Abstract
Cells decide whether to grow and divide by integrating internal and external signals. Non-autonomous cell growth and proliferation occurs when microenvironmental signals from neighboring cells, both physical and secreted, license this decision. Understanding these processes is vital to developing an accurate framework for cell–cell interactions and cellular decision-making, and is useful for advancing new therapeutic strategies to prevent dysregulated growth. Here, we review some recent examples of non-autonomous cell growth in the mammary gland and tumor cell proliferation.
Introduction
Classic studies of cell growth and proliferation have used relatively homogeneous populations of immortalized cells due to their experimental tractability. In these cases, cells seem to autonomously decide whether to grow and divide, as microenvironmental cues are dominated by media formulation and physical cues such as local cells density. These homogeneous cell cultures allow for the detailed analysis of pathways controlling the cell cycle, for example, through gain and loss of function perturbations targeted to specific gene products. However, in multicellular organisms, interactions between heterogeneous cell populations are often the primary regulators of growth during development, normal tissue function, and even in diseases such as cancer. This realization has motivated the development of organoid, co-culture, and tumor xenograft models that provide a means to study so-called non-autonomous cell growth. While all cellular decisions in metazoan are to one degree or another regulated by signals generated by neighboring cells and tissues, we define non-cell autonomous behaviors as those cases where a signal or perturbation impinging on a cell triggers a response in a neighboring cell that may not have received the signal or that may be incapable of receiving the signal. In the case of proliferative control, this means that a signal impinging on one cell type triggers growth and proliferation in a neighboring or secondary cell type. Understanding the principles and mechanisms that regulate non-autonomous cell growth will be critical for gaining deeper insight into the consequences of tumor heterogeneity, epithelial-stromal interactions, and will provide fertile ground for the discovery of new therapeutic targets and strategies. Here, we provide a summary of recent findings related to the non-autonomous regulation of cell growth and proliferation in the mammary gland, and discuss the implications of these findings for diseases such as cancer.
Non-autonomous cell growth in the mammary gland
The mammary gland is a bilayered, tubular epithelial tree. Lining the lumen are milk producing, luminal epithelial cells (LEPs) surrounded by a layer of contractile myoepithelial cells (MEPs). The MEP layer is surrounded by a basement membrane, separating the epithelium from a stroma containing numerous additional cell types such as fibroblasts, adipocytes, lymphocytes, neurons, and endothelial cells [1,2]. Within the luminal layer of the epithelium, there exists a minor population of post-mitotic, hormone sensing cells which range from 7 to 30% in the human mammary gland depending on a host of factors including age, pregnancy history, and exposure to hormones [3]. These cells are generally defined using immunohistochemical (IHC) detection of nuclear localized estrogen receptor alpha (ERα) and progesterone receptor (PR), and are frequently referred to as ER(+) cells [4]. Without dividing themselves [3,5], this minor population integrates hormonal signals produced by the ovaries and coordinates post-natal development and post-pubertal cyclical expansion/regression of the mammary gland via an intricate paracrine signaling network (Figure 1a).
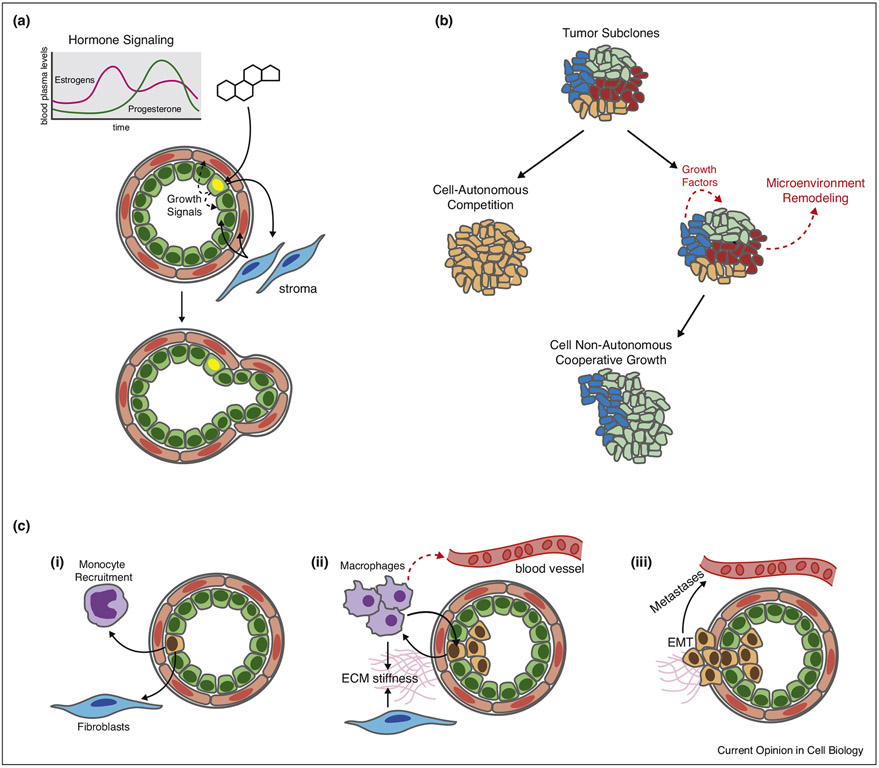
Figure 1.
Examples of non-cell autonomous growth. (a) Hormone signaling in the mammary gland. Hormone-sensing cells are restricted to the luminal epithelial lineage (green) and integrate cyclic hormonal signals, such as progesterone. These signals drive proliferation in neighboring cells via paracrine signaling both within the epithelium and through stromal–epithelial cross-talk. This key regulator of mammary gland growth is preserved but dysregulated in many breast cancers. (b) Tumor subclones can overgrow/compete (subclone sweep) with the neighbors or cooperate through secreted factors and physical remodeling of the microenvironment. (c) Monocytes and fibroblasts can be recruited to tumors by secretion of cytokines, where they mature into secretory macrophages and cancer-associated fibroblasts, which in turn can promote tumor proliferation through the secretion of additional cytokines and other forms of microenvironmental remodeling. This cross talk can enhance invasion, EMT, and ultimately, metastasis (Fig. 1a).
At three different developmental stages – prepubertal, postpurbertal, and pregnant – a distinct spectrum of hormones signal through ER(+) cells which in turn send out new signals to the microenvironment that serve to coordinate growth and morphogenesis. At the pre-pubertal stage, for example, estrogen-dependent signaling activates the sheddase activity of the ADAM17 protease to release transforming growth factor alpha (TGF-α) and amphiregulin, Epidermal Growth Factor Receptor (EGFR) ligands, to fibroblasts in the stromal compartment which then signal back to the epithelial compartment promoting ductal tree elongation [2,6,7•]. The complete mechanism of stromal signaling back to the epithelial compartment remains unclear, but depends on fibroblast growth factors 10 in the stroma and fibroblast growth factor receptors 1 and 2 in the epithelium [8,9]. At the post-pubertal stage, ERα activation is also permissive of PR expression, which when activated, results in local proliferation, primarily via secretion of paracrine signaling molecules: RANKL and Wnt4 [10-12]. Rajaram et al. used serial engraftment of mutant mammary epithelial tissue with deletions in either RANKL or Wnt4 in contralateral mouse fat pads and observed that loss of RANKL modestly reduced proliferation, primarily by a reduction in the amount of side branching. By the third serial transplant, loss of Wnt4 secretion led to a drastic decrease in proliferation, more so even than PR deletion. They went on to parse Wnt4 secretion in the perinatal as well as pubertal and adult mammary gland. Their work and others supports a model in which canonical Wnt4 signaling downstream of PR is important for cell self-renewal and drives proliferation of CD44high CK5+ luminal progenitor cells required for cyclical expansion and regression in the pubertal and adult mammary gland [11,13]. Though these experiments were conducted in mice, additional work has suggested PR is a key signaling pathway in the human mammary gland as well [10]. Finally, at the pregnant stage, prolactin receptor activation can trigger secondary effects in the epithelium beyond its typical role in stimulating the production of milk proteins such as Beta casein during pregnancy. Recent work by Tarulli et al. has demonstrated a unique response of hormone sensing/ER(+) cells to low levels of prolactin due to differences in Wip1 expression, a phosphatase that regulates Jak-Stat signaling. This difference results in hormone sensing/ER(+) cells converting prolactin signals into paracrine growth factors such as RANKL and IGF2 as opposed to milk production [14].
In these three different developmental stages, a rare population of hormone sensing cells within the luminal compartment integrate systemic signals impinging on the gland through the blood stream to drive the exclusive proliferation of their neighbors. In cancer, these and other signaling pathways are frequently co-opted by the tumor and used in a dysregulated manner to promote tumor progression [15]. In invasive breast cancer, for example, ERα signaling is active and necessary for the growth and survival of nearly two thirds of tumors. This observation has driven the development and frequent clinical use of endocrine therapies that block estrogenic signaling, either through competitive inhibitors of ER such as tamoxifen, or through blocking the local conversion of androgens to estrogens via aromatase inhibitors [16]. Strikingly, patients with tumors containing as few as 1% ER+ cells, as defined by IHC staining, still benefit from anti-endocrine therapy [17]. It is interesting that these therapies are effective while presumably targeting such a small proportion of tumor cells, and highlights how non-autonomous proliferation may act as a hub for proliferative control in both the normal and diseased mammary gland. While we still know relatively few mechanistic details governing the estrogenic paracrine signaling circuit in ER (+) breast cancers, particularly a detailed understanding of stromal–epithelial cross-talk, it is likely to be important given its role in normal gland function. It is possible, for example, that estrogenic signaling, in addition to allowing PR expression, is permissive of stromal alterations, creating a permissive microenvironment for tumor growth through mechanisms that remains to be determined. Along these lines, there is evidence that dysregulated RANKL is predictive of disease [18] and there is growing interest in targeting paracrine signaling such as by RANKL inhibition for breast cancer treatment [19,20] though specific mechanisms remain to be elucidated. Wnt4 has also been implicated as an important factor in cell line models of breast cancer, including in an endocrine resistant model, again paralleling its important role in normal physiology [21,22]. Further insight into these pathways, and the changes they promote within the stromal and epithelial compartments, will likely be fertile ground for novel therapeutic interventions [2,23].
While hormone-sensing cells and their canonical ligands are central to paracrine signaling in the mammary gland, and also play a role in estrogen-dependent breast cancers, recent studies have unveiled additional examples of divisions of labor within the mammary gland that regulate its growth and remodeling. Sonic Hedgehog pathway is a key pathway for epithelial-to-mesenchymal transition across several tissues [24]. Visbal et al. demonstrated that overexpression of smoothened, a key transducer of hedgehog signaling, in luminal cells, stimulates proliferation in neighboring luminal cells and also serves to alter the stromal microenvironment, attracting macrophages [25]. This is a phenotype frequently observed in ductal carcinoma in-situ lesions (DCIS) and is conceptually analogous to the recruitment of tumor-associated macrophages and promotion of a tumorigenic microenvironment discussed below. A follow-up study by O’Toole et al. demonstrated negative survival in a cohort of 279 patients with invasive ductal carcinoma that correlated with the level of hedgehog signaling in the epithelium. They also showed that receptors in the stromal compartment, rather than epithelium, mediated this effect [26].
Mutualism of heterogeneous tumor subclones
With the aid of next generation sequencing technologies, there is now a general appreciation that tumors comprise a heterogeneous mixture of genetically and epigenetically distinct subclones. Tumor heterogeneity can be a consequence of genomic instability and Darwinian competition, but can also arise phenotypically through dysregulated lineage hierarchies [27,28], local variations in microenvironment, and signaling feedback loops [29,30,31•]. There is particular interest in examining intra-tumoral heterogeneity and how different subclones interact via competition or cooperation in order to accurately stratify risk [32], assess metastatic potential [33], and design more precise interventions [34-37]. For example, Marusyk et al. developed a panel of 18 MDA-MB-468 subclones, each transduced to overexpress a single secreted factor known to be important for breast cancer progression and transplanted different combinations to investigate clone dynamics. They demonstrated that instead of competing to take over the tumor, a subset of subclones within a polyclonal tumor can collaborate and drive non-autonomous growth or invasion of other subclones (Fig. 1b) [38••]. This concept has been observed in other recent studies as well. Working in vitro, Todhunter et al. observed that organoids containing a minor subpopulation of RAS-transformed MCF10AT cells enhanced the growth rate of the majority, non-transformed MCF10A cells [39]. In a related experiment with modified MCF10A cells, xenografts containing mixtures of two different lines, one mildly invasive (AT1) and one very invasive (CA1d), produced larger tumors in mouse transplantation studies than a pure population of CA1d or AT1 cells alone. One potential mechanism behind this observation was that the slower growing AT1 line was a source of TGFα and TGFβ, driving expansion of CA1ds [40]. Cross-talk between heterogeneous cancer cells can operate through a number of different pathways. Zhang et al., for example, analyzed a (Lin−CD29HighC-D24Low) subclone population within a murine model of breast cancer and observed higher transcripts of secreted factors Wnt2, Wnt9a, Cxcl12, and IL-6. While these particular cells are not very proliferative themselves, the study demonstrated that blocking these secreted ligands suppressed tumor formation and frequency by inhibiting growth of neighboring cells [41]. These studies add to emerging models of the tumor as organ-like structure which, like normal tissues, have cells serving specialized roles in maintaining tumor expansion, invasion, and survival [42]. Moreover, this more granular view of the tumor will inform new strategies to block growth of heterogeneous tumors.
Cancer associated fibroblasts (CAFs) and tumor associated macrophages (TAMs)
Another mechanism of non-autonomous cell growth is epithelial-stromal cross talk. As described above using estrogen-dependent signaling in the mammary gland as an example, estrogen triggered release of amphiregulin and TGF-α to the stromal compartment is critical for regulating epithelial growth. However, this cross-talk is also a well-documented phenomenon observed during tumor progression and involves tumorigenic cells co-opting normal wound healing processes to promote a proliferative microenvironment [15,43]. This is usually achieved via the secretion of cytokines to recruit blood vessels, fibroblasts, and immune cells. Broadly, once these cells are recruited and activated, they collaborate with tumor cells to support proliferation by two general mechanisms: indirectly by promoting angiogenesis, and more directly by secreting paracrine factors and remodeling the extracellular matrix (ECM).
Tumor associated macrophages (TAMs) are recruited as inflammatory, circulating monocytes by cytokines secreted by tumor cells including CCL2 and CSF-1. Once in the tumor, they differentiate into a secretory, M2-like state [44,45]. The number of local TAMs has been shown to correlate with the proliferation index of the tumor cells as determined by Ki67 staining [46]. Recent work has uncovered numerous cytokines secreted by the TAMs that function to not only drive proliferation in nearby epithelial cells such as IL-6, CCL20, CXCL-1, EGF and IL-10 [47-50], but also extravasation and metastasis [51]. Cancer associated fibroblasts (CAFs) are similarly recruited and activated by secretions from tumor cells such as TGFβ and PDGF. They in turn secrete growth factors such as HGF, EGF, and IGF as signaling molecules which serve to drive cell division of neighboring tumor cells [52-54].
In addition to paracrine signaling, another important effect of both TAMs and CAFs is on remodeling the physical properties of the tumor microenvironment by deposition of ECM proteins. CAFs have been shown to enhance the stiffness of the microenvironment and changes to tissue mechanics have separately been shown to increase cell proliferation an invasion Fig. 1c [55]. Alternatively, mutations in macrophages themselves may be sufficient to drive aberrant microenvironmental changes and initiate tumors in other cells. Experiments by Lujambio et al. used a mouse model of a conditional p53 deletion in hepatic stellate cells, which serves to remove cell cycle inhibition, and after inducing injury in the liver, demonstrated that the enhanced production of fibrotic tissue resulted in a higher incidence of tumors but without a change in grade or size [56]. Work by Afik et al. using LC/MS-MS suggests that like CAFs, TAMs induce proliferation of epithelial cells by remodeling the microenvironment, specifically by enhancing the deposition of collagen I and XIV [57•].
TAMs and CAFs may also modulate the susceptibility of tumor cells to chemotherapeutic drugs and potentially even ionizing radiation by non-autonomous mechanisms, tilting the balance of apoptosis and proliferation back toward proliferation [58-61]. Therefore, interest is growing in targeting these microenvironmental changes directly [62-64] to fight cancer progression and resistance mechanisms. This was explored in seminal work by Straussman et al. showing that mutated BRAF melanoma cells are resistant to RAF inhibition when co-cultured with stromal cells that specifically secrete HGF and that in-vivo, stromal HGF correlates with poor prognosis [65]. These studies were extended to show that combination therapies including of HGF and RAF inhibition lead to a lower incidence of resistance by removing a key mechanism for rewiring pro-growth signals during drug treatment. In other recent study, combination therapies incorporating fibrosis inhibitors to prevent microenvironmental remodeling by CAFs along with more standard chemotherapeutics was superior at inhibiting tumor growth [66,67]. It now seems clear that factors secreted by the tumor or stroma have a widespread potential to overcome sensitivity to tumor targeted kinase inhibitors, perhaps necessitating the use of such combinatorial treatment strategies [61].
Conclusion and future work
Examples of non-autonomous cell growth, such as those described here in the mammary gland are common in normal physiology. Moreover, there is an increasing appreciation that similar mechanisms likely play an important role in the progression and therapeutic response of cancers. Generally, these pro-proliferative signals take the form of paracrine growth signals or direct modulators of the physical and chemical microenvironment, and recent studies suggest that these pathways can be manipulated pharmacologically for therapeutic benefit. Therefore, a mechanistic examination of non-autonomous cell proliferation is vital in order to enhance our understanding of the complex cellular interactions that regulate normal physiology, as well as to develop the next generation of novel, rationally designed therapeutics.
One important challenge which is beginning to be addressed is developing a better understanding of the difference between stable cell ‘types’ and dynamical cell ‘states’ [68]. Measurements at a fixed time point or tissue state may not be sufficient reveal the dynamical cell-cell interaction landscape, leading to context-dependent, rather than generalizable, findings. Another challenge is identifying the correct in vitro or in vivo model to examine non-autonomous growth control. Given the complexity of cellular interactions regulating normal and diseased tissues, along with the important differences between human and mouse physiology [69], this can be a particularly vexing problem. However, recent advances in cell culture systems – be they 2D co-cultures, 3D systems with extra cellular matrix mimetics [70], or even ‘organ on a chip’ technologies [71] – are poised to advance our understanding of complex multicellular interactions among human cells. Still, there are significant obstacles to fully deciphering the signaling pathways, both autocrine and paracrine, in these reductionist models. For example, recent work demonstrated that omitting even a single cell type from an interacting network of cells significantly changed cytokine signaling to make intestinal epithelial cells hypersensitive to TNFα [72]. Other work has demonstrated that vital signaling partners in-vitro [73] can be redundant in-vivo [74], arguing in favor of work in tractable animal model systems. However, given the differences between human and animal physiology in a number of tissues, translational studies may required new models [69]. Progress in patient derived xenografts (PDX) transplanted into immunocompromised mouse mammary fatpads offers an exciting intermediate model [75]. However, even these can omit key interactions, for example, through the differences between mouse and human stroma [69]. Thus, opportunities exist for advances in the ‘humanization’ of the murine fat pad and other tissues as well. A challenge to the field will therefore be to mitigate the various short-comings of each model by potentially utilizing multiple systems to validate the cellular interactions most relevant to human health and disease.
Acknowledgements
We are grateful for support from the Department of Defense Breast Cancer Research Program (W81XWH-10-1-1-1023 and W81XWH-13-1-0221 to Z.J. G.) and US National Institutes of Health common fund (DP2 HD080351-01 to Z.J.G). We are grateful to Samantha Liang for help making figures.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Rosen JM: On hormone action in the mammary gland. Cold Spring Harb Perspect Biol 2012, 4:a013086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brisken C, O’Malley B: Hormone action in the mammary gland. Cold Spring Harb Perspect Biol 2010, 2:a003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke RB, Howell A, Potten CS, Anderson E: Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res 1997, 57:4987–4991. [PubMed] [Google Scholar]
- 4.Allred DC: Issues and updates: evaluating estrogen receptor. Mod Pathol 2010, 23:S52–S59. [DOI] [PubMed] [Google Scholar]
- 5.Graham JD, Mote PA, Salagame U, van Dijk JH, Balleine RL, Huschtscha LI, Reddel RR, Clarke CL: DNA replication licensing and progenitor numbers are increased by progesterone in normal human breast. Endocrinology 2009, 150:3318–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sternlicht MD: Mammary ductal morphogenesis requires paracrine activation of stromal EGFR via ADAM17-dependent shedding of epithelial amphiregulin. Development 2006, 133:1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.•. Morancho B, Martínez-Barriocanal Á, Villanueva J, Arribas J: Role of ADAM17 in the non-cell autonomous effects of oncogene-induced senescence. Breast Cancer Res 2015. Highlights phenotype of ADAM17 sheddase activity and effects on non-autonomous growth and metastatic potential of neighboring cancer cells.
- 8.Pond AC, Bin X, Batts T, Roarty K, Hilsenbeck S, Rosen JM: Fibroblast growth factor receptor signaling is essential for normal mammary gland development and stem cell function. Stem Cells 2012, 31:178–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu P, Ewald AJ, Martin GR, Werb Z: Genetic mosaic analysis reveals FGF receptor 2 function in terminal end buds during mammary gland branching morphogenesis. Dev Biol 2008, 321:77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanos T, Sflomos G, Echeverria PC, Ayyanan A, Gutierrez M, Delaloye J-F, Raffoul W, Fiche M, Dougall W, Schneider P et al. : Progesterone/RANKL is a major regulatory axis in the human breast. Sci Transl Med 2013, 5:182ra55. [DOI] [PubMed] [Google Scholar]
- 11.Rajaram RD, Buric D, Caikovski M, Ayyanan A, Rougemont J, Shan J, Vainio SJ, Yalcin-Ozuysal O, Brisken C: Progesterone and Wnt4 control mammary stem cells via myoepithelial crosstalk. EMBO J 2015, 34:641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brisken C, Hess K, Jeitziner R: Progesterone and overlooked endocrine pathways in breast cancer pathogenesis. Endocrinology 2015, 156:3442–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai C, Yu QC, Jiang W, Liu W, Song W, Yu H, Zhang L, Yang Y, Zeng YA: R-spondin1 is a novel hormone mediator for mammary stem cell self-renewal. Genes Dev 2014, 28:2205–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarulli GA, De Silva D, Ho V, Kunasegaran K, Ghosh K, Tan BC, Bulavin DV, Pietersen AM: Hormone-sensing cells require Wip1 for paracrinestimulation in normal and premalignantmammary epithelium. Breast Cancer Res 2013, 15:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan D, Weinberg RA: Hallmarks of cancer: the next generation. Cell 2011, 144:646–674. [DOI] [PubMed] [Google Scholar]
- 16.Dowsett M, Howell A: Breast cancer: aromatase inhibitors take on tamoxifen. Nat Med 2002, 8:1341–1344. [DOI] [PubMed] [Google Scholar]
- 17.Hammond MEH, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M et al. : American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 2010, 28:2784–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiechl S, Schramek D, Widschwendter M, Fourkala E-O, Zaikin A, Jones A, Jaeger B, Rack B, Janni W, Scholz C et al. : Aberrant regulation of RANKL/OPG in women at high risk of developing breast cancer. Oncotarget 2016, 8:3811–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotsopoulos J, Singer C, Narod SA: Can we prevent BRCA1-associated breast cancer by RANKL inhibition? Breast Cancer Res Treat 2016. [DOI] [PubMed] [Google Scholar]
- 20.Gnant M, Pfeiler G, Dubsky PC, Hubalek M, Greil R, Jakesz R, Wette V, Balic M, Haslbauer F, Melbinger E et al. : Adjuvant denosumab in breast cancer (ABCSG-18): a multicentre, randomised, double-blind, placebo-controlled trial. Lancet 2015, 386:433–443. [DOI] [PubMed] [Google Scholar]
- 21.Okolicsanyi RK, van Wijnen AJ, Cool SM, Stein GS, Griffiths LR, Haupt LM: Heparan sulfate proteoglycans and human breast cancer epithelial cell tumorigenicity. J Cell Biochem 2014, 115:967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sikora MJ, Jacobsen BM, Levine K, Chen J, Davidson NE, Lee AV, Alexander CM, Oesterreich S: WNT4 mediates estrogen receptor signaling and endocrine resistance in invasive lobular carcinoma cell lines. Breast Cancer Res 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tornillo G, Smalley MJ: ERrrr … where are the progenitors? Hormone receptors and mammary cell heterogeneity. J Mammary Gland Biol Neoplasia 2015, 20:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Tian X-J, Xing J: Signal transduction pathways of EMT induced by TGF-β, SHH, and WNT and their crosstalks. J Clin Med 2016, 5:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visbal AP, LaMarca HL, Villanueva H, Toneff MJ, Li Y, Rosen JM, Lewis MT: Altered differentiation and paracrine stimulation of mammary epithelial cell proliferation by conditionally activated smoothened. Dev Biol 2011, 352:116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Toole SA, Machalek DA, Shearer RF, Millar EKA, Nair R, Schofield P, McLeod D, Cooper CL, McNeil CM, McFarland A et al. : Hedgehog overexpression is associated with stromal interactions and predicts for poor outcome in breast cancer. Cancer Res 2011, 71:4002–4014. [DOI] [PubMed] [Google Scholar]
- 27.Weinberger LS: A minimal fate-selection switch. Curr Opin Cell Biol 2015, 37:111–118. [DOI] [PubMed] [Google Scholar]
- 28.Germain RN: T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol 2002, 2:309–322. [DOI] [PubMed] [Google Scholar]
- 29.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA et al. : Mutational landscape and significance across 12 major cancer types. Nature 2013, 502:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gay L, Baker A-M, Graham TA: Tumour cell heterogeneity. F1000Research 2016, 5:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.•. Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed Bv, Curry WT, Martuza RL et al. : Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344:1396–1401.24925914 Used next-gen sequencing to profile the global transcriptome of single glioblastoma cells measuring the extent of intratumoral heterogeneity and the correlation of heterogeneity with poor prognosis.
- 32.Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, Ji HP, Maley CC: Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 2015, 22:105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen A, Yoshida M, Goodarzi H, Tavazoie SF: Highly variable cancer subpopulations that exhibit enhanced transcriptome variability and metastatic fitness. Nat Commun 2016, 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buckley NE, Forde C, McArt DG, Boyle DP, Mullan PB, James JA, Maxwell P, McQuaid S, Salto-Tellez M: Quantication of HER2 heterogeneity in breast cancer—implications for identi cationof sub-dominant clones forpersonalised treatment. Sci Rep 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janiszewska M, Liu L, Almendro V, Kuang Y, Paweletz C, Sakr RA, Weigelt B, Hanker AB, Chandarlapaty S, King TA et al. : Technical Reports. Nature Publishing Group; 2015:1212–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun X-X, Yu Q: Intra-tumor Heterogeneity of Cancer Cells and Its Implications for Cancer Treatment. Nature Publishing Group; 2015:1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park Y, Lim S, Nam J-W, Kim S: Measuring intratumor heterogeneity by network entropy using RNA-seq data. Sci Rep 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.••. Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K: Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 2014, 514:54–58.25079331 Mathematical framework of subclone competition and collaboration and the effects on overal tumor growth phenotypes.
- 39.Todhunter ME, Jee NY, Hughes AJ, Coyle MC, Cerchiari A, Farlow J, Garbe JC, Labarge MA, Desai TA, Gartner ZJ: Programmed synthesis of three-dimensional tissues. Nat Methods 2015, 12:975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franco OE, Tyson DR, Konvinse KC, Udyavar AR, Estrada L, Quaranta V, Crawford SE, Hayward SW: Altered TGF-α/β signaling drives cooperation between breast cancer cell populations. FASEB J 2016, 30:3441–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang M, Tsimelzon A, Chang CH, Fan C, Wolff A, Perou CM, Hilsenbeck SG, Rosen JM: Intratumoral heterogeneity in a Trp53-null mouse model of human breast cancer. Cancer Discov 2015, 5:520–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egeblad M, Nakasone ES, Werb Z: Tumors as organs: complex tissues that interface with the entire organism. Dev Cell 2010, 18:884–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanahan D, Coussens LM: Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21:309–322. [DOI] [PubMed] [Google Scholar]
- 44.Chanmee T, Ontong P, Konno K, Itano N: Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014, 6:1670–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jinushi M, Komohara Y: Biochim Biophys Acta—Rev Cancer 2015, 1855:123–130. [DOI] [PubMed] [Google Scholar]
- 46.Komohara Y, Horlad H, Ohnishi K, Fujiwara Y, Bai B, Nakagawa T, Suzu S, Nakamura H, Kuratsu J-I, Takeya M: Importance of direct macrophage—tumor cell interaction on progression of human glioma. Cancer Sci 2012, 103:2165–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfe AR, Trenton NJ, Debeb BG, Larson R, Ruffell B, Chu K, Hittelman W, Diehl M, Reuben JM, Ueno NT et al. : Mesenchymal stem cells and macrophages interact through IL-6 to promote inflammatory breast cancer in pre-clinical models. Oncotarget 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu B, Jia Y, Ma J, Wu S, Jiang H, Cao Y, Sun X, Yin X, Yan S, Shang M et al. : Tumor-associated macrophage-derived CCL20 enhances the growth and metastasis of pancreatic cancer. Acta Biochim Biophys Sin 2016. [DOI] [PubMed] [Google Scholar]
- 49.Miyake M, Hori S, Morizawa Y, Tatsumi Y, Nakai Y, Anai S, Torimoto K, Aoki K, Tanaka N, Shimada K et al. : CXCL1-mediated interaction of cancer cells with tumor-associated macrophages and cancer-associated fibroblasts promotes tumor progression in human bladder cancer. NEO 2016, 18:636–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qi L, Yu H, Zhang Y, Zhao D, Lv P, Zhong Y, Xu Y: IL-10 secreted by M2 macrophage promoted tumorigenesis through interaction with JAK2 in glioma. Oncotarget 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Cao Z, Zhang XM, Nakamura M, Sun M, Hartman J, Harris RA, Sun Y, Cao Y: Novel mechanism of macrophage-mediated metastasis revealed in a zebrafish model of tumor development. Cancer Res 2015, 75:306–315. [DOI] [PubMed] [Google Scholar]
- 52.Vassilev LT: In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303:844–848. [DOI] [PubMed] [Google Scholar]
- 53.Panni RZ, Linehan DC, DeNardo DG: Targeting tumor-infiltrating macrophages to combat cancer. Immunotherapy 2013, 5:1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM: CD4+ T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 2009, 16:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samuel MS, Lopez JI, McGhee EJ, Croft DR, Strachan D, Timpson P, Munro J, Schröder E, Zhou J, Brunton VG et al. : Actomyosin-mediated cellular tension drives increased tissue stiffness and β-catenin activation to induce epidermal hyperplasia and tumor growth. Cancer Cell 2011, 19:776–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lujambio A, Akkari L, Simon J, Grace D, Tschaharganeh DF, Bolden JE, Zhao Z, Thapar V, Joyce JA, Krizhanovsky V et al. : Non-cell-autonomous tumor suppression by p53. Cell 2013, 153:449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.•. Afik R, Zigmond E, Vugman M, Klepfish M, Shimshoni E, Pasmanik-Chor M, Shenoy A, Bassat E, Halpern Z, Geiger T et al. : Tumor macrophages are pivotal constructors of tumor collagenous matrix. J Exp Med 2016, 213:2315–2331.27697834 Highlights the effect of TAMs on reorganizing the extracellular matrix, a role more often associated with CAFs.
- 58.Marusyk A, Tabassum D, Janiszewska M, Place A, Trinh A, Rozhok AI, Pyne S, Guerriero JL, Shu S, Ekram M et al. : Spatial proximity to fibroblasts impacts molecular features and therapeutic sensitivity of breast cancer cells influencing clinical outcomes. Cancer Res 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin CH, Pelissier FA, Zhang H, Lakins J, Weaver VM, Park C, LaBarge MA: Microenvironment rigidity modulates responses to the HER2 receptor tyrosine kinase inhibitor lapatinib via YAP and TAZ transcription factors. Mol Biol Cell 2015, 26:3946–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang M, Kern AM, Hulskotter M, Greninger P, Singh A, Pan Y, Chowdhury D, Krause M, Baumann M, Benes CH et al. : EGFR-mediated chromatin condensation protects KRAS-mutant cancer cells against ionizing radiation. Cancer Res 2014, 74:2825–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J et al. : Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 2012, 487:505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eder T, Weber A, Neuwirt H, Grünbacher G, Ploner C, Klocker H, Sampson N, Eder I: Cancer-associated fibroblasts modify the response of prostate cancer cells to androgen and anti-androgens in three-dimensional spheroid culture. IJMS 2016, 17:1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xing F, Okuda H, Watabe M, Kobayashi A, Pai SK, Liu W, Pandey PR, Fukuda K, Hirota S, Sugai T et al. : Hypoxia-induced Jagged2 promotes breast cancer metastasis and self-renewal of cancer stem-like cells. Oncogene 2011, 30:4075–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joyce JA: Therapeutic targeting of the tumor microenvironment. Cancer Cell 2005, 7:513–520. [DOI] [PubMed] [Google Scholar]
- 65.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT et al. : Tumour micro-environment elicits innate resistanceto RAF inhibitors through HGF secretion. Nature 2012, 487:500–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takai K, Le A, Weaver VM, Werb Z: Targeting the cancer-associated fibroblasts as a treatment in triple-negative breast cancer. Oncotarget 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li M, Li M, Yin T, Shi H, Wen Y, Zhang B, Chen M, Xu G, Ren K, Wei Y: Targeting of cancer-associated fibroblasts enhances the efficacy of cancer chemotherapy by regulating the tumor microenvironment. Mol Med Rep 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Janes KA: Single-cell states versus single-cell atlases—two classes of heterogeneity that differ in meaning and method. Curr Opin Biotechnol 2016, 39:120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parmar H: Epithelial–stromal interactions in the mouse and human mammary gland in vivo. Endocr Relat Cancer 2004, 11:437–458. [DOI] [PubMed] [Google Scholar]
- 70.Simian M, Bissell MJ: Organoids: a historical perspective of thinking in three dimensions. J Cell Biol 2017, 216:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Skardal A, Shupe T, Atala A: Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov Today 2016, 21:1399–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lau KS, Cortez-Retamozo V, Philips SR, Pittet MJ, Lauffenburger DA, Haigis KM: Multi-scale in vivo systems analysis reveals the influence of immune cells on TNF-α-induced apoptosis in the intestinal epithelium. PLoS Biol 2012, 10:e1001393–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H: Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469:415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valenta T, Degirmenci B, Moor AE, Herr P, Zimmerli D, Moor MB, Hausmann G, Cantù C, Aguet M, Basler K: Wnt ligands secreted by subepithelial mesenchymal cells are essential for the survival of intestinal stem cells and gut homeostasis. Cell Rep 2016, 15:911–918. [DOI] [PubMed] [Google Scholar]
- 75.Dobrolecki LE, Airhart SD, Alferez DG, Aparicio S, Behbod F, Bentires-Alj M, Brisken C, Bult CJ, Cai S, Clarke RB et al. : Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]