Abstract
T cell-based immunotherapy, for example, with T cell-recruiting bispecific antibody (bsAb), has revolutionized oncological treatment. However, many patients do not respond to treatment, and long-term remissions are still rare. Several tumor immune evasion mechanisms have been reported to counteract efficiency of T cell-engaging therapeutics. Platelets largely affect cancer pathophysiology by mediating tumor invasion, metastasis, and immune evasion. On treatment of patients in a clinical trial with a PSMA×CD3 bsAb (NCT04104607), we observed profound treatment-associated platelet activation, mirrored by a decrease of total platelet count. On modeling the treatment setting, we found that platelet activation significantly reduced bsAb-mediated CD4+ and CD8+ T-cell reactivity as revealed by impaired T-cell degranulation, secretion of perforin, and ultimately, inhibition of target cell lysis. This effect occurred in a transforming growth factor beta (TGF-β)-dependent manner and was not restricted to PSMA×CD3 bsAb, but rather observed with various CD3-directed bispecific constructs, including the approved CD19×CD3 bsAb blinatumomab. BsAb-mediated T-cell reactivity could be restored by platelet inhibition and specifically by blocking the TGF-β axis. Together, our findings demonstrate that platelets undermine the efficacy of T cell-recruiting bsAb and identify modulation of platelet function as a means to reinforce the effectiveness of bsAb treatment.
Keywords: immunotherapy, t-lymphocytes, translational medical research
Background
Immunotherapeutic strategies such as T cell-recruiting bispecific antibodies (bsAbs), chimeric antigen receptor (CAR) T cells, and immune checkpoint inhibition demonstrate remarkable efficacy in cancer treatment.1–4 However, long-term remissions are still rare, and many patients do not respond at all to treatment.5 6 Various immune escape mechanisms have been identified to undermine the efficacy of T cell-engaging therapeutics.6 7 Particularly in solid tumors, immunosuppressive cells in the tumor microenvironment (TME), including cancer-associated fibroblasts, regulatory T cells, and myeloid-derived suppressor cells, have been described to suppress antitumor immunity.7 8 In addition, numerous studies demonstrated that elevated platelet counts and expression of certain platelet surface markers are associated with an advanced disease stage and worse outcome.9–11 Platelets mediate tumor cell survival and tumor growth, among others, by secretion or expression of P-selectin, platelet-derived growth factor, vascular endothelial growth factor, and several others.12 Notably, platelets may also promote tumor immune evasion by impairing natural killer (NK) and T-cell function.13–16 However, data documenting the effect of platelet–tumor–immune cell interaction in humans that elucidate the underlying mechanisms are largely lacking. We report here our clinical observation that patients treated with a bispecific PSMA×CD3 antibody in a multicenter phase I study (NCT04104607)17 18 display treatment-related platelet activation and transient decrease of platelet count. Based on this observation, we modeled the effect of platelets on bsAb-induced T-cell antitumor reactivity and demonstrate that activated platelets undermine bsAb-mediated T-cell reactivity in a transforming growth factor beta (TGF-β)-dependent manner, indicating that therapeutic modulation of platelet function may serve to reinforce efficacy of bsAb treatment.
Methods
Cell lines
The tumor cell lines LNCaP, SaOS-2, and SEM were purchased from German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). Mycoplasma contamination was excluded via a PCR-based method. Authenticity was determined by validating the respective immunophenotype described by the provider using flow cytometry.
Primary patient material
Peripheral blood mononuclear cells (PBMCs) of healthy donors were isolated by density gradient centrifugation using Biocoll cell separation solution (Biochrom, Berlin, Germany). Platelets were isolated from blood of healthy donors and patients with prostate cancer receiving PSMA×CD3 (CC-1) in our multicenter phase I study (NCT04104607) treated at the University Hospital Tuebingen. Written informed consent in accordance with the Helsinki protocol was given in all cases.
Preparation of platelets
Citrated blood was briefly centrifuged for 20 min at 120×g. After harvesting platelet-rich plasma, platelets were washed with citrate wash buffer (128 mmol/L NaCl, 11 mmol/L glucose, 7.5 mmol/L Na2HPO4, 4.8 mmol/L sodium citrate, 4.3 mmol/L NaH2PO4, 2.4 mmol/L citric acid, and 0.35% bovine serum albumin) and resuspended in Roswell Park Memorial Institute (RPMI1640) containing 10% fetal calf serum (FCS).
For platelet activation ex vivo and generation of platelet releasate (PR), 10 µM of thrombin receptor activator peptide 6 (TRAP-6) was added to the platelets. In the next step, platelet suspensions were stirred for 1 min and subsequently centrifuged for 15 min at 2200×g. The supernatant was used as PR. For platelet blocking, washed platelets were incubated with dabigatran (5 mg/mL) or indomethacin (10 µg/mL). For blocking of TGF-β receptor signaling, a combination of an ALK5 inhibitor (20 µM, SB431542; Selleckchem, Houston, Texas, USA) and anti-TGF-β antibody (2 µg/mL, clone MAB1835; R&D Systems, Minneapolis, Minnesota, USA) was used.
Platelet aggregation
Platelet aggregation was analyzed using the four-channel light transmission platelet aggregometer APACT 4004 (Elitech, Puteaux, France) according to the manufacturer’s instructions. For platelet stimulation, the bsAb PSMA×CD3 (CC-1),18 NKG2D×CD3,19 20 or CD19×CD3 (blinatumomab) were used. TRAP-6 (10 µM) served as positive control.
Analysis of cytokine secretion
To analyze cytokine secretion, supernatant of platelets and PR was collected and analyzed for interleukin (IL)-2, tumor necrosis factor, TGF-β1, interferon gamma, IL-6 and CXCL8 using LEGENDplex assays (BioLegend, San Diego, California, USA).
Immunofluorescence
For immunofluorescence staining, tumor cells, PBMCs, and platelets were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS),(pH 7.4) for 10 min at −20°C. Cells were blocked using 5% BSA, 0.2% Triton X-100, and 0.1% Tween for 60 min. Blocking buffer was aspirated and cells were washed three times with phosphate-buffered saline with Tween® detergent (PBST), (PBS+0.1% TWEEN 20). Staining was conducted using unlabeled primary antibodies against CD41 (clone HIP8; Invitrogen, Carlsbad, California, USA), CD61 (clone SJ19-09, Invitrogen), CD62P (clone AK4, Thermo Fisher Scientific, Waltham, Massachusetts, USA), tubulin (clone 11H10), and perforin (clone δG9); Alexa-Fluor 594 labeled anti-rabbit or Alexa-Fluor 488 labeled anti-mouse (both Invitrogen) were used as secondary antibodies. For staining of actin, fluorescein-conjugated phalloidin (Abcam, Cambridge, Great Britain) was used according to the manufacturer’s instructions. Slides were mounted in fluorescent-mounting medium. 4′,6-diamidino-2-phenylindole (DAPI) was used for counterstaining. Image acquisition was performed using an Olympus BX63 microscope and a DP80 camera (Olympus, Shinjuku, Japan). Images were processed using ImageJ.21
Flow cytometry
To determine their activation status, platelets were stained using the directly fluorescence-labeled mAbs CD41-PE or CD41-PECy5 (both clone HIP8) and CD62P-FITC (clone AK-4) or the corresponding isotype control (all from BD Biosciences, San Diego, California, USA). For analysis of surface expression levels, unlabeled antibodies against prostate-specific membrane antigen (PSMA), (clone 10B3),18 CD3 (clone UCHT-1; BioCell Technology, Newport Beach, California, USA), ADAM10 (clone 163003, R&D Systems), and a biotinylated NKG2D-Fc22 were used followed by a goat anti-mouse PE conjugate (Dako, Glostrup, Denmark) or a streptavidin-conjugated phycoerythrin (PE) conjugate (Life Technologies, Carlsbad, California, USA) and a CD19-PE mAb (BD Biosciences). For detection and blocking of CD40L, the CD40L antibody clone MK13A4 (Enzo Life Sciences, Lausen, Swiss) was used. For studies on T-cell activation and degranulation, the mAbs CD69-PE and CD107a-PE (BD Pharmingen) as well as CD3-APC/Fire750, CD4-APC, CD8-FITC, and CD56-BV711 (BioLegend) were used. For flow cytometric analysis of target cell lysis, LNCaP cells were loaded with 2.5 µM CellTrace Violet cell proliferation dye (Thermo Fisher Scientific) and cultured with PBMC (effector to target (E:T) ratio 2.5:1) in the presence or absence of PSMA×CD3 (200 ng/mL) and platelets (platelet to target (P:T) 100:1). Standard calibration beads (Sigma-Aldrich, St. Louis, Missouri, USA) were used to ensure the analysis of equal assay volumes and therefore to account for the number of target cells that had vanished from the culture. Dead cells were excluded from analysis by 7-AAD (BioLegend). Measurements were performed using a FACS Canto II or FACS Fortessa (BD Biosciences), and data were analyzed using the software FlowJo V.10 (FlowJo LCC, Ashland, Oregon, USA).
T-cell activation and degranulation assays
To determine activation and degranulation in the presence of target cells, tumor cells were cultured with allogenic PBMCs of healthy donors and with or without platelets or PR in the indicated E:T:P ratios for 4 and 24 hours, respectively. For analysis of degranulation after 4 hours, CD107a-PE (1:25), BD GolgiStop, and BD GolgiPlug (1:1000, both BD Biosciences) were added to the coculture. Analysis was conducted using flow cytometry.
Cytotoxicity assays
Long-term cytotoxicity analyses were performed using the IncuCyte S3 Live-Cell Analysis System (Essen BioScience, Sartorius, Göttingen). Tumor cells were cultured with PBMCs of healthy donors (E:T ratio of 5:1) in the presence or absence of PSMA×CD3 (200 ng/mL) or PR. The TGF-β axis was blocked using the combination of an ALK5 inhibitor (SB431542, 20 µM) (Selleckchem) and anti-TGF-β antibody (clone MAB1835, 2 µg/mL) (R&D Systems). Live cell imaging pictures were taken every 4 hours with ×10 magnification. To quantify living cells, confluences were normalized to the respective measurement at t=0 hour.
Statistics
Student’s t test, Mann‐Whitney U test, one‐way nalysis of variance (ANOVA) and Friedman’s test were used for continuous variables. If significant differences by ANOVA were found, groupwise comparison was done (Tukey’s multiple comparison test). If significant differences were found by Friedman’s test, Dunn’s multiple comparisons test was used. Bars and error bars represent means of results and SE of the mean, respectively. All statistical tests were considered statistically significant when the p value was below 0.05. Statistical analysis was performed using GraphPad Prism V.8.1.0. Raw data can be found in online supplemental file 2.
jitc-2021-003655supp002.xlsx (57.6KB, xlsx)
Results
Treatment with T cell-recruiting bsAbs induces platelet activation
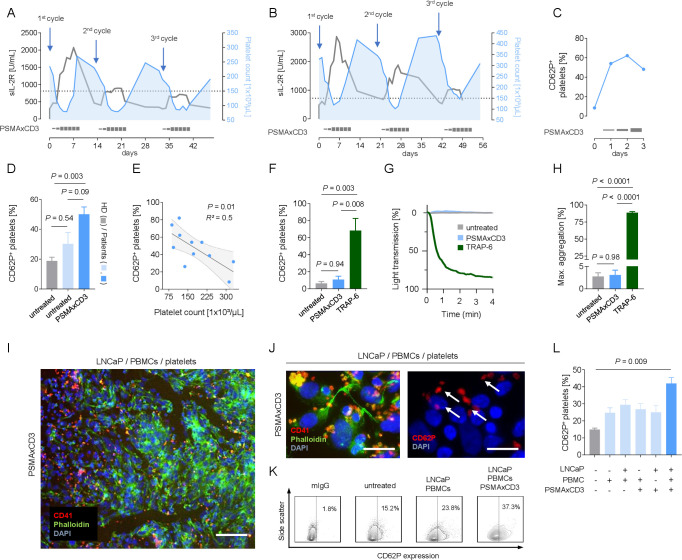
Although it has been shown that platelets, within the TME, allow for tumor immune escape,13 15 16 their role in the context of immunotherapy is largely unknown. When we monitored platelet counts in patients with prostate cancer treated with the bispecific PSMA×CD3 antibody CC-1,17 23 we observed a treatment-associated decrease in all study subjects (figure 1A, B). Platelet counts were inversely related to serum levels of soluble IL-2 receptor, indicating an association with T-cell activation induced by bsAb. Application of the PSMA×CD3 bsAb further was associated with upregulation of the activation marker P-selectin (CD62P) on platelets (figure 1C, D). Since we also observed a negative association of platelet count and platelet activation (CD62P expression) (figure 1E), we hypothesized that tumor–T cell interaction induced by the bsAb caused platelet activation, which in turn led to the decrease of total platelet count.
Figure 1.
Treatment with bispecific antibody leads to platelet activation. (A, B) Association of platelet count and sIL-2R concentration in two patients with prostate cancer receiving bispecific PSMA×CD3 (CC-1) antibody. (C) Time course of platelet activation (CD62P expression) on CC-1 treatment. (D) Platelet activation (CD62P expression) in untreated healthy donors (gray, n=5), untreated patients with prostate cancer (light blue, n=4) and in patients with prostate cancer receiving PSMA×CD3 (dark blue, n=5). Statistical significance was calculated by one-way ANOVA and Tukey’s multiple comparisons test. (E) Association of CD62P expression and total platelet count. Statistical significance was calculated using simple linear regression analysis. (F) Lack of platelet activation on PSMA×CD3 treatment (200 ng/mL) (n=4). Classical platelet agonist TRAP-6 (10 µM) served as a positive control. Statistical significance was calculated by one-way ANOVA and Tukey’s multiple comparisons test. (G) Platelet aggregation (percentage of light transmission during the indicated time interval) was studied in the presence or absence of PSMA×CD3 (200 ng/mL) and TRAP-6. (H) Maximum aggregation levels on PSMA×CD3 and TRAP-6 treatment (n=3). Statistical significance was calculated by one-way ANOVA and Tukey’s multiple comparisons test. (I) Representative immunofluorescence staining of LNCaP cells coincubated with human PBMCs, platelets and PSMA×CD3 (scale bar indicates 100 µm, original magnification ×20). (J) Representative staining for platelet activation (CD62P expression) after incubating platelets for 30 min with LNCaP cells and PBMCs (effector to target ratio of 4:1) and PSMA×CD3 (scale bar indicates 20 µm, original magnification ×63). (K, L) Platelets of healthy donors (n=3) were analyzed for CD62P expression after culturing them for 2 hours in the presence or absence of LNCaP cells alone or in combination with PSMA×CD3 and/or PBMCs of healthy donors. (K) Exemplary plots and (L) pooled data are shown. Statistical significance was calculated by one-way ANOVA and Tukey’s multiple comparisons test. ANOVA, analysis of variance; DAPI, 4′,6-diamidino-2-phenylindole; HD, healthy donor; PBMC, peripheral blood mononuclear cell; sIL-2R, soluble interleukin-2 receptor; TRAP-6, thrombin receptor activator peptide 6.
After excluding that the PSMA×CD3 bsAb alone induced platelet activation or aggregation (figure 1F–H), we established a coculture system using PSMA-expressing prostate cancer cells (LNCaP) and PBMC to mimic tumor–platelet–immune cell interactions in the presence or absence of the bsAb CC-1. As observed in patients, presence of the bsAb during tumor–platelet–immune cell interaction (figure 1I) resulted in profound platelet activation (figure 1J–L). Since platelet activation was most prominent when platelets were coincubated with stimulated immune cells (figure 1L), we hypothesized that activated T cells might trigger platelet activation in our experimental setting. In line with published data that T cells can stimulate platelets via the CD40–CD40L axis,24 we observed upregulation of CD40L preferentially on activated CD4+ T cells (online supplemental figure 2A, B), and blocking CD40–CCD40L interaction reduced platelet activation with the effect closely failing to reach statistical significance (online supplemental figure 2C). Overall, we conclude that platelets represent so far underestimated players that can affect the efficacy of T cell-engaging bsAb.
jitc-2021-003655supp001.pdf (1MB, pdf)
Platelets impair bsAb-induced T-cell activity
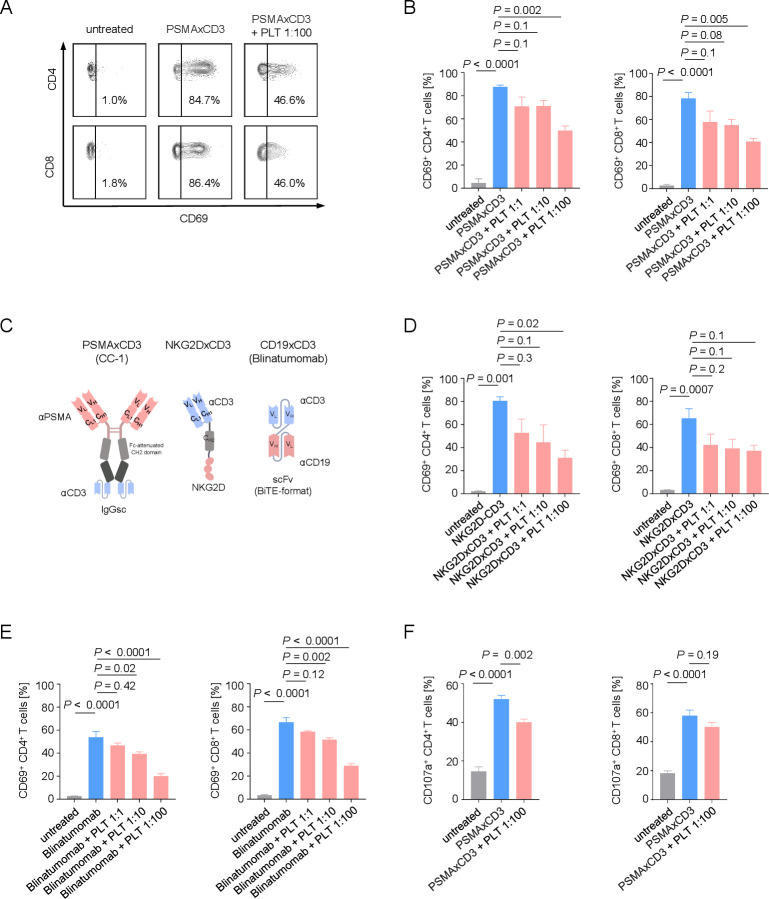
To further characterize the influence of platelets on PSMA×CD3 induced T cell reactivity, we cultured LNCaP cells with PBMCs and platelets at different platelet-immune cell ratios in the presence or absence of the PSMA×CD3 bsAb. CC-1-mediated activation of CD4+ and CD8+ T cells (CD69 expression) was inversely associated with effector to platelet ratios (figure 2A, B). To exclude that this effect exclusively occurred with the target antigen PSMA and/or specifically with the employed tumor cells, we extended our model system by using a recently described treatment model for sarcoma using a NKG2D×CD3 bispecific fusion protein (BFP)20 (figure 2D) as well as a leukemia model employing a B-ALL cell line treated with the CD19×CD3 bispecific T-cell engager (BiTE) blinatumomab (figure 2E). The inhibitory effect of platelets on CD3-mediated T-cell activation was found to be independent from the format of the T cell-recruiting compound (bsAb, BFP, and BiTE) (figure 2C), tumor entity, and target antigen (figure 2D, E), with none of the constructs inducing platelet activation or aggregation in the absence of effector cells (online supplemental figure 1A–C). Alike in patients treated with the PSMA×CD3 bsAb, a treatment-associated decrease of total platelet counts was observed in two patients with B-ALL treated with blinatumomab (online supplemental figure 3), providing further evidence that platelets might affect T-cell activation independently of tumor entity, target antigen or T cell-recruiting agent. Further analyses in our prostate cancer model using the bsAb PSMA×CD3 revealed a reduction of bsAb-induced degranulation of CD4+ and CD8+ T cells (CD107a expression) on co-incubation with platelets (figure 2F).
Figure 2.
Platelets impair the therapeutic T-cell activation capacity of bispecific antibodies. (A, B) Activation of CD4+ and CD8+ T cells was determined by analysis of CD69 expression after 24 hours. PBMCs of healthy donors were incubated with LNCaP cells (E:T 4:1) in the presence or absence of PSMA×CD3 (200 ng/mL) and with the indicated E:P ratios. (A) Exemplary flow cytometry results and (B) combined data are shown (n=3). (C) Schematic illustration of the different bispecific T cell-engaging formats used in our study. (D, E) Activation of CD4+ and CD8+ T cells was analyzed on coculture of PBMCs of healthy donors with target cells at an E:T ratio of 2.5:1.0 and the indicated E:P ratios for 24 hours. (D) The sarcoma cell line SaOS-2 was cultured with PBMCs (n=3) and NKG2DxCD3 (2.5 µg/mL). (E) The B-ALL cell line (SEM) was cultured with PBMCs (n=4) and CD19xCD3 (blinatumomab, 200 pg/mL). (F) Degranulation of CD4+ and CD8+ T cells was determined by analysis of CD107a expression after 4 hours. PBMCs of healthy donors were cultivated with LNCaP cells at an E:T ratio of 4:1 in the presence or absence of PSMA×CD3 (200 ng/mL) or platelets (E:P 1:100). Pooled data (n=5) are shown. (B, D–F) Statistical significance was calculated by one-way analysis of variance and Tukey’s multiple comparisons test. E:P, effector to platelet; E:T, effector to target; PLT, platelets; PBMC, peripheral blood mononuclear cell.
Impairment of bsAb-induced T-cell reactivity is mediated via the TGF-β axis
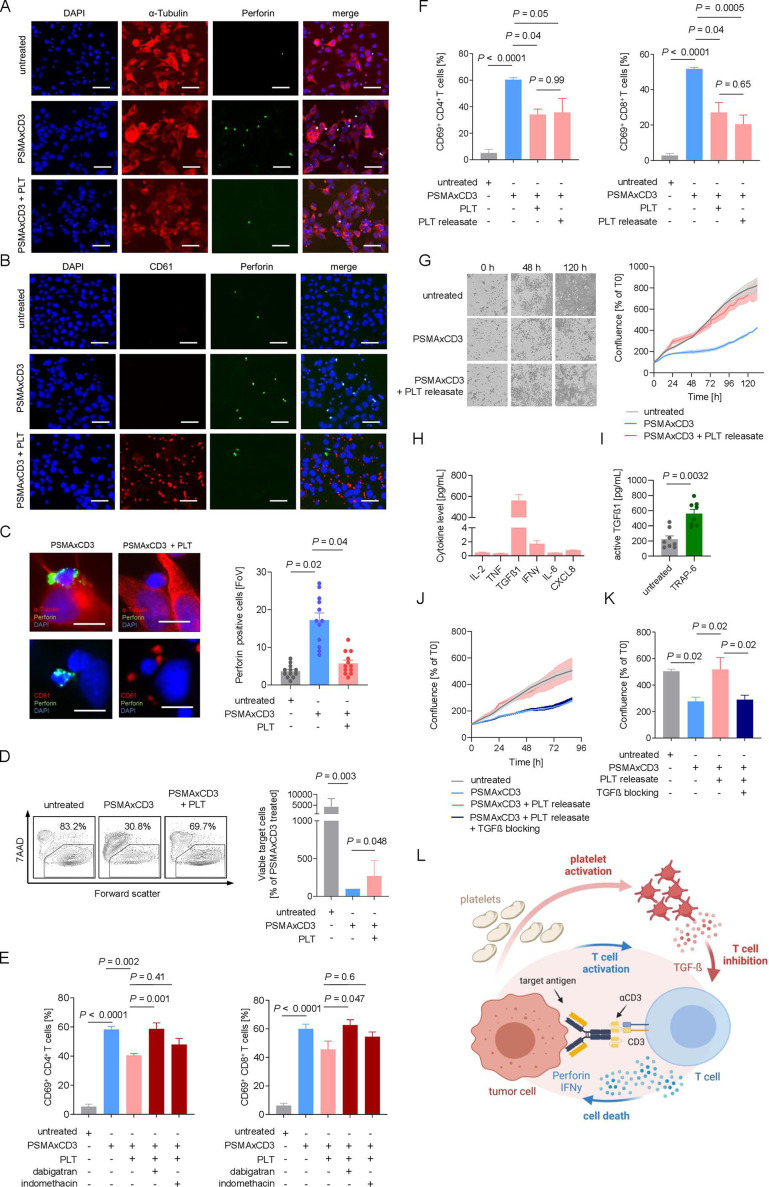
Next, we analyzed how platelet-mediated inhibition of T-cell activation and degranulation affected bsAb-induced tumor cell lysis. Treatment with the bsAb PSMA×CD3 significantly increased the number of perforin-secreting immune effector cells (figure 3A–C) and was significantly reduced by the presence of platelets. This was mirrored by profound platelet-mediated inhibition of target cell lysis quantified by flow cytometry-based cytotoxicity assays (figure 3D).
Figure 3.
Platelets undermine treatment efficacy of bsAb via the TGF-β axis. (A–C) Immunofluorescent staining after 2.5 hours of coincubation of LNCaP cells and PBMCs (E:T ratio of 4:1) in the presence of PSMA×CD3 (200 ng/mL) and platelets (E:P ratio of 10:1). Statistical significance was calculated by one-way ANOVA and Tukey’s multiple comparisons test. (A) Cells were stained for tubulin (red) and perforin (green). Nuclei were stained using DAPI (blue). Scale bar indicates 100 µm, original magnification ×20. (B) Platelets were stained for CD61 (red); nuclei were stained using DAPI (blue) and perforin (green). Scale bar indicates 100 µm, original magnification ×20. (C) Exemplary immunofluorescence staining of tumor–platelet immune cell interactions (left). Scale bar indicates 20 µm, original magnification ×63. Quantification of perforin-positive cells per FoV (n=12) (right). (D) PBMCs of healthy donors (n=7) were incubated with LNCaP cells (E:T ratio of 2.5:1.0) with and without PSMA×CD3 in the presence or absence of platelets (E:P 100:1) as indicated for 72 hours. Then target cell lysis was analyzed by flow cytometry to determine the percentage of 7-aminoactinomycin (7AAD)-positive LNCap cells. Left panel: exemplary plots obtained with a single donor; right panel: combined data obtained with seven independent donors displayed as percentage of viable target cells after bsAb treatment. Statistical significance was calculated by Kruskal-Wallis and Dunn’s multiple comparisons test. (E) Activation of CD4+ and CD8+ T cells was determined by expression of CD69 after 24 hours of treatment with PSMA×CD3 and platelets (E:T ratio of 4:1, E:P ratio of 100:1). Platelets were preincubated with dabigatran (0.5 mM) or indomethacin (0.05 mM) for 30 min before adding to the assay. Statistical significance was calculated by one-way ANOVA and Tukey’s multiple comparisons test. (F) Activation of CD4+ and CD8+ T cells was analyzed after incubation with LNCaP cells (E:T ratio of 4:1) and treatment with anti-PSMA×CD3, platelets (E:P ratio of 100:1) or PR (n=4). Statistical significance was calculated by one-way ANOVA and Tukey’s multiple comparisons test. (G) PSMA×CD3-induced cell death of LNCaP cells was determined using live cell imaging. Cells were incubated with PBMCs and platelets. In the left panel, representative pictures at 0, 48, and 120 hours are displayed. In the right panel, pooled data are shown (n=8). (H) PR has been generated as described in the Methods section, and the indicated cytokines were measured using LEGENDplex assay (n=8). (I) The concentration of active TGF-β in the platelet supernatant before and after stimulation with TRAP-6 is shown (n=8). Statistical significance was calculated using Student’s t-test. (J, K) Cytotoxicity of PBMC against LNCaP cells after treatment with PSAM×CD3, and platelet release with or without blocking of the TGF-β axis using the combination of SB431542 (20 µM) and anti-TGF-β antibody (2 µg/mL) is shown (n=8). (K) Pooled data of cell confluence measured after 88 hours of treatment. Statistical significance was calculated by one-way ANOVA and Holm-Sidak’s multiple comparisons test. (L) Schematic illustration of the proposed model of an inhibitory loop driven by platelets and platelet-released TGF-β, undermining the efficacy of T cell-recruiting bsAbs. The graphic was created by BioRender (BioRender.com, Toronto, Canada). ANOVA, analysis of variance; bsAb, bispecific antibody; DAPI, 4′,6-diamidino-2-phenylindole; E:P, effector to platelet; E:T, effector to target; FoV, field of view; PBMC, peripheral blood mononuclear cell; PR, platelet releasate; TGF-β, transforming growth factor beta.
As platelet activation results in secretion of immunomodulatory factors,15 16 we next investigated whether inhibition of platelet activation with anticoagulant drugs would prevent their inhibitory effect on bsAb-mediated T cell activation. Treatment with dabigatran, an oral anticoagulant, efficiently restored CD4+ and CD8+ T cell activation whereas indomethacin had no relevant effect (figure 3E). Next, we compared the influence of platelets and PR on T cell reactivity and found that both reduced T cell activation to the same extent (figure 3F). This indicates that soluble factors released from activated platelets impair bsAb-induced T cell reactivity, which is in line with previous observations.16 25 Remarkably, live cell imaging revealed that PR efficiently reduced bsAb-mediated tumor cell lysis (figure 3G). To unravel the factor within PR that inhibits T cell activity, we quantified several key immunomodulatory cytokines in PR preparations (figure 3H). Of all tested cytokines, active TGF-β1 was found to be most elevated and modulated on platelet activation (figure 3H, I).
To functionally validate that indeed platelet-derived TGF-β subverts the efficacy of T cell-engaging therapeutics, we determined the effect of TGF-β blockade on bsAb-induced cytotoxicity on exposure to platelet release, which resulted in restoration of T cell reactivity (figure 3J, K). We provide here a new model showing that an inhibitory loop driven by platelet-derived TGF-β limits the efficacy of T cell-recruiting bsAb (figure 3L).
Discussion
Tumor immune escape mechanisms are limiting the efficacy of immunotherapy. Especially, in solid tumors, the TME has been shown to be crucial for diminishing antitumor immune responses.26 Although it has been shown that platelets, as parts of the TME, possess the ability to modulate tumor immune escape,13 16 the importance in the context of immunotherapy is poorly investigated so far. We observed that treatment with the novel bispecific PSMA×CD3 antibody CC-117 18 leads to a significant platelet activation and a transient decrease of total platelet count in vivo. Since a similar effect was observed in patients with acute lymphocytic leukemia (ALL) treated with the CD19×D3 BiTE blinatumomab, we hypothesize that platelets represent so far underestimated players affecting the efficacy of T cell-engaging bsAb, which is in line with data that platelets impair NK cell-mediated tumor immunosurveillance and antitumor T-cell reactivity in a model of adoptive T-cell transfer.15 16
In our preclinical setting, we were able to show that bsAb-mediated tumor–immune cell interactions efficiently induce platelet activation, while the T cell-engaging therapeutics alone do not bind or activate platelets. The bsAb-induced T-cell activation was accompanied by an upregulation of CD40L preferentially on CD4+ T cells, and the induced platelet activation was partly diminished on blocking CD40–CD40L interaction. This is in line with published data that T cells stimulate platelets via the CD40–CD40L axis.24 However, further data are needed to validate the involvement of CD40–CD40L interaction.
When investigating the influence of activated platelets on the PSMA×CD3-induced effector cell reactivity, we observed a significant inhibition of T-cell reactivity. Of note, this phenomenon was not restricted to the format of the bispecific T cell-recruiting agents (IgGsc, BFP, or BiTE), the tumor entity, or target antigen (PSMA, NKG2DL, or CD19). This observation suggests that the immunomodulating effect of platelets might be a general mechanism of platelet-mediated tumor-immune escape on treatment with T cell-engaging therapies. This hypothesis is further supported by our finding that soluble factors, secreted via activated platelets, efficiently subvert bsAb induced T-cell cytotoxicity, which is in line with observations reported for adoptive T-cell therapies and immune checkpoint blockade.16 25
Our finding that only the reversible thrombin inhibitor dabigatran, but not indomethacin, abrogated the inhibitory effect of platelets is supported by data showing that dabigatran can block thrombin-induced TGF-β activation via platelet-bound glycoprotein A repetitions predominant.25 Considering that pharmacological inhibition of platelets restores efficacy of bsAb treatment, our data not only emphasize the role of platelets in tumor–immune cell interaction in general and bsAb-based immunotherapy in particular, but also indicate that blocking platelet activation with clinically already available drugs or neutralization of TGF-β may serve to increase efficacy of T cell-recruiting bsAb.
Bearing in mind that CAR T cells use a similar mechanism to induce cytotoxicity, a platelet-mediated inhibition of CAR T-cell reactivity is probable. However, further data are needed to investigate such an assumption. In conclusion, our study provides further important data supporting the evidence of an inhibitory loop driven by platelets undermining antitumor T-cell cytotoxicity and thus facilitating tumor immune escape on treatment with T cell-engaging agents.
Acknowledgments
We thank Celine Reiß, Hannah Zug, Sabrina Sauter, Michael Beller, and the Flow Cytometry Core Facility Berg of the University Hospital Tuebingen for their excellent technical support.
Footnotes
Contributors: MSL and CH designed, performed, and interpreted the experiments. MSL, CH, BK, SM, JSH, MM, and LZ provided analysis and interpretation of data. MSL, BK, MM, CH, and HRS wrote the manuscript. HRS, GJ, and CH designed the study. All authors contributed to the writing of the paper and approved the final manuscript.
Funding: This project was supported by the Deutsche Krebshilfe (70113999), the Wilhelm Sander-Stiftung (2017.100.2), and Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy (EXC 2180–39 090 067) and DFG (project number SA 1360/9–3 and 374031971 - TRR 240/project B05).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information. The datasets supporting the conclusions of this article are included within the article.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human subjects and was approved by Medical Faculty of the Eberhard Karls University and the University Hospital Tuebingen (13/2007 V). Subjects gave informed consent to participate in the study before taking part.
References
- 1.Bargou R, Leo E, Zugmaier G, et al. Tumor regression in cancer patients by very low doses of a T cell-engaging antibody. Science 2008;321:974–7. 10.1126/science.1158545 [DOI] [PubMed] [Google Scholar]
- 2.Brentjens RJ, Davila ML, Riviere I, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med 2013;5:177ra38. 10.1126/scitranslmed.3005930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996;271:1734–6. 10.1126/science.271.5256.1734 [DOI] [PubMed] [Google Scholar]
- 4.Ishida Y, Agata Y, Shibahara K, et al. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. Embo J 1992;11:3887–95. 10.1002/j.1460-2075.1992.tb05481.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kantarjian H, Stein A, Gökbuget N, et al. Blinatumomab versus chemotherapy for advanced acute lymphoblastic leukemia. N Engl J Med 2017;376:836–47. 10.1056/NEJMoa1609783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah NN, Fry TJ. Mechanisms of resistance to CAR T cell therapy. Nat Rev Clin Oncol 2019;16:372–85. 10.1038/s41571-019-0184-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh A, Dees S, Grewal IS. Overcoming the challenges associated with CD3+ T-cell redirection in cancer. Br J Cancer 2021;124:1037–48. 10.1038/s41416-020-01225-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roma-Rodrigues C, Mendes R, Baptista P, et al. Targeting tumor microenvironment for cancer therapy. Int J Mol Sci 2019;20:840. 10.3390/ijms20040840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erpenbeck L, Schön MP. Deadly allies: the fatal interplay between platelets and metastasizing cancer cells. Blood 2010;115:3427–36. 10.1182/blood-2009-10-247296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gay LJ, Felding-Habermann B. Contribution of platelets to tumour metastasis. Nat Rev Cancer 2011;11:123–34. 10.1038/nrc3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinterleitner C, Zhou Y, Tandler C, et al. Platelet-Expressed TNFRSF13B (TACI) predicts breast cancer progression. Front Oncol 2021;11:642170. 10.3389/fonc.2021.642170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leblanc R, Peyruchaud O. Metastasis: new functional implications of platelets and megakaryocytes. Blood 2016;128:24–31. 10.1182/blood-2016-01-636399 [DOI] [PubMed] [Google Scholar]
- 13.Placke T, Salih HR, Kopp H-G. GITR ligand provided by thrombopoietic cells inhibits NK cell antitumor activity. J Immunol 2012;189:154–60. 10.4049/jimmunol.1103194 [DOI] [PubMed] [Google Scholar]
- 14.Placke T, Örgel M, Schaller M, et al. Platelet-Derived MHC class I confers a pseudonormal phenotype to cancer cells that subverts the antitumor reactivity of natural killer immune cells. Cancer Res 2012;72:440–8. 10.1158/0008-5472.CAN-11-1872 [DOI] [PubMed] [Google Scholar]
- 15.Kopp H-G, Placke T, Salih HR. Platelet-Derived transforming growth factor-beta down-regulates NKG2D thereby inhibiting natural killer cell antitumor reactivity. Cancer Res 2009;69:7775–83. 10.1158/0008-5472.CAN-09-2123 [DOI] [PubMed] [Google Scholar]
- 16.Rachidi S, Metelli A, Riesenberg B, et al. Platelets subvert T cell immunity against cancer via GARP-TGFβ axis. Sci Immunol 2017;2. 10.1126/sciimmunol.aai7911. [Epub ahead of print: 05 May 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heitmann JS, Walz JS, Pflügler M, et al. Protocol of a prospective, multicentre phase I study to evaluate the safety, tolerability and preliminary efficacy of the bispecific PSMAxCD3 antibody CC-1 in patients with castration-resistant prostate carcinoma. BMJ Open 2020;10:e039639. 10.1136/bmjopen-2020-039639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zekri L, Vogt F, Osburg L, et al. An IgG‐based bispecific antibody for improved dual targeting in PSMA‐positive cancer. EMBO Mol Med 2021;13:e11902. 10.15252/emmm.201911902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Märklin M, Hagelstein I, Koerner SP, et al. Bispecific NKG2D-CD3 and NKG2D-CD16 fusion proteins for induction of NK and T cell reactivity against acute myeloid leukemia. J Immunother Cancer 2019;7:143. 10.1186/s40425-019-0606-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagelstein I, Lutz MS, Schmidt M, et al. Bispecific NKG2D-CD3 and NKG2D-CD16 fusion proteins as novel treatment option in advanced soft tissue sarcomas. Front Immunol 2021;12:653081. 10.3389/fimmu.2021.653081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider CA, Rasband WS, Eliceiri KW. Nih image to ImageJ: 25 years of image analysis. Nat Methods 2012;9:671–5. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paczulla AM, Rothfelder K, Raffel S, et al. Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature 2019;572:254–9. 10.1038/s41586-019-1410-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kauer J, Hörner S, Osburg L, et al. Tocilizumab, but not dexamethasone, prevents CRS without affecting antitumor activity of bispecific antibodies. J Immunother Cancer 2020;8:e000621. 10.1136/jitc-2020-000621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danese S, de la Motte C, Reyes BMR, et al. Cutting edge: T cells trigger CD40-dependent platelet activation and granular RANTES release: a novel pathway for immune response amplification. J Immunol 2004;172:2011–5. 10.4049/jimmunol.172.4.2011 [DOI] [PubMed] [Google Scholar]
- 25.Metelli A, Wu BX, Riesenberg B, et al. Thrombin contributes to cancer immune evasion via proteolysis of platelet-bound GARP to activate LTGF-β. Sci Transl Med 2020;12. 10.1126/scitranslmed.aay4860. [Epub ahead of print: 08 Jan 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murciano-Goroff YR, Warner AB, Wolchok JD. The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res 2020;30:507–19. 10.1038/s41422-020-0337-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-003655supp002.xlsx (57.6KB, xlsx)
jitc-2021-003655supp001.pdf (1MB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information. The datasets supporting the conclusions of this article are included within the article.