Abstract
Epidemiological evidence suggests that some methicillin-resistant Staphylococcus aureus (MRSA) strains are more prone to dissemination than others. We studied 72 MRSA strains, collected through nationwide MRSA surveillance in 1992 through 1999 and known to be either (i) sporadic, (ii) local outbreak strains spread within one hospital, or (iii) epidemic strains spread among hospitals, by antimicrobial susceptibility testing, hybridization of the mec hypervariable region (HVR), and ribotyping. Our results show that two main groups can be identified among these strains. The first group includes mainly nonepidemic, nonmultiresistant MRSA strains showing a specific mec HVR hybridization pattern, A, in combination with a variety of ribotypes. The other group includes multiresistant strains with mec HVR hybridization pattern B or C in association with closely related ribotype a or b. Sixty-four percent (9 of 14) of Finnish epidemic MRSA strains belong to the latter group. These findings support the existence of differences in epidemic potential among MRSA strains.
Certain methicillin-resistant Staphylococcus aureus (MRSA) strains have efficiently disseminated both locally and internationally. The factors affecting dissemination of resistant organisms include hospital size and type, antimicrobial policies of hospitals, frequency of patient transfer between wards and hospitals, certain risk factors predisposing patients to MRSA colonization and infection, and infection control measures (1, 2, 4, 25). Based on epidemiological evidence, it has also been suggested that some MRSA strains have properties that enable them to disseminate particularly well (19).
The search for genetic properties linked to epidemic spread can be initiated by comparing the characteristics of known epidemic and sporadic strains. The genetic background of MRSA strains is determined by the mec DNA and the original chromosome of methicillin-susceptible S. aureus to which the mec DNA has integrated. Although the genomes of the susceptible and the resistant strains have diversified since this initial integration, certain genotyping methods, such as ribotyping, can be used to delineate the genetic background of the MRSA strains (10). Ribotyping with concomitant analysis of the mec DNA thus provides information on the potential differences in the genomic background or on the mec DNA of epidemic and sporadic strains.
The 20- to 50-kb mec DNA includes the mecA gene, coding for penicillin-binding protein 2a, which is primarily responsible for methicillin resistance, and the complete or truncated regulatory region mecRI-mecI (14, 20). The mecA downstream sequences contain series of direct repeat units (dru region), an open reading frame whose product has amino acid similarity to the N terminus of Escherichia coli glycerophosphoryl diester phosphodiesterase (UpgQ), and insertion sequence IS431 (16, 21). The genomic distance between mecA and IS431 is highly variable, and this region is called the mec hypervariable region (HVR) (11, 16, 21). This region and the insertion sequences and transposons within the mec DNA can serve as hot spots, which collect antibiotic resistance genes or other markers into the MRSA genome (24).
In this study, we searched for molecular traits linked to epidemic spread using a subset of 72 MRSA isolates from the Finnish MRSA strain collection. This population-based collection represents all Finnish MRSA strains typed since 1992 and includes both epidemic and sporadic strains.
(These results have partly been presented at the 9th European Congress of Clinical Microbiology and Infectious Diseases, 21 to 24 March 1999, Berlin, Germany [S. Salmenlinna and J. Vuopio-Varkila, 9th Eur. Congr. Clin. Microbiol. Infect. Dis., poster PO112, 1999].)
MATERIALS AND METHODS
Since 1992, all clinical microbiology laboratories in Finland have sent MRSA isolates to the Hospital Infection Laboratory at the National Public Health Institute (KTL) for typing. In 1995, MRSA became a notifiable laboratory finding in Finland. Most laboratories report the findings electronically to the National Infectious Disease Register at KTL.
Routine MRSA typing scheme in 1992 through 1999.
All MRSA isolates referred to KTL were typed by phages and tested for antimicrobial susceptibility. Phage typing was performed with the universal set of phages (18) at 1× and 100× routine test dilutions, both with and without heat treatment of the bacteria (5). The antimicrobial susceptibilities were tested by the disk diffusion method according to guidelines recommended by the National Committee for Clinical Laboratory Standards (15). The antimicrobials tested were oxacillin, ampicillin, penicillin, cephalexin, cefuroxime, gentamicin, tobramycin, erythromycin, clindamycin, chloramphenicol, ciprofloxacin, rifampin, fusidic acid, and mupirocin. MICs of oxacillin were determined by the E-test according to the manufacturer's instructions (AB Biodisk, Solna, Sweden). Additional studies using pulsed-field gel electrophoresis (PFGE) and, occasionally, ribotyping were performed if the isolates met any of the following criteria: (i) they were known to be linked to a suspected epidemic based on epidemiological information obtained from the submitting hospital, (ii) they possessed an antimicrobial susceptibility pattern and a phage type similar to those for a strain identified earlier in the laboratory, and (iii) they could not be typed by phages. PFGE and ribotyping were performed as described earlier (22). Briefly, for PFGE, genomic DNA prepared in agarose blocks was digested with SmaI restriction endonuclease, and the chromosomal fragments were separated with a CHEF DR III (Bio-Rad, Richmond, Calif.) for 24 h, with initial and final switching times of 10 and 60 s, respectively. PFGE patterns were interpreted as different if more than three band differences occured. Ribotyping was performed by digesting genomic DNA with EcoRI restriction endonuclease. DNA fragments were separated by electrophoresis, transferred to a nylon membrane, and hybridized with a probe for the rrn operon of E. coli (3).
Strain definitions.
By combining the typing data and epidemiological information from hospitals, all strains were classified as either sporadic, local outbreak, or epidemic strains. A sporadic MRSA strain was defined as a strain isolated from one person only and displaying an individual antimicrobial susceptibility pattern and/or phage type. For non-phage-typeable strains, the strain relatedness was ascertained by PFGE. MRSA isolates sharing the same typing patterns and isolated from two or more persons in the same hospital were defined as representative of a locally spread outbreak strain. MRSA isolates sharing the same typing patterns and isolated from two or more persons and at two or more hospitals were defined as representative of an epidemic MRSA strain.
Selection of isolates for analysis of genomic background.
From the MRSA collection, built up by strains sent for typing since 1992, 72 isolates were selected for more detailed analysis of genomic background. These isolates included all 47 sporadic isolates from one representative year, namely, 1995, as well as one representative of each MRSA strain which has caused epidemics (n = 14) or local outbreaks (n = 11) from 1992 to 1998. Eighteen of these strains have been described previously (22).
Analysis of the HVR mec determinant.
Genomic variation within the mec determinant was analyzed by hybridization analysis. The genomic DNA from MRSA strains was digested with EcoRI and BglII restriction enzymes and hybridized separately with two different probes prepared from plasmid pBBB30 (21). HVR probe I is a 3.6-kb fragment that recognizes the HVR of the mec determinant, starting within the mecA gene up to IS431 mec. HVR probe II is a 1.5-kb fragment digested with HindIII from the 3.6-kb fragment. This probe recognizes the dru and an E. coli ugpQ-like sequence (orf145) within the mec determinant. The extraction of genomic DNA, digestion with restriction enzymes, gel electrophoresis with digoxigenin-labeled molecular weight marker III (Roche Molecular Biochemicals) as a migration control, blotting on to a nylon membrane, and labeling reactions were performed as previously described (22). The hybridization was performed according to instructions provided by the nylon membrane manufacturer (Boehringer Mannheim). Briefly, after 2 h of prehybridization in 20 ml of hybridization buffer (0.75 M NaCl, 75 mM sodium citrate, 1% blocking reagent, 0.1% N-lauroylsarcosine, 0.02% sodium dodecyl sulfate) at 60°C, about 80 ng of probe (HVR I or HVR II) was added to 20 ml of hybridization buffer, boiled for 10 min, and hybridized with DNA fragments for 20 to 24 h at 60°C.
Ribotyping.
Ribotyping with EcoRI was used to detect the phylogenetic relationship within the MRSA strains. MluI-digested Citrobacter koseri was used as the migration control (22). A difference of one band in the ribopattern, as detected by visual analysis, was considered to represent a new type. An arbitrary identification letter was given to each ribotype. Computer-assisted analysis of the ribotypes was performed by BioNumerics, version 1.0 (Applied Maths, Kortrijk, Belgium), using the Dice coefficient with a position tolerance setting of 0.9%, optimization setting of 0.8%, and unweighted pair-group method using arithmetic averages.
Statistical analysis.
Statistical associations were tested by the chi-square method using Yates' correction.
RESULTS
MRSA strains.
Since 1995, the annual number of MRSA isolations reported to the National Infectious Disease Register has varied from 89 to 211, with an incidence per population of 10,000 of 0.17 to 0.41 (Table 1). During the period from 1992 through 1999, a total of 1,299 MRSA strains were referred to the Hospital Infection Laboratory (Table 1), i.e., 118 to 225 MRSA strains per year. According to the definitions described above, 538 (41%) of the isolates were sporadic, 109 (8%) were local outbreak strains, and 652 (50%) were epidemic strains. The annual percentage of epidemic strains ranged between 30 and 77%.
TABLE 1.
MRSA strains isolated in Finland in 1992 through 1999
| Yr | No. of MRSA strains reported to the National Infectious Disease Register | Incidencea | No. of MRSA strains referred to the Hospital Infection Laboratoryc | No. of sporadic strains (%) | No. of local outbreak strains (%) | No. of epidemic strains (%) |
|---|---|---|---|---|---|---|
| 1992 | NAb | NA | 140 | 30 (21) | 2 (1) | 108 (77) |
| 1993 | NA | NA | 128 | 41 (32) | 30 (23) | 57 (45) |
| 1994 | NA | NA | 225 | 76 (34) | 12 (5) | 137 (61) |
| 1995 | 89 | 0.17 | 137 | 47 (35) | 6 (4) | 84 (61) |
| 1996 | 108 | 0.21 | 118 | 55 (46) | 2 (2) | 61 (52) |
| 1997 | 120 | 0.23 | 143 | 93 (65) | 8 (5) | 42 (30) |
| 1998 | 189 | 0.37 | 196 | 120 (61) | 11 (6) | 65 (33) |
| 1999 | 211 | 0.41 | 212 | 76 (36) | 38 (18) | 98 (46) |
| Total | NA | NA | 1,299 | 538 (41) | 109 (8) | 652 (50) |
Incidence per population of 10,000 based on MRSA cases reported to the National Infectious Disease Register.
NA, not applicable.
Mean, 162 per year.
The observation that epidemic strains represent half of all MRSA isolates in Finland led us to hypothesize that these strains may harbor characteristics favorable for epidemic spread. To test this hypothesis, we selected 72 MRSA strains for further analysis. These included all sporadic isolates from 1995 (n = 47) and one representative isolate of each epidemic (n = 14) and outbreak (n = 11) strain identified between 1992 and 1998.
Antibiotic susceptibility.
Of the 47 sporadic strains, 14 (30%) were multiresistant (i.e., resistant to more than three antibiotic groups in addition to beta-lactams), 14 (30%) were resistant to beta-lactams and one to three other antibiotic groups, and 19 (40%) were resistant to beta-lactams only. The corresponding figures for epidemic strains were 11 (79%), 1 (7%), and 2 (14%), respectively. The corresponding figures for local outbreak strains were 5 (45%), 3 (27%), and 3 (27%), respectively. Together, these results indicate that multiresistance was more common among epidemic strains (11 of 14) than among nonepidemic MRSA strains (19 of 58) (P < 0.005).
Analysis of the HVR mec determinant.
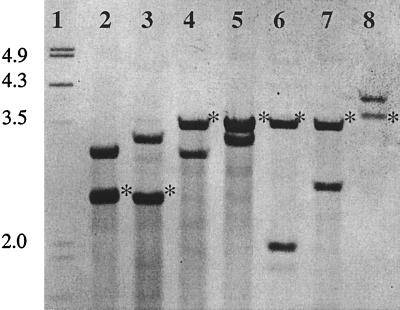
Hybridization with HVR probe I differentiated the 72 strains into 7 types: A, B, C, D, E, F, and G (Fig. 1), each of which comprised two bands. HVR probe II recognized the other of these two bands showing the position of dru-orf145 and flanking sequences with three different migration profiles. HVR types A, C, E, and F showed a dru-orf145 band of approximately 3.5 kb, HVR types B and D showed a band of approximately 2.5 kb, and HVR type G showed a band of more than 3.5 kb.
FIG. 1.
HVR probe I hybridization patterns. Lanes 1, molecular size marker (fragment sizes are indicated as kilobase pairs); 2 to 8, HVR types B, D, A, C, E, F, and G. Asterisks indicate the fragments recognized by HVR probe II (the dru-orf145 fragments).
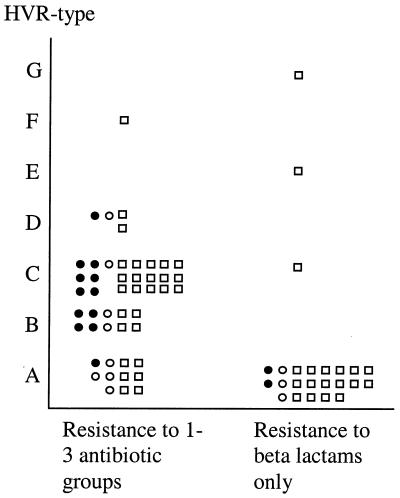
Most sporadic strains were either HVR type A (22 of 47 [47%]) or type C (16 of 47 [34%]) (Fig. 2). The epidemic strains were mostly HVR type B (4 of 14 [29%]) or type C (6 of 14 [43%]). Three epidemic strains were HVR type A, and one was type D. Of the 11 local outbreak strains, 7 (64%) were HVR type A and 4 were type B, C, or D.
FIG. 2.
HVR type in relation to antibiotic resistance patterns and epidemic spread. ●, epidemic strain; ○, local outbreak strain; □, sporadic strain.
Ribotyping.
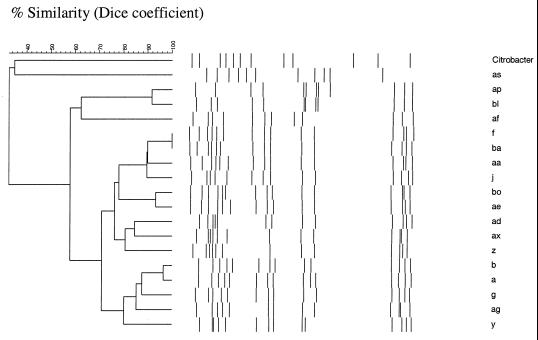
Ribotyping with EcoRI yielded 18 different ribotypes (Fig. 3). The most prevalent types were a and b (35 [49%] of 72 strains). Of the sporadic strains, 21 (45%) showed either ribotype a or ribotype b. The corresponding figures for epidemic and local outbreak strains were 9 (64%) and 5 (45%), respectively. Ribotypes a and b differed from each other by only one band. Computer-assisted analysis of ribotypes showed that a >92% similarity level corresponded to a one- to two-band difference and an 87 to 90% similarity level corresponded to a three- to four-band difference (Fig. 3).
FIG. 3.
Dendrogram of relatedness of the ribotypes.
Linkage of antibiotic susceptibility to genotype and epidemiological background.
In combined analysis, the MRSA strains clustered into two main groups (Tables 2 and 3 and Fig. 2). Thirty-two of 48 strains resistant to more than one antibiotic group, the majority of which were multiresistant, associated with ribotype a or b (32 of all 35 strains with ribotype a or b, P < 0.001), and 32 of all 33 strains associated with HVR type B or C (P < 0.001). Thirty-one strains showed both ribotype a or b and HVR type B or C. This group included 9 of the 14 (64%) epidemic strains, 3 of the 11 (27%) local outbreak strains, and 19 of the 47 (40%) sporadic strains. The remaining 16 strains, expressing resistance to antibiotic groups in addition to the beta-lactams, belonged to heterogeneous ribotypes with HVR types A, C, F, and D (Table 2).
TABLE 2.
Strains resistant to other antibiotic group(s) in addition to beta-lactams
| HVR type | No. of strains (n = 48) associated with the following ribotype:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | g | f | j | y | z | aa | ad | ae | ag | ax | ba | bo | af | bl | ap | as | |
| A | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||
| B | 6 | 4 | ||||||||||||||||
| C | 11 | 10 | 1 | |||||||||||||||
| D | 4 | |||||||||||||||||
| E | ||||||||||||||||||
| F | 1 | |||||||||||||||||
| G | ||||||||||||||||||
TABLE 3.
Strains resistant to beta-lactams only
| HVR type | No. of strains (n = 24) associated with the following ribotype:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | g | f | j | y | z | aa | ad | ae | ag | ax | ba | bo | af | bl | ap | as | |
| A | 3 | 3 | 4 | 1 | 6 | 1 | 1 | 1 | 1 | |||||||||
| B | ||||||||||||||||||
| C | 1 | |||||||||||||||||
| D | ||||||||||||||||||
| E | 1 | |||||||||||||||||
| F | ||||||||||||||||||
| G | 1 | |||||||||||||||||
In contrast, MRSA strains which were resistant to beta-lactams only (n = 24) showed remarkable heterogeneity in ribotypes (21 of 37 strains with ribotype other than a or b, P < 0.001) and were mostly HVR type A (21 of 32, P < 0.001) (Table 3). Although the resistance solely to beta-lactams did not directly relate to sporadicity, the majority of strains resistant to beta-lactams only and showing HVR type A (16 of 21 [76%]) were sporadic by nature. The remaining three strains, with HVR types C, E, and G, were all sporadic and had different ribotypes.
DISCUSSION
Based on antimicrobial susceptibility testing, ribotyping, and hybridization analysis of the mec region of 72 epidemic, local outbreak, and sporadic MRSA strains, the prevalent Finnish MRSA strains can be classified into two main groups. One group includes strains showing mec HVR hybridization pattern A combined with a variety of ribotypes and resistance to beta-lactam antibiotics only. The majority of these strains are sporadic by nature. The other group includes strains with mec HVR hybridization pattern B or C in association with ribotype a or b and resistance to other antibiotic groups in addition to beta-lactams. This group includes both epidemic and sporadic strains.
The incidence of MRSA in Finland is low, which allows efficient nationwide surveillance. Practically all MRSA isolates are routinely typed in Finland, which is also evident from convergence of figures on strains referred to the Hospital Infection Laboratory and reported to the National Infectious Disease Register. The MRSA prevention and screening policies in different health care facilities in Finland are rather uniform, which also justifies comparison of the spreading capacities of different MRSA strains. The national guidelines, from 1995, include protocols for rapid identification of MRSA, prevention of MRSA transmission, and treatment of MRSA infection and carriage, as well as actions to be undertaken when a new case of MRSA infection or carriage is recognized. All patients who have been hospitalized outside the Nordic countries are advised to be screened for MRSA carriage and treated in contact isolation until a negative culture result is obtained.
In accordance with previous reports (16, 17, 23), Finnish MRSA strains showed variability within the HVR of the mecA complex. We detected changes both in the sizes of the dru-orf145 fragment and in the fragments containing major parts of the mecRI and mecA genes. The strains with HVR type B or D, but expressing three different ribotypes, showed an approximately 1-kb-smaller dru-orf145 fragment than the other strains, suggesting that deletions may have occurred within the dru region in different MRSA lineages, as recently also reported by Oliveira et al. (17). Sequencing of the different HVR types is warranted to reveal the molecular nature of this variability.
The majority of epidemic strains belonged to a cluster expressing HVR type B or C, multiresistance, and ribotype a or b. It may be argued that this cluster represents two related clones expressing two different HVR types (17). However, this was not the case, since additional PFGE analysis determined that the majority (24 of 31) of these strains were different (data not shown). HVR types B and C may also be common among pandemic MRSA strains, as several of these Finnish epidemic MRSA strains were originally imported from abroad. Although ribotypes a and b were common among the epidemic strains, they were also found among sporadic strains but notably only among those which were resistant to not only the beta-lactams but also other drugs. Some of the HVR type C strains remained sporadic and may thus represent strains which possess the potential for epidemic spread, but depending on infection control efforts, they failed to cross-infect.
A large proportion of sporadic strains were HVR type A. These strains were typically heterogeneous, expressing a variety of different ribotypes, and were resistant to beta-lactams only. The reservoir for these sporadic, nonmultiresistant MRSA strains remains unknown. It is possible that some of them are not linked to hospitalization. Community strains of MRSA have been reported within remote communities (13), among drug abusers (12), and recently among children and healthy adults without any known risk factors (8, 9). In Finland, approximately one-third of MRSA strains are resistant to beta-lactams only. Further studies are required to investigate the epidemiology of these strains in detail.
In conclusion, we found two epidemiologically distinct groups of MRSA strains in Finland. The first group includes multiresistant, epidemic MRSA strains; the other group includes sporadic MRSA strains resistant to beta-lactams only. The mecA complex of these strains showed heterogenieity. This finding is promising in respect to future development of diagnostic tests for detection of MRSA strains with epidemic-spreading capacity.
ACKNOWLEDGMENTS
We thank Outi Lyytikäinen for the data on Infectious Disease Register and valuable advice and Elina Siren and Ritva Scotford for technical assistance.
REFERENCES
- 1.Ayliffe G A J. The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 1997;24(Suppl. 1):74–79. doi: 10.1093/clinids/24.supplement_1.s74. [DOI] [PubMed] [Google Scholar]
- 2.Boyce J M. Methicillin-resistant Staphylococcus aureus: a continuing infection control challenge. Eur J Clin Microbiol Infect Dis. 1994;13:45–49. doi: 10.1007/BF02026126. [DOI] [PubMed] [Google Scholar]
- 3.Brosius J, Ullrich A, Raker M A, Gray A, Dull T J, Gutell R R, Noller H F. Construction and fine mapping of recombinant plasmids containing the rrn B ribosomal RNA operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 4.Coello R, Glynn J R, Gaspar C, Picazo J J, Fereres J. Risk factors for developing clinical infection with methicillin-resistant Staphylococcus aureus (MRSA) amongst hospital patients initially only colonized with MRSA. J Hosp Infec. 1997;37:39–46. doi: 10.1016/s0195-6701(97)90071-2. [DOI] [PubMed] [Google Scholar]
- 5.Davies H G, Martin D R. Heat shocking as a useful adjunct to routine phage typing of Staphylococcus aureus. J Hosp Infect. 1987;10:4–9. doi: 10.1016/0195-6701(87)90026-0. [DOI] [PubMed] [Google Scholar]
- 6.Dobbeling B N. The epidemiology of methicillin-resistant Staphylococcus aureus colonization and infection. J Chemother. 1995;7(Suppl. 3):99–103. [PubMed] [Google Scholar]
- 7.Dubin D T, Matthews P R, Chikramane S G, Stewart P R. Physical mapping of the mec region of an American methicillin-resistant Staphylococcus aureus strain. Antimicrob Agents Chemother. 1991;35:1661–1665. doi: 10.1128/aac.35.8.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorak E J, Yamada S M, Brown J D. Community-acquired methicillin-resistant Staphylococcus aureus in hospitalized adults and children without known risk factors. Clin Infect Dis. 1999;29:797–800. doi: 10.1086/520437. [DOI] [PubMed] [Google Scholar]
- 9.Herold B C, Immergluck L C, Maranam M C, Lauderdale D S, Gaskin R E, Boyle-Vavra S, Leitch C D, Daum R S. Community acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA. 1998;279:593–595. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 10.Hiramatsu K, Kondo N, Ito T. Genetic basis for molecular epidemiology of MRSA. J Infect Chemother. 1996;2:117–129. doi: 10.3412/jsb.52.417. [DOI] [PubMed] [Google Scholar]
- 11.Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine D, Cushing R, Jui J, Brown W. Community-acquired methicillin-resistant Staphylococcus aureus endocarditis in the Detroit Medical Center. Ann Intern Med. 1982;97:330–338. doi: 10.7326/0003-4819-97-3-330. [DOI] [PubMed] [Google Scholar]
- 13.Maguire G P, Arthur A D, Boustead P J, Dwyer B J, Currie B J. Clinical experience and outcome of community-acquired and nosocomial methicillin-resistant Staphylococcus aureus in a northern Australian hospital. J Hosp Infect. 1998;38:273–281. doi: 10.1016/s0195-6701(98)90076-7. [DOI] [PubMed] [Google Scholar]
- 14.Matthews P R, Reed K C, Stewart P R. The cloning of chromosomal DNA associated with methicillin and other resistances in Staphylococcus aureus. J Gen Microbiol. 1987;133:1919–1929. doi: 10.1099/00221287-133-7-1919. [DOI] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2–A6. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 16.Nishi J I, Miyanohara H, Nakajima T, Kitajima J, Yoshinaga M, Maruyama I, Miyata K. Molecular typing of the methicillin-resistance determinant (mec) of clinical strains of Staphylococcus aureus based on mec hypervariable region length polymorphisms. J Lab Clin Med. 1995;126:29–35. [PubMed] [Google Scholar]
- 17.Oliveira D C, Wu S W, de Lencastre H. Genetic organization of the downstream region of the mecA element in methicillin-resistant Staphylococcus aureus isolates carrying different polymorphisms of this region. Antimicrob Agents Chemother. 2000;44:1906–1910. doi: 10.1128/aac.44.7.1906-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker M T. Phage typing of Staphylococcus aureus. Methods Microbiol. 1972;7B:1–28. [Google Scholar]
- 19.Phillips I. Epidemic potential and pathogenicity in outbreaks with EMRSA and EMREC. J Hosp Infect. 1991;18(Suppl. A):197–201. doi: 10.1016/0195-6701(91)90024-3. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds P E, Fuller C. Methicillin-resistant Staphylococcus aureus: presence of identical additional penicillin binding protein in all strains examined. FEMS Microbiol Lett. 1986;33:251–254. [Google Scholar]
- 21.Ryffel C, Bucher R, Kayser F H, Berger-Bächi B. The Staphylococcus aureus mec determinant comprises an unusual cluster of direct repeats and codes for a gene product similar to the Escherichia coli sn-glycerophosphoryl diester phosphodiesterase. J Bacteriol. 1991;173:7416–7422. doi: 10.1128/jb.173.23.7416-7422.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmenlinna S, Lyytikäinen O, Kotilainen P, Scotford R, Siren E, Vuopio-Varkila J. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Finland. Eur J Clin Microbiol Infect Dis. 2000;19:101–107. doi: 10.1007/s100960050438. [DOI] [PubMed] [Google Scholar]
- 23.Schmitz F-J, Steiert M, Tichy H-V, Hofmann B, Verhoef J, Heinz H-P, Köhrer K, Jones W E. Typing of methicillin-resistant Staphylococcus aureus isolates from Düsseldorf by six genotypic methods. J Med Microbiol. 1998;47:341–351. doi: 10.1099/00222615-47-4-341. [DOI] [PubMed] [Google Scholar]
- 24.Stewart P R, Dubin D T, Chikramane S G, Inglis B, Matthews P R, Poston S M. IS257 and small plasmid insertions in mec-region of the chromosome of Staphylococcus aureus. Plasmid. 1994;31:12. doi: 10.1006/plas.1994.1002. [DOI] [PubMed] [Google Scholar]
- 25.Wagenvoort J H T. Dutch measures to control MRSA and the expanding European union. Eurosurveillance. 2000;5:26–28. doi: 10.2807/esm.05.03.00031-en. [DOI] [PubMed] [Google Scholar]