Abstract
Phenotypic integration is an important metric that describes the degree of covariation among traits in a population, and is hypothesized to arise due to selection for shared functional processes. Our ability to identify the genetic and/or developmental underpinnings of integration is marred by temporally overlapping cell-, tissue-, and structure-level processes that serve to continually ‘overwrite’ the structure of covariation among traits through ontogeny. Here we examine whether traits that are integrated at the phenotypic level, also exhibit a shared genetic basis (e.g., pleiotropy). We micro-CT scanned two hard tissue traits, and two soft tissue traits (mandible, pectoral girdle, atrium, and ventricle respectively) from an F5 hybrid population of Lake Malawi cichlids, and used geometric morphometrics to extract 3D shape information from each trait. Given the large degree of asymmetric variation that may reflect developmental instability, we separated symmetric- from asymmetric-components of shape variation. We then performed quantitative trait loci (QTL) analysis to determine the degree of genetic overlap between shapes. While we found ubiquitous associations among traits at the phenotypic level, except for a handful of notable exceptions, our QTL analysis revealed few overlapping genetic regions. Taken together, this indicates developmental interactions can play a large role in determining the degree of phenotypic integration among traits, and likely obfuscate the genotype to phenotype map, limiting our ability to gain a comprehensive picture of the genetic contributors responsible for phenotypic divergence.
Keywords: Heart, bone, phenotypic integration, development, geometric morphometrics
INTRODUCTION
Phenotypic integration (i.e., the degree of covariation among traits) is an important and ubiquitous feature of multicellular organisms that shapes functionality within and among structures. Integration can permit or limit the accumulation of morphological variation by partitioning traits into distinct subunits (i.e., modules) and then varying the degree of association among these subunits (Klingenberg 2008; Goswami et al. 2014). Recent research has demonstrated that differences among populations in the pattern and magnitude of phenotypic integration can have major consequences for taxonomic diversification, rates of morphological evolution, and the ability of a population to respond to selection (Smith et al. 2015; Hu et al. 2016; Conith et al. 2018a, 2020). However, in spite of being an important intrinsic feature of animals, the origins of integration are unclear. Indeed, the question of what actually determines the pattern and magnitude of trait covariation has become a central line of inquiry since Olson and Miller placed phenotypic integration into a statistical framework (Olson and Miller 1958). The difficultly in characterizing phenotypic integration stems from the overlapping influences of genetics, development, and the environment that together form the adult phenotype (Hallgrímsson et al. 2009; Jamniczky et al. 2010). Untangling these roles has proved difficult as each factor can temporally and spatially vary in their relative contributions to the phenotype.
Theory predicts that traits with similar functions should evolve shared genetic control (i.e., genetic integration), while traits with divergent functions should evolve independent genetic control (i.e., genetic parcellation). Such genetic organization would optimize the evolutionary potential of a complex anatomical system; however, the hierarchical natural of development makes it challenging to detect instances of genetic integration, or parcellation (Young and Hallgrímsson 2005; Hallgrimsson et al. 2019). It is often assumed that the connection between genotype and phenotypes is linear, and therefore covariation in phenotype should be reflected in the genotype (Figure 1a); however, it is becoming increasingly apparent that this is an oversimplification, and that in most instances genetic (co)variation will be masked by the interactions between gene products, including cells, tissues, and structures (Figure 1b–c). Indeed, while genome-wide association studies (GWAS) are commonly used to understand the association between genotype and phenotype, these techniques typically highlight only a handful of causative loci and explain only a relatively small proportion of the total phenotypic variation, likely due to the role of pleiotropy, epistasis, and development (Liu et al. 2012; Hallgrimsson et al. 2014; Adhikari et al. 2016). What we are measuring when we characterize phenotypic variation may better reflect developmental interactions, resulting in a link between genotype and phenotype that is more tenuous (Hallgrimsson et al. 2014), or at least biased such that interactions that occur above the level of the genome will distill any measureable genetic signal to those genes centered on regulating the most pertinent developmental interactions. Therefore, depending on the strength and level of interactions, the adult phenotype could differentially reflect genetic, cellular, tissue, or structural level processes.
Figure 1.
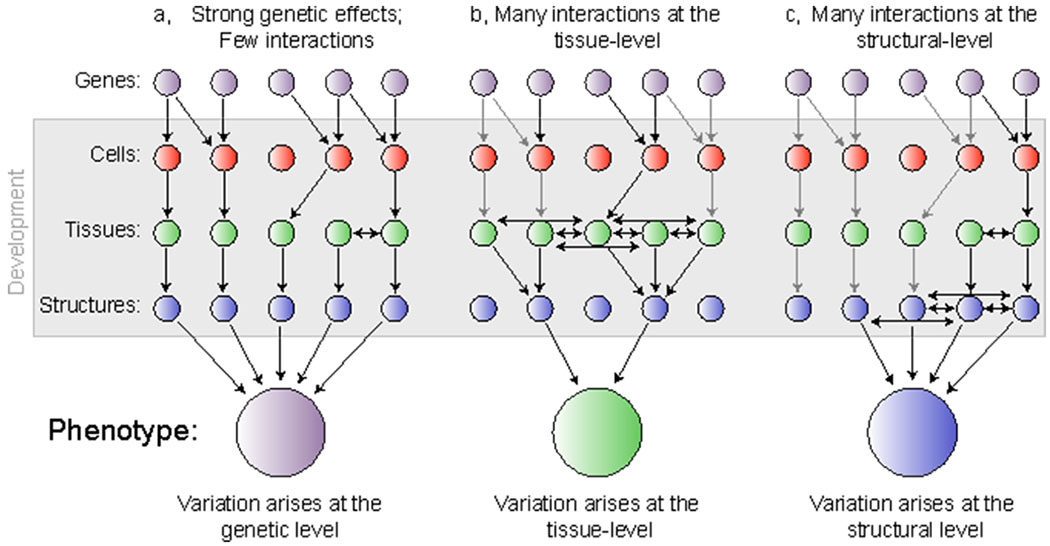
The phenotype is shape by a multitude of genetic and developmental processes. a, Few interactions among developmental processes impact the eventual phenotype leading to a clear picture of how genotype patterns phenotype. b, Developmental interactions occurring among tissues obscure our ability to fully trace the underlying genetic basis resulting in a phenotype that mostly reflects tissue-level processes. c, Developmental interactions occurring among structures can obscure our ability to understand both the genetic basis for a given trait, and the cellular and tissue level processes that may have impacted the shape of each structure. Gray arrows refer to interactions between levels that will go undetected due to the hierarchical nature of developmental processes.
Waddington was the first to present the idea that processes occurring above the level of the genome can impact the phenotype (Waddington 1942b, 1957). Among his key findings, Waddington demonstrated that certain developmental mechanisms existed to suppress phenotypic variation both within individuals (i.e., developmental stability) and between individuals (i.e., canalization), resulting in a more integrated phenotype across a population (Waddington 1942a, 1953). Like Waddington, we are defining development as processes that are occurring above the level of the genome, including but not limited to coordinated growth and kinematics that can occur at any point throughout ontogeny. Indeed, we are taking ‘development’ to refer to the totality of those processes, from patterning events that occur in the embryo to remodeling events that occur in the adult. While numerous traits that are functionally integrated arise from different embryonic sources, all should be subject to spatially and temporally interacting cell, tissue, and organ level developmental processes. More recently, Hallgrímsson et al. (2009) expanded upon these ideas by using the metaphor of a palimpsest, an ancient writing tablet that can be re-used many times but retains a faint imprint of what came before it, to describe how a structure such as the skull is subject to temporally overlapping developmental processes that leave an ‘imprint’ on the phenotype. The Hallgrímsson palimpsest model makes two key predictions that we use to frame our own questions: (1) interactions among cells, tissues, and/or organs occurring at different times in development are important in determining structural geometry across the skull, and (2) many of the interactions that contribute to adult shape variation operate above the level of the genome (Figure 1). Recent studies have demonstrated that craniofacial structures are subjected to an array of overlapping morphogenic fields through development, and soft and hard tissue are constantly remodeling in response to biomechanical, molecular, and cellular interactions (Jamniczky et al. 2010; Yang et al. 2010; Choe and Crump 2014; Conith et al. 2019a).
Here we took advantage of a genetic mapping population to disentangle the relative contributions of genetics and development on phenotypic integration. In particular, we assess phenotypic integration and developmental stability (i.e., fluctuating asymmetry) in the craniofacial region of Lake Malawi cichlid fishes to examine if or how these two features are reflected in the genotype-phenotype map. We characterize two bone complexes responsible for independent functional tasks: the mandible, responsible for food capture and processing, and the pectoral girdle, responsible for housing the musculature of the pharyngeal jaw and the pectoral fin. We compare these structures to two components of the teleost heart: the ventricle and atrium. We selected these four anatomical units as they conduct functionally disparate tasks (i.e., locomotion, respiration, and feeding), have diverse developmental origins (i.e., mandible = neural crest cell (NCC); pectoral girdle = mesoderm; heart = NCC and mesoderm), but are all located within the cranial region, and therefore should be subject to a suite of overlapping morphogenic fields that may impact their development, growth, and overall shape. Furthermore, given the divergence in eco-morphology between the two parental species used for this genetic cross (Parsons et al. 2016), this system represents an excellent opportunity to begin to parse the genetic and non-genetic factors that underlie phenotypic integration of structures likely under divergent selective pressures between taxa.
To assess the first prediction of the palimpsest model, that integration of structures is evident across the skull, we examine covariation among and between our hard and soft tissue traits (i.e., mandible, pectoral girdle, atrium, and ventricle), in our hybrid cichlids. While hybridization can affect the strength of integration among traits (Parsons et al. 2011b), hybridization is a common occurrence in wild populations, and gene flow is prominent among Lake Malawi cichlids (Malinsky et al. 2018). Thus, a hybrid genomic background is not an artificial state in this system. Further, using a recombinant hybrid population can prove useful in assessing the relative roles of genetics and development in trait integration, as it allows us to decouple these processes in a way that cannot be achieved in “natural” populations. We use 3D geometric morphometrics to characterize shape in these structures, statistically separating the asymmetric from symmetric components of landmark variation from all structures. We then conduct a suite of correlations among the structures using the symmetric and asymmetric configurations. By removing the asymmetric component of variation, the remaining symmetric component should reflect a shape that is mostly free from developmental noise (Klingenberg 2019). In structures that form within overlapping developmental fields, we may predict that these traits will exhibit a greater degree of correlation in the asymmetric component of shape, because any perturbation in the system (i.e., developmental noise) may be propagated to both traits. Evidence for correlations among structures suggests traits can be integrated regardless of spatial and temporal differences or developmental origin. Evidence for a lack of correlation among structures suggests development within a shared morphogenetic environment can still elicit independent phenotypic patterning.
We then address the second prediction of the palimpsest model, that patterns of phenotypic (co)variation are strongly influenced by interactions operating above the level of the genome. In particular, we use a quantitative trait loci (QTL) analysis to reveal those regions of the genome responsible for how variation in trait geometry is established, and assess the degree to which disparate traits that phenotypically covary map to similar genomic regions. Overlapping QTL would suggest that shared genetic control, and not developmental interactions, is important in producing an integrated craniofacial structure. This scenario could arise from multiple, non-mutually exclusive, events, including: (1) a very large genetic effect that shapes multiple traits (e.g., Eda in stickleback bony armor plates and lateral line system (Archambeault et al. 2020)), (2) an early acting genetic effect that is steadily amplified by development through ontogeny in a way that preserves the signal through ontogeny (e.g., maturation of distinct heart chambers via Notch1b expression and muscular contraction (Samsa et al. 2015)), or (3) an accumulation of many relatively minor genetic effects of a single locus at multiple points throughout ontogeny (e.g., ptch1 in cichlid craniofacial development (Hu and Albertson 2014, 2017; Navon et al. 2020)). On the other hand, observing the relative independence of QTL among structures that are phenotypically integrated would suggest that these associations manifest as a result of developmental interactions that occur above the level of the genome. This scenario does not preclude roles for genetics, but rather suggests that developmental interactions have “overwritten” early genetic signals, obscuring the G-P map (e.g., the Palimpsest model (Hallgrímsson et al. 2009, 2012)).
MATERIALS AND METHODS
Hybrid Population
We generated an F5 hybrid population from two, wild-caught African cichlids from Lake Malawi: Labeotropheus fuelleborni (LF) and Tropheops ‘red cheek’ (TRC). Further details of the pedigree mapping are provided in the supplement. While LF and TRC are both members of the mbuna clade (Malinsky et al. 2018), they exhibit differences in craniofacial anatomy, feeding behavior, and microhabitat. LF is considered a highly specialized algae scraper, and exhibits traits consistent with this behavior: a steep craniofacial profile, wide and short oral jaws, and a generally robust feeding apparatus. Indeed, one striking specialization that arose in the Labeotropheus genus is a pronounced snout that permits the ripping of algae from rocks by using the snout as a fulcrum to lever filamentous algae away from the rocks (Conith et al. 2018b). As a result, Labeotropheus rarely venture far from rocks, avoiding the more turbulent waters present in their rocky shore habitats. TRC are considered more generalist feeders relative to LF, exhibiting nipping, sifting, and suction feeding behaviors to consume algae and small invertebrates. As a result, TRC have a more shallow craniofacial profile, narrower and longer oral jaws, and a more gracile skull shape. While the diet of TRC is similar to LF, TRC plucks filamentous algae using a twisting and jerking method that is presumably more energetically expensive, and means they are often subjected to the turbulent waters surrounding the rocks (Konings 2007; Conith et al. 2019b).
In crossing LF with TRC we gained a range of craniofacial traits for morphological analysis that reflected the full range of phenotypes between these two species, and even included transgressive phenotypes (Parsons et al. 2011a). Our F5 hybrid population comprised 140 individuals that were housed in identical environments in a recirculating system and fed a diet of algae and egg yolk flakes, thus limiting environmental variation. Hybrid animals were collected at ~5 months. Our sample also included four LF and four TRC individuals to examine how traits in the hybrids are distributed relative to the parentals.
Morphological Analysis
We used 3D geometric morphometrics to extract the shape of two bones, the mandible and pectoral girdle, and two major components of the heart, the atrium and ventricle from μCT scanned specimens (Figure 2). For further X-ray scanning information, see the supplement. We used a combination of fixed-, semi-, and surface- landmarks (LMs) to characterize shape information in each structure and mirrored LM positions on both the left and right sides in all our structures, which allowed us to separate the symmetric from asymmetric components of shape variation (Figure 3; Table S1). We placed a series of LMs at key functional and developmental positions (i.e., processes, muscle insertion points, sutures etc.) on the mandible and pectoral girdle. We used a total of 20 fixed LMs on the mandible, and 46 fixed landmarks along with 90 sliding semi-landmarks on the pectoral girdle. To generate a detailed digital representation of the heart we combined human and semi-automated landmarking procedures. We first placed 2 fixed LMs and 97 sliding semi-landmarks on the atrium, alongside 2 fixed LMs and 62 sliding semi-landmarks on the ventricle for all hybrids and parentals. We then combined this with a semi-automated procedure to distribute surface semi-landmarks across the atrium and ventricle to generate a high-resolution anatomical characterization of the heart. This involved constructing two 3D templates using computer aided design (CAD) software (FreeCAD v.0.16.6712) that broadly mimicked atrium and ventricle morphology. The atrium was modeled as a cylindrical segment, and the ventricle was modeled as a cone (Figure S1). Fixed and sliding semi-landmarks were then placed on each of the CAD models that corresponded to their position in the real structures, alongside surface semi-landmarks (atrium, 240 surface LMs; ventricle, 160 surface LMs). We then used the R package Morpho to map the surface semi-landmarks from the templates to each atrium and ventricle specimen using the placePatch function (Schlager 2017, 2018). All human-digitized landmarks were placed using Landmark Editor (Wiley et al. 2005).
Figure 2.
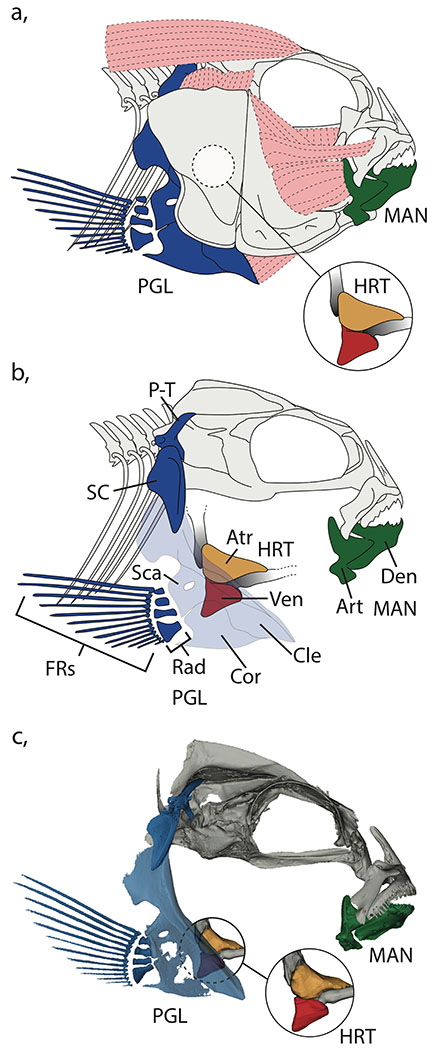
Positioning of the mandible, pectoral girdle, atrium, and ventricle in the cichlid head. a, Schematic demonstrating the full compliment of muscles and bones in the head. b, Schematic of the cichlid head with the suspensorium removed to clearly reveal the heart, mandible, and pectoral girdle. c, μCT scan of the head region including models of the heart reconstructed from contrast enhanced images. MAN, mandible (dark green); PGL, pectoral girdle (dark blue); HRT, heart, Atr, atrium (orange); Ven, ventricle (red); Den, dentary; Art, articular; P-T, post-temporal; SC, supra-cleitrum; Cle, cleithrum; Sca, scapula; Cor, coronoid; Rad, radials; FRs, fin rays.
Figure 3.
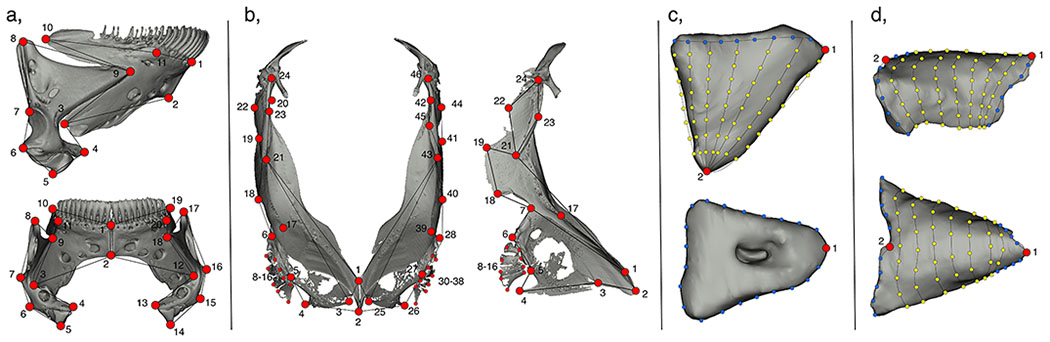
Geometric morphometric schematics for the placement of all landmarks. a, mandible; b, pectoral girdle; c, ventricle; d, atrium. Fixed landmarks, red circles; semi-landmark curves, blue circles; patch landmarks, yellow circles.
We used general least squares Procrustes superimposition (GPA) to remove the effects of size, translation, and rotation from each structure among all our specimens with the gpagen geomorph function (Rohlf 1998; Adams et al. 2017). We then performed a Procrustes ANOVA between the geometric size of the skull and shape of each structure using the procD.lm geomorph function (Adams et al. 2017) and found a significant effect of allometry in all cases (mandible, r2 = 0.032, F = 4.49, p = 0.002; pectoral girdle, r2 = 0.048, F = 7.36, p = 0.001; atrium, r2 = 0.024, F = 3.31, p = 0.001; ventricle, r2 = 0.016, F = 2.25, p = 0.009). To remove the allometric component of shape variation, we extracted the landmark residuals from this Procrustes ANOVA model to obtain landmark data sets for all four traits for use in subsequent analyses.
To assess the degree of fluctuating asymmetry in our allometrically corrected mandible, pectoral girdle, atrium, and ventricle landmark configurations we used the bilat.symmetry geomorph function. Given our traits can be traced embryologically to initially developing with a separate left and right sides, we treat all traits as paired structures for the purpose of our symmetry analyses. All traits fuse their midlines at a similar embryonic stage (Kratochwil et al. 2015). While the heart then loops and jogs, it still originated as a paired structure (Desgrange et al. 2018). All structures were landmarked for both left and right sides separated by their midlines, allowing us to assess object symmetry. First, the landmarks from one side are reflected onto the other and undergo Procrustes superimposition. The variation among the reflected and original landmark configurations for all comprises the symmetric component of shape variation. The asymmetric component of variation was calculated through quantifying the deviation of the original landmark configuration from the symmetric consensus of the original and mirrored configuration (Klingenberg et al. 2002). The degree of asymmetry was then statistically evaluated using Procrustes ANOVA using residual randomization permutation procedures (Collyer et al. 2015). All structures were found to exhibit significant degrees of directional asymmetry (mandible, r2 = 0.042, F = 26.99, p = 0.001; pectoral girdle, r2 = 0.030, F = 15.76, p = 0.001; atrium, r2 = 0.201, F = 78.02, p = 0.001; ventricle, r2 = 0.641, F = 552.44, p = 0.001). We therefore extracted landmark configurations from all structures that reflected the symmetric and asymmetric components of variation for use in future analyses. This resulted in three different landmark configurations for each trait: landmark configurations that reflect the allometrically corrected data prior to symmetry analysis (termed ‘original configuration’), landmark configurations of the symmetric component of shape variation (termed ‘symmetric configuration’), and landmark configurations of the asymmetric component of shape variation (termed ‘asymmetric configuration’). Isolating the symmetric and asymmetric components of variation allows us to reduce the amount of developmental noise present in our morphological data, and should increase the power to detect a genetic signal.
We then performed a principal component (PC) analysis on the original, symmetric, and asymmetric configurations to quantify variation in shape for all four traits (Table S2). We initially excluded all parentals in our PC analysis and then exported the first two PC axes that typically represented between 20-40% of the total shape variation for each structure, and saved those PC scores for to use as traits in the mapping analyses (there are currently no methods available to assess shape data in a multivariate framework). We then included parentals in the PC analysis and plotted the first two PC axes for each trait to visualize the major differences in shape variation among individuals.
We assessed the degree of correlation among all traits within their respective landmark configurations (i.e., original, symmetric, asymmetric). We used the two.b.pls geomorph function (Adams et al. 2017), which performs a partial least squares analysis between two sets of Procrustes-aligned coordinates to assess the degree of association (Rohlf and Corti 2000; Adams and Collyer 2016).
Quantitative Trait Loci Analysis
To determine the relative roles of genetics and development in shaping phenotypic integration we began by constructing a F5 genetic map of cichlids. For full information on map construction, see the supplement. We then performed a QTL analysis on the PC scores extracted from all our structures that reflected the symmetric, asymmetric, and the original components of variation. We ran each PC axis as a separate trait through our QTL analysis, and identified regions of the genome that represented a possible association between phenotype and genotype. This approach would enable the separation of loci responsible for overall geometric change from loci responsible for asymmetric change. The genetic regions highlighted in each case could be compared to the original datasets to reveal how important the effects of directional asymmetry are in each structure. Additionally, considering the effects of symmetry and asymmetry independently may aid in identifying cryptic genetic variation that are only revealed when asymmetry is removed.
While the approach outlined above uses PC scores as its data source that reflect maximum variation within a trait, these data are subtlety different to those observed in the phenotypic integration analysis, which fixes the PC axes to vary with maximum covariation between two traits. As a result, we also assessed where the trait data derived from the covariation between our traits localized to on our QTL map. We mapped covariation between all possible symmetric trait configurations and performed a QTL analysis in an attempt to derive a map that retains consistency in the traits displayed in the integration analyses, and in the QTL analyses. We discuss these results in the supplementary information section, but note we found broad similarity in QTL localization between the variation- and covariation-derived trait data.
We began searching for putative loci using the multiple QTL mapping (MQM) approach (Broman and Sen 2009) implemented in the R package r/qtl (Broman and Wu 2020). Following an initial QTL scan using the scanone function, we progressively added cofactors to the model and cofactor fit was determined via maximum-likelihood backward elimination in the mqmscan function. We continued to add cofactors to the model until the logarithm of odds (LOD) score was maximized. We quantitatively assessed QTL marker significance using the mqmpermutation function. The mqmpermutation function reshuffles the phenotypic data relative to genotypic data 1000 times, disassociating any possible relationship between genotype and phenotype, to generate a null distribution (Arends et al. 2010). QTL marker LOD scores that exceeded the 5% LOD threshold level are deemed significant. Once a significant QTL peak was determined, we used the function bayesint to calculate an approximate Bayesian credible interval in which a potential candidate locus would reside within.
Once we had identified the approximate position of a candidate marker, we then fine mapped that linkage group to further refine candidate marker position. This involved using the Maylandia zebra (MZ) genome to anchor QTL intervals to particular stretches of physical sequence (Conte and Kocher 2015). We matched the QTL marker range revealed by the Bayesian credible interval analysis to corresponding stretches of the MZ genome. We then identified additional RAD-seq SNPs across the linkage group of interest and genotyped them in the F5. These were selected to span the linkage group with an average spacing of around one marker every 150kb. This provided a greater resolution of genotypes, to assess genotype-phenotype correlations across the credible intervals. We then examined the difference in the average trait values between individuals with the LF allele and TRC allele at every marker position using the effectsplot function. We then performed a permutation test to determine fine map marker significance, which involved randomizing the trait data relative to genotypic data 100 times to create a null distribution.
RESULTS
Quantification of shape variation in hard and soft tissue traits
We found that the major axes of morphological variation in the mandible and pectoral girdle reflects a spectrum from more LF-like to more-TRC like shapes, and are largely consistent with the major axes of variation present across the rock dwelling cichlids in Lake Malawi (Cooper et al. 2010; Hulsey et al. 2018). Morphological variation in the atrium and ventricle also spans a spectrum from LF-like morphologies to TRC-like morphologies, and change along these axes mimic broad differences observed in other lineages, such as cavefish (Tang et al. 2018). Notably, variation in heart structures reflect changes characteristic of differences in foraging behavior and energetics (Figure 4; Figure S2), from the energetically demanding nip-and-twist mode of TRC, to the more efficient and highly specialize mode of LF, whereby animals use their hypertrophied snout pads to help leverage algae from the substrate (Konings 2007).
Figure 4.
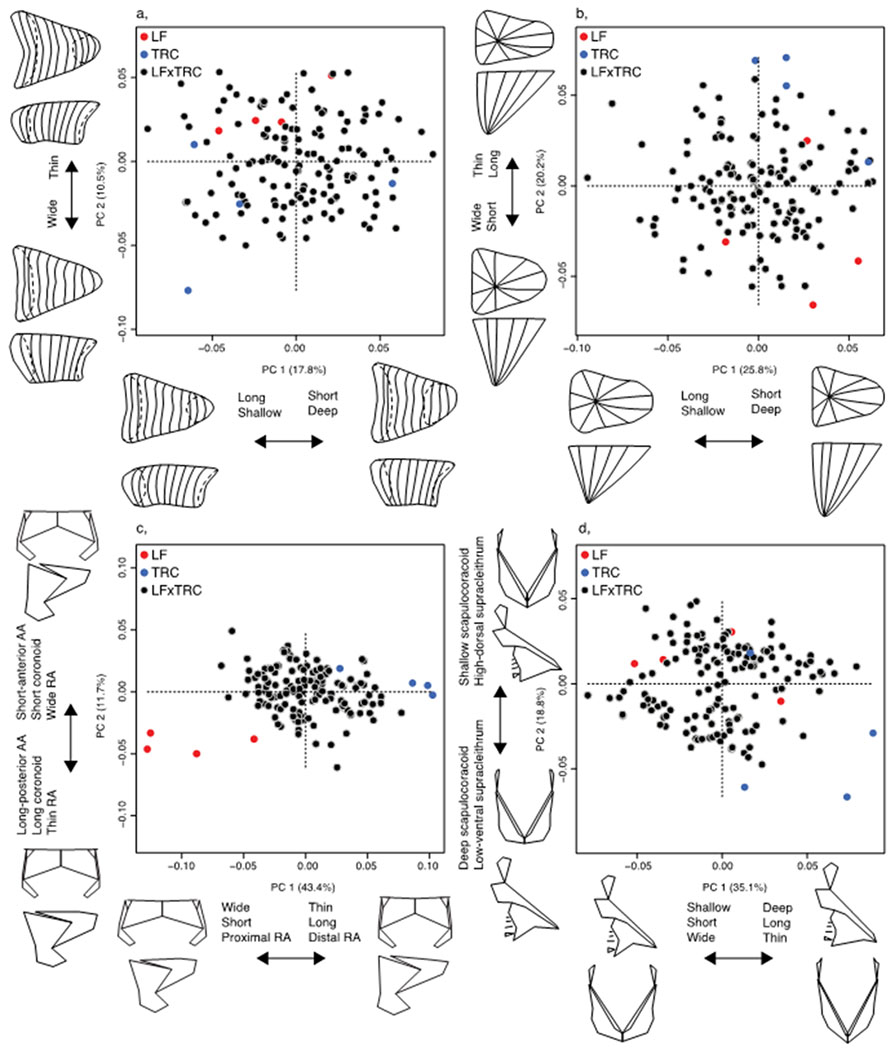
Symmetric configuration principal component morphospaces for all hybrid structures. A, atrium; b, ventricle; c, mandible; d, pectoral girdle. Hybrids, black circles; LF, red circles; TRC, blue circles.
Atrium PC1 reflects elements of asymmetry and length (18.4%), and PC2 represents depth and the size of the connection between the atrium and sinus venosus (9.4%). When assessing the symmetric component of atrium variation, PC1 reflects length, depth, and the size of the connection between the atrium and sinus venosus (17.8%), and PC2 represents width (10.5%). When assessing the asymmetric component of atrium variation, PC1 reflects a left-right skew of the posterior surface (36.1%), and PC2 represents left-right skew of the anterior surface (9.2%). Ventricle PC1 reflects elements of asymmetry and depth (17.8%), and PC2 represents length and width (14.1%). When assessing the symmetric component of ventricle variation, PC1 reflects depth and length (26.5%), and PC2 represents width and length (18.6%). When assessing the asymmetric component of ventricle variation, PC1 reflects a left-right extension of the posterior surface (35.5%), and PC2 represents left-right invagination of the medial surface (19.2%).
Mandible PC1 reflects width, length, and a proximal-medial shift of the RA (30.4%), and PC2 represents coronoid length, retroarticlar process (RA) width, and ascending arm (AA) length and anterior-posterior position (9.9%). When assessing the symmetric component of mandible variation, PC1 reflects similar changes in the RA, mainly differences in width, length, and proximal to distal shift in positioning (38.9%). PC2 still describes variation in AA length, but now also describes differences in coronoid length, and RA width (12.6%). When assessing the asymmetric component of mandible variation, PC1 reflects a left-right change in width and RA position (20.9%), and PC2 represents a left-right shift in the height of the AA and depth of the RA (17.3%). Pectoral girdle PC1 reflects over all depth, length, and width of the structure (26.2%), and PC2 represents scapulocoracoid depth, a left-right shift in the height of the scapulocoracoid, supracleithrum depth and width, and a left-right shift in anterior-posterior positioning of the supracleithrum (14.3%). When assessing the symmetric component of pectoral girdle variation, PC1 again reflects depth, length, and width (35.5%), and PC2 represents scapulocoracoid depth and supracleithrum height (18.4%). When assessing the asymmetric component of pectoral girdle variation, PC1 reflects a left-right skew in the height of the whole pectoral girdle (39.0%), and PC2 represents a left-right shift in the anterior-posterior positioning of the supracleithrum, and a left-right shift in the height of the scapulocoracoid and radials (8.8%).
Correlations among and within hard and soft tissue traits
We find evidence for moderate associations in shape among our hard tissue configurations (Table 1; Figure 5–6). The mandible and pectoral girdle are tightly correlated across symmetric, asymmetric, and original landmark configurations. This indicates tight integration can occur between traits with diverse functional roles (locomotion and feeding), different developmental origins (NCC derived and mesodermally derived), and spatially distant positions. The correlation between asymmetric configurations is interesting as it suggests a common origin of developmental noise in these distinct anatomical components.
Table 1.
Associations among hard and soft tissue traits. a, associations among traits using the original landmark configurations. b, associations among traits using the symmetric component of shape variation. c, associations among traits using the asymmetric component of shape variation. Left of diagonal, p-values; right of diagonal, r2 values.
| a Original | |||
|---|---|---|---|
| Atrium | 0.392 | 0.264 | 0.288 |
| 0.001 | Ventricle | 0.175 | 0.267 |
| 0.013 | 0.040 | Mandible | 0.202 |
| 0.004 | 0.001 | 0.006 | Cleithrum |
| b Symmetry | |||
| Atrium | 0.253 | 0.233 | 0.286 |
| 0.004 | Ventricle | 0.151 | 0.240 |
| 0.006 | 0.014 | Mandible | 0.184 |
| 0.001 | 0.001 | 0.002 | Cleithrum |
| c Asymmetry | |||
| Atrium | 0.359 | 0.070 | 0.109 0.228* |
| 0.001 | Ventricle | 0.084 | 0.135 0.121* |
| 0.864 | 0.745 | Mandible | 0.170 |
| 0.115 0.002* |
0.026 0.152* |
0.016 | Cleithrum |
indicates outliers removed.
Figure 5.
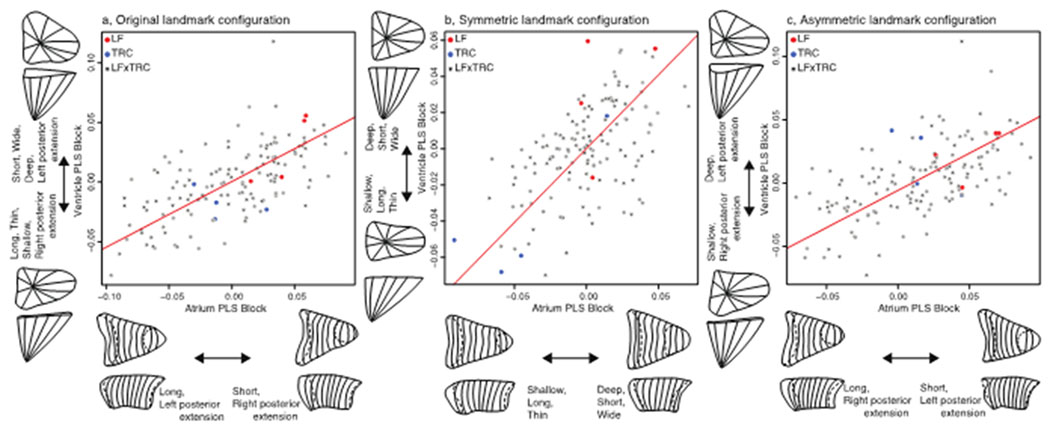
Two-block partial least squares analysis to assess association between ventricle and atrium for each landmark configuration. a, original landmark configuration; b, symmetric component of shape variation; c, asymmetric components of shape variation. Hybrids, black circles; LF, red circles; TRC, blue circles.
Figure 6.
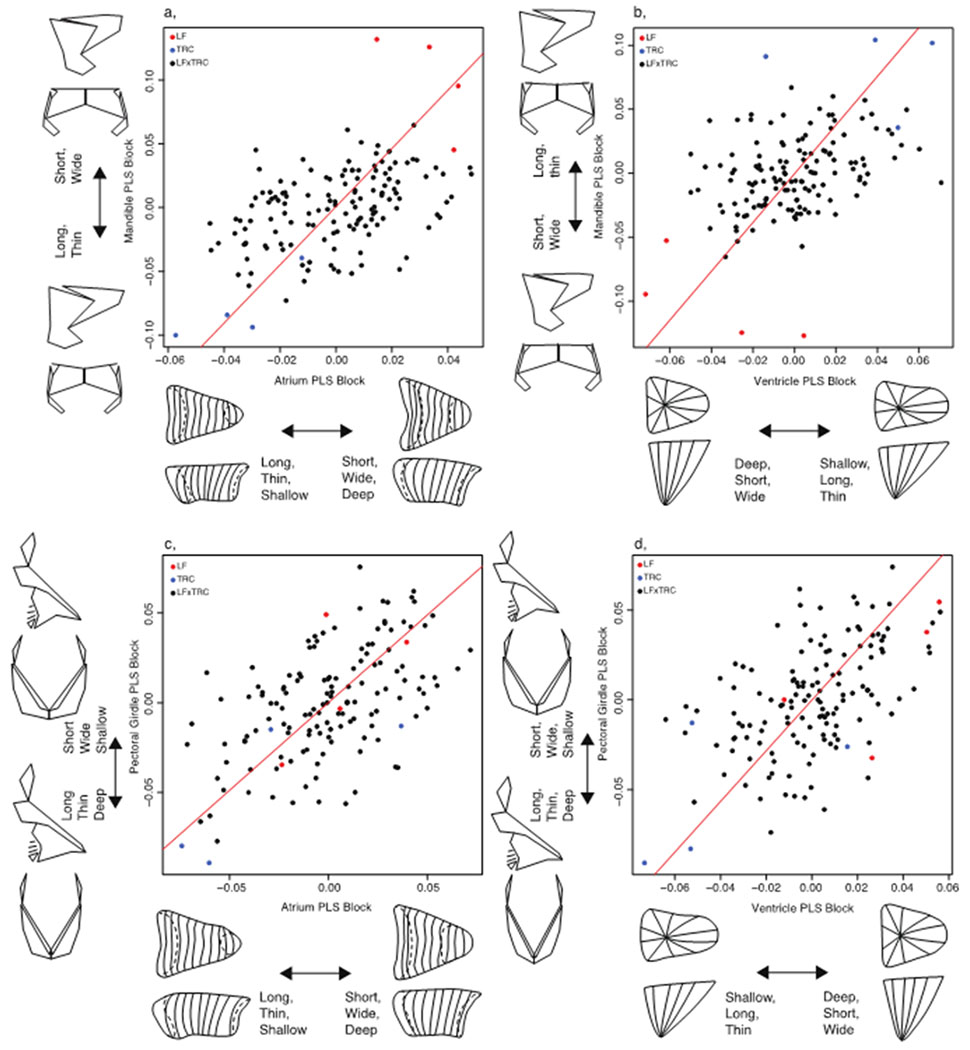
Two-block partial least squares analysis to assess association between heart and bony tissues using the symmetric component of shape variation. A, association between atrium and mandible; b, association between ventricle and mandible; c, association between atrium and the pectoral girdle; d, association between ventricle and the pectoral girdle. Hybrids, black circles; LF, red circles; TRC, blue circles.
We also found evidence for moderate shape associations among our soft tissue traits. The atrium and ventricle exhibit tight integration when comparing symmetric, asymmetric, and original landmark configurations. The correlation between asymmetric configurations in different compartments of the heart is likely due to common asymmetric patterning events (Desgrange et al. 2018). When comparing the correlations based on the original and symmetric configurations, the positions of the parental species are more divergent in the symmetric configurations (Figure 5), possibly because the original configurations contain asymmetric variation due to developmental noise (Figure 5c).
Notably, we also observe associations between hard and soft tissues (Figure 6; Figure S3). When comparing both the symmetric and original configurations between hard and soft tissues we observe tight integration across tissues, which suggests that species-specific (i.e., LF-TRC) aspects of structural variation are developing together across tissue types (i.e., bone or cardiac muscle). However, correlations between tissues are absent when comparing asymmetric correlations, which may be due to different origins for asymmetry in hard and soft tissue – e.g., developmental noise in bones, asymmetric gene expression in hearts. Additionally, parental species are not placed at the extremes of the distributions for asymmetric configurations. This appears to reflect a departure from normal, directional asymmetric patterning of the heart observed in the parental species. In particular, both parental species are characterized by heart shapes with posterior extensions toward the left. Hybrids, on the other hand, possess both left- and right-facing hearts extensions, which indicates greater fluctuating asymmetry (i.e., developmental instability) in the hybrid population. We also note that comparing the asymmetric components of variation in the pectoral girdle to the atrium and ventricle is made more challenging by the presence of two outliers (Table 1c), as their removal can change the outcome of the correlation by either reducing the significance of the correlation (i.e., pectoral girdle-ventricle,) or increasing significance (i.e., pectoral girdle-atrium).
Genetic mapping provides limited evidence for genetic pleiotropy
Our QTL analysis reveals multiple regions of the genome that underlie variation in mandible, pectoral girdle, and heart shapes (Table S3). We found a total of 42 significant QTL (30 at the <0.05 genome-wide level, and a further 12 at the <0.10 level), which may be partitioned into symmetric (9 at <0.05 and 5 at <0.10), asymmetric (10 at <0.05 and 2 at <0.10), and total components of shape variation (11 at <0.05 and 5 at <0.10). Each structure exhibited between 3-5 QTL depending on configuration. We typically recovered fewer QTL for the asymmetric components (3 QTL), compared to the symmetric (3-4 QTL), or total shape components (3-5 QTL). We identified significant LOD scores on 17 of 25 linkage groups (Table S4). All significant QTL explained between 8%-18% of the variation in the F5 population. Modes of inheritance for traits included additive, dominant, and overdominant effects.
We found minimal evidence for shared genetic architecture (i.e., pleiotropy) between traits based on the lack of overlapping QTL regions (Figure 7; Figure S4; Table S5). We found a single instance where QTL for symmetric traits localize to the same linkage group across structures (LG 22), no instances of QTL overlap between asymmetric traits, and two instances of overlap among asymmetric-symmetric traits (LGs 5, 22). When we considered overlap among the symmetric-asymmetric configurations and the originals, we noticed several instances in which QTL derived from the original configurations are conspicuously isolated from their symmetric and/or asymmetric components of variation (atrium, LG12; ventricle, LG4, LG17, LG19; mandible, LG15; pectoral girdle, LG14). Data derived from the original configurations typically exhibited more QTL per trait, likely because these configurations combined symmetric and asymmetric aspects of the morphology. The QTL regions that are unique to the original configurations are intriguing and suggest unique regions of the genome may control the interaction between symmetric and asymmetric morphological variation.
Figure 7.
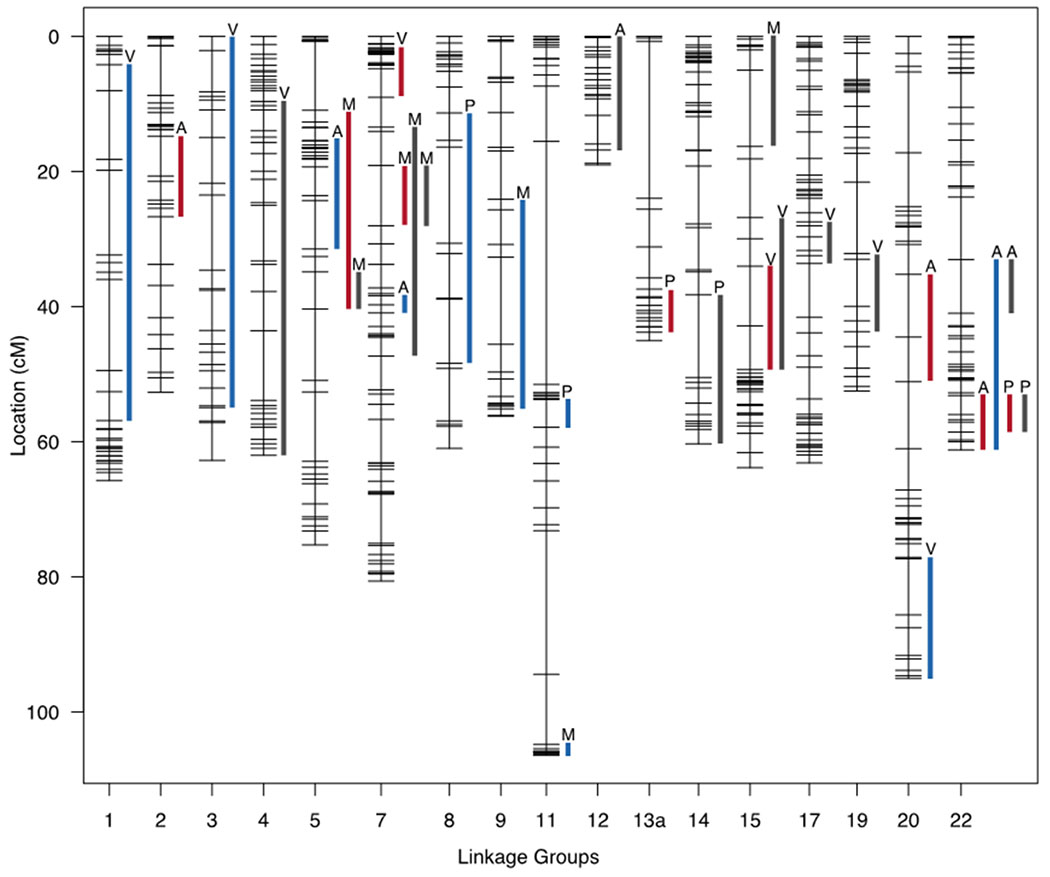
QTL map for all structures and all components of variation. Lines reflect significant QTL regions at the 95% level for the original configurations (dark gray), symmetric components of variation (red), and asymmetric components of variation (blue). Letters on top of the lines indicate structure identity: Atrium (A), ventricle (V), mandible (M), pectoral girdle (P).
Given the very large genomic regions revealed by our QTL intervals, any perceived overlap among structures could be due to separate, but linked loci. Since an F5 mapping pedigree provides a large number of recombination events, we employed a fine mapping technique to refine our QTL intervals. These analyses focused on three linkage groups (LG7, LG11, LG22), and revealed a modest to large number of shared significant markers between two structures (Figure 8; Table S6). For the symmetric comparisons, the mandible and ventricle overlapped across a physical stretch of 18Mb on LG7, and the pectoral girdle and atrium overlapped along a 9Mb stretch on LG22. For the asymmetric comparison, the mandible and pectoral girdle overlapped for around 2Mb on LG11. The fine mapping approach served to narrowed interval size for these QTL, and identified additional markers that share a significant genotype-phenotype relationship for distinct traits. For these loci, we cannot reject the hypothesis of shared genetic control between tissues, and a number of genes within these overlapping regions represent potential candidates for controlling global shape change across multiple tissue types across the head (Table S7).
Figure 8.
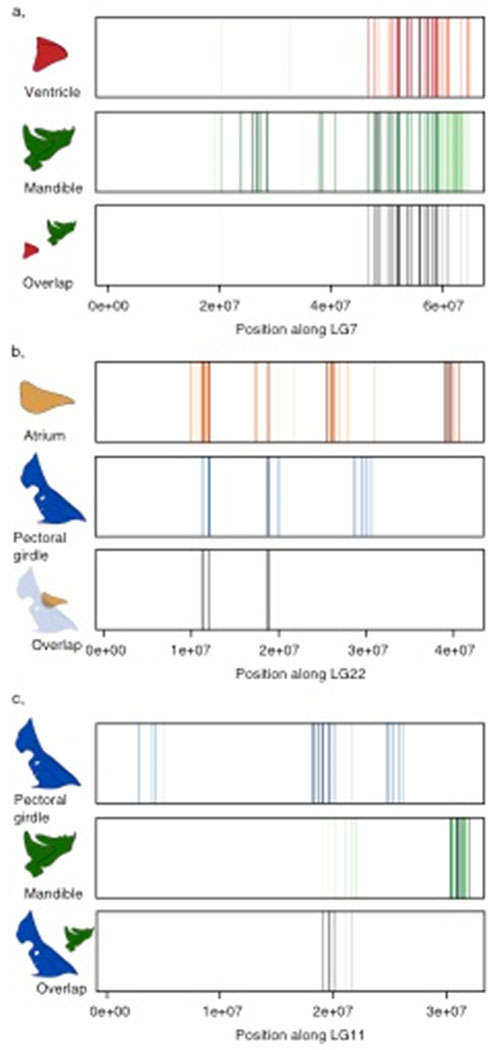
Fine maps depicting significant QTL markers across a linkage group and the degree of overlap among those QTL markers. Each line reflects a significant marker, and the opacity of a line indicates significance with solid lines representing highly significant markers, while translucent lines reflect less significant markers. Black lines reflect overlapping markers between traits with their opacity determined by their shared significance values. a, comparing the symmetric component of variation between ventricle and mandible. b, comparing the symmetric component of variation between atrium and pectoral girdle. c, comparing the asymmetric component of variation between pectoral girdle and mandible.
DISCUSSION
Phenotypic integration is present within and between disparate structures
The cranial region of vertebrates is a highly complex, multifunctional structure housing the major sensory and feeding structures, and in teleost fishes, this anatomical region is physically associated with the heart, as well as the appendicular and rostral axial skeleton. Skull development is tightly regulated at the genetic level, especially at early stages (e.g., patterning), but involves inputs from diverse developmental sources, acting at the cellular, tissue, and structural levels (e.g., neural crest cell migration and differentiation, cellular condensation, organ growth, and muscle bone interactions (Hallgrímsson et al. 2009)). Waddington discussed the importance of such extrinsic forces that act upon the initial genetic variation to shape the eventual phenotype at levels acting above the genome (Waddington 1957). Here we examined the impact of developmental processes, such as phenotypic integration and developmental stability (i.e., fluctuating asymmetry), on the adult phenotype and examine how development can influence our ability to detect a genetic signal (Figure 1). We found strong support for our first prediction, that phenotypic integration is evident across the skull. Indeed, we found evidence for shape correlations among all structures indicating that the cranial region, as a whole, reflects a fairly well integrated trait complex, despite comparing disparate tissue types (i.e., bone, cardiac muscle).
It is perhaps unsurprising to find general associations among structures in the head given how traits are likely subjected to, and influenced by, spatially and temporally overlapping developmental fields, alongside shared cellular origins and genetic signals. To our knowledge, no formal assessment of cardiac shape integration exists, although different heart structures appear to be associated with body size (Hu et al. 2000; Shifatu et al. 2018). Recent studies have also demonstrated that patterns of trait covariation can span tissues with vastly different developmental origins (e.g., Martínez-Vargas et al. 2017; Conith et al. 2019; Yamamoto et al. 2020). Taken together, it appears that patterns of integration can encompass multiple tissue types, even extending beyond structures in close physical proximity, to act as a global mechanism permitting the skull to operate as a single unit.
We also found a surprising number of correlations among our structures when using the asymmetric components of shape variation. However, these correlations are typically confined to comparisons within a specific tissue type (mandible-pectoral girdle and atrium-ventricle). These tissue-specific associations may result from differences in the capacity of each tissue to remodel; bone tissue takes longer to remodel relative to cardiac tissue, and our heart structures exhibited the highest levels of fluctuating asymmetry, a trend not unusual for soft tissue traits (Żelaźniewicz and Pawłowski 2018; Hedrick et al. 2019). Additionally, the heart is asymmetrically patterned early in development, and thus asymmetric gene expression may contribute to shared laterality detected in our analyses (Desgrange et al. 2018; Guerra et al. 2018), whereas asymmetry in our bony structures is more likely due to developmental instabilities that arise over broad windows of ontogeny. Fluctuating asymmetry can vary among traits due to functional and structural (i.e., cellular composition) differences, and the strength of selection acting upon a trait (Aparicio and Bonal 2002). We note that there is some evidence to suggest asymmetry may be under genetic control in both the heart (Guerra et al. 2018) and mandible (Albertson and Yelick 2005; Stewart and Albertson 2010) of fish, which could also contribute to (co)variation in our asymmetric components of shape.
Developmental processes limit our ability to link genotype to phenotype
Waddington predicted that developmental and other external environmental processes operating above the level of the genome could have large effects on the eventual phenotype, producing a range of shapes that are not directly genetically determined, but are the result of interacting cellular, tissue, or organ level processes (Waddington 1957; Jamniczky et al. 2010). Our second prediction, that phenotypic (co)variation is largely shaped by processes and interactions operating above the level of the genome, echoes those of Waddington, and is largely supported. We observed that phenotypic associations (i.e., integration) among traits rarely result in genetic associations among traits. Indeed, despite widespread phenotypic correlations among our skeletal and cardiac structures, we found little evidence for overlapping QTL regions that would likely signify shared genetic control. The lack of genetic overlap in our QTL analysis applies to both asymmetric and symmetric traits. Within symmetric components of shape, a lack of overlap suggests that either, 1) developmental processes have obfuscated the G-P map, limiting our ability to gain a clear picture of the major genetic contributors to phenotypic differences among our traits (e.g., Figure 1), or 2) that pleiotropy does not play a large role in determining covariation of traits across the head, as noted elsewhere (i.e., Navon et al. 2017). Within asymmetric structures, a lack of overlap suggests different genomic regions are responsible for producing asymmetric variation among structures, demonstrating a large number of genes maybe responsible for regulating fluctuating asymmetry and not a core subset. We also found little genetic overlap between symmetric and asymmetric traits for a given structure, suggesting independence between the genes that control overall geometry and those that influence left-right symmetry and/or developmental noise.
The Hallgrímsson et al. (2009) palimpsest model takes Waddington’s ideas further, suggesting developmental processes build on each other, constantly rewriting the pattern of covariation. As a result, the phenotype could differentially reflect specific developmental processes depending on the strength of interactions through ontogeny (Figure 1b–c). Recent, empirical evidence suggests that a number of developmental factors can obscure the G-P signal even further, such as non-linear developmental processes, behavioral modulation, and biophysical feedback (Hallgrimsson et al. 2019). For example, Hu and Albertson (2017) demonstrated how variation in gaping behavior during the first two weeks of cichlid development can elicit drastically different rates of bone deposition due to the mechanical environment in which the mandible develops. Notably, the phenotypic effects on bone length induced by manipulating this behavior were equal to those predicted by QTL or induced by molecular manipulations. Thus, even within larval stages of development, structural interactions have the potential to induce phenotypic variation of sufficient magnitude to “rewrite” variation encoded in the genome. Additionally, biophysical processes operating during ventricle development appear to influence the shape of the ventricle by altering the timing of cardiac differentiation based on hemodynamics (Sedmera et al. 2003; Auman et al. 2007; Jensen et al. 2012). Taken together, these examples highlight how variation in developmental regulation can operate on existing genetic variation to further shape phenotypic variation, while simultaneously obscuring the relationship between genotype and phenotype.
Phenotypic integration may be governed by pleiotropy, shared development origins, overlapping developmental fields, and coordinated growth
While overlapping regions among structures were rare overall, our fine mapping data reveals a handful of shared markers between structures, and these regions reveal potential candidate genes involved in important regulatory roles (e.g., Notch, Wnt). We found a substantial overlapping region on LG7 (18Mb), between the mandible (symmetric PC2, length and width), and ventricle (symmetric PC2, length and width). A number of genes involved in cardiac and bone development reside within this, albeit large, region including notch1a, camk2b2, and nrg1. All three genes have been implicated in regulating development of tissues across the head and have multiple roles in patterning both hard, and soft tissue traits (Rothschild et al. 2009; Samsa et al. 2015; Liang et al. 2019).
In spite of these few regions of genetic overlap, the mandible, pectoral girdle, atrium, and ventricle are phenotypically correlated to a degree that is far greater than what can be explained by pleiotropy. This suggests outsized roles for factors operating above the level of the genome, including common cell origins, cross-talk between developmental fields, and/or the need to function in concert with neighboring structures. Neural crest cells (NCCs) are responsible for patterning much of the cranial region in fishes, producing most of the cartilage, bone, dentine, and connective tissues present in the head. In addition, NCCs contribute to cardiac tissues, and unlike mammals and birds, where these cells only give rise to the outflow track, fish NCCs contribute to all chambers of the heart (Sato and Yost 2003). Thus, the heart and mandible in fishes share a common cellular origin, which may contribute to the integration between these distinct tissues. In addition, structures developing in close proximity to each other are more likely to experience a similar developmental environment, and may depend on signals derived from a common cell population (e.g., heart and pharyngeal skeleton (Choe and Crump 2014)).
There is also mounting evidence to suggest biomechanical forces acting across structures are critical to ‘normal’ development and growth, and can lead to tight patterns of covariation. Indeed, the shapes of various bony structures across the head are strongly associated with cranial skeletal muscles, and these associations exist beyond simply origin-insertion points on a given bone (Conith et al. 2019a; Yamamoto et al. 2020). Such muscle-bone associations likely arise from a biomechanical environment that is shared over ontogeny (Hallgrímsson et al. 2009), and this idea may even extend to the heart, which also depends on biomechanical inputs for proper development and function (Sedmera et al. 2003).
Morphological variation among hybrids mimics variation observed across cichlids
The major axes of divergence among our structures reflect characteristic changes that occur across lake Malawi cichlids (Albertson and Kocher 2006; Cooper et al. 2010; Hulsey et al. 2018). Differences in mandible shape reflect a spectrum from wide, short, and robust jaws to thin, long, and slender jaws, features that are linked to differences in diet and biomechanics (Albertson et al. 2003; Hu and Albertson 2014). Patterns of variation and covariation in cichlid jaw shape have been well studied at the phenotypic and genetic levels (Albertson et al. 2005; Albertson and Kocher 2006; Hu et al. 2014). The pectoral girdle, on the other hand, is an important but understudied element of fish anatomy. It supports musculoskeletal components of both the pectoral and pelvic fins, and it provides attachment sites for muscles that power the lower pharyngeal jaw and hyoid apparatus. Thus, as a single bony complex, it participates in multiple functions, including lower jaw depression (via the urohyal), prey processing (via the pharyngeal jaw), and locomotion. Differences in pectoral girdle morphology among hybrids represent shape change from deep, long, and thin to shallow, short, and wide; shape differences that are characteristic of divergence in foraging and locomotory behavior across cichlid species (Hulsey et al. 2018).
Another novel aspect of this study, is that it documents variation in heart shape, which ranged from short, deep, and wide reflecting an LF-like morphotype to shallow, long and thin reflecting a TRC-like morphotype. Deeper ventricle shapes are typically associated with more active and athletic fish that require greater blood pressure and stroke work (Claireaux et al. 2005). LF therefore appear to develop a more athletic heart, running counter to our initial expectation that LF is a more efficient forager and should therefore expend less overall energy while feeding, compared to TRC (Conith et al. 2019b). Both species are territorial, with males vigorously defending breeding territories (Ribbink et al. 1983), and a recent study examining activity levels across cichlid species found that LF and TRC exhibit high and indistinguishable levels of activity relative to other species (Lloyd et al. 2020). LF do occupy a greater depth range relative to TRC, foraging up to depths of 18 meters while TRC often occupy depths up to 6 meters (Ribbink et al. 1983). Fish residing at greater depths experience greater hydrostatic pressure and lower levels of dissolved oxygen necessitating higher blood pressure and stroke, which may explain the deeper ventricle in LF. Indeed, dissolved oxygen concentrations decline rapidly with depth in Lake Malawi, dropping by around 30% from the surface to 10 meters, and then becoming highly variable beyond 10 meters with oxygen concentrations dropping by as much as 60% relative to the surface waters (Martin et al. 1998). However, it is unknown whether this drop in dissolved oxygen concentration is necessary and sufficient to elicit differences in ventricle shape between species. Given that the heart develops adjacent to the pectoral girdle, it is also possible that differences in heart shape arise due to conformational changes in the surrounding skeleton, which limits the shape of the pericardial cavity. Regardless of its specific origins, given that heart shape can be a reliable predictor of habitat and activity, and that it appears to have a tractable genetic basis, we suggest that this aspect of cichlid anatomy will be a fruitful area of future research.
CONCLUSIONS
Phenotypic integration is an important concept in biology (Felice and Goswami 2018; Watanabe et al. 2019), but how it arises and persists within populations is unclear, but likely involves multiple and hierarchical inputs from genetics, development and kinematics. As a result, understanding the link between genotype and phenotype can be greatly complicated by these emergent properties arising above the genomic level. By assessing the degree of association among four structures with vastly different genetic and developmental origins (mandible, pectoral girdle, atrium, ventricle) in the head of cichlid fish we found evidence for integration among all structures. We then assessed the degree of genetic association among these traits and found few instances where a phenotypic association resulted in a genetic association. These dichotomous results strongly support the ideas first laid out by Waddington (1957) and refined by Hallgrimmson et al. (2009). In addition, we assert that they frame an important challenge for the field moving forward: How to describe the full range of genetic effects that yield coordinated development and growth of the phenotype. For early developmental events, classic mutational analysis will remain a useful approach, as single genetic lesions can lead to primary defects across disparate traits (Mašek and Andersson 2017). To decipher the genotype-phenotype relationship over extended periods of the ontogeny, transgenic systems in model organisms that allow gene effects to be assessed in a time-specific manner may also be useful (Wehner et al. 2014; Mishra et al. 2020). However, such an approach would be limited to only a handful of model systems, and work arising from these models may not be representative of wider patterns. As an alternative, genetic mapping studies at multiple life-history stages or in different environments may be used to understand the extent to which the genotype-phenotype map changes over ontogeny. The few studies that have taken such an approach suggest that these studies will be highly informative (Erickson et al. 2014; Parsons et al. 2016). Taken together, these are exciting times when conceptual and technological advances have the potential to substantially improve the resolution at which we understand the phenotype.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Greg Lin at Harvard for μCT access, Daniel Pulaski for assistance with segmentation software, Ryan Felice for providing R scripts to perform automated landmark placement, and members of the Albertson lab for comments on key elements of this project. This work was funded by the National Institutes of Health #R01DE026446 to RCA.
Footnotes
DATA ACCESSIBILITY STATEMENT
Scripts and data produced by this study are available in the supporting information section and on Dryad (doi: 10.5061/dryad.dr7sqv9wx).
REFERENCES
- Adams DC, and Collyer ML. 2016. On the comparison of the strength of morphological integration across morphometric datasets. Evolution (N. Y). 70:2623–2631. John Wiley & Sons, Ltd (10.1111). [DOI] [PubMed] [Google Scholar]
- Adams DC, Collyer ML, Otarola-Castillo E, and Sherratt E. 2017. Geomorph: Software for geometric morphometric analyses. CRAN. [Google Scholar]
- Adhikari K, Fuentes-Guajardo M, Quinto-Sánchez M, Mendoza-Revilla J, Camilo Chacón-Duque J, Acuña-Alonzo V, Jaramillo C, Arias W, Lozano RB, Pérez GM, Gómez-Valdés J, Villamil-Ramírez H, Hunemeier T, Ramallo V, Silva de Cerqueira CC, Hurtado M, Villegas V, Granja V, Gallo C, Poletti G, Schuler-Faccini L, Salzano FM, Bortolini M-C, Canizales-Quinteros S, Cheeseman M, Rosique J, Bedoya G, Rothhammer F, Headon D, González-José R, Balding D, and Ruiz-Linares A. 2016. A genome-wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation. Nat. Commun. 7:11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, and Kocher TD. 2006. Genetic and developmental basis of cichlid trophic diversity. Heredity (Edinb). 97:211–221. [DOI] [PubMed] [Google Scholar]
- Albertson RC, Streelman JT, and Kocher TD. 2003. Directional selection has shaped the oral jaws of Lake Malawi cichlid fishes. Proc. Natl. Acad. Sci. 100:5252–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, Streelman JT, Kocher TD, and Yelick PC. 2005. Integration and evolution of the cichlid mandible: the molecular basis of alternate feeding strategies. Proc. Natl. Acad. Sci. U. S. A. 102:16287–16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, and Yelick PC. 2005. Roles for fgf8 signaling in left–right patterning of the visceral organs and craniofacial skeleton. Dev. Biol. 283:310–321. [DOI] [PubMed] [Google Scholar]
- Aparicio JM, and Bonal R. 2002. Why do some traits show higher fluctuating asymmetry than others? A test of hypotheses with tail feathers of birds. Heredity (Edinb). 89:139–144. [DOI] [PubMed] [Google Scholar]
- Archambeault SL, Bärtschi LR, Merminod AD, and Peichel CL. 2020. Adaptation via pleiotropy and linkage: Association mapping reveals a complex genetic architecture within the stickleback Eda locus. Evol. Lett. 4:282–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends D, Prins P, Jansen RC, and Broman KW. 2010. R/qtl: high-throughput multiple QTL mapping. Bioinformatics 26:2990–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auman HJ, Coleman H, Riley HE, Olale F, Tsai H-J, and Yelon D. 2007. Functional Modulation of Cardiac Form through Regionally Confined Cell Shape Changes. PLoS Biol. 5:e53. Public Library of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, and Sen S. 2009. A guide to QTL mapping with R/qtl. Springer-Verlag, New York, NY. [Google Scholar]
- Broman KW, and Wu H. 2020. QTL: Tools for Analyzing QTL Experiments. CRAN. [Google Scholar]
- Choe CP, and Crump JG. 2014. Tbx1 controls the morphogenesis of pharyngeal pouch epithelia through mesodermal Wnt11r and Fgf8a. Development 141:3583–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claireaux G, McKenzie DJ, Genge AG, Chatelier A, Aubin J, and Farrell AP. 2005. Linking swimming performance, cardiac pumping ability and cardiac anatomy in rainbow trout. J. Exp. Biol. 208:1775–1784. [DOI] [PubMed] [Google Scholar]
- Collyer ML, Sekora DJ, and Adams DC. 2015. A method for analysis of phenotypic change for phenotypes described by high-dimensional data. Heredity (Edinb). 115:357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conith AJ, Kidd MR, Kocher TD, and Albertson RC. 2020. Ecomorphological divergence and habitat lability in the context of robust patterns of modularity in the cichlid feeding apparatus. BMC Evol. Biol. 20:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conith AJ, Lam DT, and Albertson RC. 2019a. Muscle-induced loading as an important source of variation in craniofacial skeletal shape. genesis 57:e23263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conith AJ, Meagher MA, and Dumont ER. 2018a. The Influence of Climatic Variability on Morphological Integration, Evolutionary Rates, and Disparity in the Carnivora. Am. Nat. 191:000–000. [DOI] [PubMed] [Google Scholar]
- Conith MR, Conith AJ, and Albertson RC. 2019b. Evolution of a soft-tissue foraging adaptation in African cichlids: Roles for novelty, convergence, and constraint. Evolution (N. Y). 73:2072–2084. John Wiley & Sons, Ltd. [DOI] [PubMed] [Google Scholar]
- Conith MR, Hu Y, Conith AJ, Maginnis MA, Webb JF, and Albertson RC. 2018b. Genetic and developmental origins of a unique foraging adaptation in a Lake Malawi cichlid genus. Proc. Natl. Acad. Sci. 115:7063–7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte MA, and Kocher TD. 2015. An improved genome reference for the African cichlid, Metriaclima zebra. BMC Genomics 16:724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper WJ, Parsons K, McIntyre A, Kern B, McGee-Moore A, and Albertson RC. 2010. Bentho-pelagic divergence of cichlid feeding architecture was prodigious and consistent during multiple adaptive radiations within African rift-lakes. PLoS One 5:e9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgrange A, Le Garrec J-F, and Meilhac SM. 2018. Left-right asymmetry in heart development and disease: forming the right loop. Development 145:dev162776. [DOI] [PubMed] [Google Scholar]
- Erickson PA, Glazer AM, Cleves PA, Smith AS, and Miller CT. 2014. Two developmentally temporal quantitative trait loci underlie convergent evolution of increased branchial bone length in sticklebacks. Proc. R. Soc. B Biol. Sci. 281:20140822. Royal Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felice RN, and Goswami A. 2018. Developmental origins of mosaic evolution in the avian cranium. Proc. Natl. Acad. Sci. 115:555–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami A, Smaers JB, Soligo C, and Polly PD. 2014. The macroevolutionary consequences of phenotypic integration : from development to deep time. Philos. Trans. R. Soc. B Biol. Sci. 369:20130254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra A, V Germano RF, Stone O, and Arnaout R. 2018. Distinct myocardial lineages break atrial symmetry during cardiogenesis in zebrafish. Elife 7:e32833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrimsson B, Green RM, Katz DC, Fish JL, Bernier FP, Roseman CC, Young NM, Cheverud JM, and Marcucio RS. 2019. The developmental-genetics of canalization. Semin. Cell Dev. Biol. 88:67–79. Elsevier Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrímsson B, Jamniczky HA, Martínez-Abadías N, Young NM, and Marcucio RS. 2012. The Phenogenomics of Craniofacial Shape. FASEB J. 26:337.4–337.4. [Google Scholar]
- Hallgrímsson B, Jamniczky H, Young NM, Rolian C, Parsons TE, Boughner JC, and Marcucio RS. 2009. Deciphering the Palimpsest: Studying the Relationship Between Morphological Integration and Phenotypic Covariation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrimsson B, Mio W, Marcucio RS, and Spritz R. 2014. Let’s Face It—Complex Traits Are Just Not That Simple. PLoS Genet. 10:10–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick BP, Antalek-Schrag P, Conith AJ, Natanson LJ, and Brennan PLR. 2019. Variability and asymmetry in the shape of the spiny dogfish vagina revealed by 2D and 3D geometric morphometrics. J. Zool. 308:16–27. John Wiley & Sons, Ltd. [Google Scholar]
- Hu N, Sedmera D, Yost HJ, and Clark EB. 2000. Structure and function of the developing zebrafish heart. Anat. Rec. 260:148–157. John Wiley & Sons, Ltd. [DOI] [PubMed] [Google Scholar]
- Hu Y, and Albertson RC. 2017. Baby fish working out: An epigenetic source of adaptive variation in the cichlid jaw. Proc. R. Soc. B Biol. Sci. 284:20171018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, and Albertson RC. 2014. Hedgehog signaling mediates adaptive variation in a dynamic functional system in the cichlid feeding apparatus. Proc. Natl. Acad. Sci. 11:8530–8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Ghigliotti L, Vacchi M, Pisano E, Detrich HW, and Albertson RC. 2016. Evolution in an extreme environment: developmental biases and phenotypic integration in the adaptive radiation of antarctic notothenioids. BMC Evol. Biol. 16:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Parsons KJ, and Albertson RC. 2014. Evolvability of the Cichlid Jaw: New Tools Provide Insights into the Genetic Basis of Phenotypic Integration. Evol. Biol. 41:145–153. Springer US. [Google Scholar]
- Hulsey CD, Holzman R, and Meyer A. 2018. Dissecting a potential spandrel of adaptive radiation: Body depth and pectoral fin ecomorphology coevolve in Lake Malawi cichlid fishes. Ecol. Evol. 8:11945–11953. John Wiley & Sons, Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamniczky HA, Boughner JC, Rolian C, Gonzalez PN, Powell CD, Schmidt EJ, Parsons TE, Bookstein FL, and Hallgrímsson B. 2010. Rediscovering Waddington in the post-genomic age: Operationalising Waddington’s epigenetics reveals new ways to investigate the generation and modulation of phenotypic variation. BioEssays 32:553–558. [DOI] [PubMed] [Google Scholar]
- Jensen B, Boukens BJD, V Postma A, Gunst QD, van den Hoff MJB, Moorman AFM, Wang T, and Christoffels VM. 2012. Identifying the Evolutionary Building Blocks of the Cardiac Conduction System. PLoS One 7:e44231. Public Library of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP 2008. Morphological Integration and Developmental Modularity. Annu. Rev. Ecol. Evol. Syst. 39:115–132. [Google Scholar]
- Klingenberg CP 2019. Phenotypic Plasticity, Developmental Instability, and Robustness: The Concepts and How They Are Connected Phenotype Noise n. Front. Ecol. Evol. 7:56. [Google Scholar]
- Klingenberg CP, Barluenga M, and Meyer A. 2002. SHAPE ANALYSIS OF SYMMETRIC STRUCTURES: QUANTIFYING VARIATION AMONG INDIVIDUALS AND ASYMMETRY. Evolution (N. Y). 56:1909–1920. John Wiley & Sons, Ltd (10.1111). [DOI] [PubMed] [Google Scholar]
- Konings A 2007. Malawi Cichlids in Their Natural Habitat. 4th ed. Cichlid Press. [Google Scholar]
- Kratochwil CF, Sefton MM, and Meyer A. 2015. Embryonic and larval development in the Midas cichlid fish species flock (Amphilophus spp.): a new evo-devo model for the investigation of adaptive novelties and species differences. BMC Dev. Biol. 15:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S-T, Chen J-R, Tsai J-J, Lai Y-H, and Hsiao C-D. 2019. Overexpression of Notch Signaling Induces Hyperosteogeny in Zebrafish. Int. J. Mol. Sci. 20:3613. MDPI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Van Der Lijn F, Schurmann C, Zhu G, Chakravarty MM, Hysi PG, Wollstein A, Lao O, De Bruijne M, Ikram MA, Van Der Lugt A, Uitterlinden G, Hofman A, Niessen WJ, Rivadeneira F, Homuth G, De Zubicaray G, Mcmahon KL, Thompson PM, Daboul A, Puls R, Hegenscheid K, Bevan L, Pausova Z, Medland SE, Montgomery GW, Wright MJ, Wicking C, Boehringer S, Spector TD, Martin NG, Biffar R, and Kayser M. 2012. A Genome-Wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans. PLoS Genet. 8:e1002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd E, Chhouk B, Keene AC, and Albertson RC. 2020. Diversity in rest-activity patterns among Lake Malawi cichlid fishes suggests novel axis of habitat partitioning. bioRxiv 2020.07.14.203505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky M, Svardal H, Tyers AM, Miska EA, Genner MJ, Turner GF, and Durbin R. 2018. Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nat. Ecol. Evol. 2:1940–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P, Granina L, Martens K, and Goddeeris B. 1998. Oxygen concentration profiles in sediments of two ancient lakes: Lake Baikal (Siberia, Russia) and Lake Malawi (East Africa). Hydrobiologia 367:163–174. [Google Scholar]
- Martínez-Vargas J, Ventura J, Machuca Á, Muñoz-Muñoz F, Fernández MC, Soto-Navarrete MT, Durán AC, and Fernández B. 2017. Cardiac, mandibular and thymic phenotypical association indicates that cranial neural crest underlies bicuspid aortic valve formation in hamsters. PLoS One 12:e0183556. Public Library of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mašek J, and Andersson ER. 2017. The developmental biology of genetic Notch disorders. Development 144:1743 LP–1763. [DOI] [PubMed] [Google Scholar]
- Mishra R, Sehring I, Cederlund M, Mulaw M, and Weidinger G. 2020. NF-κB Signaling Negatively Regulates Osteoblast Dedifferentiation during Zebrafish Bone Regeneration. Dev. Cell 52:167–182.e7. [DOI] [PubMed] [Google Scholar]
- Navon D, Male I, Tetrault ER, Aaronson B, Karlstrom RO, and Albertson RC. 2020. Hedgehog signaling is necessary and sufficient to mediate craniofacial plasticity in teleosts. Proc. Natl. Acad. Sci. 117:19321–19327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navon D, Olearczyk N, and Albertson RC. 2017. Genetic and developmental basis for fin shape variation in African cichlid fishes. Mol. Ecol. 26:291–303. John Wiley & Sons, Ltd (10.1111). [DOI] [PubMed] [Google Scholar]
- Olson EC, and Miller RL. 1958. Morphological Integration. University of Chicago Press, Chicago, IL. [Google Scholar]
- Parsons KJ, Concannon M, Navon D, Wang J, Ea I, Groveas K, Campbell C, and Albertson RC. 2016. Foraging environment determines the genetic architecture and evolutionary potential of trophic morphology in cichlid fishes. Mol. Ecol. 25:6012–6023. [DOI] [PubMed] [Google Scholar]
- Parsons KJ, Cooper WJ, and Albertson RC. 2011a. Modularity of the Oral Jaws Is Linked to Repeated Changes in the Craniofacial Shape of African Cichlids. Int. J. Evol. Biol. 2011:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons KJ, Son YH, and Albertson RC. 2011b. Hybridization Promotes Evolvability in African Cichlids: Connections Between Transgressive Segregation and Phenotypic Integration. Evol. Biol. 38:306–315. [Google Scholar]
- Ribbink AJ, Marsh BA, Marsh AC, Ribbink AC, and Sharp BJ. 1983. A preliminary survey of the cichlid fishes of rocky habitats in Lake Malawi. South African J. Zool. 18:149–309. [Google Scholar]
- Rohlf FJ 1998. On applications of geometric morphometrics to studies of ontogeny and phylogeny. Syst. Biol. 47:147–167. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ, and Corti M. 2000. Use of Two-Block Partial Least-Squares to Study Covariation in Shape. Syst. Biol. 49:740–753. [DOI] [PubMed] [Google Scholar]
- Rothschild SC, Easley CA, Francescatto L, Lister JA, Garrity DM, and Tombes RM. 2009. Tbx5-mediated expression of Ca2+/calmodulin-dependent protein kinase II is necessary for zebrafish cardiac and pectoral fin morphogenesis. Dev. Biol. 330:175–184. [DOI] [PubMed] [Google Scholar]
- Samsa LA, Givens C, Tzima E, Stainier DYR, Qian L, and Liu J. 2015. Cardiac contraction activates endocardial Notch signaling to modulate chamber maturation in zebrafish. Development 142:4080 LP–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager S 2018. Morpho: Calculations and Visualisations Related to Geometric Morphometrics. CRAN. [Google Scholar]
- Schlager S 2017. Morpho and Rvcg - Shape Analysis in R. Pp. 217–256 in Zheng G, Li S, and Szekely G, eds. Statistical Shape and Deformation Analysis. [Google Scholar]
- Sedmera D, Reckova M, DeAlmeida A, Sedmerova M, Biermann M, Volejnik J, Sarre A, Raddatz E, McCarthy RA, Gourdie RG, and Thompson RP. 2003. Functional and morphological evidence for a ventricular conduction system in zebrafish and Xenopus hearts. Am. J. Physiol. Circ. Physiol. 284:H1152–H1160. American Physiological Society. [DOI] [PubMed] [Google Scholar]
- Shifatu O, Glasshagel-Chilson S, Nelson HM, Patel P, Tomamichel W, Higginbotham C, Evans PK, Lafontant GS, Burns AR, and Lafontant PJ. 2018. Heart Development, Coronary Vascularization and Ventricular Maturation in a Giant Danio (Devario malabaricus). J. Dev. Biol. 6:19. MDPI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AJ, Nelson-Maney N, Parsons KJ, James Cooper W, and Craig Albertson R. 2015. Body Shape Evolution in Sunfishes: Divergent Paths to Accelerated Rates of Speciation in the Centrarchidae. Evol. Biol. 42:283–295. Springer US. [Google Scholar]
- Stewart TA, and Albertson RC. 2010. Evolution of a unique predatory feeding apparatus: functional anatomy, development and a genetic locus for jaw laterality in Lake Tanganyika scale-eating cichlids. BMC Biol. 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JLY, Guo Y, Stockdale WT, Rana K, Killen AC, Mommersteeg MTM, and Yamamoto Y. 2018. The developmental origin of heart size and shape differences in Astyanax mexicanus populations. Dev. Biol. 441:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH 1942a. Canalization of development and the inheritance of acquired characters. Nature 150:563–565. [DOI] [PubMed] [Google Scholar]
- Waddington CH 1953. GENETIC ASSIMILATION OF AN ACQUIRED CHARACTER. Evolution (N. Y). 7:118–126. John Wiley & Sons, Ltd (10.1111). [Google Scholar]
- Waddington CH 1942b. The Epigenotype. Endeavour 1:18–20. [Google Scholar]
- Waddington CH 1957. The Strategy of the Genes. George Allen & Unwin, London, UK. [Google Scholar]
- Watanabe A, Fabre A-C, Felice RN, Maisano JA, Müller J, Herrel A, and Goswami A. 2019. Ecomorphological diversification in squamates from conserved pattern of cranial integration. Proc. Natl. Acad. Sci. 116:14688 LP–14697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner D, Cizelsky W, Vasudevaro MD, Özhan G, Haase C, Kagermeier-Schenk B, Röder A, Dorsky RI, Moro E, Argenton F, Kühl M, and Weidinger G. 2014. Wnt/β-Catenin Signaling Defines Organizing Centers that Orchestrate Growth and Differentiation of the Regenerating Zebrafish Caudal Fin. Cell Rep. 6:467–481. [DOI] [PubMed] [Google Scholar]
- Wiley DF, Amenta N, Alcantara DA, Ghosh D, Kil YJ, and et al. 2005. Evolutionary Morphing. Pp. 431–438 in Proceedings of IEEE Visualization. [Google Scholar]
- Yamamoto M, Takada H, Ishizuka S, Kitamura K, Jeong J, Sato M, Hinata N, and Abe S. 2020. Morphological association between the muscles and bones in the craniofacial region. PLoS One 15:e0227301. Public Library of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kilgallen S, Andreeva V, Spicer DB, Pinz I, and Friesel R. 2010. Conditional expression of Spry1 in neural crest causes craniofacial and cardiac defects. BMC Dev. Biol. 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NM, and Hallgrímsson B. 2005. Serial homology and the evolution of mammalian limb covariation structure. Evolution (N. Y). 59:2691–2704. [PubMed] [Google Scholar]
- Żelaźniewicz A, and Pawłowski B. 2018. Maternal breast and body symmetry in pregnancy and offspring condition. Am. J. Phys. Anthropol. 166:127–138. John Wiley & Sons, Ltd. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


