Abstract
Conserved from yeast to humans and composed of six core subunits (Elp1–Elp6), Elongator is a multiprotein complex that catalyzes the modification of the anticodon loop of transfer RNAs (tRNAs) and in turn regulates messenger RNA decoding efficiency. Previous studies showed that yeast Elongator consists of two subassemblies (yElp1/2/3 and yElp4/5/6) and adopts an asymmetric overall architecture. Yet, much less is known about the structural properties of the orthologous human Elongator. Furthermore, the order in which the different Elongator subunits come together to form the full assembly as well as how they coordinate with one another to catalyze tRNA modification is not fully understood. Here, we purified recombinant human Elongator subunits and subassemblies and examined them by single-particle electron microscopy. We found that the human Elongator complex is assembled from two subcomplexes that share similar overall morphologies as their yeast counterparts. Complementary co-purification and pulldown assays revealed that the scaffolding subunit human ELP1 (hELP1) has stabilizing effects on the human ELP3 catalytic subunit. Furthermore, the peripheral hELP2 subunit appears to enhance the integrity and substrate-binding ability of the dimeric hELP1/2/3. Lastly, we found that hELP4/5/6 is recruited to hELP1/2/3 via hELP3. Collectively, our work generated insights into the assembly process of core human Elongator and the coordination of different subunits within this complex.
Introduction
As a key adaptor molecule in translation, transfer RNAs (tRNAs) undergo a broad range of enzyme-catalyzed post-transcriptional modifications. A hotspot of tRNA modification is position 34 or the first position of the anticodon.1 This tRNA nucleoside interacts with the third position of a messenger RNA (mRNA) codon and mediates non-Watson–Crick pairing, which allows the ribosome to accommodate different “wobble pairs” in the decoding process.2 tRNA modifications at position 34 regulate wobble pairing and enhance decoding efficiency by altering the conformation of the anticodon.3,4 A key enzyme in tRNA post-transcriptional modification is Elongator, which is a multiprotein enzyme complex that specifically modifies uridine bases at position 34 (U34).5 In particular, Elongator introduces a carboxymethyl group to the C5-position of this uridine (cm5U), priming this nucleoside for subsequent conversion to 5-carbamoylmethyl (ncm5U), 5-methoxycarbonylmethyl (mcm5U), or 5-methoxy-carbonyl-methyl-2-thio (mcm5s2U) groups by other partner enzymes in the cascade.6
Conserved from yeast to humans, Elongator was originally discovered as a component of the elongating RNA Polymerase II ternary complex.7−11 However, further studies in fission yeast Schizosaccharomyces pombe and budding yeast Saccharomyces cerevisiae suggested that the most likely physiological function of Elongator is in post-transcriptional modification of tRNA.12−16 Subsequent tandem affinity purification analyses in yeast Saccharomyces cerevisiae revealed that Elongator is an ∼650 kDa multiprotein complex composed of two copies of six subunits (Elp1, Elp2, Elp3, Elp4, Elp5, and Elp6). These proteins are arranged into two discrete subassemblies: Elp1-Elp2-Elp3 (hereafter Elp1/2/3) and Elp4-Elp5-Elp6 (hereafter Elp4/5/6).8,9 Often referred to as core Elongator, Elp1/2/3 contains main catalytic subunit Elp3 and two structural proteins Elp1 and Elp2. Elp1 and Elp2, through their WD40 beta propeller domains, generate a scaffold that houses Elp3 as well as mediates substrate interaction.17 X-ray crystallographic analysis of human ELP1 (hELP1) and yeast Elp1 (yElp1) further showed the tetratricopeptide domain at the C-terminus of this protein mediates dimerization required for full complex assembly.18 The recent crystal structure of a bacterial orthologue of Elp3 showed that this catalytic subunit contains a catalytically inactive lysine acetyltransferase domain, an iron–sulfur (Fe–S) cluster binding site, and a radical S-adenosylmethionine domain that mediates the chemical modification.19 For Elp4/5/6, biochemical and structural characterization of yeast Elp4/5/6 (yElp4/5/6) showed that these proteins form a hexameric RecA ATPase-like assembly that is capable of both hydrolyzing ATP and binding tRNA.20,21
The overall architecture and subunit arrangement of full yeast Elongator were recently resolved through negative stain single-particle electron microscopy (EM) analysis of core Elongator and full Elongator isolated from yeast S. cerevisiae.22,23 Notably, yeast core Elongator was observed to adopt a symmetrical bi-lobal architecture. This symmetry was broken in full Elongator because of the yElp4/5/6 subcomplex binding to one of two lobes. More recently, high-resolution cryo-EM analysis of yeast core Elongator (yElp1/2/3) generated further molecular details on interaction interface between different subunits as well as how yeast core Elongator engages in interaction with its tRNA substrate.24 Despite these recent advances, many fundamental questions on Elongator remain unanswered. For example, the precise roles of the different subunits in supporting Elongator function are not fully understood. Furthermore, the proposed mechanism of substrate carboxymethylation by Elongator derived from experiments using archaeal Elp3 has yet to be validated in the context of core and full Elongator.25
Because of its critical role in tRNA modification, Elongator dysfunction has been shown to affect proteome homeostasis by causing an increase in +1 frameshifting and a reduction in codon-dependent translation speed.26,27 In yeast, deletion mutants of individual “Elp” genes show growth defects in the presence of stressors such as high salt, caffeine, and 6-azouracil.8,9,11,28 Phenotypes of Elongator deficient mutants in higher eukaryotes are more severe but are tissue-dependent. In mice, aberrations in Elongator function within germ cells, neuronal cells, and retinal cells were shown to cause infertility, neurodegeneration, and blindness, respectively.29−32 In humans, an exon-skipping mutation of the gene encoding hELP1 causes Familial Dysautonomia, a congenital neurodevelopmental disease featuring an unusually low number of neurons in the sensory and autonomic nervous systems, which result in symptoms such as pain insensitivity, gastrointestinal dysfunction, and scoliosis.33−35 Gene mutations in other human Elongator subunits have also been associated with Amyotrophic lateral sclerosis, Rolandic epilepsy, and intellectual disability.36−45 More recently, germline loss-of-function mutation of the hELP1 gene was found to be linked to the malignant brain cancer medulloblastoma.39
Despite growing evidence supporting the direct relevance of Elongator in different human pathologies, the effects of disease-associated mutations on human Elongator are not fully understood. This is due in part to the fact that much of the current biochemical and structural knowledge on Elongator and its components is derived from yeast Elongator and bacterial orthologues of Elp3. Here, we reconstituted and characterized the biochemical and structural properties of human Elongator subunits/subassemblies and core human Elongator (hELP1/2/3). Negative stain EM analysis of human core Elongator and hELP4/5/6 revealed a similar overall architecture and subunit arrangement to yeast core Elongator and yElp4/5/6. These structural data, together with results from pulldown assays, indicated that hELP1 stabilizes hELP3 while hELP2 rigidifies the hELP1/3 subassembly. Complementary tRNA binding assays showed that substrate interaction is enhanced upon core Elongator assembly. Collectively, our work sheds light on the assembly pathway of human Elongator and potential role of hELP2 in supporting Elongator function.
Results
Conserved Architecture of Human Core Elongator
To gain insights into the structural properties of human Elongator, we attempted to reconstitute the full 6-subunit human Elongator using the baculovirus-insect cell-based system. We selected the biGBac series of plasmids, which offers a simple procedure for generating a co-expression construct containing multiple genes.40 In short, we first cloned the six genes encoding full-length hELP1 to hELP6 into pLIB entry vectors (with a 3xFLAG tag engineered to the C-terminus of hELP1 and a His tag engineered to the N-terminus of hELP2) and then used Gibson assembly to link all six Elongator genes into a multigene cassette within a pBIG2 vector. Baculovirus generated from bacmid derived from this co-expression construct was used to infect Sf9 cells for recombinant protein expression. We conducted anti-FLAG affinity chromatography on lysates prepared from these infected cells. We found that the eluted fractions showed high levels of hELP1, hELP2, and hELP3 but a complete absence of hELP4, hELP5, and hELP6. In agreement with this observation, we were unable to overexpress nor purify the hELP4/5/6 subcomplex from Sf9 cells using a separate co-expression construct that contains only these three genes (data not shown). Nevertheless, these observations demonstrated that similar to yeast Elongator, human core Elongator (hELP1/2/3) can form a stable assembly in the absence of hELP4/5/6.
To characterize the structural properties of human core Elongator, we first applied size-exclusion chromatography or gradient fixation (GraFix) to improve sample purity by removing excess FLAG-tagged hELP1 captured in the initial anti-FLAG affinity step. The extra polishing step enabled us to obtain human core Elongator with the three subunits present at near stoichiometric level (Figure 1A). We next subjected GraFix purified human core Elongator to negative stain EM analysis. Raw images showed the presence of bi-lobal particles reminiscent to yeast core Elongator (Figure 1B) as well smaller particles, which may represent alternate views, dissociated subunits, or “monomeric” hELP1/2/3. Reference-free 2D classification showed that 51% of the particles make up the dominant “moth”-shaped class averages that resembled the 2D averages obtained from previous studies of yeast core Elongator (Figures 1B, S2A), while 34% of the particles produced averages reminiscent of a single lobe of yElp1/2/3. These results suggest that yeast and human core Elongator share a conserved overall architecture. Similarly, we observed high variability in the distance between the two hELP1/2/3 lobes, as was observed for yElp1/2/3.22 However, unlike our previously determined 2D averages of yeast core Elongator, the experimental 2D averages of human core Elongator lacks sufficient details to discern intersubunit and domain boundaries, indicating that there is a higher degree of intrasubunit conformational flexibility compared to yeast core Elongator.22 This flexibility precluded determination of a 3D reconstruction of the hELP1/2/3 complex as well as high-resolution analysis of this complex by cryo-EM.
Figure 1.
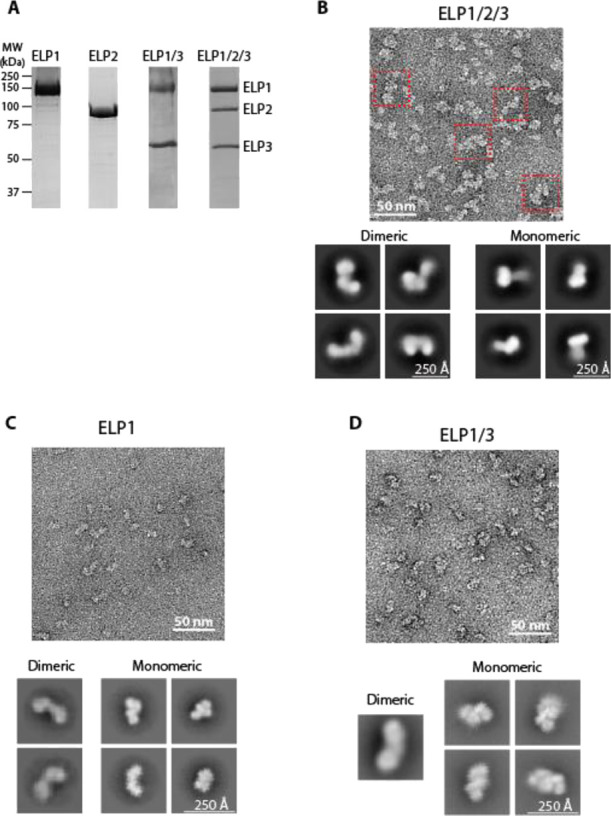
Purification and architecture of human core Elongator and its subassemblies. (A) SDS-PAGE of human core Elongator subunits purified by size-exclusion chromatography. Gel was stained with Coomassie brilliant blue. Migration of molecular weight markers is indicated on the left and expected migration of each subunit is indicated on the right. (B) Top: Representative segment of an electron micrograph of negatively stained hELP1/2/3 purified by GraFix. Red squares correspond to intact core Elongator. Bottom: Representative 2D class averages of hELP1/2/3 particles. Averages that clearly resemble a bi-lobal dimeric shape are on the left, and averages that are single lobes are shown as monomeric on the right. Uncropped images of all 2D averages are shown in Figure S2A. (C) Representative segment of an electron micrograph of negatively stained hELP1 purified by GraFix. Bottom: Representative 2D class averages from reference-free classification of hELP1 particles generating monomeric and dimeric averages. Uncropped images of all 2D averages are shown in Figure S3A. (D) Top: Representative segment of an electron micrograph of negatively stained hELP1/3 purified by GraFix. Bottom: Representative 2D class averages of best-resolved hELP1/3 particles. Uncropped images of all 2D averages are shown in Figure S3B.
hELP2 Locks hELP1/3 into a More Rigid Dimeric Assembly
To examine the subunit organization of human core Elongator, we next used the baculovirus-insect cell system to produce individual Elongator subunits and subassemblies. We first focused on the largest subunit, hELP1, which forms the main scaffold that mediates dimerization and substrate binding. We were able to produce full-length ELP1 alone as well as hELP1 in complex with hELP3 (hELP1/3) (Figure 1A). We next characterized these samples by negative stain EM. Raw images showed that hELP1 is structurally heterogeneous (Figure 1C). 2D class averages obtained from reference-free classification show overall morphologies that appear to correspond to the monomeric and dimeric states of the protein (Figure 1C). Although only ∼10% of the class averages represent dimeric hELP1 compared to 87% for monomeric hELP1, we believe the actual dimer-to-monomer ratio is higher in solution. Notably, the asymmetry of the hELP1 dimer can lead to one half of the dimer being averaged out in the 2D image analysis procedure. Indeed, our size-exclusion chromatography coupled to multiangle light scattering (SEC-MALS) analysis suggested that hELP1 is predominantly dimeric (Figure S4). Systematic comparison of experimental 2D class averages corresponding to monomeric hELP1 with the 3D model of monomeric hELP1 (generated from the core yeast Elongator cryo-EM structure) showed remarkable conservation in the overall morphology (Figure 2A). The region corresponding to the hELP1 C-terminal dimerization domain (hereafter “CTD”) was blurry in all class averages, indicating that the CTD is more conformationally flexible compared to the N-terminal WD40 domains of this protein.
Figure 2.
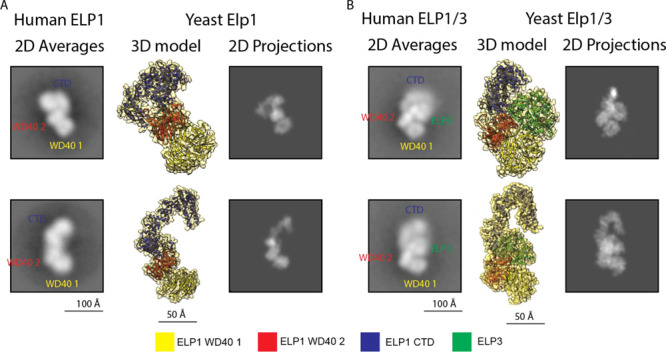
Structural comparison of hELP1 and ELP1/3 with their yeast orthologues. (A) Left: Representative 2D class averages of hELP1 particles with labels denoting the approximate location of hELP1 domains. Centre: 3D model of monomeric yElp1 (PDB: 6QK7) in orientations corresponding to the 2D averages to the left. Right: 2D projections generated from the 3D models in the center. (B) Left: Representative 2D class averages of hELP1/3 particles with labels denoting the approximate location of hELP1 domains and hELP3. Centre: 3D model of monomeric yElp1/3 (PDB: 6QK7) in orientations corresponding to the 2D averages to the left. Right: 2D projections generated from the 3D models in the center. A color legend corresponding to the 3D models is shown at the bottom.
For hELP1/3, while we clearly observed particles resembling dimeric hELP1/3 in raw EM images (Figure 1D), the majority of class averages obtained from 2D analysis have overall morphology resembling single-lobe “monomeric” yELP1/3 (2% dimeric, 96% monomeric) (Figure 1D). Like hELP1 alone, the CTD of hELP1 was blurry in the hELP1/3 2D class averages, indicating that hELP3 does not have stabilizing effects on the hELP1 CTD. Comparison of hELP1/3 class averages to the 3D model of yElp1/3 suggests that the orientation of hELP3 relative to both WD40 domains of hELP1 is conserved from yeast to humans (Figure 2B). Much like hELP1, the limited number of 2D averages corresponding to dimeric hELP1/3 could be attributed to conformational flexibility that prevents this complex from adopting a symmetrical dimer, leading to one half of the hELP1/3 dimer being averaged out in the 2D image analysis procedure. The fact that a significantly higher proportion of 2D class average resembles the dimeric state in our earlier 2D negative stain EM analysis of hELP1/2/3 indicated that hELP2 binding likely reduces overall flexibility of hELP1/3 by locking the two lobes in a more rigid symmetrical conformation.
hELP3 Stability Is Enhanced by Complex Formation with hELP1
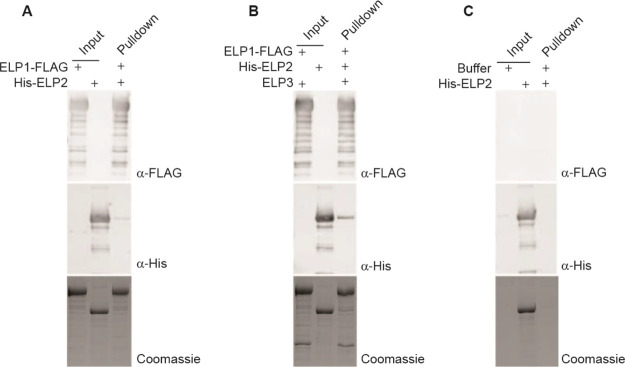
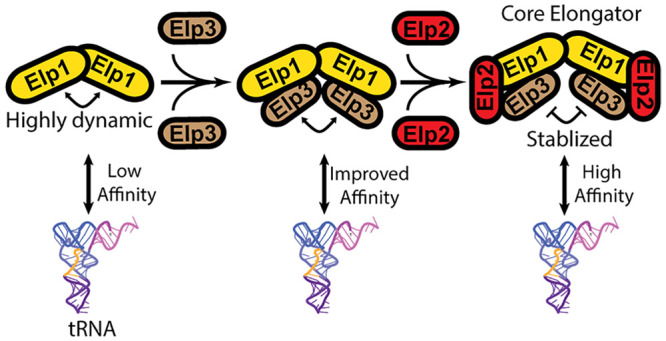
Despite trying different methods to optimize expression, we were unable to purify human ELP3 alone using our system. This was not unexpected as previous studies on archaeal and bacterial Elp3 necessitate the use of anaerobic conditions to preserve the integrity of the Fe–S cluster present in this protein. Interestingly and as detailed earlier, we were able to reconstitute an hELP1/3 subcomplex, indicating that hELP1 stabilizes hELP3 and potentially prevents hELP3 from aggregating and/or sequesters it from harmful effects in an aerobic environment. Based on this observation, we hypothesized that the assembly of human core Elongator likely begins with the formation of the hELP1-hELP3 subcomplex followed by the recruitment and binding of hELP2. To test this hypothesis, we carried out in vitro co-immunoprecipitation assays using the purified human core Elongator subunits and subassemblies. We found that a very low amount of hELP2 was pulled down by immobilized hELP1, indicating that hELP1 alone interacts weakly with hELP2 (Figure 3A). By contrast, the hELP1/3 subassembly showed stronger interaction with hELP2, as shown by an increase in the signal for hELP2 in the pulldown (Figure 3B). Interestingly, the assembly of hELP2 into core Elongator appears to be limited by the amount of hELP3, as demonstrated by the amount of hELP2 corresponding to the lesser amount of hELP3 present, despite an excess of hELP1. In summary, our pulldown results support an assembly pathway whereby hELP1 and hELP3 form a structural platform for hELP2 to bind and further stabilize the complex.
Figure 3.
Intersubunit assembly of human core Elongator proteins. (A) In vitro pulldown assay using FLAG-tagged hELP1 immobilized on M2-FLAG resin as prey and his-tagged hELP2 as bait. Input and pulldown fractions were analyzed by SDS-PAGE followed by an anti-FLAG (top) or anti-His (middle) Western blot or by Coomassie blue staining (bottom). (B) In vitro pulldown assay using FLAG-tagged hELP1/3 immobilized on M2-FLAG resin as prey and his-tagged hELP2 as bait. Input and pulldown fractions were analyzed by SDS-PAGE followed by an anti-FLAG (top) or anti-His (middle) Western blot or by Coomassie staining (bottom). (C) Negative control pulldown assay using unloaded M2-FLAG resin as bait and purified His-hELP2 as bait. Input and pulldown fractions were analyzed by SDS-PAGE followed by an anti-FLAG (top) or anti-His (middle) Western blot or by Coomassie blue staining (bottom).
Integration of hELP2 Enhances Human Core Elongator Interaction with tRNA
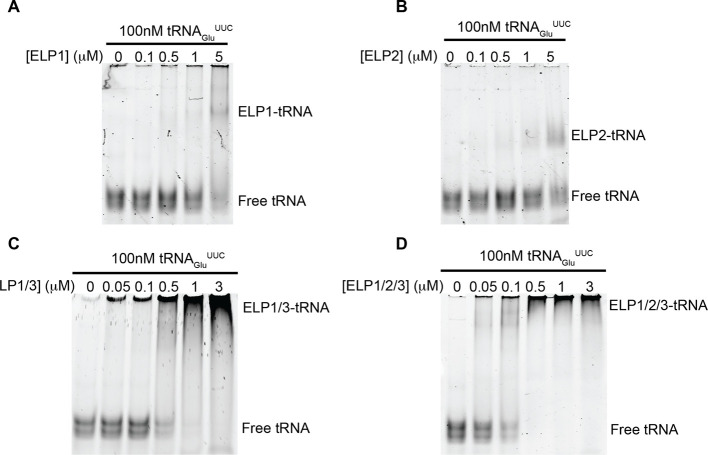
We next evaluated the contribution of different core Elongator subunits toward substrate recognition and binding. To this end, we generated a putative substrate tRNA of Elongator, tRNAGluUUC, by in vitro transcription and used this reagent for electrophoretic mobility shift assays (EMSAs) to assess binding. We observed minimal binding of hELP1 alone to tRNA, requiring a 10- to 50-fold molar excess of the protein to cause a shift in migration of the tRNA within the gel (Figures 4A, S1A). Similarly, the observed affinity of hELP2 for tRNAs within our study was quite weak, requiring a larger molar excess of hELP2 for the formation of a ternary hELP2-tRNA complex (Figures 4B, S1B). Significant binding of Elongator proteins to tRNAGluUUC was observed when we carried out the EMSA using the hELP1/3 subassembly, where we could detect binding even at submolar ratios of hELP1/3:tRNA (Figures 4C, S1C). Despite very minimal direct binding between hELP2 and tRNAGluUUC, we observed an increase in substrate binding when using the fully assembled core Elongator (hELP1/2/3) compared to hELP1/3 (Figures 4D, S1D), further demonstrating a role for hELP2 in stabilizing this assembly. This mode of substrate interaction is consistent with the recent cryo-EM structure of yeast core Elongator in complex with tRNAAlaUGC, where direct interactions between the substrate and both hELP1 and hELP3 were observed.24
Figure 4.
Hierarchical interactions of human core Elongator subassemblies with tRNA. EMSA demonstrating the interaction between recombinant tRNAGluUUC and Elongator subassemblies. 100 nM tRNA was incubated with the indicated amount of hELP1 (A), hELP2 (B), hELP1/3(C), or hELP1/2/3 (D) and run on a 5% native polyacrylamide gel. Nucleic acids within the gel were then stained and visualized using SYBR Gold and visualized. Migration of the various species of tRNA and tRNA:protein complexes are indicated to the right of each gel.
Human ELP4/5/6 Subcomplex Forms a Hexameric ATPase Ring
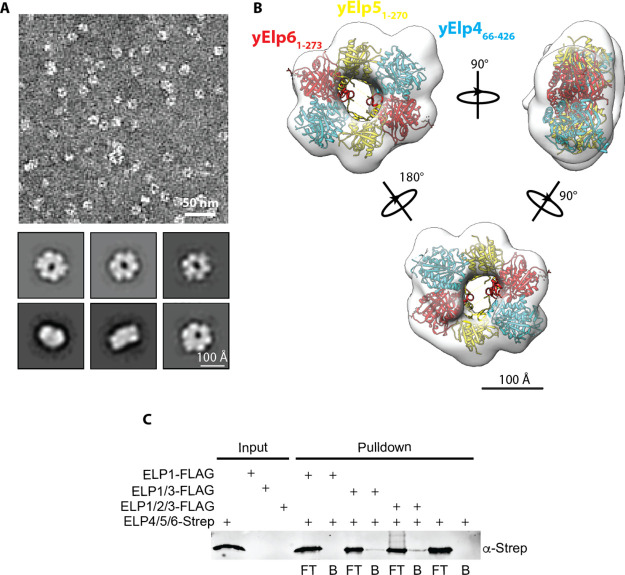
As we were unable to express hELP4/5/6 using the baculovirus-insect cell system, we tested the ability of a bacterial expression system to produce recombinant hELP4/5/6. We first cloned the genes encoding each of the three subunits to a pQLink vector and then assembled a multigene, multipromoter co-expression plasmid by ligase independent cloning.41 Despite low levels of expression, we were able to recover a small amount of hELP4/5/6 using a combination of nickel-based affinity chromatography, ion exchange chromatography, and gel filtration (typical yield of ∼1 μg/L of E. coli expression culture). We subjected purified hELP4/5/6 to negative stain EM analysis. Raw micrographs revealed ring-shaped particles resembling the morphology of yElp4/5/6 (Figure 5A). 2D classification and subsequent 3D reconstruction generated a density map in which we were able to fit the crystal structure of the truncated yElp4/5/6 subcomplex (Figure 5B). However, the limited resolution of our EM density map precluded precise subunit assignment to each of the lobes. Interestingly, we observed unaccounted density after this fitting. This density might correspond to the missing structural elements resulting from truncation of the yElp4 subunit in the previous crystallized assembly.
Figure 5.
Molecular architecture and assembly of the hELP4/5/6 subcomplex. (A) (Top) Representative segment of a negative stained micrograph generated using purified hELP4/5/6. (Bottom) Representative 2D class averages of hELP4/5/6 generated using Relion v3.0. Uncropped image of all 100 class averages are shown in Figure S2B. (B) 3D reconstruction of hELP4/5/6 generated using ab initio reconstruction in cryoSPARC v2.14. Crystal structure of the truncated yElp4/5/6 subcomplex (PDB ID: 4A8J) was docked into the 3D model and individual subunits are denoted based on color. (C) In vitro pulldown assay using FLAG-tagged hELP1, hELP1/3, or hELP1/2/3 immobilized on M2-FLAG resin as prey and Strep-tagged hELP4/5/6 as bait. Flow through fractions (FT) were collected to detect unbound protein and any bound proteins (B) were eluted from the resin using 3xFLAG peptide. Input and pulldown fractions were analyzed by SDS-PAGE followed by an anti-Strep Western blot.
We next investigated how hELP4/5/6 interacts with human core Elongator. We carried out in vitro pulldown assays using hELP4/5/6 as prey against FLAG-tagged hELP1, hELP1/3, or hELP1/2/3 purified from insect cells and immobilized on anti-FLAG resin. Contrary to previous reports, we were unable to detect interaction between hELP1 alone and hELP4/5/6 (Figure 5C), suggesting the previously observed co-immunoprecipitation of hELP1 with hELP6 was likely through a bridging protein.18 We found that both hELP1/3 and hELP1/2/3 complexes were able to pulldown hELP6 (Figure 4C), indicating that hELP3 is the primary interface for the hELP4/5/6 subcomplex binding to core Elongator. Because of very limited yield of purified hELP4/5/6, we were unable to carry out experiments to test the effects of this complex on human core Elongator-tRNA substrate interaction. Based on the sequence and structural similarities between the yeast and human homologs, we expect the observed high affinity of yElp4/5/6 to tRNAGluUUC (Figure S1E,F) to be conserved in humans as well.
Discussion
Over the past decade, structural analyses of yeast Elongator and Elongator subunits/subassemblies from yeast and other bacterial species have advanced our understanding of the overall architecture, subunit organization, and substrate interaction of this tRNA modifying complex.17−19,21−24 However, the extent to which this knowledge can be applied to human Elongator remains unclear as molecular studies on this orthologous complex, apart from human ELP1, have lagged behind due in part to challenges in obtaining pure human Elongator. Results from our negative stain EM analyses of human ELP1/2/3 and human ELP4/5/6 not only enable visualization of these two major subassemblies of human Elongator for the first time but also demonstrated that their overall architecture is conserved compared to those of the well-characterized yeast Elongator. However, we were unable to capture images of full human Elongator as challenges in expressing hELP4/5/6 in insect, bacterial, and human cells (data not shown) have precluded us from reconstituting the full 6-subunit assembly. Examination of different available databases showed that the levels of mRNA of hELP4, hELP5, and hELP6 are lower than that of hELP1, hELP2, and hELP3 across different human tissues and cell types, a scenario that mirrored what was observed in yeast cells.22,46 We postulate that much like in yeast, the low level of hELP4/5/6 in the cytoplasm of human cells would restrict loading of this subassembly onto hELP1/2/3, resulting in an overall asymmetric architecture of human full Elongator. An early experimental evidence supporting this hypothesis is that the original multistep purification showed stoichiometric level of six subunits in the purified human Elongator which can only be achieved if only one hELP4/5/6 ring is loaded onto hELP1/2/3.47
It is generally accepted that full Elongator involves the assembly of two discrete subcomplexes (hELP4/5/6 and hELP1/2/3) that were formed independently. Yet, little is known how the different subunits come together to form each subcomplex because of previous studies focusing primarily on characterizing preassembled core Elongator or full Elongator from yeast cells. The ability to produce individual human Elongator subunits and distinct subassemblies offered us the opportunity to investigate this poorly understood aspect of Elongator biology. Through examining the biochemical and structural features of these subunits and subassemblies, we were able to gain insights into the mechanism of Elongator assembly. By validating that full-length hELP1 can form a dimer in the absence of other Elongator subunits (Figure S4), we confirmed that this largest subunit forms the scaffold for complex assembly. Our subsequent and serendipitous discovery that hELP3 can only be purified when co-expressed with hELP1 indicated that the hELP1 scaffold stabilizes hELP3 and that our reconstituted hELP1/3 represents an assembly intermediate. The observation that hELP2 binds more strongly to hELP1/3 compared to hELP1 indicated that the binding of two copies of hELP2 to the periphery of hELP1/3 likely represents the final step in the assembly of core Elongator. Furthermore, the integration of this last subunit also rigidifies the core assembly, likely priming it for tRNA substrate binding and the recruitment of hELP4/5/6. Deletion mutants of yElp4, yElp5, and yElp6 share the same phenotypes as mutants carrying deletions of core Elongator genes (yElp1, yElp2, and yElp3). This suggested that, despite apparently being less stable, hELP4/5/6 is an indispensable component of Elongator. How hELP4/5/6 precisely modulates the activity and/or alters the conformation of core Elongator would require acquisition of high-resolution structural data on apo-full Elongator and full Elongator in complex with the tRNA substrate.
Materials and Methods
Plasmids and Cloning
The cDNAs encoding hELP1 (isoform 1) with a C-terminal 3xFLAG tag and hELP2 (isoform 2) were codon optimized for expression in S. frugiperda (Sf9) cells and synthesized as 5 (hELP1) or 2 (hELP2) GeneArt fragments (Thermo Fisher). The fragments were then assembled into a pLIB (hELP1) or pLIB-10xHis (hELP2) vector via Gibson assembly. cDNAs encoding hELP3, hELP4, hELP5, and hELP6 were purchased from the DNASU repository then subcloned into pLIB (hELP3, hELP4, hELP5, and hELP6), pQLinkH (hELP4), pQLinkN (hELP5) or pQLink-TwinStrepII (hELP6) vectors. hELP1-3xFLAG, 10xHis-hELP2, and hELP3 CDS were combined into one pBIG1a, and hELP4, hELP5, and hELP6 CDS were combined into one pBIG1b vector by Gibson assembly following the published protocol,40 and 6xHis-hELP4, hELP5, and TwinStrepII-hELP6 CDS were combined into one pQLink vector by ligation-independent cloning, following the published protocol.41 A table summarizing all plasmids used in this study is provided as Table S1.
Baculovirus Generation and Protein Expression
The insect cell expression plasmid was transformed into DH10EMBacY cells for bacmid production. Bacmids were isolated using an isopropanol precipitation protocol and the presence of all cDNAs was confirmed via PCR. Then, 2.5 μg of bacmid was combined with 5 μL of TransIT transfecting reagent (Mirus) in 200 μL of Grace’s media. The combined mixture was used to infect Sf9 cells freshly seeded in 6-well plates at a density of 0.8 × 106 cells/mL and the infection was allowed to proceed for 72 h. The supernatant containing the baculovirus (P1) was harvested, and the virus was propagated for another generation in 6-well plates (P2). P3 virus was produced by infecting a 50 mL suspension culture grown in ESF921 media (Expression Systems) with 500 μL of P2 virus for 72 h and harvesting the virus-containing supernatant. For large-scale protein expression, 250 or 500 mL cultures of Sf9 cells were infected at a density between 2 and 3 × 106 cells/mL with P3 virus at a ratio of 1 mL/100 × 106 cells. Cell pellets were harvested by centrifugation after 66–72 h of expression, washed with PBS, then flash-frozen in liquid nitrogen.
The pQLink-hELP4/5/6 plasmid was transformed into T7 Express E. colicells via heat shock. A single colony was used to inoculate a 5 mL overnight culture and grown for 16–18 h. One liter of expression cultures were inoculated with 1 mL of the overnight culture and grown at 37 °C until reaching an OD600 of 0.6. The expression was induced by adding IPTG to a final concentration of 1 mM and allowed to continue for 18 h at 16 °C. Following protein expression, cells were harvested by centrifugation at 4,000 rpm in a JLA-9.1 rotor (Beckman), pellets were washed with PBS, then flash-frozen in liquid nitrogen.
Protein Purification
Cell pellets of hELP1, hELP2, hELP1/3, hELP1/2/3, or hELP4/5/6 were all resuspended in lysis buffer (40 mM HEPES-NaOH pH 7.4, 300 mM NaCl, 10% (v/v) glycerol, and 0.01% DDM) containing protease inhibitors (1 mM PMSF, 1× Protease Inhibitor cocktail EDTA-free (Roche), 25 μg/mL benzamidine, and 0.25 U/mL benzonase). Cells were lysed by sonication on ice using a Branson Sonifier 450 set to 50% duty cycle and output control 5 for 60 s followed by 120 s of cooling, repeated six times. The lysate was cleared by centrifugation for 30 min at 18,000 rpm (JA-25.50 rotor, Beckman). The supernatant was then incubated with M2-FLAG resin (hELP1, hELP1/3, hELP1/2/3) (Sigma) or Ni-NTA beads (hELP2, hELP4/5/6) (Thermo Fisher) for 1 h at 4 °C while rocking. The mixtures were then passed over 5 mL Pierce centrifuge columns (Thermo Fisher) and the flow through discarded. The settled resin was washed with six washes of 10CV lysis buffer alone (M2-FLAG) or lysis buffer supplemented with 50 mM imidazole (NiNTA). Proteins were eluted from the columns by incubation with 4 × 1 CV lysis buffer supplemented with 0.5 mg/mL 3xFLAG peptide (M2-FLAG) or with 250 mM imidazole (NiNTA). Elution fractions were then concentrated to 500 μL using a 100 k MWCO concentrator (Millipore). For use in pulldown assays or EMSAs, the proteins were loaded onto a Superdex200 (hELP1, hELP2, hELP4/5/6) or a Superose 6 (hELP1/3, hELP1/2/3) gel filtration column (Cytiva) equilibrated with SEC buffer (40 mM HEPES-NaOH pH 7.4, 150 mM NaCl, 5% glycerol, 0.01% DDM). For use in EM studies, the proteins were loaded on a 15–30% glycerol gradient with 0–0.1% glutaraldehyde, and centrifuged for 16 h at 40,000 rpm (SW-55 rotor, Beckman). Elution peak fractions were analyzed by SDS-PAGE for purity. Protein concentrations were measured by a Bradford assay in 250 μL reactions, following the manufacturer’s instructions (BioRad). hELP1 and hELP2 proteins were flash-frozen in liquid nitrogen and stored at −70 °C until use in pulldowns or EMSAs. hELP1/3, hELP1/2/3, and hELP4/5/6 samples were used fresh for all experiments.
Negative Stain EM and Imaging Processing
Purified hELP1, hELP1/3, hELP1/2/3, and hELP4/5/6 were adsorbed to glow discharged carbon-coated grids and stained with uranyl formate. Stained hELP1, hELP1/3, or hELP1/2/3 specimens were examined using a Talos L120C transmission electron microscope (Thermo Fisher Scientific) operated at an acceleration voltage of 120 kV and equipped with a Ceta camera. hELP4/5/6 samples were imaged using a Tecnai Spirit transmission electron microscope (FEI Company) operated at an acceleration voltage of 120 kV and equipped with an Eagle 4 K CCD camera.
For hELP1, 220 micrographs were acquired at a nominal magnification of 45,000× at a defocus of ∼1.2 μm at a pixel size of 2.228 Å/pixel. Contrast transfer function (CTF) estimation for each micrograph was carried out using CTFFIND4.42 Two hundred particles were manually picked then aligned to generate 2D class averages for template-based autopicking in Relion 3.0. A total of 81,007 particles were autopicked and extracted with a box size of 200 pixels. Particles were then exported to cryoSPARC v3.243 and subjected to 2D classification. A total of 61,968 distinct particles were subjected to another round of 2D classification into 50 classes (Figure S3A), from which particle distribution was calculated and the figures were generated.
For hELP1/3, 150 micrographs were acquired at a nominal magnification of 45,000× at a defocus of ∼1.2 μm at a pixel size of 2.228 Å/pixel. CTF estimation for each micrograph was carried out using CTFFIND4. A total of 200 particles were manually picked then aligned to generate 2D class averages for template-based autopicking in Relion 3.0.44 A total of 109,093 particles were autopicked and extracted with a box size of 200 pixels. Particles were then exported to cryoSPARC v3.2 and subjected to 2D classification into 50 classes. A total of 75,859 particles classified into discrete classes were subjected to a second round of 2D classification (Figure S3B), and representative class averages were selected for figure generation.
For hELP1/2/3, 100 micrographs were acquired at a nominal magnification of 45,000× at a defocus of ∼1.2 μm and binned twice to obtain a pixel size of 4.53 Å/pixel. CTF estimation for each micrograph was carried out using CTFFind4. Two hundred particles were manually picked and then aligned to generate 2D class averages for template-based autopicking in Relion 3.0. A total of 71,732 particles were autopicked and extracted with a box size of 100 pixels. Particles were then exported to cryoSPARC v3.2 and subjected to 2D classification, where 38,529 particles were found to classify well and were subjected to a second round of 2D classification (Figure S2A). The distribution of “dimeric” and “monomeric” particles was calculated from these averages.
For hELP4/5/6, 85 micrographs were acquired at a nominal magnification of 49,000× at a defocus of ∼1 μm and binned twice to obtain a pixel size of 4.67 Å/pixel. CTF estimation for each micrograph was carried out using CTFFind4. A total of 286 particles were manually picked and then aligned to generate 2D class averages for template-based autopicking in Relion 3.0. A total of 35,272 particles were autopicked and extracted with a box size of 60 pixels. Particles were then subjected to 2D classification into 100 classes (Figure S2B). For 3D reconstruction, the micrographs were processed in cryoSPARC v2.14, where CTF estimation was carried out using CTFFind4; 50 particles were manually picked and averaged into 2D classes for use as templates for autopicking, which yielded 64,734 particles. The particles were iteratively classified by 2D averaging twice, and the remaining particles were used for ab initio reconstruction of two 3D models; the more complete model was used for subsequent docking using the crystal structure of yElp4/5/6 (PDB ID: 4A8J).
tRNA Substrate Preparation
cDNA encoding human tRNAGluUUC was synthesized and placed downstream of a T7 promoter in a pMA-T cloning vector (Thermo Fisher). The plasmid was linearized in a 50 μL reaction by incubation with 2 μL of SwaI for 1 h at 25 °C, then purified by phenol–chloroform extraction. The linearized plasmid was then used as a template for transcription using T7 RNA polymerase, following the manufacturer’s protocol (NEB Quick High Yield RNA Synthesis Kit). The completed reaction was incubated with 2 μL DNAseI for 15 min at 37 °C to digest the template DNA, and the remaining tRNA was purified by phenol–chloroform extraction followed by ethanol precipitation. Next, tRNA refolding was carried out by incubating the purified tRNA at 85 °C for 1 min then cooled to 25 °C over 60 min. Correctly folded tRNAs were separated from unfolded or misfolded species by size-exclusion chromatography using a Superdex 75 column (Cytiva). Samples from every step of the procedure were analyzed by agarose gel electrophoresis to ensure purity and correct size.
Electrophoretic Mobility Shift Assay
All EMSAs were carried out using 5% native polyacrylamide gels made from 40% acrylamide/bis-acrylamide, 37.5:1 (BioRad), which were prerun in 0.5× TBE alone for 1 h at 150 V, at 4 °C. Then, 100 nM tRNAGluUUC was incubated with 0–5 μM hELP1, hELP1/3, hELP1/2/3, or yyElp4/5/6 and diluted to 9 μL reactions in EMSA buffer (40 mM HEPES pH 7.4, 100 mM NaCl, 1 mM MgCl2). Reactions were incubated on ice for 1 h, and then 1 μL of 50% glycerol was added for a final concentration of 5% (v/v). Samples were then loaded on native PAGE gels and subjected to electrophoresis for 1 h at 150 V at 4 °C. tRNA and tRNA–protein complexes were stained on the gels by incubating with 50 mL of 1× SYBR Gold for 30 min under rotation. The gels were subsequently imaged using a ChemiDoc MP imaging system (BioRad). The gels were then stained for proteins using PAGE blue following the manufacturer’s instructions (Thermo Scientific) and imaged using the ChemiDoc MP.
Pulldown Assays
First, 1 μM hELP1-3xFLAG or hELP1-3xFLAG/hELP3 was combined with 1 μM 10xHis-hELP2 in 200 μL reactions and incubated with 50 mL of M2-FLAG resin for 1 h at 4 °C under rotation. The M2-FLAG resin was isolated by centrifugation for 2 min at 5000 × g, and the supernatant was collected to analyze unbound protein. The resin was then washed three times with 500 μL of pulldown buffer by resuspending the slurry in buffer and collecting the resin by centrifugation. Bound proteins were the eluted by incubation with 200 μL of 0.5 mg/mL 3xFLAG peptide for 30 min at 4 °C (“Pulldown”). Then, 1 μM hELP1, hELP1/3, or hELP2 alone (“Input”), along with FLAG elution fractions from each pulldown were loaded evenly across 3 SDS-PAGE gels. Two gels were then transferred to nitrocellulose membranes by wet transfer for 1.5 h at 100 V, and the third gel was stained using Coomassie blue. Membranes were blocked for 1 h at RT with a 5% skimmed milk solution then incubated with a 1:2000 dilution of mouse anti-FLAG antibody (Sigma) or mouse anti-His antibody for 1 h at RT. Blots were then washed and incubated with a 1:10,000 dilution of anti-mouse IRDye680 (Li-Cor BioSciences) secondary antibody. Blots and stained gels were then imaged using a ChemiDoc MP.
Acknowledgments
This work was supported a Discovery Grant from the Natural Science and Engineering Research Council of Canada (RGPIN-2018-03951) and a Foundation Grant from the Canadian Institutes of Health Research (FDN-143228) to C.K.Y. This research project was supported in part by the UBC High Resolution Macromolecular Cryo-Electron Microscopy Facility (HRMEM).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c05719.
Plasmids used in this study, interactions of Elongator Subassemblies with tRNA, gallery of negative stain EM class averages of human Elongator subcomplexes, gallery of negative stain EM class averages of hELP1 and hELP1/3 assemblies, and biophysical characterization of the oligomeric state of hELP1 (PDF)
Accession Codes
Human ELP1: O95163. Human ELP2: Q6IA86. Human ELP3: Q9H9T3. Human ELP4: Q96EB1. Human ELP5: Q8TE02. Human ELP6: Q0PNE2. Yeast Elp4: Q02884. Yeast Elp5: P38874. Yeast Elp6: Q04868.
Author Contributions
U.D. was involved in conception and design, acquisition of structural and biochemical data, analysis and interpretation of data, and drafting or revising the article; D.M. and F.Z. were involved in the acquisition of biochemical data and analysis and interpretation of data. C.K.Y. was involved in conception and design, coordinating the project, analysis, and interpretation of data, and drafting or revising the article.
The authors declare no competing financial interest.
Notes
All data described in this study are available upon request to the corresponding author.
Supplementary Material
References
- Machnicka M. A.; Olchowik A.; Grosjean H.; Bujnicki J. M. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 2014, 11, 1619–1629. 10.4161/15476286.2014.992273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. J. O.; Esberg A.; Huang B.; Björk G. R.; Byström A. S. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol. 2008, 28, 3301–3312. 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant P. C.; Bajji A. C.; Sundaram M.; Kumar R. K.; Davis D. R. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 2005, 44, 8078–8089. 10.1021/bi050343f. [DOI] [PubMed] [Google Scholar]
- Vendeix F. A. P.; Murphy F. V.; Cantara W. A.; Leszczyńska G.; Gustilo E. M.; Sproat B.; Malkiewicz A.; Agris P. F. Human tRNA(Lys3)(UUU) is pre-structured by natural modifications for cognate and wobble codon binding through keto-enol tautomerism. J. Mol. Biol. 2012, 416, 467–485. 10.1016/j.jmb.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalwadi U.; Yip C. K. Structural insights into the function of Elongator. Cell. Mol. Life Sci. 2018, 75, 1613–1622. 10.1007/s00018-018-2747-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. J. O.; Xu F.; Byström A. S. Elongator-a tRNA modifying complex that promotes efficient translational decoding. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 401–408. 10.1016/j.bbagrm.2017.11.006. [DOI] [PubMed] [Google Scholar]
- Otero G.; Fellows J.; Li Y.; de Bizemont T.; Dirac A. M.; Gustafsson C. M.; Erdjument-Bromage H.; Tempst P.; Svejstrup J. Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 1999, 3, 109–118. 10.1016/S1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- Krogan N. J.; Greenblatt J. F. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 8203–8212. 10.1128/MCB.21.23.8203-8212.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler G. S.; Petrakis T. G.; Ethelberg S.; Tokunaga M.; Erdjument-Bromage H.; Tempst P.; Svejstrup J. Q. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 2001, 276, 32743–32749. 10.1074/jbc.M105303200. [DOI] [PubMed] [Google Scholar]
- Li Y.; Takagi Y.; Jiang Y.; Tokunaga M.; Erdjument-Bromage H.; Tempst P.; Kornberg R. D. A multiprotein complex that interacts with RNA polymerase II elongator. J. Biol. Chem. 2001, 276, 29628–29631. 10.1074/jbc.C100274200. [DOI] [PubMed] [Google Scholar]
- Wittschieben B. Ø.; Otero G.; de Bizemont T.; Fellows J.; Erdjument-Bromage H.; Ohba R.; Li Y.; Allis C. D.; Tempst P.; Svejstrup J. Q. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 1999, 4, 123–128. 10.1016/S1097-2765(00)80194-X. [DOI] [PubMed] [Google Scholar]
- Huang B.; Johansson M. J. O.; Byström A. S. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 2005, 11, 424–436. 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esberg A.; Huang B.; Johansson M. J. O.; Byström A. S. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell 2006, 24, 139–148. 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Lu J.; Huang B.; Esberg A.; Johansson M. J. O.; Byström A. S. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA 2005, 11, 1648–1654. 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W. D.; Thuriaux P.; Kohli J.; Ebert P.; Kersten H.; Gehrke C.; Kuo K. C.; Agris P. F. An antisuppressor mutation of Schizosaccharomyces pombe affects the post-transcriptional modification of the “wobble” base in the anticodon of tRNAs. J. Biol. Chem. 1984, 259, 2856–2862. 10.1016/S0021-9258(17)43226-1. [DOI] [PubMed] [Google Scholar]
- Grossenbacher A. M.; Stadelmann B.; Heyer W. D.; Thuriaux P.; Kohli J.; Smith C.; Agris P. F.; Kuo K. C.; Gehrke C. Antisuppressor mutations and sulfur-carrying nucleosides in transfer RNAs of Schizosaccharomyces pombe. J. Biol. Chem. 1986, 261, 16351–16355. 10.1016/S0021-9258(18)66573-1. [DOI] [PubMed] [Google Scholar]
- Dong C.; Lin Z.; Diao W.; Li D.; Chu X.; Wang Z.; Zhou H.; Xie Z.; Shen Y.; Long J. The yElp2 subunit is essential for elongator complex assembly and functional regulation. Structure 2015, 23, 1078–1086. 10.1016/j.str.2015.03.018. [DOI] [PubMed] [Google Scholar]
- Xu H.; Lin Z.; Li F.; Diao W.; Dong C.; Zhou H.; Xie X.; Wang Z.; Shen Y.; Long J. Dimerization of elongator protein 1 is essential for Elongator complex assembly. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 10697–10702. 10.1073/pnas.1502597112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.-Y.; Abbassi N. E. H.; Zakrzewski K.; Chramiec-Głąbik A.; Jemioła-Rzemińska M.; Różycki J.; Glatt S. The Elongator subunit yElp3 is a non-canonical tRNA acetyltransferase. Nat. Commun. 2019, 10, 625. 10.1038/s41467-019-08579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt S.; Létoquart J.; Faux C.; Taylor N. M. I.; Séraphin B.; Müller C. W. The Elongator subcomplex yElp456 is a hexameric RecA-like ATPase. Nat. Struct. Mol. Biol. 2012, 19, 314–320. 10.1038/nsmb.2234. [DOI] [PubMed] [Google Scholar]
- Lin Z.; Zhao W.; Diao W.; Xie X.; Wang Z.; Zhang J.; Shen Y.; Long J. Crystal structure of elongator subcomplex yElp4-6. J. Biol. Chem. 2012, 287, 21501–21508. 10.1074/jbc.M112.341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiaputra D. T.; Cheng D. T.; Lu S.; Hansen J. M.; Dalwadi U.; Lam C. H.; To J. L.; Dong M.-Q.; Yip C. K. Molecular architecture of the yeast Elongator complex reveals an unexpected asymmetric subunit arrangement. EMBO Rep. 2017, 18, 280–291. 10.15252/embr.201642548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauden M. I.; Kosinski J.; Kolaj-Robin O.; Desfosses A.; Ori A.; Faux C.; Hoffmann N. A.; Onuma O. F.; Breunig K. D.; Beck M.; Sachse C.; Séraphin B.; Glatt S.; Müller C. W. Architecture of the yeast Elongator complex. EMBO Rep. 2017, 18, 264–279. 10.15252/embr.201643353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauden M. I.; Jaciuk M.; Weis F.; Lin T.-Y.; Kleindienst C.; Abbassi N. E. H.; Khatter H.; Krutyhołowa R.; Breunig K. D.; Kosinski J.; Müller C. W.; Glatt S. Molecular basis of tRNA recognition by the Elongator complex. Sci. Adv. 2019, 5, eaaw2326 10.1126/sciadv.aaw2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvadurai K.; Wang P.; Seimetz J.; Huang R. H. Archaeal yElp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat. Chem. Biol. 2014, 10, 810–812. 10.1038/nchembio.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tükenmez H.; Xu H.; Esberg A.; Byström A. S. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res. 2015, 43, 9489–9499. 10.1093/nar/gkv832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffena J.; Lefcort F.; Zhang Y.; Lehrmann E.; Chaverra M.; Felig J.; Walters J.; Buksch R.; Becker K. G.; George L. Elongator and codon bias regulate protein levels in mammalian peripheral neurons. Nat. Commun. 2018, 9, 889. 10.1038/s41467-018-03221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohloff F.; Fichtner L.; Jablonowski D.; Breunig K. D.; Schaffrath R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 2001, 20, 1993–2003. 10.1093/emboj/20.8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F.-J.; Shen L.; Jang C.-W.; Falnes P. Ø.; Zhang Y. Ikbkap/yElp1 deficiency causes male infertility by disrupting meiotic progression. PLoS Genet. 2013, 9, e1003516 10.1371/journal.pgen.1003516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K.-T.; Inoue A.; Lee Y.-J.; Jiang C.-L.; Lin F.-J. Loss of Ikbkap/yElp1 in mouse oocytes causes spindle disorganization, developmental defects in preimplantation embryos and impaired female fertility. Sci. Rep. 2019, 9, 18875. 10.1038/s41598-019-55090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M. Z.; Gruner K. A.; Qin C.; Tourtellotte W. G. A neuron autonomous role for the familial dysautonomia gene hELP1 in sympathetic and sensory target tissue innervation. Development 2014, 141, 2452–2461. 10.1242/dev.107797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki Y.; Ramirez G.; Salcedo E.; Stabio M. E.; Lefcort F. Loss of ikbkap causes slow, progressive retinal degeneration in a mouse model of familial dysautonomia. eNeuro 2016, 3, ENEURO.0143-16.2016 10.1523/ENEURO.0143-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaugenhaupt S. A.; Blumenfeld A.; Gill S. P.; Leyne M.; Mull J.; Cuajungco M. P.; Liebert C. B.; Chadwick B.; Idelson M.; Reznik L.; Robbins C.; Makalowska I.; Brownstein M.; Krappmann D.; Scheidereit C.; Maayan C.; Axelrod F. B.; Gusella J. F. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 2001, 68, 598–605. 10.1086/318810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S. L.; Coli R.; Daly I. W.; Kichula E. A.; Rork M. J.; Volpi S. A.; Ekstein J.; Rubin B. Y. Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 2001, 68, 753–758. 10.1086/318808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold-von Simson G.; Axelrod F. B. Familial dysautonomia: update and recent advances. Curr. Probl. Pediatr. Adolesc. Health Care 2006, 36, 218–237. 10.1016/j.cppeds.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Simpson C. L.; Lemmens R.; Miskiewicz K.; Broom W. J.; Hansen V. K.; van Vught P. W. J.; Landers J. E.; Sapp P.; van den Bosch L.; Knight J.; Neale B. M.; Turner M. R.; Veldink J. H.; Ophoff R. A.; Tripathi V. B.; Beleza A.; Shah M. N.; Proitsi P.; van Hoecke A.; Carmeliet P.; Horvitz H. R.; Leigh P. N.; Shaw C. E.; van den Berg L. H.; Sham P. C.; Powell J. F.; Verstreken P.; Brown R. H.; Robberecht W.; al-Chalabi A. Variants of the elongator protein 3 (hELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet. 2009, 18, 472–481. 10.1093/hmg/ddn375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strug L. J.; Clarke T.; Chiang T.; Chien M.; Baskurt Z.; Li W.; Dorfman R.; Bali B.; Wirrell E.; Kugler S. L.; Mandelbaum D. E.; Wolf S. M.; McGoldrick P.; Hardison H.; Novotny E. J.; Ju J.; Greenberg D. A.; Russo J. J.; Pal D. K. Centrotemporal sharp wave EEG trait in rolandic epilepsy maps to Elongator Protein Complex 4 (hELP4). Eur. J. Hum. Genet. 2009, 17, 1171–1181. 10.1038/ejhg.2008.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. S.; Srivastava S.; Farwell K. D.; Lu H.-M.; Zeng W.; Lu H.; Chao E. C.; Fatemi A. hELP2 is a novel gene implicated in neurodevelopmental disabilities. Am. J. Med. Genet. A 2015, 167, 1391–1395. 10.1002/ajmg.a.36935. [DOI] [PubMed] [Google Scholar]
- Kojic M.; Gawda T.; Gaik M.; Begg A.; Salerno-Kochan A.; Kurniawan N. D.; Jones A.; Drożdżyk K.; Kościelniak A.; Chramiec-Głąbik A.; Hediyeh-Zadeh S.; Kasherman M.; Shim W. J.; Sinniah E.; Genovesi L. A.; Abrahamsen R. K.; Fenger C. D.; Madsen C. G.; Cohen J. S.; Fatemi A.; Stark Z.; Lunke S.; Lee J.; Hansen J. K.; Boxill M. F.; Keren B.; Marey I.; Saenz M. S.; Brown K.; Alexander S. A.; Mureev S.; Batzilla A.; Davis M. J.; Piper M.; Bodén M.; Burne T. H. J.; Palpant N. J.; Møller R. S.; Glatt S.; Wainwright B. J. Elp2 mutations perturb the epitranscriptome and lead to a complex neurodevelopmental phenotype. Nat. Commun. 2021, 12, 2678. 10.1038/s41467-021-22888-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszak S. M.; Robinson G. W.; Gudenas B. L.; Smith K. S.; Forget A.; Kojic M.; Garcia-Lopez J.; Hadley J.; Hamilton K. V.; Indersie E.; Buchhalter I.; Kerssemakers J.; Jäger N.; Sharma T.; Rausch T.; Kool M.; Sturm D.; Jones D. T. W.; Vasilyeva A.; Tatevossian R. G.; Neale G.; Lombard B.; Loew D.; Nakitandwe J.; Rusch M.; Bowers D. C.; Bendel A.; Partap S.; Chintagumpala M.; Crawford J.; Gottardo N. G.; Smith A.; Dufour C.; Rutkowski S.; Eggen T.; Wesenberg F.; Kjaerheim K.; Feychting M.; Lannering B.; Schüz J.; Johansen C.; Andersen T. V.; Rösli M.; Kuehni C. E.; Grotzer M.; Remke M.; Puget S.; Pajtler K. W.; Milde T.; Witt O.; Ryzhova M.; Korshunov A.; Orr B. A.; Ellison D. W.; Brugieres L.; Lichter P.; Nichols K. E.; Gajjar A.; Wainwright B. J.; Ayrault O.; Korbel J. O.; Northcott P. A.; Pfister S. M. Germline Elongator mutations in Sonic Hedgehog medulloblastoma. Nature 2020, 580, 396–401. 10.1038/s41586-020-2164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann F.; Peters J.-M. Expressing Multi-subunit Complexes Using biGBac. Methods Mol. Biol. 2018, 1764, 329–343. 10.1007/978-1-4939-7759-8_21. [DOI] [PubMed] [Google Scholar]
- Scheich C.; Kummel D.; Soumailakakis D.; Heinemann U.; Bussow K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007, 35, e43 10.1093/nar/gkm067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A.; Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015, 192, 216–221. 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A.; Rubinstein J. L.; Fleet D. J.; Brubaker M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Zivanov J.; Nakane T.; Forsberg B. O.; Kimanius D.; Hagen W. J.; Lindahl E.; Scheres S. H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 2018, 7, e42166 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatheodorou I.; Moreno P.; Manning J.; Fuentes A. M.-P.; George N.; Fexova S.; Fonseca N. A.; Füllgrabe A.; Green M.; Huang N.; Huerta L.; Iqbal H.; Jianu M.; Mohammed S.; Zhao L.; Jarnuczak A. F.; Jupp S.; Marioni J.; Meyer K.; Petryszak R.; Prada Medina C. A.; Talavera-López C.; Teichmann S.; Vizcaino J. A.; Brazma A. Expression Atlas update: from tissues to single cells. Nucleic Acids Res. 2019, 48, D77–D83. 10.1093/nar/gkz947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes N. A.; Otero G.; Winkler G. S.; Marshall N.; Dahmus M. E.; Krappmann D.; Scheidereit C.; Thomas C. L.; Schiavo G.; Erdjument-Bromage H.; Tempst P.; Svejstrup J. Q. Purification and characterization of the human elongator complex. J. Biol. Chem. 2002, 277, 3047–3052. 10.1074/jbc.M110445200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.