Abstract
We evaluated two automated systems, MagNA Pure (Roche Molecular Biochemicals, Indianapolis, Ind.) and BioRobot 9604 (Qiagen, Inc., Chatsworth, Calif.) as effective replacements for the manual IsoQuick method (Orca Research, Inc., Bothell, Wash.) for extraction of herpes simplex virus (HSV) DNA from dermal and genital tract specimens prior to analysis by LightCycler PCR. Of 198 specimens (152 genital, 46 dermal), 92 (46.2%) were positive for HSV DNA by LightCycler PCR after automated extraction of specimens with either the MagNA Pure or BioRobot 9604 instrument. The manual IsoQuick method yielded HSV DNA (total n = 95) from three additional specimens that were negative by the automated method (P = 0.25, sign test). Although the mean numbers of LightCycler PCR cycles required to reach positivity differed statistically significantly among all three of the methods of extraction, the estimated means differed by no more than 1.5 cycles (P < 0.05). Seventy (76%) of the 92 specimens that were LightCycler PCR positive by all three extraction methods were also positive by shell vial cell culture assay. HSV DNA was detected by a lower LightCycler PCR cycle number (26.1 cycles) in specimens culture positive for the virus than in culture-negative samples (33.3 cycles) (P < 0.0001). The manual IsoQuick and automated MagNA Pure and BioRobot 9604 methods provide standardized, reproducible extraction of HSV DNA for LightCycler PCR. The decision to implement a manual versus an automated procedure depends on factors such as costs related to the number of specimens processed rather than on the minimal differences in the technical efficiency of extraction of nucleic acids among these methods.
The LightCycler (Roche Molecular Biochemicals, Indianapolis, Ind.) is an automated instrument which can monitor the development of amplified target nucleic acid by fluorescence resonance energy transfer after each amplification cycle. The instrument provides rapid (30 to 40 min) PCR results by precise air-controlled temperature cycling; most importantly, the amplification and detection of an amplified product occur in a closed system, which virtually eliminates the likelihood of carryover contamination. In three evaluations that involved 877 dermal and genital and a few ocular specimens and yielded 285 herpes simplex virus (HSV)- and 44 varicella-zoster virus (VZV)-positive results, LightCycler PCR produced a greater sensitivity for the detection of HSV (22%) and VZV (91%) than did the shell vial cell culture assay for the laboratory diagnosis of these viral infections (5, 6, 7). Based on these performance characteristics, together with comparable cost analysis for LightCycler PCR and shell vial cell culture, we implemented the molecular amplification procedure in May 2000 for the routine diagnosis of HSV dermal and genital and VZV dermal infections.
Preliminary to LightCycler PCR, we extracted nucleic acids from dermal and genital specimens by the manual IsoQuick (Orca Research, Inc., Bothell, Wash.) procedure. In the present study, we evaluated two automated systems, MagNA Pure (Roche Molecular Biochemicals, Indianapolis, Ind.) and BioRobot 9604 (Qiagen, Inc., Chatsworth, Calif.) as effective replacements for the manual IsoQuick method with the goal of implementing a cost-efficient, standardized system for the processing of clinical specimens.
MATERIALS AND METHODS
Specimens and shell vial assay.
Genital (n = 152) and dermal (n = 46) swab specimens from patients suspected of having HSV infections were extracted into 2-ml volumes of serum-free medium; the specimen extracts were then divided into four equal aliquots. Each of two shell vial MRC-5 cell cultures received 200 μl of inoculum from one aliquot. The vials were centrifuged, incubated overnight at 36°C, and stained by the indirect immunofluorescence test as previously described (10). Nucleic acids were extracted from the remaining aliquots by three different extraction techniques and processed for amplification of HSV DNA by LightCycler PCR. Nucleic acid extracts obtained by each method were stored at 4°C for a maximum of 2 weeks before PCR amplification.
IsoQuick nucleic acid extraction.
Nucleic acids were extracted manually from a 200-μl volume of serum-free extract of genital or dermal swab specimens by the IsoQuick procedure (Orca Research, Inc.), which utilizes guanidine thiocyanate and a noncorrosive extraction reagent, in accordance with the manufacturer's instructions (4, 7).
MagNA Pure nucleic acid extraction.
A second aliquot (200 μl) was extracted by the MagNA Pure LC automated extractor (Roche Molecular Biochemicals) by using the DNA isolation extraction kit produced by the same manufacturer.
Qiagen BioRobot 9604 nucleic acid extraction.
Another 200-μl aliquot of each specimen extract was extracted in accordance with the manufacturer's instructions by using the QIAamp 96 DNA blood BioRobot kit and the Qiagen BioRobot 9604 system (Qiagen, Inc.).
LightCycler PCR.
A 5-μl aliquot of each extracted specimen was added to 15 μl of a master mixture for amplification. LightCycler PCR (Roche Molecular Biochemicals) was performed as previously described (6). Melting curve features of the LightCycler software were used to distinguish between the two genotypes of HSV (6).
Statistical analysis.
This experiment was a two-factor repeated-measures design. One factor, automated nucleic acid extraction, is a repeated measure with three levels, one for each of the three extraction methods. It is a repeated measure because material from the same sample was tested by each of the three methods. The other factor, shell vial culture positivity, is not a repeated factor.
The response variable for this experiment was the number of LightCycler amplification cycles required to detect HSV DNA. The distribution of the number of cycles was sufficiently Gaussian to allow the mean number of cycles to be analyzed with a two-factor repeated-measures analysis of variance (15). In this analysis, an F test is performed for each of the two main effect terms, extraction method and shell vial positivity. In addition, an F test is performed for the interaction of the extraction method and shell vial positivity; i.e., a test of whether the effect of the method of extraction depends on the shell vial culture result. In order to determine which pairs of mean numbers of cycles were significantly different from one another for the three combinations of two methods of extraction, a Student-Newman-Keuls multiple-comparison test was performed (1).
All specimens were detected by LightCycler PCR within 45 cycles; however, three specimens extracted by the two automated methods were not detected within the same number of cycles. Therefore, in order to avoid biasing the analysis in favor of the two automated extraction methods, a value of 45 cycles was used for these methods for each of the three specimens.
Ninety-five percent confidence intervals were calculated for each mean number of cycles to HSV positivity. P values of less than 0.05 were considered to be statistically significant.
RESULTS
Of 198 specimens (genital, n = 152; dermal, n = 46), 92 (46.5%) were positive for HSV DNA by LightCycler PCR after automated extraction of specimens with the MagNA Pure and BioRobot 9604 instruments. All three methods gave identical results for 195 (92 positive and 103 negative) specimens. The manual IsoQuick method yielded HSV DNA (total n = 95) from three additional specimens (cycle 37, n = 1; cycle 40, n = 2) that were negative by MagNA Pure or BioRobot 9604 (P = 0.25, sign test) (Table 1). Of the total of 95 HSV DNA-positive specimens detected by LightCycler PCR, 25 were not positive by the shell vial cell culture detection method. Of these 25 specimens, all were positive for HSV DNA by LightCycler PCR when extracted by IsoQuick (n = 25) and 22 were positive when extracted by the automated nucleic acid extraction methods (MagNA Pure and BioRobot 9604).
TABLE 1.
Numbers of LightCycler PCR cycles necessary to detect HSV DNA with regard to the extraction method used and the shell vial assay results
| Shell vial assay result | Mean no. of cycles (SD); 95% CIa
|
|||
|---|---|---|---|---|
| IsoQuick | MagNA Pure | BioRobot 9604 | Total | |
| Negative (n = 25) | 32.6 (4.4); 30.6–34.2 | 33.5 (6.1); 31.0–36.0 | 34.1 (5.5); 31.9–36.4 | 33.3 (5.3); 32.1–34.6 |
| Positive (n = 70) | 25.6 (3.8); 24.7–26.5 | 26.0 (4.0); 25.0–27.0 | 26.9 (4.0); 25.9–27.8 | 26.1 (4.0); 25.6–26.7 |
CI, confidence interval.
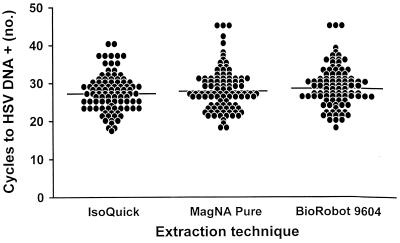
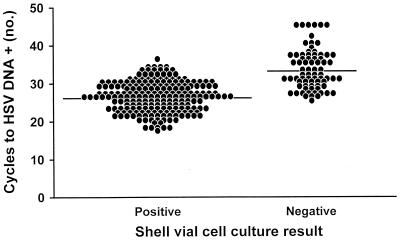
The mean numbers of LightCycler PCR cycles necessary to detect HSV DNA, with regard to the method of extraction used and shell vial assay positivity, are shown in Table 1. Although the mean numbers of cycles differed significantly among the three methods of extraction (P value, <0.05; Student-Newman-Keuls test), the estimated means differed by no more than 1.5 cycles (Fig. 1). LightCycler PCR was positive in all instances in which the shell vial assay yielded HSV; in addition, LightCycler PCR was exclusively positive for 75 specimens. The PCR cycle number of the total specimens positive by LightCycler PCR and the shell vial assay (n = 210) was compared to that of the specimens that were detected by LightCycler PCR but not by the shell vial cell culture method (n = 75) (Table 1). The mean number of cycles necessary to detect HSV was significantly greater for specimens negative by the shell vial assay, 33.3 cycles, than for specimens positive by the shell vial assay, 26.1 cycles (P value, <0.0001; F test) (Fig. 2).
FIG. 1.
Numbers of LightCycler PCR cycles required for the detection of HSV DNA by three nucleic acid extraction methods.
FIG. 2.
Numbers of LightCycler PCR cycles required for the detection of HSV DNA by the IsoQuick, MagNA Pure, and BioRobot 9604 extraction methods with regard to shell vial cell culture results. There were shell vial cell culture-positive samples.
DISCUSSION
Our laboratory has performed extensive evaluations focused on optimizing the performance characteristics of LightCycler PCR for the routine detection of viral DNA from dermal (HSV and VZV) and genital tract (HSV) specimens (5, 6, 7). For these assays, we initially implemented the LightCycler PCR assay by using IsoQuick, a commercially available manual method for the nucleic acid extraction step. We have had substantial experience with this manual technique, since IsoQuick has been used routinely in our laboratory as part of several molecular assays, including extraction of DNA from over 18,000 cerebrospinal fluid specimens prior to conventional PCR amplification of HSV DNA (2, 11, 14). Nevertheless, the initial front-end step of molecular diagnostic testing, that is, extraction of nucleic acid from clinical specimens, has not been critically optimized, as is now possible with the amplification and detection steps of LightCycler PCR. Based on our experience with LightCycler PCR for HSV and VZV and the characteristics of this closed, automated, rapid, real-time amplification detection system, we have implemented this assay in our laboratory (800 to 900 specimens/month) and anticipate the extension of this technology to many nucleic acid targets for pathogens identified throughout our clinical microbiology laboratory.
Two major concerns are relevant to a full-scale transition to real-time PCR in the clinical microbiology laboratory. First, an automated method is needed for the extraction of nucleic acids. Second, the costs and efficiency of the processing steps associated with automated methods must be critically evaluated.
To address the first question, we evaluated two automated methods for nucleic acid extraction: MagNA Pure, a system designed by the manufacturer of LightCycler PCR, and BioRobot 9604, a system designed by Qiagen for high-volume specimen processing preparatory to PCR detection (3, 8, 9, 12, 13). We compared the performance of these two automated methods with that of the manual IsoQuick system. Of 198 specimens processed by all three methods, 195 yielded identical results; 3 specimens were positive after IsoQuick extraction only but were positive only after 37 cycles of LightCycler amplification (P = 0.25, sign test). Nevertheless, because of the large sample size of the study and the extremely high precision of the LightCycler PCR, a difference of one or two amplification cycles between the extraction methods provided enough statistical power to demonstrate significant differences.
Overall, based on technical performance, we considered all three methods (IsoQuick, MagNA Pure, and BioRobot 9604) appropriate for optimal and clinically equivalent DNA extraction from genital and dermal specimens as front-end nucleic acid extraction procedures prior to LightCycler PCR for HSV DNA. In this study, we also demonstrated increased positivity of LightCycler PCR, regardless of the extraction procedure used, compared to the conventional shell vial assay (P < 0.001; sign test). These results agree with our previous reports demonstrating higher target DNA copy numbers in those specimens containing sufficient active virus to be detected by cell culture than in specimens negative by that assay (5, 6, 7).
The second consideration for the selection of an automated or manual nucleic acid extraction method relates to workload units and, ultimately, the cost per test. For example, the manual IsoQuick method required 1.98 min and the MagNA Pure method required 1.4 min for nucleic acid extraction per sample of the total LightCycler PCR test time (extraction and analysis) of 8.5 min according to workload recording information. MagNA Pure contains an eight-nozzle pipette head that can process a variable number of samples (1 to 32) in one run that requires 1.5 h; after initial aliquotting of specimens into the instrument, the extraction proceeds automatically. The unit list cost of MagNA Pure extraction is $1.10, compared to $1.71 for IsoQuick. Nevertheless, the numbers of College of American Pathology workload recording units were almost identical for the IsoQuick and MagNA pure methods. Thus, based on equivalent technical performance and workload costs, the choice of the manual IsoQuick or the automated MagNA Pure method can be somewhat optional for a laboratory, depending on the number of specimens that require nucleic acid extraction. For 28 samples, only 14 to 16 min of hands-on time is required for processing by MagNA Pure, compared with 56 to 60 min by the manual IsoQuick extraction methods. The BioRobot 9604 and Qiagen extraction method can process 96 samples in a single run; however, the hands-on time is approximately 88 min.
Our primary goal in this investigation was to compare the technical efficiency, cost, and processing capabilities of the MagNA Pure nucleic acid extraction method with those of the manual IsoQuick procedure preliminary to LightCycler PCR for the routine laboratory diagnosis of HSV infections. In addition, we tested aliquots of specimens after DNA extraction by the BioRobot 9604 system and assayed them for HSV target DNA by LightCycler PCR. The BioRobot 9604 system was optimal for this comparison because of its attractive automation technologies and the selective binding properties of Qiagen QIAamp silica-based membrane in a high-throughput 96-well format. The instrument has a microtiter diluting and dispensing workstation utilizing a tube bar code reader and liquid level-sensing pipette tips to ensure accurate dispensing of specimens and reagents. Cross contamination of nucleic acids in specimens is effectively controlled by using seal tapes on microtiter plates during centrifugation. This centrifugation is a hands-on step that prevents the BioRobot 9604 from being completely automated. The potential for batch mode processing of up to 96 samples ($1.73/sample) within a 2-h period affords a high throughput capacity for the clinical laboratory. Nevertheless, the BioRobot 9604 system has not yet been implemented in our laboratory for routine processing of specimens.
Overall, we feel that the two automated nucleic acid extraction methods (MagNA Pure and BioRobot 9604) are equivalent for the nucleic acid extraction of specimens preparatory to LightCycler PCR in laboratories processing high numbers of specimens since the numbers of amplification cycles required for HSV detection differed by a maximum of fewer than 1.5 cycles. Thus, the decision to implement a manual (IsoQuick) versus an automated (MagNA Pure or BioRobot 9604) procedure prior to LightCycler PCR depends on factors such as cost and number of specimens rather than on manual differences in technical efficiency of extraction between these methods. Our results indicate that the MagNA Pure and BioRobot 9604 automated nucleic acid extraction methods provide standardized, reproducible, and cost-effective methods for the processing of high numbers of dermal and genital specimens for HSV LightCycler PCR.
REFERENCES
- 1.Bancroft T A. Topics in intermediate statistical methods. 2nd ed. Vol. 1. Ames, Iowa: The Iowa State University Press; 1968. pp. 100–113. [Google Scholar]
- 2.Casas I, Powell L, Klapper P E, Cleator G M. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J Virol Methods. 1995;53:25–36. doi: 10.1016/0166-0934(94)00173-e. [DOI] [PubMed] [Google Scholar]
- 3.Cloud J L, Carroll K C, Pixton P, Erali M, Hillyard D R. Detection of Legionella species in respiratory specimens using PCR with sequencing confirmation. J Clin Microbiol. 2000;38:1709–1712. doi: 10.1128/jcm.38.5.1709-1712.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espy M J, Patel R, Paya C V, Smith T F. Comparison of three methods for extraction of viral nucleic acids from blood cultures. J Clin Microbiol. 1995;33:41–44. doi: 10.1128/jcm.33.1.41-44.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espy M J, Ross T K, Teo R, Svien K A, Wold A D, Uhl J R, Smith T F. Evaluation of LightCycler PCR for implementation of laboratory diagnosis of herpes simplex virus infections. J Clin Microbiol. 2000;38:3116–3118. doi: 10.1128/jcm.38.8.3116-3118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espy M J, Teo R, Ross T K, Svien K A, Wold A D, Uhl J R, Smith T F. Diagnosis of varicella-zoster virus infections in the clinical laboratory by LightCycler PCR. J Clin Microbiol. 2000;38:3187–3189. doi: 10.1128/jcm.38.9.3187-3189.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espy M J, Uhl J R, Mitchell P S, Thorvilson J N, Svien K A, Wold A D, Smith T F. Diagnosis of herpes simplex virus infections in the clinical laboratory by LightCycler PCR. J Clin Microbiol. 2000;38:795–799. doi: 10.1128/jcm.38.2.795-799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer M, Huber W, Kallivroussis A, Ott P, Opravil M, Luthy R, Weber R, Cone R W. Highly sensitive methods for quantitation of human immunodeficiency virus type 1 RNA from plasma, cells, and tissues. J Clin Microbiol. 1999;37:1260–1264. doi: 10.1128/jcm.37.5.1260-1264.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fransen K, Mortier D, Heyndrickx L, Verhofstede C, Janssens W, van der Groen G. Isolation of HIV-1 RNA from plasma: evaluation of seven different methods for extraction (part two) J Virol Methods. 1998;76:153–157. doi: 10.1016/s0166-0934(98)00115-3. [DOI] [PubMed] [Google Scholar]
- 10.Gleaves C A, Wilson D J, Wold A D, Smith T F. Detection and serotyping of herpes simplex virus in MRC-5 cells by use of centrifugation and monoclonal antibodies 16 h postinoculation. J Clin Microbiol. 1985;21:29–32. doi: 10.1128/jcm.21.1.29-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell P S, Espy M J, Smith T F, Toal D R, Rys P N, Berbari E F, Osmon D R, Persing D H. Laboratory diagnosis of central nervous system infections with herpes simplex virus by PCR performed with cerebrospinal fluid specimens. J Clin Microbiol. 1997;35:2873–2877. doi: 10.1128/jcm.35.11.2873-2877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niubo J, Li W, Henry K, Erice A. Recovery and analysis of human immunodeficiency virus type 1 (HIV) RNA sequences from plasma samples with low HIV RNA levels. J Clin Microbiol. 2000;38:309–312. doi: 10.1128/jcm.38.1.309-312.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shafer R W, Levee D J, Winters M A, Richmond K L, Huang D, Merigan T C. Comparison of QIAamp HCV kit spin columns, silica beads, and phenol-chloroform for recovering human immunodeficiency virus type 1 RNA from plasma. J Clin Microbiol. 1997;35:520–522. doi: 10.1128/jcm.35.2.520-522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang Y-W, Mitchell P S, Espy M J, Smith T F, Persing D H. Molecular diagnosis of herpes simplex virus infections in the central nervous system. J Clin Microbiol. 1999;37:2127–2136. doi: 10.1128/jcm.37.7.2127-2136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winer B J. Statistical principles in experimental design. 2nd ed. New York, N.Y: McGraw-Hill Book Company; 1971. pp. 518–539. [Google Scholar]