Conversely, when the bound biprobe melts from the target sequence, a decrease in light emission from Cy5 is observed. By continually monitoring the emission peak of Cy5 (675 nm) during the melt cycle, the temperature at which the biprobe detaches from the single-stranded PCR product is determined and visualized as a defined melting peak. The melting peak data are characteristic of a particular probe-target sequence because mismatches between probe and target affect the kinetics of melting, producing different melting peaks for each species of interest. In the present study we developed an assay that would identify Campylobacter species both from field isolates and from DNA extracted from fecal samples.
MATERIALS AND METHODS
Bacterial strains and clinical fecal samples.
Reference strains used for assay development and assessment of biprobe specificities are listed in Table 1. To evaluate the assay, 46 C. jejuni and 13 C. coli Penner serotype reference strains were tested. In addition, 110 field isolates already identified to species level by phenotype were tested; these were blind-coded and comprised 26 C. jejuni, 12 C. coli, 13 C. lari, 24 C. upsaliensis, 13 C. fetus, 11 C. helveticus, 10 C. hyointestinalis, and 1 C. lanienae isolate.
TABLE 1.
Reference strains of Campylobacter, Arcobacter, and Helicobacter species
| Species | Sourcea |
|---|---|
| Campylobacter spp. | |
| C. jejuni subsp. jejuni | NCTC 11351T |
| C. jejuni subsp. doylei | NCTC 11951T |
| C. coli | NCTC 11350 |
| C. lari | NCTC 11352T |
| C. upsaliensis | NCTC 11541T |
| C. helveticus | NCTC 12470T |
| C. fetus subsp. fetus | NCTC 10842T |
| C. fetus subsp. venerealis | NCTC 10354T |
| C. hyointestinalis subsp. hyointestinalis | NCTC 11608T |
| C. sputorum subsp. sputorum | NCTC 11528T |
| C. sputorum subsp. faecalis | NCTC 11415T |
| C. sputorum subsp. bubulus | NCTC 11367T |
| C. gracilis | NCTC 12738T |
| C. concisus | NCTC 11485T |
| C. rectus | NCTC 11489T |
| C. curvus | NCTC 11649T |
| C. showae | NCTC 12843T |
| C. mucosalis | NCTC 11001 |
| C. lanienae | NCTC 13004T |
| [Bacteroides] ureolyticusb | NCTC 10941T |
| Helicobacter spp. | |
| H. canis | NCTC 12739T |
| H. pullorum | NCTC 12824T |
| H. rappini | NCTC 12461T |
| H. acinonyx | NCTC 12686T |
| H. cinaedi | NCTC 12423T |
| H. fennelliae | NCTC 11612T |
| H. mustelae | NCTC 12198T |
| H. pylori | NCTC 11637T |
| H. hepaticus | ATCC 51448T |
| H. muridarum | NCTC 12714T |
| H. pamatensis | ATCC 51478T |
| H. nemestriae | NCTC 12491T |
| Arcobacter spp. | |
| A. cryaerophilus | NCTC 11885T |
| A. skirrowii | NCTC 12713T |
| A. butzleri | NCTC 12481T |
| A. nitrofigilis | NCTC 12551T |
| Wollinella succinogenes | NCTC 11488T |
NCTC, National Collection of Type Cultures; ATCC, American Type Culture Collection.
Species incertae sedis which is genotypically Campylobacter (28).
Thirty-eight fecal samples from gastroenteritis patients from a large-scale study (13) were tested. From 24 of these samples, campylobacters had been identified to the species level by PCR-ELISA (15 C. jejuni, 2 C. coli, 2 C. hyointestinalis, and 5 C. upsaliensis isolates). From 19 of these 24, Campylobacter had been identified to the genus level by selective culture. The remaining 14 clinical samples were negative both by Campylobacter PCR-ELISA and culture: in 9 samples no pathogenic organisms were detected, while 1 sample was positive for Salmonella, 1 for Shigella, 1 for Escherichia coli, 1 for Cryptosporidium, and 1 for Giardia.
Sample preparation.
A boiled colony suspension in sterile water was prepared from 24- to 48-h blood agar plate cultures. Archived DNA extracts stored at −20°C were tested retrospectively; DNA was extracted from these fecal specimens as previously described (4, 12).
PCR and biprobe melting peak analysis.
Symmetric PCR was performed on a RoboCycler (Stratagene, La Jolla, Calif.) using previously described Campylobacter genus-specific primers (16) C412F (GGA TGA CAC TTT TCG GAG C) and C1228R (CAT TGT AGC ACG TGT GTC).
Heminested asymmetric PCR and melting peak analysis were performed in glass capillaries in a LightCycler apparatus (Bio/Gene Ltd., Kimbolton, United Kingdom) (34, 35) using 1 μl of first-round PCR product in a total volume of 10 μl. Asymmetric PCR is used to produce excess copies of the strand complementary to the biprobes. The reaction mixture contained 2.5 pmol of the single primer C690F (AGATACCCTGGTAGTCCACG), 50 mM Tris-HCl (pH 8.3), 5 mM MgCl2, 0.5 mg of bovine serum albumin/ml, 200 μM each deoxynucleoside triphosphate, 0.4 U of Platinum Taq DNA polymerase (Life Technologies Ltd., Paisley, United Kingdom), SYBR Green 1 (Bio/Gene Ltd.) at 0.01%, and 5 pmol of biprobe labeled at the 5′ end with Cy5 and at the 3′ end with biotin. This type of nucleic acid detection system is subject to a patent held by the Defence Evaluation Research Agency, Farnborough, United Kingdom and Bio/Gene Ltd. (3). Primers and probes were HPSF (highly purified salt-free) grade, synthesized by MWG-Biotech UK Ltd., Milton Keynes, England. Thermal cycling and fluorescence acquisition conditions comprised an initial denaturation cycle at 95°C for 10 s and 40 amplification cycles (with a temperature transition rate of 20°C/s) of 95°C for 0 s; annealing at 60°C for 10 s, and extension at 74°C for 30 s. Fluorescence readings were taken after each cycle following the extension step. This was followed by melting analysis of the probe-PCR product duplex consisting of 95°C for 0 s, then cooling to 40°C before the temperature was raised to 99°C at a rate of 0.1°C/s with continuous fluorescence acquisition.
RESULTS
Design of biprobes and assay development.
The available EMBL DNA sequences coding for the 16S rRNA gene of Campylobacter were aligned using the Clustal multiple alignment program (10). A series of probes for species of interest were then designed, targeting variable regions between nucleotide positions 690 and 1228. The probes were 20 to 25 bases in length. There were four design parameters: hybridization to sites away from either end of the PCR product, melting temperatures above that of the primer, mismatching of bases central to the probe itself, and minimum hairpin loop formation. Probes were designed to have up to four base mismatches with the species (sequence) of interest.
The exact effect of mismatched bases on the melting behavior of a given probe cannot be predicted from the sequence alone, due to secondary structure and other steric effects. Performance was therefore assessed against a panel of reference strain sample DNAs (Table 1) to identify probes which gave discrete melting peaks, with good separation of the melting temperatures for the species in question. No single probe produced discrete melting peaks discriminatory for all the species of interest. However, combining data for three probes allowed identification of C. jejuni/C. coli, C. lari, C. upsaliensis, C. helveticus, C. hyointestinalis, C. fetus, and C. lanienae.
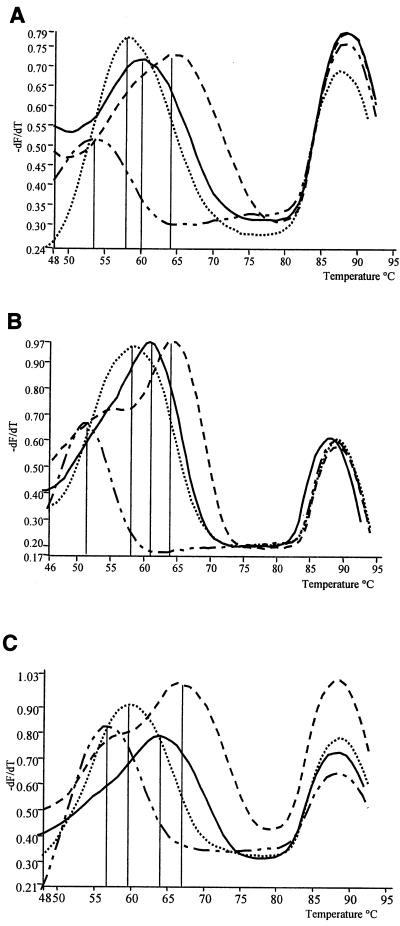
The sequence and labeling of the chosen combination of probes were as follows: biprobe A, Cy5-GCA CCC CAA CAA CTA GTG TAC A-biotin; biprobe B, Cy5-CAG CAC CTG TCA CTA ATT TCT TG-biotin; biprobe C, Cy5-GCA AGC TAG CAC TCT CTT ATC TCT-biotin. During end point analysis of the melting kinetics of a given biprobe/target, a decrease in light emission from Cy5 was observed, and this was recorded by continually monitoring the emission peak of Cy5 during the melt cycle. Data collected were presented in the form of a negative first-derivative plot of fluorescence/temperature versus temperature (−dF/dT versus T), calculated by LightCycler software. This plot converts the data into discrete melting-peak graphs. The secondary peak observed on such plots corresponds to residual double-stranded PCR product, since Cy5 has a spectral overlap with SYBR Green 1. Such plots, as produced by each probe with the range of reference strain sample DNAs, are shown in Fig. 1. The combined melting-peak profiles constitute an identification scheme as presented in Table 2. Six species (C. lari, C. upsaliensis, C. helveticus, C. hyointestinalis, C. fetus, and C. lanienae) produced unique combined melting-peak profiles. However, C. jejuni and C. coli could not be differentiated at the species level. The melting peaks of all strains of C. hyointestinalis with biprobe B and the melting peaks of all strains of C. fetus with biprobe C showed a “shoulder effect” at the lower temperature range (Fig. 1B and C).
FIG. 1.
Biprobe fluorescence data. Shown are negative first-derivative plots, reflecting the change in fluorescence divided by the change in temperature (−dF/dT). The melting peaks seen in the range from 48 to 75°C correspond to the decrease in FRET and hence the decrease in light emitted at the Cy5 wavelength, as the biprobe melts from the single-stranded target DNA. The secondary peak seen at 86°C acts as a positive amplification control and corresponds to residual double-stranded PCR product. (A) Melting peaks obtained with biprobe A. C. jejuni/C. coli gave a peak at 64°C (dashed line), C. jejuni gave a peak at 58°C (dotted line), C. lari/C. upsaliensis gave a peak at 60°C (solid line), and C. helveticus gave a peak at 54°C (irregularly dashed line). (B) Melting peaks obtained with biprobe B. C. hyointestinalis gave a peak at 64°C (dashed line), C. jejuni/C. coli/C. lari/C. helveticus gave a peak at 62°C (solid line), C. fetus/C. lanienae gave a peak at 58°C (dotted line), and C. upsaliensis gave a peak at 51°C (irregularly dashed line), (C) Melting peaks obtained with biprobe C. C. fetus gave a peak at 67°C (dashed line), C. jejuni/C. coli/C. lanienae gave a peak at 64°C (solid line), C. jejuni/C. coli/C. lari/C. helveticus gave a peak at 60°C (dotted line), and C. upsaliensis gave a peak at 56°C (irregularly dashed line).
TABLE 2.
Campylobacter sp. identification by combined biprobe melting-peak profiles
| Campylobacter sp. (no. of strains tested)a | Melting peaks (°C) for biprobe:
|
Penner serotype reference strain profilec | ||
|---|---|---|---|---|
| A | B | C | ||
| C. lari (14) | 60 | 62 | 60 | |
| C. upsaliensis (25) | 60 | 51 | 56 | |
| C. helveticus (12) | 54 | 62 | 60 | |
| C. hyointestinalis (11) | — | 64 | — | |
| C. fetus (15) | — | 58 | 67 | |
| C. lanienae (2) | — | 58 | 64 | |
| C. jejuni/C. coli (72) | 64 | 62 | 64 | C. jejuni HS 1–6, 10, 11, 13, 15–18, 22, 31, 37, 40, 44, 50, 52, 62, 64 |
| C. coli HS 5, 14, 25, 26, 28, 30, 34, 39, 47, 48, 51, 54 | ||||
| C. jejuni/C. coli (8) | 64 | 62 | 60 | C. jejuni HS 23, 35, 43, 63 |
| C. coli HS 20 | ||||
| C. jejuni (20) | 58 | 62 | 60 | C. jejuni HS 7–9, 12, 19, 21, 27, 29, 32, 33, 36, 38, 41, 42, 43, 53, 55, 57, 58, 60 |
Reference and field isolates.
—, no melting peak.
Heat-stable (HS) Penner serotype designations apply only to C. jejuni and C. coli.
The 46 Penner heat-stable (HS) serotype reference strains of C. jejuni produced one of two melting peaks with biprobe A (64 or 58°C) and with biprobe C (64 or 60°C) but a single peak (62°C) with biprobe B (Fig. 1; Table 2). The 13 C. coli HS reference strains produced a single melting peak with biprobe A, identical to one of the C. jejuni peaks (64°C). With biprobes B and C, C. coli strains produced melting peaks identical to those of the C. jejuni strains (Fig. 1; Table 2). Combining these results yielded the three possible profiles shown in Table 2. For example, A/B/C peaks of 64:62:64 or 60 could denote C. jejuni or C. coli. Peaks of 58:62:60 denoted only C. jejuni. The distribution of each profile with respect to the HS serotypes is shown in Table 2.
Evaluation of Campylobacter field isolates.
One hundred ten field isolates were blind tested. The profiles obtained with the three biprobes above were identified according to the scheme outlined in Table 2. Controls for each melting peak were included in each run to remove run-to-run variation. Thirteen C. lari, 24 C. upsaliensis, 13 C. fetus, 11 C. helveticus, 10 C. hyointestinalis, and 1 C. lanienae isolate were all identified by the PCR-biprobe assay and had the characteristic species profiles described in Table 2. Twenty-six C. jejuni and 12 C. coli field isolates were identified by the assay as C. jejuni/C. coli. Twenty-five C. jejuni field isolates and all 12 C. coli field isolates gave melting-peak profiles of 64:62:64 (A/B/C). One C. jejuni field isolate gave the A/B/C profile 64:62:60. PCR-biprobe identification therefore showed 100% correlation with phenotypic identification to species level for all tested field isolates.
Evaluation of fecal DNA extracts.
Thirty-eight archived fecal DNA extracts were tested. The combined melting-peak profiles of these extracted DNAs were compared with corresponding results for the samples obtained by phenotypic identification to the genus level (selective culture) and by PCR-ELISA of the extracted DNA (13). As can been seen from Table 3, identifications by the PCR-biprobe assay and the PCR-ELISA were the same. There were five samples where culture was negative but both the PCR-biprobe and PCR-ELISA procedures detected a Campylobacter isolate (one C. hyointestinalis and four C. upsaliensis isolates). Extracts from clinical samples containing no enteropathogens or other enteropathogens such as Salmonella or Cryptosporidium spp. were negative by all three methods.
TABLE 3.
Comparison of Campylobacter sp. identification from fecal DNA extracts by three methods: phenotypic (genus level), PCR-ELISA, and PCR-biprobe assay
| No. of samples | Identification by:
|
PCR-biprobe result
|
||
|---|---|---|---|---|
| Culture | PCR-ELISA | A/B/C profile (°C)a | Identification | |
| 15 | Campylobacter sp. | C. jejuni/C. coli | 64:62:64 | C. jejuni/C. coli |
| 2 | Campylobacter sp. | C. jejuni/C. coli | 58:62:60 | C. jejuni/C. coli |
| 1 | Campylobacter sp. | C. hyointestinalis | —:64:— | C. hyointestinalis |
| 1 | Negative | C. hyointestinalis | —:64:— | C. hyointestinalis |
| 1 | Campylobacter sp. | C. upsaliensis | 60:51:56 | C. upsaliensis |
| 4 | Negative | C. upsaliensis | 60:51:56 | C. upsaliensis |
| 9 | Negative | Negative | Negative profile | Negative |
| 1 | Salmonella | Negative | Negative profile | Negative |
| 1 | Shigella | Negative | Negative profile | Negative |
| 1 | Escherichia coli | Negative | Negative profile | Negative |
| 1 | Cryptosporidium | Negative | Negative profile | Negative |
| 1 | Giardia | Negative | Negative profile | Negative |
—, no melting peak.
DISCUSSION
We describe here a PCR-biprobe assay for detection and identification to the species level of campylobacters from isolates and DNA extracts of fecal samples. The multicopy 16S rRNA gene, which comprises conserved regions interspersed with variable regions, was used as the target for this DNA sequence-based species differentiation. Conserved regions of the gene were targeted by Campylobacter genus-specific oligonucleotide primers, while variable regions suitable for identification of individual taxa were targeted for biprobe hybridization. Thus, the oligonucleotides C412F–C1228R (first-round PCR) and C690F (asymmetric second-round PCR) were used as primers to generate the genus-specific amplicon, and this was subsequently identified by its combined melting-peak profiles with the three biprobes. The assay showed no cross-reaction with other Campylobacter species or with species of Helicobacter or Arcobacter. Under test conditions it correctly identified 110 field isolates according to previous phenotypic identification to the species level. It was robust enough to identify Campylobacter species directly in DNA extracted from fecal specimens.
Six taxa were differentiated to the species level, namely, C. lari, C. upsaliensis, C. helveticus, C. hyointestinalis, C. fetus, and C. lanienae. C. jejuni and C. coli were identified as a single taxon. This was a limitation but was not unexpected, since previous 16S ribosomal DNA (rDNA) sequencing of a 467-nucleotide region of 52 Penner serotype reference strains showed that all belonged to one of three different cross-species groups, between which sequences varied at just three nucleotide positions (20). This is a particular feature of the 16S rDNA of these two species and has not been found for any other pair of Campylobacter species. Comparison of the 16S sequences of C. jejuni and C. coli with that of biprobe A indicated that a perfect match of target to probe produced a melting peak at 64°C, while a melting peak at 58°C represented a single-base change from G to A in the target sequence. Similarly, for biprobe C, melting peaks at 64 and 60°C represented a single-base change from A to T in the target sequence. The distribution of the biprobe melting-peak profiles showed complete congruence with the sequence data and confirmed that there are no clear-cut species-specific differences between the 16S rRNA gene sequences of C. jejuni and C. coli. The shoulder effect was seen with the melting peaks of C. hyointestinalis and C. fetus analyzed with biprobes B and C, respectively. This is possibly a result of hybridization of the probe to the 16S rDNA target being destabilized in the lower temperature range, by DNA sequences remote from the probe binding site that form secondary structures. Indeed, this shoulder phenomenon has also been observed by other workers (7).
The assay was validated for use with clinical (fecal) samples. Here, PCR-biprobe analysis of 38 fecal DNA extracts was more sensitive than phenotypic identification of isolates to the genus level, in that the former was 100% concordant with identification by PCR-ELISA (13). All the culture-positive and PCR-ELISA-positive samples were also positive by the PCR-biprobe assay (17 C. jejuni/C. coli, 1 C. hyointestinalis, and 1 C. upsaliensis isolate). All the culture-negative, PCR-ELISA-negative fecal samples or those containing other enteropathogens were negative by the PCR-biprobe assay. In five diarrheic samples negative by culture, both the PCR-biprobe assay and the PCR-ELISA detected 1 C. hyointestinalis and 4 C. upsaliensis isolates. These results are explained by the fact that the culture conditions typically used for isolation of campylobacters in human gastroenteritis (C. jejuni, C. coli, and C. lari) are inhibitory for other campylobacters. Hence the “rare” species detected here by the PCR-biprobe assay (C. hyointestinalis and C. upsaliensis) would not have been isolated by routine laboratory culture.
Previous studies that have used PCR and/or hybridization for identification of Campylobacter to the species level based on 16S rDNA (8, 13, 14, 16, 18, 27, 31), 23S rDNA (5, 6), flagellin genes (22, 32, 33), or genes encoding a protein involved in siderophore transport (9), hippuricase (15), or aspartokinase (15) can distinguish only between C. jejuni and C. coli. The gene encoding the GTP-binding protein (29, 30) and the glyA gene, which encodes serine hydroxymethyltransferase (1), can identify four Campylobacter species, but neither has yet been applied to clinical samples. We have previously described a PCR-ELISA with first- and second-round pangenus PCRs that eliminated the need for multiple single-species reactions and then achieved identification by probe hybridization (20). The present work applies a PCR multiple biprobe assay directly to identification of Campylobacter spp.. Direct PCR amplification of fecal DNA extracts removes the need for isolation and culture of campylobacters, as demonstrated in earlier PCR applications (12, 22, 32). The biprobe assay can be completed within 1 h of first-round conventional PCR, and data analysis is quick and accurate. Further development of assay-specific software would allow for automatic peak assignment and comparison. Compared to PCR-ELISA, the risk of contamination is reduced by performing the second-round PCR and product identification in a closed-tube system. The assay is flexible and can be tailored by incorporating additional biprobes to identify further species of interest. Our study employed an Idaho Technology LightCycler with the capacity for two-channel detection (SYBR Green and Cy5). Second-generation real-time PCR platforms can perform three- to four-channel detection, which may allow monitoring of all three biprobes described in this report in a single reaction. All such systems have the potential for automated handling and a capacity for higher throughput. PCR-biprobe assays can play a role both in the diagnostic laboratory and in molecular ecology studies of the prevalence and significance of campylobacters in human disease, especially with regard to those species which are not amenable to routine culture and whose role in human disease is inadequately understood.
ACKNOWLEDGMENTS
This work was partially funded by a grant from the Department of Health, London, United Kingdom (DH220B).
We thank the Campylobacter Reference Unit, Central Public Health Laboratory, London, United Kingdom, and the Campylobacter Collaborating Unit, Preston Public Health Laboratory, for the provision of field isolates.
REFERENCES
- 1.Al Rashid S T, Dakuna I, Louie H, et al. Identification of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, Arcobacter butzleri, and A. butzleri-like species based on the glyA gene. J Clin Microbiol. 2000;38:1488–1494. doi: 10.1128/jcm.38.4.1488-1494.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. A report of the study of infectious intestinal disease in England. London, United Kingdom: The Stationery Office; 2000. [Google Scholar]
- 3.Bio/Gene Ltd.The Secretary of State for Defence. 21 July 1999. Nucleic acid detection system. Great Britain patent GB2333359A.
- 4.Boom R, Sol C J, Salimans M M, Jansen C L, Wertheim-van Dillen P M, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eyers M, Chapelle S, van Camp G, Goossens H, de Wachter R. Discrimination among thermophilic Campylobacter species by polymerase chain reaction amplification of 23S rRNA gene fragments. J Clin Microbiol. 1993;31:3340–3343. doi: 10.1128/jcm.31.12.3340-3343.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fermer C, Engvall E O. Specific PCR identification and differentiation of the thermophilic campylobacters, Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis. J Clin Microbiol. 1999;37:3370–3373. doi: 10.1128/jcm.37.10.3370-3373.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson J R, Saunders N A, Burke B, Owen R J. Novel method for rapid determination of clarithromycin sensitivity in Helicobacter pylori. J Clin Microbiol. 1999;37:3746–3748. doi: 10.1128/jcm.37.11.3746-3748.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giesendorf B A, Quint W G. Detection and identification of Campylobacter spp. using the polymerase chain reaction. Cell Mol Biol. 1995;41:625–638. [PubMed] [Google Scholar]
- 9.Gonzalez I, Grant K A, Richardson P T, Park S F, Collins M D. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J Clin Microbiol. 1997;35:759–763. doi: 10.1128/jcm.35.3.759-763.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 11.Kendall E J, Tanner E I. Campylobacter enteritis in general practice. J Hyg. 1982;88:155–163. doi: 10.1017/s0022172400070030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawson A J, Linton D, Stanley J, Owen R J. Polymerase chain reaction detection and speciation of Campylobacter upsaliensis and C. helveticus in human faeces and comparison with culture techniques. J Appl Microbiol. 1997;83:375–380. doi: 10.1046/j.1365-2672.1997.00240.x. [DOI] [PubMed] [Google Scholar]
- 13.Lawson A J, Logan J M, O'Neill G L, Desai M, Stanley J. Large-scale survey of Campylobacter species in human gastroenteritis by PCR and PCR-enzyme-linked immunosorbent assay. J Clin Microbiol. 1999;37:3860–3864. doi: 10.1128/jcm.37.12.3860-3864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawson A J, Shafi M S, Pathak K, Stanley J. Detection of campylobacter in gastroenteritis: comparison of direct PCR assay of faecal samples with selective culture. Epidemiol Infect. 1998;121:547–553. doi: 10.1017/s0950268898001630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linton D, Lawson A J, Owen R J, Stanley J. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J Clin Microbiol. 1997;35:2568–2572. doi: 10.1128/jcm.35.10.2568-2572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linton D, Owen R J, Stanley J. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res Microbiol. 1996;147:707–718. doi: 10.1016/s0923-2508(97)85118-2. [DOI] [PubMed] [Google Scholar]
- 17.Livak K J, Flood S J, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 18.Marshall S M, Melito P L, Woodward D L, Johnson W M, Rodgers F G, Mulvey M R. Rapid identification of Campylobacter, Arcobacter, and Helicobacter isolates by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene. J Clin Microbiol. 1999;37:4158–4160. doi: 10.1128/jcm.37.12.4158-4160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mead P S, Slutsker L, Dietz V, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metherell L A, Logan J M, Stanley J. PCR-enzyme-linked immunosorbent assay for detection and identification of Campylobacter species: application to isolates and stool samples. J Clin Microbiol. 1999;37:433–435. doi: 10.1128/jcm.37.2.433-435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.On S L. Identification methods for campylobacters, helicobacters, and related organisms. Clin Microbiol Rev. 1996;9:405–422. doi: 10.1128/cmr.9.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oyofo B A, Thornton S A, Burr D H, Trust T J, Pavlovskis O R, Guerry P. Specific detection of Campylobacter jejuni and Campylobacter coli by using polymerase chain reaction. J Clin Microbiol. 1992;30:2613–2619. doi: 10.1128/jcm.30.10.2613-2619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selvin P R. Fluorescence resonance energy transfer. Methods Enzymol. 1995;246:300–334. doi: 10.1016/0076-6879(95)46015-2. [DOI] [PubMed] [Google Scholar]
- 24.Stanley J, Burnens A P, Linton D, On S L, Costas M, Owen R J. Campylobacter helveticus sp. nov., a new thermophilic species from domestic animals: characterization, and cloning of a species-specific DNA probe. J Gen Microbiol. 1992;138:2293–2303. doi: 10.1099/00221287-138-11-2293. [DOI] [PubMed] [Google Scholar]
- 25.Tauxe R V. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 9–19. [Google Scholar]
- 26.Tyagi S, Kramer F R. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 27.van Camp G, Fierens H, Vandamme P, Goossens H, Huyghebaert A, de Wachter R. Identification of enteropathogenic Campylobacter species by oligonucleotide probes and polymerase chain reaction based on 16S rRNA genes. Syst Appl Microbiol. 1993;16:30–36. [Google Scholar]
- 28.Vandamme P, Daneshvar M I, Dewhirst F E, et al. Chemotaxonomic analyses of Bacteroides gracilis and Bacteroides ureolyticus and reclassification of B. gracilis as Campylobacter gracilis comb. nov. Int J Syst Bacteriol. 1995;45:145–152. doi: 10.1099/00207713-45-1-145. [DOI] [PubMed] [Google Scholar]
- 29.van Doorn L J, Giesendorf B A, Bax R, van der Zeijst B A, Vandamme P, Quint W G. Molecular discrimination between Campylobacter jejuni, Campylobacter coli, Campylobacter lari and Campylobacter upsaliensis by polymerase chain reaction based on a novel putative GTPase gene. Mol Cell Probes. 1997;11:177–185. doi: 10.1006/mcpr.1997.0100. [DOI] [PubMed] [Google Scholar]
- 30.van Doorn L J, Verschuuren-van Haperen A, Burnens A, et al. Rapid identification of thermotolerant Campylobacter jejuni, Campylobacter coli, Campylobacter lari, and Campylobacter upsaliensis from various geographic locations by a GTPase-based PCR-reverse hybridization assay. J Clin Microbiol. 1999;37:1790–1796. doi: 10.1128/jcm.37.6.1790-1796.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanniasinkam T, Lanser J A, Barton M D. PCR for the detection of Campylobacter spp. in clinical specimens. Lett Appl Microbiol. 1999;28:52–56. doi: 10.1046/j.1365-2672.1999.00474.x. [DOI] [PubMed] [Google Scholar]
- 32.Waegel A, Nachamkin I. Detection and molecular typing of Campylobacter jejuni in fecal samples by polymerase chain reaction. Mol Cell Probes. 1996;10:75–80. doi: 10.1006/mcpr.1996.0011. [DOI] [PubMed] [Google Scholar]
- 33.Wassenaar T M, Newell D G. Genotyping of Campylobacter spp. Appl Environ Microbiol. 2000;66:1–9. doi: 10.1128/aem.66.1.1-9.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wittwer C T, Herrmann M G, Moss A A, Rasmussen R P. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques. 1997;22:130–131. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]
- 35.Wittwer C T, Ririe K M, Andrew R V, David D A, Gundry R A, Balis U J. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques. 1997;22:176–181. doi: 10.2144/97221pf02. [DOI] [PubMed] [Google Scholar]