Abstract
MRI allows accurate and reliable organ delineation for many disease sites in radiation therapy because MRI is able to offer superb soft-tissue contrast. Manual organ-at-risk (OAR) delineation is labor-intensive and time-consuming. This study aims to develop a deep learning-based automated multi-organ segmentation method to release the labor and accelerate the treatment planning process for head-and-neck (HN) cancer radiotherapy. A novel regional convolutional neural network (R-CNN) architecture, namely, mask scoring R-CNN has been developed in this study. In the proposed model, a deep attention feature pyramid network is used as backbone to extract the coarse features given MRI, followed by the feature refinement using R-CNN. The final segmentation is obtained through mask and mask scoring networks taking those refined feature maps as input. With the mask scoring mechanism incorporated into conventional mask supervision, the classification error can be highly minimized in conventional mask R-CNN architecture. A cohort of 60 HN cancer patients receiving external beam radiation therapy was used for experimental validation. Five-fold cross validation was performed for the assessment of our proposed method. The Dice similarity coefficients of brain stem, left/right cochlea, left/right eye, larynx, left/right lens, mandible, optic chiasm, left/right optic nerve, oral cavity, left/right parotid, pharynx, and spinal cord are 0.89±0.06, 0.68±0.14/0.68±0.18, 0.89±0.07/0.89±0.05, 0.90±0.07, 0.67±0.18/0.67±0.10, 0.82±0.10, 0.61±0.14, 0.67±0.11/0.68±0.11, 0.92±0.07, 0.85±0.06/0.86±0.05, 0.80±0.13, and 0.77±0.15, respectively. After the model training, all OARs can be segmented within 1 minute.
Keywords: Image segmentation, magnetic resonance imaging, MRI, radiation therapy, deep learning, mask R-CNN
1. INTRODUCTION
Radiation therapy is a common treatment approach for head and neck (HN) cancer which occur in the regions including oral cavity, pharynx, larynx, salivary glands, paranasal sinuses, and nasal cavity with an approximate rate of 4% among all cancers in the United States.(Ratko et al., 2014; Siegel et al., 2020) In general, HN tumors are very close to organs at risk (OARs), therefore, steep-dose gradients are expected in modern radiotherapy modalities(Webb, 2015; Otto, 2008; Lomax et al., 2001). In radiation therapy treatment planning, dose distribution is optimized and evaluated using the spatial information of target and OAR delineation, thus, the accuracy of target and OAR delineation can directly affect the quality of treatment plan.(Aliotta et al., 2019; Dai et al., 2021b) Current radiation therapy uses CT images for treatment planning. Attribute to its superb soft-tissue contrast, MRI becomes more and more common in radiation therapy as an extra imaging modality facilitating target and OAR delineation.(Khoo and Joon, 2006; Chandarana et al., 2018) For instance, the complexity of head and neck anatomy brings difficulties to accurately contour target and OARs using only CT images, while the utility of MRI can improve the accuracy of both tumor and OAR delineations.(Metcalfe et al., 2013; Prestwich et al., 2012; Chang et al., 2005)
Manual contouring is current clinical practice in the radiotherapy treatment planning workflow.(Jonsson et al., 2019) However, it is rather time-consuming especially for HN cases where the anatomy is complex and many surrounding OARs have to be contoured.(Vorwerk et al., 2014) Meanwhile, manual delineation highly relies on the physician’s knowledge and expertise, therefore, a high intra- and inter-observer variability is often observed.(Brouwer et al., 2012; Vinod et al., 2016) Automated approaches for contouring OARs are highly desired, yet existing automatic delineation methods are still limited.(van Dijk et al., 2020) At present, two main types of approaches have been investigated for organ segmentation in MRI. The first type is called atlas-based method, in which, organ segmentations are obtained through registering the input images to the atlas images that have been annotated and validated by experts.(Bondiau et al., 2005; Wardman et al., 2016; Bevilacqua et al., 2010) Atlas-based methods are in general repeatable and reliable, yet the trade-off between the robustness and accuracy has to be carefully made in order to achieve clinical applicable results.(Bondiau et al., 2005) The atlas suffers from lack of case-by-case variability which not only comes from natural anatomical differences but also from pathological factors, which may deteriorate the perform of atlas-based methods.(Mlynarski et al., 2019) By adding more atlases in the database could improve the performance but still limited due to the impossibility of building a database containing every variability.(Van de Velde et al., 2016; Peressutti et al., 2016) Moreover, atlas-based methods often underperform for these organs with small volumes.(Teguh et al., 2011) Finally, atlas-based methods rely on the registration between different anatomies where the accuracy of deformation can highly affect the delineation accuracy.(Zhong et al., 2010) The second type of automatic segmentation methods based on machine learning, especially, deep learning algorithms have been investigated recently and achieved encouraging initial results.(Tong et al., 2019; Sharp et al., 2014; Yang et al., 2014; Chen et al., 2018; Sahiner et al., 2019; Mlynarski et al., 2019) Both traditional machine learning methods such as random forests,(Damopoulos et al., 2019; Dong et al., 2019) support vector machine,(Yang et al., 2014) and modern convolutional neural networks and their variations(Chen et al., 2018; Mlynarski et al., 2019; Tong et al., 2019; Dai et al., 2020) obtain segmentations by extracting abstract and complex features from images, which show better adaptation to the natural diversity of subjects than atlas-based approaches.(Meyer et al., 2018) Compared to traditional machine learning methods that rely on handcraft features, deep learning-based method can learn deep features which can be directly used to represent the learning-target, emerging as a promising method for improving the performance of automated organ contouring.(Sahiner et al., 2019; van Dijk et al., 2020; Meyer et al., 2018) While encouraging, at present, to our knowledge, few investigations have been carried out on simultaneously segmenting multiple organs in complex structures on MRI images such as head and neck cases.(Mlynarski et al., 2019)
Mask R-CNN has been recently proposed as a new generation of image segmentation method from computer vision community.(He et al., 2017a) Compared to traditional image segmentation approaches using U-net and its variations(Ronneberger et al., 2015), mask R-CNN has been developed for instance segmentation(Chiao et al., 2019), where, the classification, localization and labeling of individual object are done simultaneously. The output of U-net is multi-channel equal sized binary masks which each channel represents one segmentation. While R-CNN does not perform segmentation directly, the R-CNN is used to predict the position of each organ via bounding boxes. The output of R-CNN is two aspects, one is a vector representing the bounding box index of organ, the other one is a vector indicating the class of the organ within the bounding box. Attribute to its superior performance, mask R-CNN has been investigated for medical image segmentation, and initial inspiring results have been achieved in either tumor(Zhang et al., 2019; Chiao et al., 2019; Lei et al., 2020c) or organ(Liu et al., 2018; Lei et al., 2020e) segmentation for CT and MRI images, and so forth. Mask R-CNN introduces a simple and flexible way to improve the accuracy of regions-of-interest (ROIs) localizations and reduce the dependency on localization accuracy.(He et al., 2017b) In mask R-CNN, the score of the instance mask is predicted by a classifier applied on the proposal feature, where the quality of mask is measured using classification confidence. The classification confidence and mask quality are not always correlated. For example, high mask score but poor mask quality may exist. Therefore, when classifying multiple organs at once, mask R-CNN is prone to artifacts because mask quality is not always well correlated with label accuracy.(Lei et al., 2020d; Lei et al., 2020a; Lei et al., 2020b; Harms et al., 2021)
In this study, we implemented a regional convolutional neural network (R-CNN) architecture, namely, mask scoring R-CNN, a variation of mask R-CNN which was introduced in computer vision area, for the task of multi-organ auto-delineation in HN MRI. In mask scoring R-CNN, a correlation between the mask quality and regional class is built through independent trainable mask and mask scoring networks. Compared to original mask scoring R-CNN architecture, here, a deep attention feature pyramid network is used as backbone to extract the coarse features from input MRI. The coarse feature maps are then refined using R-CNN. The final segmentation is obtained through mask and mask scoring networks taking those refined feature maps as input. With the mask scoring mechanism incorporated into conventional mask supervision, the classification error can be highly minimized in conventional mask R-CNN architecture. Manual contours from MRI were used as ground truth contours to assess the performance of the proposed method. The proposed method aims to offer a solution for fast and accurate OAR contouring in HN radiation therapy.
2. MATERIALS AND METHODS
2.A. Method overview
The workflow is schematically shown in Fig. 1. The model consists of five major steps. In the first step, a backbone was used to extract coarse feature map from the MRI image patch, followed by a regional proposal network (RPN) to compute those region-of-interest (ROI) candidates. In the second step, the coarse feature map was cropped by those ROI candidates (ROI alignment), followed by informative highlighting via attention gate (AG). In the third step, the refined feature map was obtained using a regional convolutional neural network (R-CNN), meanwhile, ROI for each organ was also extracted. In this fourth step, mask network was applied on the refined feature map cropped by the ROI for each organ to compute the initial segmentations. In the last step, the final segmentation was obtained through weighted averaging fusing those initial segmentations according to the mask scores offered by mask scoring network. By using paired MRI and contour (ground truth), the model was trained under the supervision of four types of loss functions, ROI loss for backbone and RPN, ROI loss for R-CNN, segmentation loss, and mask scoring loss. Once the optimal parameters of the model obtained through training, OAR contours were derived given MRI within a minute. The detail of each component will be discussed in the following sections.
FIG. 1.
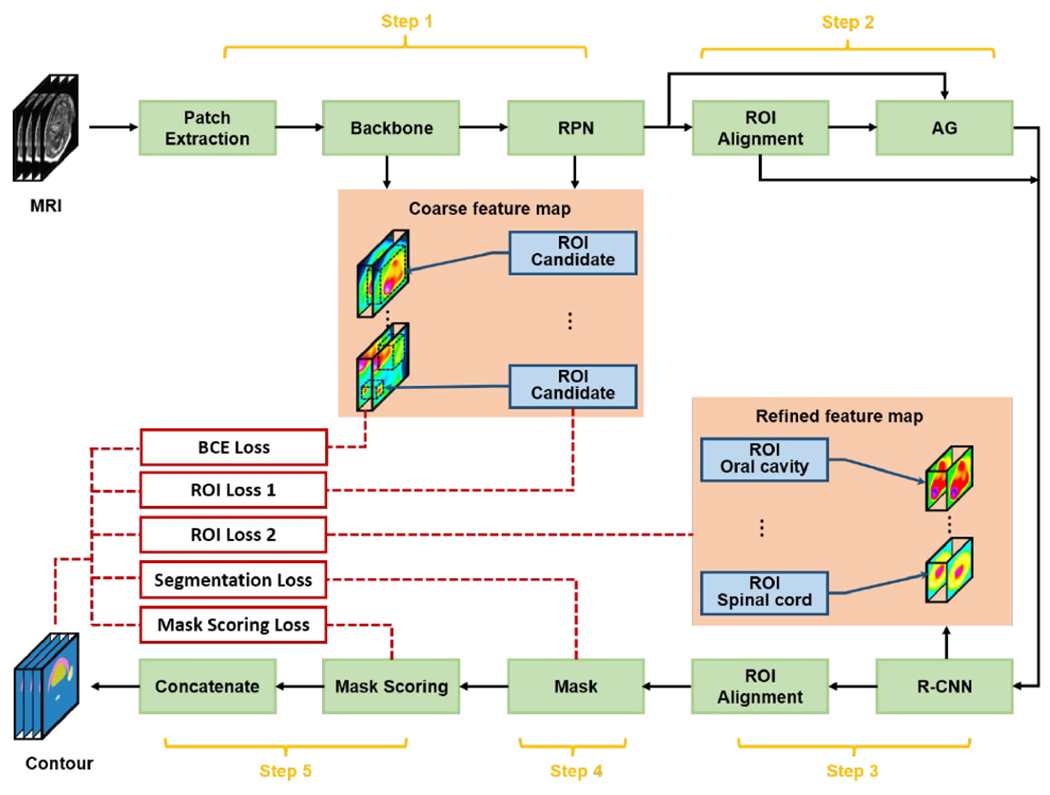
The schematic flow diagram of the proposed method. RPN, regional proposal network; ROI, region-of-interest; AG, attention gate; R-CNN, regional convolutional neural network; BCE, binary cross entropy.
2.B. Backbone
Feature pyramid network (FPN)(Lin et al., 2017) like architecture was used as backbone as shown in Fig. 2. FPN has been proposed to collect pyramid representations that are fundamental components in recognition system at different scales to improve the performance of deep learning-based algorithms for object detection. The FPN in this study consists of two primary pathways, encoding path and decoding path. The encoding pathway was constructed by a series of convolutional operations that computes hierarchical feature maps at several scales, while the decoding pathway was composed by deconvolutions. In the decoding path, higher pyramid levels are spatially coarser but semantically stronger. Those features in the decoding pathway are enhanced by the lateral connections which combine the feature maps from the encoding and the decoding pathways that have the same spatial resolutions. To supervise the backbone, the binary cross entropy (BCE) which was introduced in our previous work (Lei et al., 2019) is used as the loss function. In this work, the loss of backbone network is calculated between the ground truth and predicted contours from the output of the last layer in the backbone network.
FIG. 2.
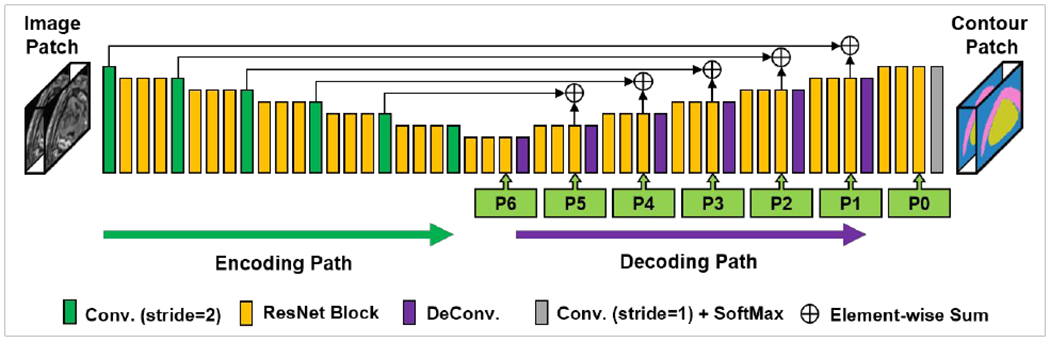
The network architecture of backbone. Conv., convolution; DeConv., deconvolution; P0-P6, pyramidal feature in hierarchic resolution levels.
2.C. RPN and R-CNN
As shown in Fig. 3, in our model, RPN (Fig. 3(a)) and R-CNN (Fig. 3(b)) are very similar. RPN was originally constructed in Faster R-CNN as a fully convolutional network which has the capability of simultaneously predicting object bounds and scores at each position(Ren et al., 2015). R-CNN was originally designed to improve the efficiency of object detection through applying convolutional networks on each ROI independently(Girshick et al., 2014). In our model, by taking the coarse feature map from the output of the backbone, RPN predicts rough locations for all the OARs. In particular, RPN estimates binary classes for all the ROIs and predicts their corresponding centers and boundaries. As shown in Fig. 3, RPN was constructed by two pathways, anchor classification and anchor regression, which have identical components including a series of convolutional operations and a fully connected layer. The final location and class of each ROI candidate was then predicted through a proposal layer combining the output from anchor classification and regression subnetworks. Assuming RPN predicts nanchors ROI candidates, for each rough ROI, the binary class can be represented by Ci ∈ R2 · nanchors, and its center (xc,yc,zc) and bound (Δw,Δh,Δd) can be represented by Bi ∈ R6 · nanchors. To supervise RPN, the classification loss of Ci and regression loss of Bi are used, which is defined as Eq. (1) in the following section 2.E. Once the rough ROIs were predicted by RPN, coarse feature maps from the backbone were cropped within rough ROIs via Bi ∈ R6·nanchors, and re-scaled via ROI alignment to share a same size, which is uniform for feeding into following R-CNN head. As shown in Fig. 3, R-CNN has similar network architecture with RPN, but with different leamable parameters. Since in the first step, RPN identifies rough ROIs from the useless background regions, R-CNN is expected to further adjust the location of these rough ROIs and classify these ROIs. Therefore, the output of R-CNN can be recognized as fine ROIs. The similar loss as RPN was used to supervise R-CNN.
FIG. 3.
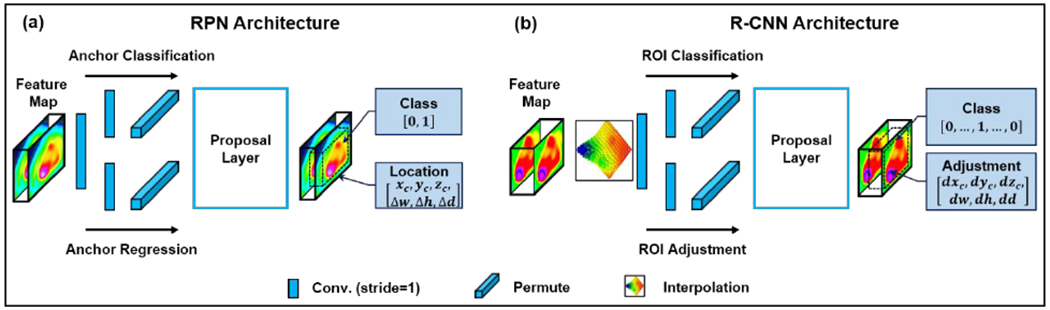
The network architecture of RPN (a) and R-CNN (b). Conv., convolution; ROI, region-of-interest.
2.D. Mask scoring and Mask
Mask head is used to segment binary mask of contours within fine ROIs. The mask head is implemented by several convolution layers as shown in Figure 4. The segmentation loss, which is a BCE loss, is used to supervise the mask head. This loss is introduced in Eq. (5) in the following section 2.E. The key point to improve the final segmentation accuracy in mask R-CNN is that the class of the predicted ROI should be correlated to the organ segmentation. However, the misclassification often happens in conventional mask R-CNN. To prevent this misclassification, in this work, we used a mask scoring head to build a correlation between the output of R-CNN and the output of mask head, and utilized the output of mask head to further guide the classification result of R-CNN. We define Smusk as the score of the segmented binary mask, where the ideal Smask is equal to the intersection over union (IoU) between the segmented binary mask and the corresponding ground truth mask. The ideal Smask should only take a positive value if the segmented mask matches the ground truth class and should be zero for all other classes since a mask can only belong to one class. This requires the mask scoring head to perform two tasks well: structure classification and mask regression. A single, two-component, objective function is used to train the network to perform both tasks, and this can be denoted as Smask = Scls · SIoU where Scls is the classification score, and SIoU is the regression (IoU) score. The goal of Scls is to classify each ROI type, i.e. how likely the current segmented structure is to be the structure of interest. Since the preceding R-CNN is designed solely for classification, each ROI has a pre-determined classification score, which can be directly fed to the mask scoring (MS) subnetwork. This direct link better enforces a correlation between mask quality and classification during training. SIoU is predicted by a fully connected layer which is supervised by the sigmoid cross entropy between the predicted SIoU and the ground truth SIoU. The output of mask score head is a vector whose length matches the number of classes. The loss to supervise mask score subnetwork is introduced in Eq. (6) in the following section 2.E.
FIG. 4.
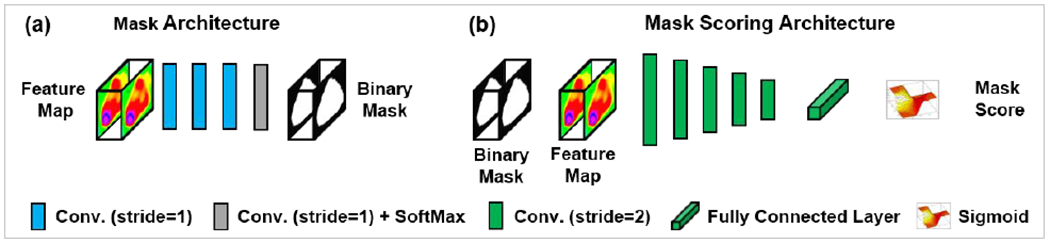
The network architecture of mask (a) and mask scoring (b). Conv., convolution.
As shown in Fig. 4(a), the mask network consists of three convolutional operation layers and a convolution plus softmax layer. The stride size in each convolutional operation was 1×1×1. Taking the refined feature map from R-CNN as an input, the mask network predicts the corresponding binary mask for each ROE Mask scoring (Fig. 4(b)) was constructed by five convolutional operation layers followed by a fully connected layer and a sigmoid layer. The mask score will be predicted by the mask scoring network taking both refined feature map and binary mask for each ROI as inputs. The mask score (output of the mask scoring network) was a vector that has an equal length to the number of ROI classes. Ideally, the mask score was expected to be the intersection over union (IoU) between predicted binary mask and its corresponding ground truth mask and to work well for classifying the mask to a proper category and regressing the score for a foreground object category. In reality, it is difficult to work out both tasks by using one single objective function. In this study, for simplicity, we separated these two tasks, mask classification and IoU regression. The mask scoring network was designed to be mainly focused on the IoU regression task. For mask classification, we directly took the results from R-CNN output.
2.E. Loss functions and model training
As shown in Fig. 1, the model was trained under the supervision of five types of loss functions. The first loss function is the BCE loss which was discussed in the above section 2.B. The second loss function can be indicated as ROI loss 1, which is a foreground/background loss LRPN can be defined as:
| (1) |
| (2) |
| (3) |
| (4) |
where, IoU is intersection over union, IROI denotes the ground truth ROI of an organ, is the predicted ROI, (xCi, yCi, zCi) and are respectively the central coordinate of ground truth and predicted ROIs, wi, hi, and di are the dimensions of ground truth ROI along x, y, and z directions, and , , are that for the predicted ROI.
The third type of loss function is ROI loss 2, which has identical definition as ROI loss 1 (Eqs. (1)–(4)). The difference is that ROI loss 2 is applied for each organ instead of the virtual classes in ROI loss 1.
The fourth type of loss function is called segmentation loss. The segmentation loss was defined as a binary cross entropy:
| (5) |
where, and are respectively the ground truth and predicted binary mask (segmentation).
The last type of loss function is the mask scoring loss, which is an IoU loss. The mask scoring loss was utilized to supervise the prediction of mask score taking binary mask and feature map as inputs. The mask scoring loss can be expressed as:
| (6) |
where, IoU and respectively indicate the ground truth IoU and the predicted IoU.
2.F. Image data acquisition, implementation and experimental setting
A retrospective study was conducted on a cohort of 60 HN cancer patients receiving radiation therapy. These patients were all treated in our department. Each dataset includes a set MRI and a set of multi-organ contours. The MRIs were acquired by a Siemens Aera 1.5T scanner as T1-weigted maps. For validation of our proposed method, MRI and contour images were re-sampled to a uniformed voxel size of 0.9766 mm × 0.9766 mm × 1.0 mm using commercial medical image processing software approved for clinical use (Velocity AI 3.2.1, Varian Medical Systems, Palo Alto, CA). The manual contours were firstly made by one junior radiation oncologist followed by the verification and editing by another senior radiation oncologist in our clinic, which were used as the ground truth in the study. Our proposed method was implemented using python 3.6.8 and Tensorflow 1.8.0. Besides, toolboxes including Pydicom, NumPy, and Batchgenerator were also respectively used for reading/writing DICOM files, random number generation and linear algebra, and data augmentation. The model was trained applying Adam optimizer with a learning rate of 2e-4 for 400 epochs. And the batch size was set to be 20. For all the experiments, five-fold cros-svalidation was performed.
Moreover, to demonstrate the advantages of the proposed mask scoring mechanism, we conducted all the experiments on the proposed, the conventional mask R-CNN based, and U-net based methods using the same in-house collected dataset. Here, patch-based method was used in the mask R-CNN and proposed method. The patch size is 128×128×64. And the overlap between two neighbor patches is 112×112×48. The final segmentation was obtained by taking average of the segments in all the patches. And the bounding boxes were set to be 64×64×32, 32×32×16, and 16×16×8, respectively. The stride size for these bounding boxes were set to be 32×32×16, 16×16×8, and 8×8×4, respectively. While for U-net, the input image size was fixed to be 256×256×256 through centralized cropping and padding.
3. RESULTS
3.A. Overall performance
Figure 5 shows the results from one representative patient. Five transverse slices are respectively shown in rows from top to bottom. The leftmost two columns are respectively the original MRI and the ground truth images. The third, fourth, and fifth columns show the predicted segmentations by our proposed, mask R-CNN, and U-net based methods, respectively. Qualitatively investigating on those images, we can see, that, our proposed method achieved less discrepancies referring to the ground truth in majority of the organs including brain stem, optic chiasm, left/right cochlea, left/right eye, mandible, left/right lens, left/right optic nerve, oral cavity, left/right parotid, and spinal cord compared to the other two methods.
FIG. 5.
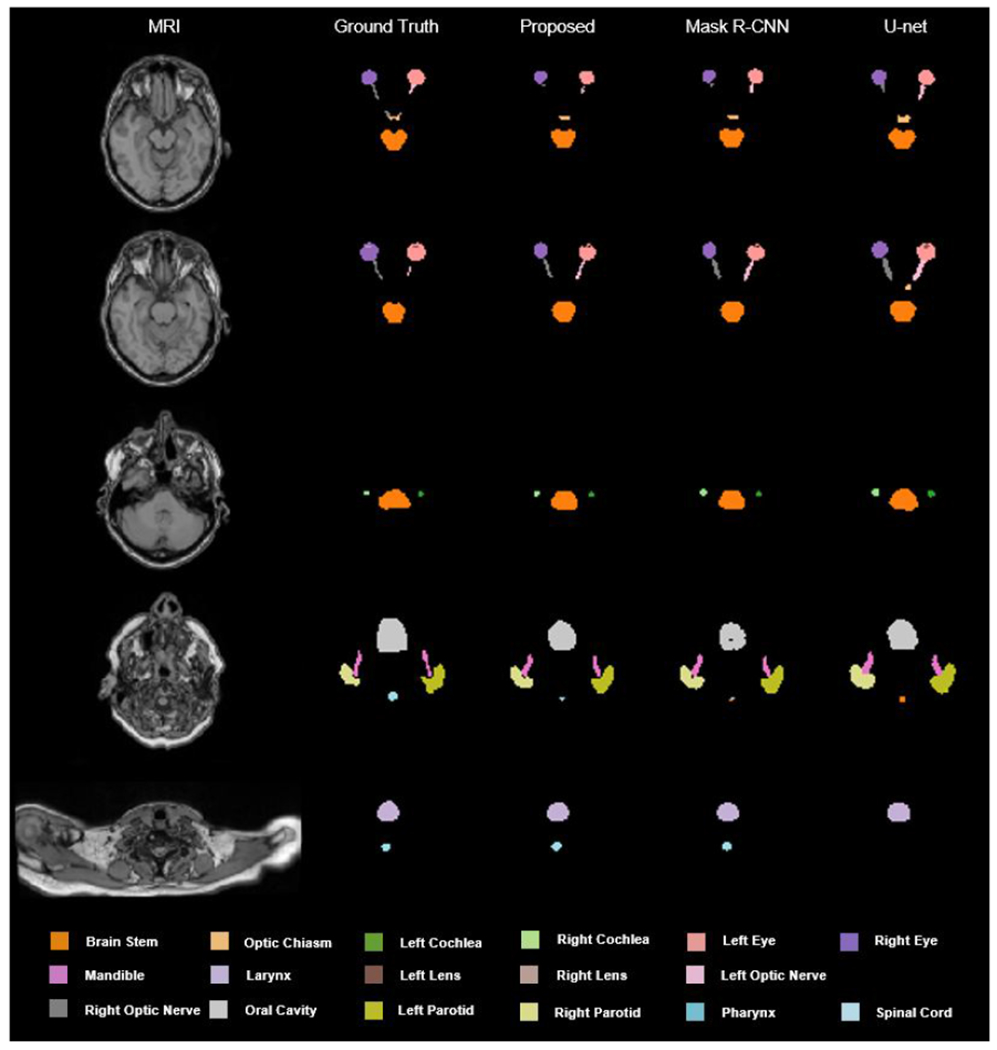
An illustrative example of the segmentation achieved by our proposed method and mask R-CNN based algorithm.
3.B. Quantitative results
To quantitatively demonstrate the performance of our proposed method, commonly used metrics (Dai et al., 2021a) including Dice similarity coefficient (DSC), 95th percentile Hausdorff distance (HD95), mean surface distance (MSD), and residual mean square distance (RMS) were calculated. The ablation studies of the proposed method were conducted by comparisons among the proposed method, the model without mask score subnetwork (Mask R-CNN method), the model without attention gate (No-AG method). The results were summarized in Table 1, where, the mean and standard deviation of these metrics are listed for each organ. Our proposed method achieved the mean DSC value of 0.78 over 17 organs, outperforming the mask R-CNN based method (mean DSC of 0.75) and No-AG method (mean DSC of 0.73). We also conducted the paired student t-test for all the metrics to analyze the statistical significance of the improvements by our proposed method. The results were summarized in Table 2.
TABLE 1.
Overall quantitative results achieved by the proposed, the mask R-CNN, and the No-AG methods.
| Organ | Method | DSC | HD95 (mm) | MSD (mm) | RMS (mm) |
|---|---|---|---|---|---|
| Brain Stem | Proposed | 0.89±0.06 | 2.91±1.35 | 0.98±0.57 | 1.43±0.70 |
| Mask R-CNN | 0.88±0.06 | 3.07±1.36 | 1.11±0.57 | 1.54±0.70 | |
| No-AG | 0.86±0.06 | 3.91±1.45 | 1.37±0.58 | 1.88±0.75 | |
| Left Cochlea | Proposed | 0.68±0.14 | 1.80±0.76 | 0.68±0.39 | 0.96±0.41 |
| Mask R-CNN | 0.66±0.14 | 1.88±0.79 | 0.73±0.37 | 1.00±0.40 | |
| No-AG | 0.64±0.14 | 2.66±1.10 | 1.01±0.44 | 1.34±0.49 | |
| Right Cochlea | Proposed | 0.68±0.18 | 1.91±0.85 | 0.71±0.41 | 1.02±0.47 |
| Mask R-CNN | 0.63±0.17 | 2.14±0.79 | 0.83±0.36 | 1.13±0.41 | |
| No-AG | 0.61±0.17 | 2.92±0.96 | 1.05±0.39 | 1.46±0.53 | |
| Left Eye | Proposed | 0.89±0.07 | 2.08±1.06 | 0.79±0.44 | 1.10±0.52 |
| Mask R-CNN | 0.88±0.06 | 2.13±0.99 | 0.86±0.41 | 1.15±0.49 | |
| No-AG | 0.86±0.06 | 3.00±1.16 | 1.06±0.44 | 1.45±0.58 | |
| Right Eye | Proposed | 0.89±0.05 | 2.04±0.74 | 0.80±0.30 | 1.10±0.36 |
| Mask R-CNN | 0.88±0.05 | 2.00±0.69 | 0.83±0.30 | 1.11±0.34 | |
| No-AG | 0.86±0.05 | 2.95±1.06 | 1.08±0.34 | 1.53±0.46 | |
| Larynx | Proposed | 0.90±0.07 | 3.73±1.84 | 1.13±0.71 | 1.72±0.91 |
| Mask R-CNN | 0.88±0.07 | 3.99±1.88 | 1.34±0.73 | 1.92±0.92 | |
| No-AG | 0.86±0.07 | 4.76±1.99 | 1.56±0.73 | 2.26±0.99 | |
| Left Lens | Proposed | 0.67±0.18 | 1.75±0.83 | 0.68±0.46 | 0.96±0.48 |
| Mask R-CNN | 0.65±0.16 | 1.84±0.76 | 0.74±0.39 | 1.00±0.42 | |
| No-AG | 0.63±0.16 | 2.52±1.02 | 0.98±0.41 | 1.40±0.51 | |
| Right Lens | Proposed | 0.67±0.10 | 1.70±0.46 | 0.62±0.18 | 0.90±0.19 |
| Mask R-CNN | 0.66±0.10 | 1.77±0.47 | 0.67±0.19 | 0.94±0.20 | |
| No-AG | 0.64±0.10 | 2.50±0.80 | 0.80±0.22 | 1.22±0.28 | |
| Mandible | Proposed | 0.82±0.10 | 3.62±2.26 | 1.08±0.60 | 1.84±1.02 |
| Mask R-CNN | 0.79±0.11 | 4.02±2.48 | 1.30±0.67 | 2.05±1.09 | |
| No-AG | 0.77±0.11 | 4.93±2.53 | 1.50±0.67 | 2.39±1.21 | |
| Optic Chiasm | Proposed | 0.61±0.14 | 3.94±2.03 | 1.06±0.63 | 1.72±0.89 |
| Mask R-CNN | 0.56±0.15 | 4.21±2.13 | 1.21±0.68 | 1.87±0.94 | |
| No-AG | 0.54±0.15 | 4.96±2.23 | 1.47±0.66 | 2.24±1.00 | |
| Left Optic Nerve | Proposed | 0.67±0.11 | 3.25±2.11 | 0.91±0.38 | 1.51±0.69 |
| Mask R-CNN | 0.62±0.10 | 3.37±1.83 | 1.04±0.35 | 1.57±0.61 | |
| No-AG | 0.60±0.10 | 4.14±1.87 | 1.28±0.41 | 1.96±0.65 | |
| Right Optic Nerve | Proposed | 0.68±0.11 | 2.96±1.32 | 0.87±0.29 | 1.50±0.50 |
| Mask R-CNN | 0.61±0.10 | 2.97±1.10 | 1.02±0.27 | 1.56±0.45 | |
| No-AG | 0.61±0.09 | 4.14±3.43 | 1.30±0.48 | 1.96±0.99 | |
| Oral Cavity | Proposed | 0.92±0.07 | 4.56±3.89 | 1.40±1.45 | 2.14±1.94 |
| Mask R-CNN | 0.90±0.07 | 5.10±3.90 | 1.74±1.49 | 2.47±1.98 | |
| No-AG | 0.88±0.07 | 5.87±3.95 | 2.03±1.47 | 2.77±2.02 | |
| Left Parotid | Proposed | 0.85±0.06 | 3.75±1.15 | 1.35±0.50 | 1.88±0.62 |
| Mask R-CNN | 0.83±0.06 | 3.90±1.08 | 1.53±0.47 | 2.00±0.57 | |
| No-AG | 0.81±0.06 | 4.70±1.24 | 1.75±0.50 | 2.31±0.62 | |
| Right Parotid | Proposed | 0.86±0.05 | 3.86±1.42 | 1.35±0.52 | 2.12±1.43 |
| Mask R-CNN | 0.83±0.05 | 4.10±1.41 | 1.58±0.51 | 2.26±1.36 | |
| No-AG | 0.81±0.05 | 4.86±1.55 | 1.79±0.53 | 2.60±1.43 | |
| Pharynx | Proposed | 0.80±0.13 | 3.16±2.52 | 0.91±0.87 | 1.54±1.23 |
| Mask R-CNN | 0.76±0.13 | 3.36±2.56 | 1.05±0.89 | 1.66±1.24 | |
| No-AG | 0.74±0.13 | 4.41±2.45 | 1.24±0.90 | 1.90±1.29 | |
| Spinal Cord | Proposed | 0.77±0.15 | 8.18±8.27 | 1.78±1.39 | 3.37±2.92 |
| Mask R-CNN | 0.73±0.15 | 8.41±8.11 | 1.97±1.39 | 3.51±2.86 | |
| No-AG | 0.72±0.15 | 9.13±8.08 | 2.18±1.40 | 3.74±2.89 |
TABLE 2.
Statistical comparisons between the proposed method and conventional mask R-CNN, the proposed and No-AG, respectively.
| Organ | Method | DSC | HD95 (mm) | MSD (mm) | RMS (mm) |
|---|---|---|---|---|---|
| Brain Stem | Proposed vs. Mask R-CNN | <0.01 | <0.01 | <0.01 | <0.01 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 | |
| Left Cochlea | Proposed vs. Mask R-CNN | <0.01 | 0.04 | <0.01 | 0.01 |
| Proposed vs. No-AG | 0.17 | <0.01 | <0.01 | <0.01 | |
| Right Cochlea | Proposed vs. Mask R-CNN | <0.01 | 0.01 | <0.01 | <0.01 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 | |
| Left Eye | Proposed vs. Mask R-CNN | <0.01 | 0.25 | <0.01 | <0.01 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 | |
| Right Eye | Proposed vs. Mask R-CNN | <0.01 | 0.24 | <0.01 | <0.01 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 | |
| Larynx | Proposed vs. Mask R-CNN | <0.01 | <0.01 | <0.01 | <0.01 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 | |
| Left Lens | Proposed vs. Mask R-CNN | <0.01 | 0.05 | <0.01 | 0.02 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 | |
| Right Lens | Proposed vs. Mask R-CNN | <0.01 | 0.06 | <0.01 | 0.02 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 | |
| Mandible | Proposed vs. Mask R-CNN | <0.01 | <0.01 | <0.01 | <0.01 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 | |
| Optic Chiasm | Proposed vs. Mask R-CNN | <0.01 | <0.01 | <0.01 | <0.01 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 | |
| Left Optic Nerve | Proposed vs. Mask R-CNN | <0.01 | 0.25 | <0.01 | 0.04 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 | |
| Right Optic Nerve | Proposed vs. Mask R-CNN | <0.01 | 0.96 | <0.01 | 0.05 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | 0.02 | |
| Oral Cavity | Proposed vs. Mask R-CNN | <0.01 | <0.01 | <0.01 | <0.01 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 | |
| Left Parotid | Proposed vs. Mask R-CNN | <0.01 | <0.01 | <0.01 | <0.01 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 | |
| Right Parotid | Proposed vs. Mask R-CNN | <0.01 | <0.01 | <0.01 | <0.01 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 | |
| Pharynx | Proposed vs. Mask R-CNN | <0.01 | <0.01 | <0.01 | <0.01 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 | |
| Spinal Cord | Proposed vs. Mask R-CNN | <0.01 | <0.01 | <0.01 | <0.01 |
| Proposed vs. No-AG | <0.01 | <0.01 | <0.01 | <0.01 |
To further demonstrate our proposed method, we implemented several advanced deep learning-based multiorgan segmentation methods including nnU-Net(Isensee et al., 2021), AnatomyNet(Zhu et al., 2019), and U-Faster-RCNN(Lei et al., 2020e). Same validation experiments were conducted using our proposed method and those advanced algorithms. Table 3 shows the quantitative DSC values that were achieved for each organ-of-interest by these methods. We also did statistical analysis using the paired student t-test. As shown in Table 3, we can see that our proposed method achieves a statistically significant improvement (p < 0.05) for all the organs in term of DSC compared to those competing methods.
TABLE 3.
Dice similarity coefficient (DSC) comparison of the proposed, the nnU-Net, the AnatomyNet, and the U-Faster-RCNN methods.
| Organ | I. Proposed | II. nnU-Net | III. Anatomy Net | IV. U-Faster-RCNN | V. Original Mask R-CNN | p value (I vs. II) | p value (I vs. III) | p value (I vs. IV) | p value (I vs. V) |
|---|---|---|---|---|---|---|---|---|---|
| Brain Stem | 0.89±0.06 | 0.87±0.06 | 0.86±0.06 | 0.86±0.07 | 0.86±0.06 | <0.01 | <0.01 | <0.01 | <0.01 |
| Left Cochlea | 0.68±0.14 | 0.65±0.14 | 0.64±0.14 | 0.63±0.14 | 0.64±0.14 | <0.01 | <0.01 | <0.01 | <0.01 |
| Right Cochlea | 0.68±0.18 | 0.62±0.17 | 0.61±0.17 | 0.61±0.16 | 0.61±0.17 | <0.01 | <0.01 | <0.01 | <0.01 |
| Left Eye | 0.89±0.07 | 0.86±0.06 | 0.86±0.06 | 0.85±0.07 | 0.86±0.06 | <0.01 | <0.01 | <0.01 | <0.01 |
| Right Eye | 0.89±0.05 | 0.87±0.05 | 0.87±0.04 | 0.85±0.04 | 0.87±0.05 | <0.01 | <0.01 | <0.01 | <0.01 |
| Larynx | 0.90±0.07 | 0.86±0.07 | 0.86±0.07 | 0.85±0.07 | 0.85±0.07 | <0.01 | <0.01 | <0.01 | <0.01 |
| Left Lens | 0.67±0.18 | 0.64±0.16 | 0.63±0.16 | 0.63±0.16 | 0.63±0.16 | <0.01 | <0.01 | <0.01 | <0.01 |
| Right Lens | 0.67±0.10 | 0.65±0.10 | 0.64±0.10 | 0.64±0.10 | 0.64±0.10 | <0.01 | <0.01 | <0.01 | <0.01 |
| Mandible | 0.82±0.10 | 0.78±0.11 | 0.77±0.11 | 0.77±0.11 | 0.77±0.11 | <0.01 | <0.01 | <0.01 | <0.01 |
| Optic Chiasm | 0.61±0.14 | 0.55±0.15 | 0.54±0.15 | 0.53±0.15 | 0.54±0.15 | <0.01 | <0.01 | <0.01 | <0.01 |
| Left Optic Nerve | 0.67±0.11 | 0.60±0.10 | 0.60±0.10 | 0.59±0.10 | 0.60±0.10 | <0.01 | <0.01 | <0.01 | <0.01 |
| Right Optic Nerve | 0.68±0.11 | 0.61±0.11 | 0.61±0.10 | 0.61±0.10 | 0.58±0.12 | 0.02 | <0.01 | <0.01 | <0.01 |
| Oral Cavity | 0.92±0.07 | 0.89±0.07 | 0.88±0.08 | 0.87±0.08 | 0.88±0.08 | <0.01 | <0.01 | <0.01 | <0.01 |
| Left Parotid | 0.85±0.06 | 0.82±0.06 | 0.81±0.06 | 0.80±0.06 | 0.81±0.06 | <0.01 | <0.01 | <0.01 | <0.01 |
| Right Parotid | 0.86±0.05 | 0.82±0.05 | 0.81±0.05 | 0.81±0.05 | 0.81±0.05 | <0.01 | <0.01 | <0.01 | <0.01 |
| Pharynx | 0.80±0.13 | 0.75±0.13 | 0.74±0.13 | 0.73±0.13 | 0.74±0.13 | <0.01 | <0.01 | <0.01 | <0.01 |
| Spinal Cord | 0.77±0.15 | 0.72±0.15 | 0.72±0.15 | 0.71±0.15 | 0.71±0.15 | <0.01 | <0.01 | <0.01 | <0.01 |
4. DISCUSSION AND CONCLUSIONS
Organ-at-risk delineation is a necessary and fundamental step in radiation therapy treatment planning. The proposed method, namely mask scoring R-CNN, provides a solution for automated delineation that is expected to accelerate the contouring process and release the labor without deteriorating the quality of contours. By integrating mask R-CNN framework into the segmentation task, our method can avoid the effect of useless features that are learned from the background region of MRI. By integrating mask scoring subnetwork into mask R-CNN framework, our method can diminish the misclassification issue for this multi-OAR segmentation task. The experimental results (Fig. 5 and Table 1) show that our proposed method achieved better performance compared to its strong competitor mask R-CNN. Mask R-CNN has been demonstrated as an efficient method for instance segmentation.(He et al., 2017a) Compared to the widely studied medical image segmentation algorithm U-net(Ronneberger et al., 2015), mask R-CNN achieves promising results for tumor and multi-organ segmentation.(Liu et al., 2018; Chiao et al., 2019; Zhang et al., 2019; Lei et al., 2020e) Our experimental results (Fig. 5 and Table 1) also show that mask R-CNN achieves higher accuracy than U-net based method. Based on the fundamental idea in mask R-CNN, our proposed method aims to minimize the misalignment between the mask score and the quality of mask through a scoring mechanism. The quantitative results listed in Table 1 show that our proposed method slightly outperformed mask R-CNN. For example, the increases of the mean DSC are within the range of 0.01-0.07. However, the statistical results (Table 2) show that our proposed method has significant improvement over mask R-CNN based method in the majority of these organs-at-risk in HN patients. From the p-values listed in Table 2, we can see that our proposed method achieves a statistically significant improvement (p < 0.05) for all the organs in DSC, and with very few p values larger than 0.05 in other metrics. For these p values larger than 0.05 in other metrics, it is not safe to come to the conclusion that our proposed method achieves significant improvements in these organs such as left/right eye, due to the limitations of a single metric for assessing the segmentation performance (Taha and Hanbury, 2015). Especially, for organs including brain stem, left/right cochlea, larynx, mandible, optic chiasm, oral cavity, left/right parotid, pharynx, and spinal cord, our proposed method achieved significant (p < 0.05) improvement over conventional mask R-CNN method in all the four metrics. Even with the improvement by our proposed mask scoring R-CNN method, the DSC for some organs such as left/right cochlea, left/right lens, optic chiasm, and left/right optic nerve are relatively low. These organs are challenging not only due to their low contrast on CT images but also their tiny size. A discrepancy of only a few pixels can be relatively large for these structures, which may lead to a low DSC. While further investigations have to be carried out to improve the segmentation accuracy, another possible reason might be the variations of manual contours (ground truth) of these organs with very small volumes. Through the ablation studies (Tables 1 & 2), we can see that by adding the attention gate and mask score subnetworks, the proposed method achieved significant improvements. The advantage of using attention gate lies on suppressing irrelevant regions in an input image and highlighting salient features that are useful for segmenting a specific organ. While the advantage of using mask score subnetwork is that it can build a relationship between segmentation shape and the class of organ, which is useful for differentiating small organs and reducing the risk of misclassification.
One limitation of our study is that the manual contours were recognized as the ground truth to train and evaluate our proposed method. As discussed above, uncertainty always exists in the manual contours. In other words, the ground truth might not be the ‘real’ ground truth. However, the ground truth was obtained by two experienced radiation oncologists with consensus reached. Thus, the uncertainty was minimized in our study. Yet the uncertainty still exists, future studies have to be carried out to further minimize the uncertainty. One possible way for investigating this uncertainty is to use the standard phantom. This type of phantom study could be very useful for traditional segmentation method such as atlas-based method. However, for deep learning-based methods, the anatomical variations of the standard phantom are not enough to train a usable model. Several other possible solutions can be applied. The first pathway is to gather contours from the consensus of a large group of physicians under the same guideline to reduce the inter-variables. Another way is to build a large benchmark database to train the model. More inspiring, explicit uncertainty formulation incorporating into the model itself can be another way to tackle the challenge of uncertainty.(Kendall and Gal, 2017) Careful investigations are required to perform to verify these pathways, which can guide our future studies.
In summary, in this study, a deep learning-based head-and-neck MRI multi-organ segmentation method has been implemented and validated. The proposed method offers a strategic solution for automated OAR delineation which can be integrated into daily clinical workflow to release the labor from tedious manual contouring in radiation therapy treatment planning.
Supplementary Material
ACKNOWLEDGMENTS
This research is supported in part by the National Cancer Institute of the National Institutes of Health under Award Number R01CA215718 (XY), the Department of Defense (DoD) Prostate Cancer Research Program (PCRP) Award W81XWH-17-1-0438 (TL) and W81XWH-19-1-0567 (XD), and Emory Winship Cancer Institute pilot grant (XY).
Footnotes
DISCLOSURES
The authors declare no conflicts of interest.
REFERENCES
- Aliotta E, Nourzadeh H, Siebers J J P i M and Biology 2019. Quantifying the dosimetric impact of organ-at-risk delineation variability in head and neck radiation therapy in the context of patient setup uncertainty 64 135020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua V, Piazzolla A and Stofella P Year Atlas-based segmentation of organs at risk in radiotherapy in head mris by means of a novel active contour framework International Conference on Intelligent Computing,2010), vol. Series): Springer; ) pp 350–9 [Google Scholar]
- Bondiau P-Y, Malandain G, Chanalet S, Marcy P-Y, Habrand J-L, Fauchon F, Paquis P, Courdi A, Commowick O and Rutten IJ I J o R O B P 2005. Atlas-based automatic segmentation of MR images: validation study on the brainstem in radiotherapy context 61 289–98 [DOI] [PubMed] [Google Scholar]
- Brouwer CL, Steenbakkers RJ, van den Heuvel E, Duppen JC, Navran A, Bijl HP, Chouvalova O, Burlage FR, Meertens H and Langendijk JAJRO 2012. 3D Variation in delineation of head and neck organs at risk 7 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandarana H, Wang H, Tijssen R and Das I J J J o M R I 2018. Emerging role of MRI in radiation therapy 48 1468–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT-C, Lin C-Y, Chen T-M, Kang C-J, Ng S-H, Chen I-H, Wang H-M, Cheng A-J and Liao C-T J I J o R O B P 2005. Nasopharyngeal carcinoma with cranial nerve palsy: the importance of MRI for radiotherapy 63 1354–60 [DOI] [PubMed] [Google Scholar]
- Chen L, Bentley P, Mori K, Misawa K, Fujiwara M and Rueckert D J I t o m i 2018. DRINet for medical image segmentation 37 2453–62 [DOI] [PubMed] [Google Scholar]
- Chiao J-Y, Chen K-Y, Liao KY-K, Hsieh P-H, Zhang G and Huang T-CJM 2019. Detection and classification the breast tumors using mask R-CNN on sonograms 98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Lei Y, Wang T, Dhabaan AH, McDonald M, Beitler JJ, Curran WJ, Zhou J, Liu T and Yang X 2021a. Head-and-neck organs-at-risk auto-delineation using dual pyramid networks for CBCT-guided adaptive radiotherapy Physics in Medicine Biology 66 045021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Lei Y, Wang T, Dhabaan AH, McDonald M, Beitler JJ, Curran WJ, Zhou J, Liu T, Yang X J P i M and Biology 2021b. Head-and-neck organs-at-risk auto-delineation using dual pyramid networks for CBCT-guided adaptive radiotherapy 66 045021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Lei Y, Zhang Y, Qiu RL, Wang T, Dresser SA, Curran WJ, Patel P, Liu T and Yang XJMP 2020. Automatic Multi - Catheter Detection using Deeply Supervised Convolutional Neural Network in MRI - guided HDR Prostate Brachytherapy [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damopoulos D, Lerch TD, Schmaranzer F, Tannast M, Chênes C, Zheng G, Schmid J J I j o c a r and surgery 2019. Segmentation of the proximal femur in radial MR scans using a random forest classifier and deformable model registration 14 545–61 [DOI] [PubMed] [Google Scholar]
- Dong X, Lei Y, Tian S, Liu Y, Wang T, Liu T, Curran WJ, Mao H, Shu H-K and Yang X J a p a 2019. Air, bone and soft-tissue Segmentation on 3D brain MRI Using Semantic Classification Random Forest with Auto-Context Model [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girshick R, Donahue J, Darrell T and Malik J Year Rich feature hierarchies for accurate object detection and semantic segmentation Proceedings of the IEEE conference on computer vision and pattern recognition,2014), vol. Series) pp 580–7 [Google Scholar]
- Harms J, Lei Y, Tian S, McCall NS, Higgins KA, Bradley JD, Curran WJ, Liu T and Yang XJMP 2021. Automatic delineation of cardiac substructures using a region - based fully convolutional network [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Gkioxari G, Dollár P and Girshick R Year Mask r-cnn Proceedings of the IEEE international conference on computer vision, 2017a), vol. Series) pp 2961–9 [Google Scholar]
- He K, Gkioxari G, Dollár P and Girshick RB 2017b. Mask R-CNN IEEE International Conference on Computer Vision 2980–8 [Google Scholar]
- Isensee F, Jaeger PF, Kohl SA, Petersen J and Maier-Hein K H J N m 2021. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation 18 203–11 [DOI] [PubMed] [Google Scholar]
- Jonsson J, Nyholm T, Söderkvist KJC and Oncology TR 2019. The rationale for MR-only treatment planning for external radiotherapy 18 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall A and Gal Y Year What uncertainties do we need in bayesian deep learning for computer vision? Adv. Neural Inf Process. Syst,2017), vol. Series) pp 5574–84 [Google Scholar]
- Khoo V and Joon D J T B j o r 2006. New developments in MRI for target volume delineation in radiotherapy 79 S2–S15 [DOI] [PubMed] [Google Scholar]
- Lei Y, Harms J, Dong X, Wang T, Tang X, Yu D, Beitler J, Curran W, Liu T and Yang X 2020a. Organ-at-Risk (OAR) segmentation in head and neck CT using U-RCNN vol 11314: SPIE; ) [Google Scholar]
- Lei Y, Tian S, He X, Wang T, Wang B, Patel P, Jani A, Mao H, Curran WJ, Liu T and Yang X 2019. Ultrasound prostate segmentation based on multidirectional deeply supervised V-Net Med Phys 46 3194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Tian Z, Kahn S, Curran W, Liu T and Yang X 2020b. Automatic detection of brain metastases using 3D mask R-CNNfor stereotactic radiosurgery vol 11314: SPIE; ) [Google Scholar]
- Lei Y, Tian Z, Kahn S, Curran WJ, Liu T and Yang X Year Automatic detection of brain metastases using 3D mask R-CNN for stereotactic radiosurgery Medical Imaging 2020: Computer-Aided Diagnosis, 2020c), vol. Series 11314): International Society for Optics and Photonics; ) p 113142X [Google Scholar]
- Lei Y, Zhou J, Dong X, Wang T, Mao H, McDonald M, Curran W, Liu T and Yang X 2020d. Multi-organ segmentation in head and neck MRI using U-Faster-RCNN vol 11313: SPIE; ) [Google Scholar]
- Lei Y, Zhou J, Dong X, Wang T, Mao H, McDonald M, Curran WJ, Liu T and Yang X Year Multi-organ segmentation in head and neck MRI using U-Faster-RCNN Medical Imaging 2020: Image Processing,2020e), vol. Series 11313): International Society for Optics and Photonics; ) p 113133A [Google Scholar]
- Lin T-Y, Dollár P, Girshick R, He K, Hariharan B and Belongie S Year Feature pyramid networks for object detection Proceedings of the IEEE conference on computer vision and pattern recognition,2017), vol. Series) pp 2117–25 [Google Scholar]
- Liu M, Dong J, Dong X, Yu H and Qi L Year Segmentation of lung nodule in CT images based on mask R-CNN 2018 9th International Conference on Awareness Science and Technology (iCAST),2018), vol. Series): IEEE; ) pp 1–6 [Google Scholar]
- Lomax AJ, Boehringer T, Coray A, Egger E, Goitein G, Grossmann M, Juelke P, Lin S, Pedroni E, Rohrer B, Roser W, Rossi B, Siegenthaler B, Stadelmann O, Stauble H, Vetter C and Wisser L 2001. Intensity modulated proton therapy: a clinical example Med. Phys 28 317–24 [DOI] [PubMed] [Google Scholar]
- Metcalfe P, Liney G, Holloway L, Walker A, Barton M, Delaney G, Vinod S, Tome WJT i c r and treatment 2013. The potential for an enhanced role for MRI in radiation-therapy treatment planning 12 429–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Noblet V, Mazzara C, Lallement AJC i b and medicine 2018. Survey on deep learning for radiotherapy 98 126–46 [DOI] [PubMed] [Google Scholar]
- Mlynarski P, Delingette H, Alghamdi H, Bondiau P-Y and Ayache NJapa 2019. Anatomically Consistent Segmentation of Organs at Risk in MRI with Convolutional Neural Networks [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto K 2008. Volumetric modulated arc therapy: IMRT in a single gantry arc Med. Phys 35 310–7 [DOI] [PubMed] [Google Scholar]
- Peressutti D, Schipaanboord B, van Soest J, Lustberg T, van Elmpt W, Kadir T, Dekker A and Gooding MJM p 2016. TU - AB - 202 - 10: How Effective Are Current Atlas Selection Methods for Atlas - Based Auto - Contouring in Radiotherapy Planning? 43 3738–9 [Google Scholar]
- Prestwich R, Sykes J, Carey B, Sen M, Dyker K and Scarsbrook AJCO 2012. Improving target definition for head and neck radiotherapy: a place for magnetic resonance imaging and 18-fluoride fluorodeoxyglucose positron emission tomography? 24 577–89 [DOI] [PubMed] [Google Scholar]
- Ratko TA, Douglas G, De Souza J A, Belinson SE and Aronson N 2014. Radiotherapy treatments for head and neck cancer update [PubMed] [Google Scholar]
- Ren S, He K, Girshick R and Sun J Year Faster r-enn: Towards real-time object detection with region proposal networks Adv Neural Inf. Process. Syst,2015), vol. Series) pp 91–9 [DOI] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P and Brox T Year U-net: Convolutional networks for biomedical image segmentation International Conference on Medical image computing and computer-assisted intervention, 2015), vol. Series): Springer; ) pp 234–41 [Google Scholar]
- Sahiner B, Pezeshk A, Hadjiiski LM, Wang X, Drukker K, Cha KH, Summers RM and Giger MLJMp2019. Deep learning in medical imaging and radiation therapy 46 e1–e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp G, Fritscher KD, Pekar V, Peroni M, Shusharina N, Veeraraghavan H and Yang J J M p 2014. Vision 20/20: perspectives on automated image segmentation for radiotherapy 41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD and Jemal A 2020. Cancer statistics, 2020 CA Cancer J. Clin 707–30 [DOI] [PubMed] [Google Scholar]
- Taha AA and Hanbury A J B m i 2015. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool 15 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teguh DN, Levendag PC, Voet PW, Al-Mamgani A, Han X, Wolf TK, Hibbard LS, Nowak P, Akhiat H and Dirkx M L J I J o R O B P 2011. Clinical validation of atlas-based auto-segmentation of multiple target volumes and normal tissue (swallowing/mastication) structures in the head and neck 81 950–7 [DOI] [PubMed] [Google Scholar]
- Tong N, Gou S, Yang S, Cao M and Sheng K 2019. Shape constrained fully convolutional DenseNet with adversarial training for multiorgan segmentation on head and neck CT and low - field MR images Med Phys. 46 2669–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde J, Wouters J, Vercauteren T, De Gersem W, Achten E, De Neve W and Van Hoof T J R o 2016. Optimal number of atlases and label fusion for automatic multi-atlas-based brachial plexus contouring in radiotherapy treatment planning 11 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk L V, Van den Bosch L, Aljabar P, Peressutti D, Both S, Steenbakkers RJ, Langendijk JA, Gooding MJ, Brouwer CL J R and Oncology 2020. Improving automatic delineation for head and neck organs at risk by Deep Learning Contouring 142 115–23 [DOI] [PubMed] [Google Scholar]
- Vinod SK, Jameson MG, Min M, Holloway LC J R and Oncology 2016. Uncertainties in volume delineation in radiation oncology: a systematic review and recommendations for future studies 121 169–79 [DOI] [PubMed] [Google Scholar]
- Vorwerk H, Zink K, Schiller R, Budach V, Böhmer D, Kampfer S, Popp W, Sack H and Engenhart-Cabillic R J S u O 2014. Protection of quality and innovation in radiation oncology: the prospective multicenter trial the German Society of Radiation Oncology (DEGRO-QUIRO study) 190 433–43 [DOI] [PubMed] [Google Scholar]
- Wardman K, Prestwich RJ, Gooding MJ and Speight R J J J o a c m p 2016. The feasibility of atlas - based automatic segmentation of MRI for H&N radiotherapy planning 17 146–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb S 2015. Intensity-modulated radiation therapy. CRC Press; ) [Google Scholar]
- Yang X, Wu N, Cheng G, Zhou Z, David SY, Beitler JJ, Curran WJ and Liu T J I J o R O B P 2014. Automated segmentation of the parotid gland based on atlas registration and machine learning: a longitudinal MRI study in head-and-neck radiation therapy 90 1225–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Cheng C, Zhao X and Li X J M i 2019. Multiscale Mask R-CNN–Based Lung Tumor Detection Using PET Imaging 18 1536012119863531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Kim J and Chetty I J J M p 2010. Analysis of deformable image registration accuracy using computational modeling 37 970–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Huang Y, Zeng L, Chen X, Liu Y, Qian Z, Du N, Fan W and Xie X J M p 2019. AnatomyNet: deep learning for fast and fully automated whole - volume segmentation of head and neck anatomy 46 576–89 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


