Abstract
Genetic variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continue to dramatically alter the landscape of the coronavirus disease 2019 (COVID-19) pandemic. The recently described variant of concern designated Omicron (B.1.1.529) has rapidly spread worldwide and is now responsible for the majority of COVID-19 cases in many countries. Because Omicron was recognized recently, many knowledge gaps exist about its epidemiology, clinical severity, and disease course. A genome sequencing study of SARS-CoV-2 in the Houston Methodist health care system identified 4468 symptomatic patients with infections caused by Omicron from late November 2021 through January 5, 2022. Omicron rapidly increased in only 3 weeks to cause 90% of all new COVID-19 cases, and at the end of the study period caused 98% of new cases. Compared with patients infected with either Alpha or Delta variants in our health care system, Omicron patients were significantly younger, had significantly increased vaccine breakthrough rates, and were significantly less likely to be hospitalized. Omicron patients required less intense respiratory support and had a shorter length of hospital stay, consistent with on average decreased disease severity. Two patients with Omicron stealth sublineage BA.2 also were identified. The data document the unusually rapid spread and increased occurrence of COVID-19 caused by the Omicron variant in metropolitan Houston, Texas, and address the lack of information about disease character among US patients.
Over the past 14 months, the Alpha and Delta variants of concern (VOCs) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have caused two distinct coronavirus disease 2019 (COVID-19) surges in the United States, Southeast Asia, Europe, and elsewhere (https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html, last accessed December 30, 2021; https://www.gov.uk/government/collections/new-sars-cov-2-variant, last accessed December 30, 2021), and remodeled the landscape of human behavior and many societies. Delta replaced the Alpha variant as the cause of virtually all COVID-19 in many countries (https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---13-july-2021, last accessed August 18, 2021; https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/bulletins/coronaviruscovid19infectionsurveypilot/9july2021, last accessed August 18, 2021).
At the start of the pandemic almost 2 years ago, the Houston Methodist health care system instituted a comprehensive and integrated population genomics project designed to sequence all SARS-CoV-2 samples causing COVID-19 in patients cared for at our facilities, which include eight hospitals located throughout the metroplex. The project was implemented when the initial Houston Methodist COVID-19 case was diagnosed at the end of February 2020, and has continued unabated.1, 2, 3, 4, 5, 6, 7 This project was facilitated by the existence of a single large diagnostic laboratory that serves the entire system and is seamlessly integrated with a research institute with extensive genomics expertise and capacity. A key goal was to comprehensively map the population genomics, trajectory, and other features of the pandemic in metropolitan Houston, Texas, with a population size of approximately 7.2 million. Houston is the fourth largest city in the United States, is the most ethnically diverse metropolitan area in the country, and is a major port of entry. To date, SARS-CoV-2 genomes have been sequenced from >70,000 patient samples. Many features of four distinct SARS-CoV-2 waves in Houston have been described.2, 3, 4, 5, 6
The successes of rapid SARS-CoV-2 vaccine development and documented efficacy, coupled with the significant downturn of the disease wave caused by Delta in Houston and elsewhere in fall 2021,6 suggested that the pandemic was abating. However, the identification of a new VOC designated B.1.1.529 and known as Omicron that has spread rapidly in South Africa and the United Kingdom has tempered this optimism.8, 9, 10 Inasmuch as Omicron was recognized recently, and much is not known about its epidemiology and clinical characteristics and course, we used our integrated infrastructure in an effort to address the lack of information available for US Omicron patients. Genome sequencing identified 4468 COVID-19 patients with symptomatic disease caused by Omicron in the Houston Methodist health care system beginning in late November 2021 and ending January 5, 2022. In 3 weeks, Omicron spread throughout the Houston metropolitan region to become the cause of 90% of new COVID-19 cases, and at the end of the study period caused 98% of all new cases. Compared with patients infected with either Alpha or Delta variant and cared for in our system, significantly fewer Omicron patients were hospitalized, and those who were hospitalized required significantly less intense respiratory support and had a shorter length of stay. Our findings are consistent with decreased disease severity among Houston Methodist Omicron patients. Many factors undoubtedly have contributed, including but not limited to increased vaccination uptake, population immunity, and patient demographics, such as younger age. The extent to which our findings translate to other cities and other patient populations, including children, is unknown. These data expand on our initial Omicron work7 and address the lack of information about disease character among US patients with COVID-19 caused by this VOC.
Materials and Methods
Patient Specimens
Specimens were obtained from patients registered at Houston Methodist facilities (eg, hospitals and urgent care centers) and institutions in the Houston metropolitan region that use our laboratory services. The great majority of individuals had signs or symptoms consistent with COVID-19 disease. For analyses focusing on patients with COVID-19 caused by the Omicron variant, samples obtained from November 27, 2021, through January 5, 2022, were used. This time frame was chosen because it represents the period during which an Omicron variant was first identified in our health care system and the last date of specimen collection used to generate genome sequence data for this article. Note that the genome data were generated for two distinct sampling periods. The first period included November 27, 2021, through December 23, 2021; and the second period included samples obtained between December 30, 2021, and January 5, 2022. This discontinuous sampling strategy was used in an effort to obtain the most up-to-date data available for inclusion in this study. Because of the substantial number of positive specimens obtained daily in the December 24, 2021, to December 29, 2021, period (sometimes exceeding 1500), it was not possible to sequence most of the samples collected during this period for inclusion in the study.
For analyses comparing features of patients infected with the Omicron VOC and Alpha and Delta VOCs, all patients documented to be infected with these variants in the Houston Methodist system were studied. The study included 40,991 unique patients identified in this time frame for whom we had SARS-CoV-2 genome sequences. The work was approved by the Houston Methodist Research Institute Institutional Review Board (IRB1010-0199).
SARS-CoV-2 Molecular Diagnostic Testing
Specimens obtained from symptomatic patients with a suspicion for COVID-19 disease were tested in the Molecular Diagnostics Laboratory at Houston Methodist Hospital using assays granted Emergency Use Authorization from the US Food and Drug Administration (https://www.fda.gov/medical-devices/emergency-situations-medical-devices/faqs-diagnostic-testing-sars-cov-2#offeringtests, last accessed June 7, 2021). Multiple molecular testing platforms were used, including the COVID-19 test or RP2.1 test with BioFire Film Array instruments (BioFire Diagnostics, Salt Lake City, UT), the Xpert Xpress SARS-CoV-2 test using Cepheid GeneXpert Infinity or Cepheid GeneXpert Xpress IV instruments (Cepheid, Sunnyvale, CA), the Cobas SARS-CoV-2 and Influenza A/B Assay using the Roche Liat system (Roche Diagnostics, Indianapolis, IN), the SARS-CoV-2 Assay using the Hologic Panther instrument (Hologic, Marlborough, MA), the Aptima SARS-CoV-2 Assay using the Hologic Panther Fusion system (Hologic), the Cobas SARS-CoV-2 test using the Roche 6800 system, and the SARS-CoV-2 assay using Abbott Alinity m instruments (Abbott Molecular, Des Plaines, IL). Virtually all tests were performed on material obtained from nasopharyngeal swabs immersed in universal transport media; oropharyngeal or nasal swabs, bronchoalveolar lavage fluid, or sputum treated with dithiothreitol were sometimes used. Standardized specimen collection methods were used (https://vimeo.com/396996468/2228335d56, last accessed June 7, 2021).
SARS-CoV-2 Genome Sequencing, Genome Analysis, and Identification of Variants
We sequenced the SARS-CoV-2 genome of >90% of all positive cases in the Houston Methodist health care system during the two sampling periods studied. Libraries for whole SARS-CoV-2 genome sequencing were prepared according to version 4 (https://community.artic.network/t/sars-cov-2-version-4-scheme-release/312, last accessed August 19, 2021) of the ARTIC nCoV-2019 sequencing protocol. The semi-automated workflow used has been described previously.2, 3, 4, 5, 6 Sequence reads were generated with an Illumina (San Diego, CA) NovaSeq 6000 instrument.
Viral genomes were assembled with the BV-BRC SARS-Cov2 assembly service (https://www.bv-brc.org/app/ComprehensiveSARS2Analysis, last accessed June 7, 2021, requires registration). The pipeline currently uses seqtk version 1.3-r117 for sequence trimming (https://github.com/lh3/seqtk.git, last accessed December 30, 2021) and minimap version 2.17 for aligning reads against the Wuhan-Hu-1 (NC_045512.2) reference genome. Samtools version 1.11 was used for sequence and file manipulation, where maximum depth and minimum depth parameters in mpileup were set to 8000 and 3, respectively. iVar version 1.3.1 was used for primer trimming and variant calling. Genetic lineages, VOCs, and variants of interest were identified on the basis of genome sequence data and designated by Pangolin version 3.1.17 with pangoLEARN module 2021-12-06 (https://cov-lineages.org/resources/pangolin.html, last accessed December 12, 2021). Genome data used in this study have been deposited to Global Initiative on Sharing Avian Influenza Data (GISAID; www.gisaid.org) (Supplemental Table S1).
S-Gene Target-Failure Assay
An S-gene target-failure (SGTF) assay (TaqPath COVID-19 Combo Kit; Thermo Fisher, Inc., Waltham, MA) was used as a surrogate marker for the Omicron VOC for some specimens collected between December 18, 2021, and January 5, 2022. From November 1, 2021, onward, only Delta and Omicron were documented to be circulating in metropolitan Houston, based on whole-genome sequence data. Patient samples were first tested in the clinical Molecular Diagnostics Laboratory using a RT-PCR assay with an Emergency Use Authorization, as described above. The SARS-CoV-2–positive samples were then tested with the SGTF assay, according to the manufacturer's instructions to infer an Omicron or not-Omicron lineage. That is, the SGTF assay was only performed on samples known to be positive for SARS-CoV-2. Samples yielding amplification of the S-gene were classified as a Delta variant. The SGTF data were validated on the basis of comparing the results with our extensive genome sequence data.
Patient Metadata and Geospatial Analysis
Patient metadata were acquired from the electronic medical record by standard informatics methods. Figures showing geospatial distribution of spread for Omicron were generated with Tableau version 2021.2.7 (Tableau Software, LLC, Seattle, WA) using patient home address zip codes. A vaccination breakthrough case was defined as a PCR-positive sample from a patient obtained >14 days after full vaccination (eg, both doses of the Pfizer or Moderna mRNA vaccines) was completed. A booster vaccination breakthrough case was defined as a PCR-positive sample from a patient obtained >14 days after receiving a third vaccine dose. For some cases, manual chart review was conducted to resolve discrepancies or clarify ambiguities.
Results
Omicron Epidemiologic Wave
The first Houston Methodist patient infected with the Omicron variant was identified at the end of November 2021, a time when the Delta VOC was responsible for all COVID-19 cases in metropolitan Houston.6 During this period, the metropolitan area was experiencing a steady decrease in total number of new COVID-19 cases (Figures 1 and 2).
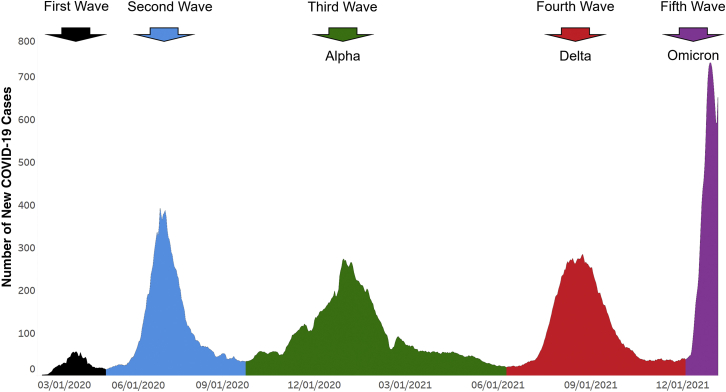
Figure 1.
Epidemiologic curve showing five COVID-19 disease waves in Houston Methodist patients. Number of new COVID-19 cases (y axis) totals are shown as a ±3-day moving average. Each of the five waves is shown in a different color. The first and second waves were composed of a heterogeneous array of SARS-CoV-2 genotypes. The Alpha variant of concern (VOC) shown in the third wave, the Delta VOC shown in the fourth wave, and the Omicron VOC shown in the fifth wave indicate their numeric prominence in those waves. The figure should not be interpreted to mean that all cases in the third, fourth, and fifth waves were caused by Alpha, Delta, and Omicron VOCs, respectively. Rather, they are the dominant single VOCs causing disease in Houston Methodist system patients in those waves. The fifth wave shown includes data through January 5, 2022. The figure was generated with Tableau version 2021.2.7 (Tableau Software, LLC, Seattle, WA), and is adapted with permission from the version presented in Christensen et al.6 The curve is essentially superimposable on COVID-19 activity in all metropolitan Houston, Texas.
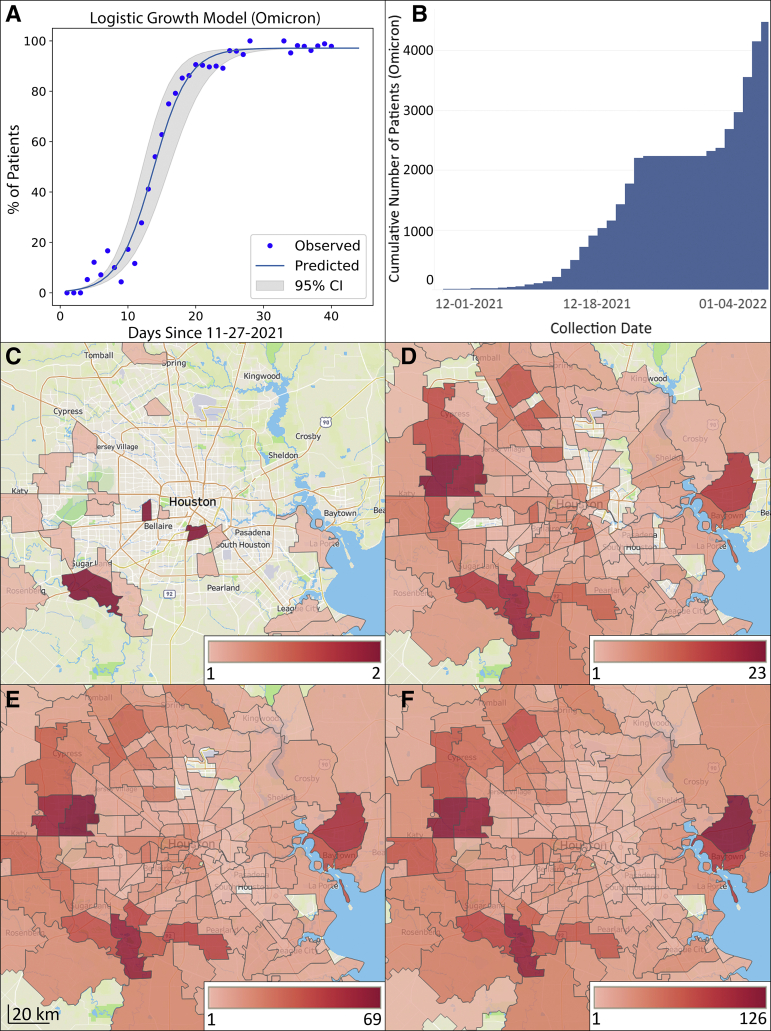
Figure 2.
Increase in Omicron frequency over time and distribution in metropolitan Houston, Texas. The study time frame was November 27, 2021, through January 5, 2022. A: Omicron logistic growth model. The estimated case doubling time is 1.8 days. B: Cumulative increase in Omicron during the study period; y axis is the cumulative number of new COVID-19 Omicron cases. At the end of the study period, Omicron caused 98% of all COVID-19 cases. The plateau between December 24, 2021, and December 30, 2021, exists because samples obtained during this period were not sequenced due to the massive number of daily positive specimens, as described in Materials and Methods. C–F: Geospatial distribution of Omicron based on home address zip code for each patient. C: November 27 to December 6. D: November 27 to December 16. E: November 27 to December 26. F: November 27 to January 5. Note differences in heat map scale for each panel. Figures were generated using Tableau version 2021.2.7. (Tableau Software, LLC, Seattle, WA).
Omicron increased in frequency unusually rapidly over a 3-week period in December (Figures 1 and 2). By December 23, the genome sequence data showed that Omicron accounted for >90% of all new COVID-19 cases in our health care system (Figure 2). The estimated case doubling time during this 3-week period was approximately 1.8 days (Figure 2), which means that Omicron increased in relative frequency approximately three times faster than Delta had increased in our area,6 an unprecedented trajectory for SARS-CoV-2 infections. By January 5, 2022, the Omicron variant caused 98% of all new COVID-19 cases diagnosed in our health care system (Figure 2). This represents the fifth wave of COVID-19 cases in metropolitan Houston (Figure 1).
Consistent with extensive infections caused by Omicron in southern Africa and elsewhere (https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html, last accessed December 28, 2021; https://www.gov.uk/government/collections/new-sars-cov-2-variant, last accessed December 28, 2021), several patients had recent travel histories to countries with a high prevalence of this VOC, suggesting acquisition of virus by some cases from abroad and importation into Houston. However, the vast majority of Omicron patients had no documented travel outside the United States and undoubtedly acquired the infection domestically, either in Houston or elsewhere.
To understand the geospatial distribution of Omicron in metropolitan Houston, patient metadata were acquired from the electronic medical record by standard informatics methods, and home address zip codes were used to visualize virus spread (Figure 2). The 4468 Houston Methodist patients infected with Omicron during this period were distributed widely throughout metropolitan Houston, with 259 different zip codes represented (Figure 2). The widespread distribution of Omicron in the Houston metroplex in an extremely short period of time reflects the ability of this variant to spread unusually rapidly and effectively between individuals and cause symptomatic disease.
Comparison of Omicron, Alpha, and Delta COVID-19 Cases
There is a considerable lack of detailed information about patients with COVID-19 caused by the Omicron VOC, and data are especially lacking for US patients. Available metadata for all Houston Methodist patients infected with Omicron, Alpha, and Delta VOCs (Tables 1 and 2) were compared. The populations differed significantly in many characteristics, including median age, hospital admission rates, maximum respiratory support, rate of vaccine breakthrough, and median length of stay (Tables 1 and 2).
Table 1.
Summary of Pertinent Patient Metadata for 7617 Unique Patients Infected with Omicron or Alpha Variant
| Variable | Omicron variant | Alpha variant | Total | Statistical analysis |
|---|---|---|---|---|
| Patients with data | 4468 (58.7) | 3149 (41.3) | 7617 | |
| Patient characteristics | ||||
| Median age, years | 44.3 | 50.0 | 47.2 |
P < 0.0001 (U-test) |
| Female | 2584 (57.8) | 1617 (51.3) | 4201 (55.2) |
P < 0.0001 (Fisher exact test) |
| Male | 1884 (42.2) | 1532 (48.7) | 3416 (44.8) | |
| Race or ethnicity | ||||
| White | 1627 (36.4) | 1240 (39.4) | 2867 (37.6) |
P < 0.0001 (χ2 test) |
| Hispanic or Latino | 992 (22.2) | 942 (29.9) | 1934 (25.4) | |
| Black | 1376 (30.8) | 729 (23.2) | 2105 (27.6) | |
| Asian | 203 (4.5) | 122 (3.9) | 325 (4.3) | |
| Other | 29 (0.6) | 32 (1.0) | 61 (0.8) | |
| Unavailable | 241 (5.4) | 84 (2.7) | 325 (4.3) | |
| BMI | ||||
| Median BMI, kg/m2 | 29.0 | 30.5 | 29.6 |
P < 0.0001 (U-test) |
| Admission data | ||||
| Admitted | 884 (19.8) | 1719 (54.6) | 2603 (34.2) |
P < 0.0001 (Fisher exact test) Odds ratio: 0.205 (95% CI, 0.185–0.227) |
| Not admitted | 3584 (80.2) | 1430 (45.4) | 5014 (65.8) | |
| Median LOS (discharged patients only), days | 3.2 | 5.1 | 4.7 |
P < 0.0001 (U-test) |
| Max respiratory support | ||||
| ECMO | 1 (0.1) | 7 (0.4) | 8 (0.3) |
P < 0.0001 (χ2 test) |
| Mechanical ventilation | 49 (5.5) | 144 (8.4) | 193 (7.4) | |
| Noninvasive ventilation | 63 (7.1) | 163 (9.5) | 226 (8.7) | |
| High-flow oxygen | 72 (8.1) | 364 (21.2) | 436 (16.7) | |
| Low-flow oxygen | 314 (35.5) | 722 (42.0) | 1036 (39.8) | |
| Room air | 385 (43.6) | 319 (18.6) | 704 (27.0) | |
| Mortality | ||||
| Alive | 4430 (99.1) | 2979 (94.6) | 7409 (97.3) |
P < 0.0001 (Fisher exact test) Odds ratio: 0.150 (95% CI, 0.105–0.214) |
| Deceased | 38 (0.9) | 170 (5.4) | 208 (2.7) | |
| Median PCR CT | ||||
| Abbott Alinity | 20.8 (n = 1961) |
22.4 (n = 1049) |
n = 3010 |
P = 0.0001 (U-test) |
| Hologic Panther | 22.7 (n = 476) |
24.2 (n = 355) |
n = 831 |
P = 0.0745 (U-test) |
| Vaccine | ||||
| Not fully vaccinated | 1971 (44.1) | 3048 (96.8) | 5019 (65.9) |
P < 0.0001 (Fisher exact test) Odds ratio: 38.232 (95% CI, 31.088–47.017) |
| Fully vaccinated | 2497 (55.9) | 101 (3.2) | 2598 (34.1) | |
Data are given as number (percentage) of patients, unless otherwise indicated.
BMI, body mass index; ECMO, extracorporeal membrane oxygenation; LOS, length of stay; Max, maximum.
Table 2.
Summary of Pertinent Patient Metadata for 20,196 Unique Patients Infected with Omicron or Delta Variant
| Variable | Omicron variant | Delta variant | Total | Statistical analysis |
|---|---|---|---|---|
| Patients with data | 4468 (22.1) | 15,728 (77.9) | 20,196 | |
| Patient characteristics | ||||
| Median age, years | 44.3 | 48.3 | 47.6 |
P < 0.0001 (U-test) |
| Female | 2584 (57.8) | 8123 (51.6) | 10,707 (53.0) |
P < 0.0001 (Fisher exact test) |
| Male | 1884 (42.2) | 7605 (48.4) | 9489 (47.0) | |
| Race or ethnicity | ||||
| White | 1627 (36.4) | 6903 (43.9) | 8530 (42.2) |
P < 0.0001 (χ2 test) |
| Hispanic or Latino | 992 (22.2) | 4179 (26.6) | 5171 (25.6) | |
| Black | 1376 (30.8) | 3450 (21.9) | 4826 (23.9) | |
| Asian | 203 (4.5) | 531 (3.4) | 734 (3.6) | |
| Other | 29 (0.6) | 112 (0.7) | 141 (0.7) | |
| Unavailable | 241 (5.4) | 553 (3.5) | 794 (3.9) | |
| BMI | ||||
| Median BMI, kg/m2 | 29.0 | 29.6 | 29.4 |
P < 0.0001 (U-test) |
| Admission data | ||||
| Admitted | 884 (19.8) | 6779 (43.1) | 7663 (37.9) |
P < 0.0001 (Fisher exact test) Odds ratio: 0.326 (95% CI, 0.301–0.353) |
| Not admitted | 3584 (80.2) | 8949 (56.9) | 12,533 (62.1) | |
| Median LOS (discharged patients only), days | 3.2 | 5.4 | 5.2 |
P < 0.0001 (U-test) |
| Max respiratory support | ||||
| ECMO | 1 (0.1) | 19 (0.3) | 20 (0.3) |
P < 0.0001 (χ2 test) |
| Mechanical ventilation | 49 (5.5) | 727 (10.7) | 776 (10.1) | |
| Noninvasive ventilation | 63 (7.1) | 641 (9.5) | 704 (9.2) | |
| High-flow oxygen | 72 (8.1) | 1796 (26.5) | 1868 (24.4) | |
| Low-flow oxygen | 314 (35.5) | 2290 (33.8) | 2604 (34.0) | |
| Room air | 385 (43.6) | 1306 (19.3) | 1691 (22.1) | |
| Mortality | ||||
| Alive | 4430 (99.1) | 14,889 (94.7) | 19,319 (95.7) |
P < 0.0001 (Fisher exact test) Odds ratio: 0.152 (95% CI, 0.110–0.211) |
| Deceased | 38 (0.9) | 839 (5.3) | 877 (4.3) | |
| Median PCR CT | ||||
| Abbott Alinity | 20.8 (n = 1961) |
21.5 (n = 5122) |
n = 7083 |
P < 0.0001 (U-test) |
| Hologic Panther | 22.7 (n = 476) |
22.6 (n = 1298) |
n = 1774 |
P = 0.1606 (U-test) |
| Vaccine | ||||
| No vaccine | 1815 (40.6) | 11,415 (72.6) | 13,230 (65.5) |
P < 0.0001 (χ2 test) |
| >7 Days past first vaccine | 156 (3.5) | 494 (3.1) | 650 (3.2) | |
| >14 Days past second vaccine | 1786 (40.0) | 3679 (23.4) | 5465 (27.1) | |
| >14 Days past third vaccine | 711 (15.9) | 140 (0.9) | 851 (4.2) | |
Data are given as number (percentage) of patients, unless otherwise indicated.
BMI, body mass index; ECMO, extracorporeal membrane oxygenation; LOS, length of stay; Max, maximum.
Patients infected with Omicron were significantly younger than patients infected with Alpha and Delta (Tables 1 and 2). More importantly, Omicron patients were hospitalized significantly less frequently than patients infected with either the Alpha or Delta variant, and had a significantly shorter median hospital length of stay (Tables 1 and 2).
Omicron vaccine breakthrough cases (Tables 1 and 2) were analyzed next. Of the 4468 total Omicron patients for whom whole genome sequence data were available, 2497 (55.9%) met the CDC definition of vaccine breakthrough cases (Tables 1 and 2). There was no simple relationship between the time elapsed since administration of the second vaccination dose and the date of vaccination breakthrough. These 2497 patients received two doses of either the Pfizer-BioNTech BNT162b2 (n = 1828; 73%) or the Moderna mRNA-1273 (n = 553; 22%) or one dose of J&J/Janssen JNJ-78436735 (n = 115; 5%) vaccine. Vaccine type was not specified for one individual. This distribution reflects the majority use of BNT162b2 vaccination doses in our health system. Compared with patients with either Alpha or Delta VOC, a significantly greater percentage of patients with breakthrough cases had the Omicron VOC (55.9% compared with 3.2% and 24.3% for Alpha and Delta VOCs, respectively) (Tables 1 and 2). Individuals with breakthrough cases after receiving a third (booster) dose of either the Pfizer-BioNTech BNT162b2 or the Moderna mRNA-1273 vaccine were analyzed next. Of the 4468 patients with the Omicron VOC, 711 (15.9%) met this criterion. Consistent with Omicron causing a significantly increased number of vaccine breakthrough cases, many studies have reported that this variant has reduced sensitivity to antibody neutralization in vitro, likely in large part due to the extensive number of amino acid and other structural changes occurring in Omicron spike protein.11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34
Spike-Gene Target-Failure Assay
To estimate Omicron variant frequency in patient samples not yet sequenced, the TaqPath COVID-19 Combo Kit assay (Thermo Fisher) was performed on 1216 samples collected from symptomatic patients between December 18, 2021, and January 5, 2022. In total, 1093 (90%) of patient samples yielded an RT-PCR result with S-gene target-failure indicative of the Omicron variant. These data are consistent with the increasing frequency of new cases of COVID-19 caused by Omicron in our population (Figure 2).
Discovery of Omicron Stealth Sublineage BA.2 in Houston
The Omicron sublineage BA.2 was first identified in November 2021 in Australia in a patient who had traveled to South Africa (https://github.com/cov-lineages/pango-designation/issues/359, last accessed December 30, 2021). This sublineage does not have the full set of polymorphisms characteristic of BA.1 (B.1.1.529) and has additional mutations unique to it (https://github.com/cov-lineages/pango-designation/issues/361, last accessed December 30, 2021). One important difference is that sublineage BA.2 lacks the spike gene deletion in the region encoding amino acid 69/70, which means that it will not be detected by the SGTF assay. As a consequence, it is sometimes referred to as the Omicron stealth variant. All full genome sequences present in our large database were inspected, including specimens obtained from symptomatic patients and asymptomatic individuals, and only two members of the BA.2 sublineage were discovered in Houston COVID-19 patients.
Discussion
This work was conducted to address the relative lack of information about disease character among US patients with COVID-19 caused by the Omicron VOC, and to compare the findings with data available for patients in the Houston Methodist system who had disease caused by the Alpha and Delta VOCs. Information relevant to the massive Omicron wave in metropolitan Houston is described here. In 3 weeks (December 1, 2021, through December 23, 2021), Omicron was first identified in our population and rapidly increased to cause 90% of all new COVID-19 cases, with an unusually fast case doubling time of 1.8 days. Analysis of samples obtained from December 30, 2021, to January 5, 2022, found that at the end of the sampling period, Omicron caused 98% of all new COVID-19 cases in our health care system.
The study was based on genome sequence analysis of 4468 Omicron samples taken from socioeconomically, geographically, and ethnically diverse symptomatic patients. Several key findings were made, including the following: i) the Omicron VOC rapidly increased as a cause of COVID-19 and spread throughout the metroplex in an unusually short period of time, far faster than any other SARS-CoV-2 variant; ii) Omicron caused significantly more vaccine breakthrough cases than the Alpha or Delta VOC; iii) Omicron patients were significantly younger than Alpha or Delta patients; iv) significantly fewer Omicron patients required hospitalization compared with Alpha and Delta patients; and v) the median length of stay for hospitalized Omicron patients was significantly shorter than for Alpha and Delta patients, and consistent with this observation, on average the maximum respiratory support required for Omicron patients was significantly less than for Alpha or Delta patients. These findings are largely consistent with many aspects of Omicron data reported from the United Kingdom, South Africa, and Canada,8, 9, 10,35, 36, 37, 38 and are consistent with experimental animal infection data suggesting that Omicron causes less severe disease in mice and hamsters.39, 40, 41, 42, 43 This study was facilitated by a comprehensive and integrated population genomics and epidemiology project2, 3, 4, 5, 6 implemented at the end of February 2020, when the initial COVID-19 case was diagnosed in the Houston Methodist health care system.
Several questions arise from these findings (namely, the underlying causes for the differences observed in Omicron compared with Alpha and Delta patients). Increased vaccine breakthrough cases may occur due to serologic and structural differences in Omicron relative to Alpha and Delta. It is also possible that waning of immunity is a contributing factor as well. Currently, no serologic or other data are available that could address this possibility in our patients. As noted above, ample in vitro and animal infection model data have accumulated, suggesting that Omicron is less virulent than Delta or Alpha VOC. We speculate that the lower age of Omicron patients may be attributable to a disproportionately greater likelihood of risky behaviors in the younger population (eg, less mask wearing and less social distancing). Regardless, additional studies are required to gain more information about factors contributing to the differences between Alpha, Delta, and Omicron patients that were identified in this study.
Because the genome of approximately 90% of SARS-CoV-2 causing COVID-19 in our diverse Houston Methodist patient population is sequenced, and has been done for almost 2 years, the composition of this virus is continuously being monitored in a major US metroplex. This affords the opportunity to rapidly assess changes in SARS-CoV-2 population genomic structure in the fourth largest city in the United States. However, this study has several limitations. Although genomes of SARS-CoV-2 causing 90% of all Houston Methodist COVID-19 cases in the study period were sequenced, this sample represents only approximately 5% of cases reported in the metropolitan region. This patient population underrepresents some demographic groups (eg, homeless individuals and pediatric patients). The samples sequenced in this study were obtained from symptomatic individuals, which means that it is possible that Omicron subvariants or features preferentially represented in asymptomatic individuals were not identified. It is likely that this study included some patients where Omicron was detected on hospital admission but was incidental to the primary cause of admission.
The identification of two asymptomatic individuals with the Omicron stealth sublineage BA.2 is potentially concerning and stresses the importance of using whole-genome sequencing to study patient samples. This sublineage lacks the spike gene deletion corresponding to amino acids 69 and 70 and is not detected by some commonly used assays. Sublineage BA.2 now accounts for approximately 5% of COVID-19 in the United Kingdom, which means that it has the ability to successfully transmit and cause disease.44 It will be important to determine if this SARS-CoV-2 genotype increases in frequency in metropolitan Houston as additional genome sequencing is conducted on samples from our patient population.
In the aggregate, these data add critical new information to features of Omicron genomic epidemiology and patient characteristics in the United States. Furthermore, the present study highlights the importance of analyzing SARS-CoV-2 genome data integrated with patient metadata and stresses the need to continue to do this in near real time as the Omicron surge continues, the virus evolves, and new variants with potentially altered fitness and biomedically relevant phenotypes are generated. Analyses of this type are also important in the context of vaccine formulation and long COVID-19, an increasing health and economic problem globally. Finally, the strategy we have used in this and previous studies2, 3, 4, 5, 6 is readily applicable to future infectious disease problems that warrant special attention.
Acknowledgments
We thank Drs. Marc Boom and Dirk Sostman for ongoing support; and Dr. Sasha M. Pejerrey for editorial contributions.
Footnotes
Supported by the Houston Methodist Academic Institute Infectious Diseases Fund; and in part with funds from the National Institute of Allergy and Infectious Diseases, NIH, Department of Health and Human Services, under contract 75N93019C00076 (J.J.D.).
P.A.C., R.J.O., and S.W.L. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2022.01.007.
Author Contributions
P.A.C., R.J.O., S.W.L., and J.M.M. had full access to all study data and take responsibility for the integrity of the data and the accuracy of data analysis; J.M.M., P.A.C., R.J.O., and S.W.L. conceived and designed the study; all authors acquired, analyzed, or interpreted data and wrote the manuscript; P.A.C. analyzed data; J.M.M. and J.J.D. obtained funding; J.M.M. provided overall supervision; P.A.C., R.J.O., and S.W.L. contributed equally and are co–first authors.
Supplemental Data
References
- 1.Dhar M.S., Marwal R., Vs R., Ponnusamy K., Jolly B., Bhoyar R.C., et al. Genomic characterization and epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. Science. 2021;374:995–999. doi: 10.1126/science.abj9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long S.W., Olsen R.J., Christensen P.A., Bernard D.W., Davis J.J., Shukla M., Nguyen M., Saavedra M.O., Yerramilli P., Pruitt L., Subedi S., Kuo H.C., Hendrickson H., Eskandari G., Nguyen H.A.T., Long J.H., Kumaraswami M., Goike J., Boutz D., Gollihar J., McLellan J.S., Chou C.W., Javanmardi K., Finkelstein I.J., Musser J.M. Molecular architecture of early dissemination and massive second wave of the SARS-CoV-2 virus in a major metropolitan area. mBio. 2020;11:6. doi: 10.1128/mBio.02707-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musser J.M., Olsen R.J., Christensen P.A., Long S.W., Subedi S., Davis J.J., Gollihar J. Rapid, widespread, and preferential increase of SARS-CoV-2 B.1.1.7 variant in Houston, TX, revealed by 8,857 genome sequences. medRxiv. 2021 [Preprint]. doi: 10.1101/2021.03.16.21253753. [Google Scholar]
- 4.Olsen R.J., Christensen P.A., Long S.W., Subedi S., Hodjat P., Olson R., Nguyen M., Davis J.J., Yerramilli P., Saavedra M.O., Pruitt L., Reppond K., Shyer M.N., Cambric J., Gadd R., Thakur R.M., Batajoo A., Finkelstein I.J., Gollihar J., Musser J.M. Trajectory of growth of severe acute respiratory (SARS-CoV-2) syndrome coronavirus 2 variants in Houston, Texas, January through May 2021, based on 12,476 genome sequences. Am J Pathol. 2021;191:1754–1773. doi: 10.1016/j.ajpath.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long S.W., Olsen R.J., Christensen P.A., Subedi S., Olson R., Davis J.J., Saavedra M.O., Yerramilli P., Pruitt L., Reppond K., Shyer M.N., Cambric J., Finkelstein I.J., Gollihar J., Musser J.M. Sequence analysis of 20,453 SARS-CoV-2 genomes from the Houston metropolitan area identifies the emergence and widespread distribution of multiple isolates of all major variants of concern. Am J Pathol. 2021;191:983–992. doi: 10.1016/j.ajpath.2021.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen P.A., Olsen R.J., Long S.W., Subedi S., Davis J.J., Hodjat P., Walley D.R., Kinskey J.C., Saavedra M.O., Pruitt L., Reppond K., Shyer M.N., Cambric J., Gadd R., Thakur R.M., Batajoo A., Mangham R., Pena S., Trinh T., Yerramilli P., Nguyen M., Olson R., Snehal R., Gollihar J., Musser J.M. Delta variants of SARS-CoV-2 cause significantly increased vaccine breakthrough COVID-19 cases in Houston, Texas. Am J Pathol. 2022;192:320–331. doi: 10.1016/j.ajpath.2021.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen P.A., Olsen R.J., Long S.W., Snehal R., Davis J.J., Saavedra M.O., Reppond K., Shyer M.N., Cambric J., Gadd R., Thakur R.M., Batajoo A., Mangham R., Pena S., Trinh T., Kinskey J.C., Williams G., Olson R., Gollihar J., Musser J.M. Signals of significantly increased vaccine breakthrough, decreased hospitalization rates, and less severe disease in patients with coronavirus disease 2019 caused by the Omicron variant of severe acute respiratory syndrome coronavirus 2 in Houston, Texas. Am J Pathol. 2022 doi: 10.1016/j.ajpath.2022.01.007. [Epub Ahead of Print] doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. medRxiv. 2021 doi: 10.1101/2021.12.19.21268028. [Preprint]. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott P., Bodinier B., Eales O., Wang H., Haw D., Elliott J., Whitaker M., Jonnerby J., Tang D., Walters C.E., Atchison C., Diggle P.J., Page A.J., Trotter A.J., Ashby D., Barclay W., Taylor G., Ward H., Darzi A., Cooke G.S., Chadeau-Hyam M., Donnelly C.A. Rapid increase in Omicron infections in England during December 2021: REACT-1 study. Science. 2022 doi: 10.1126/science.abn8347. [Epub ahead of print]. doi: 10.1126/science.abn8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheikh A., Kerr S., Woolhouse M., McMenamin J., Robertson C. University of Edinburgh; Edinburgh, Scotland: 2021. Severity of Omicron Variant of Concern and Vaccine Effectiveness against Symptomatic Disease: National Cohort with Nested Test Negative Design Study in Scotland. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng B., Ferreira I., Abdullahi A., Kemp S.A., Goonawardane N., Papa G., Fatihi S., Charles O., Collier D., Collaboration C.-N.B.C.-, Consortium TgtP. J., Choi J., Hyeon Lee J., Mlcochova P., James L., Doffinger R., Thukral L., Sato K., Gupta R.K. SARS-CoV-2 Omicron spike mediated immune escape, infectivity and cell-cell fusion. bioRxiv. 2021 [Preprint]. doi: 10.1101/2021.12.17.473248. [Google Scholar]
- 12.Zeng C., Evans J.P., Qu P., Faraone J., Zheng Y.-M., Carlin C., Bednash J.S., Zhou T., Lozanski G., Mallampalli R., Saif L.J., Oltz E.M., Mohler P., Xu K., Gumina R.J., Liu S.-L. Neutralization and stability of SARS-CoV-2 Omicron variant. bioRxiv. 2021 [Preprint]. doi: 10.1101/2021.12.16.472934. [Google Scholar]
- 13.Jacobsen H., Strengert M., Maass H., Ynga Durand M.A., Kessel B., Harries M., Rand U., Abassi L., Kim Y., Lueddecke T., Hernandez P., Ortmann J., Heise J.-K., Castell S., Gornyk D., Gloeckner S., Melhorn V., Lange B., Dulovic A., Haering J., Junker D., Schneiderhan-Marra N., Poehlmann S., Hoffmann M., Krause G., Cicin-Sain L. Diminished neutralization responses towards SARS-CoV-2 Omicron VoC after mRNA or vector-based COVID-19 vaccinations. medRxiv. 2021 doi: 10.1038/s41598-022-22552-y. [Preprint]. doi: 10.1101/2021.12.21.21267898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggink D., Andeweg S.P., Vennema H., van Maarseveen N., Vermaas K., Vlaemynck B., Schepers R., van Gageldonk-Lafeber A.B., van den Hof S., Reusken C.B.E.M., Knol M.J. Increased risk of infection with SARS-CoV-2 Omicron compared to Delta in vaccinated and previously infected individuals, the Netherlands, 22 November to 19 December 2021. medRxiv. 2021 doi: 10.2807/1560-7917.ES.2022.27.4.2101196. [Preprint]. doi: 10.1101/2021.12.20.21268121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edara V.-V., Manning K.E., Ellis M., Lai L., Moore K.M., Foster S.L., Floyd K., Davis-Gardner M.E., Mantus G., Nyhoff L.E., Bechnack S., Alaaeddine G., Naji A., Samaha H., Lee M., Bristow L., Hussaini L., Ciric C.R., Nguyen P.-V., Gagne M., Roberts-Torres J., Henry A.R., Godbole S., Grakoui A., Sexton M., Piantadosi A., Waggoner J.J., Douek D.C., Anderson E.J., Rouphael N., Wrammert J., Suthar M.S. mRNA-1273 and BNT162b2 mRNA vaccines have reduced neutralizing activity against the SARS-CoV-2 Omicron variant. Cell Medicine Reports. 2022;3:100529. doi: 10.1016/j.xcrm.2022.100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou j, Xia H., Xie X., Kurhade C., Machado R.R., Weaver S.C., Ren P., Shi P.-Y. Neutralization against Omicron SARS-CoV-2 from previous non-Omicron infection. bioRxiv. 2021 doi: 10.1038/s41467-022-28544-w. [Preprint]. doi: 2021.2012.2020.473584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikemura N., Hoshino A., Higuchi Y., Taminishi S., Inaba T., Matoba S. SARS-CoV-2 Omicron variant escapes neutralization by vaccinated and convalescent sera and therapeutic monoclonal antibodies. medRxiv. 2021 [Preprint]. doi: 10.1101/2021.12.13.21267761. [Google Scholar]
- 18.Dejnirattisai W., Shaw R.H., Supasa P., Liu C., Stuart A.S., Pollard A.J., Liu X., Lambe T., Crook D., Stuart D.I., Mongkolsapaya J., Nguyen-Van-Tam J.S., Snape M.D., Screaton G.R. Reduced neutralisation of SARS-CoV-2 Omicron B.1.1.529 variant by post-immunisation serum. Lancet. 2022;399:234–236. doi: 10.1016/S0140-6736(21)02844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cameroni E., Saliba C., Bowen J.E., Rosen L.E., Culap K., Pinto D., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L., Iketani S., Guo Y., Chan J.F.-W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., Yu J., Chik K.K.-H., Yuen T.T.-T., Yoon C., To K.K.-W., Chen H., Yin M.T., Sobieszczyk M.E., Huang Y., Wang H.H., Sheng Z., Yuen K.-Y., Ho D.D. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 21.Planas D., Saunders N., Maes P., Benhassine F.G., Planchais C., Porrot F., Staropoli I., Lemoine F., Pere H., Veyer D., Puech J., Rodary J., Bolland W.H., Buchrieser J., Baele G., Dellicour S., Raymenants J., Gorissen S., Geenen C., Vanmechelen B., Wawina T., Marti J., Cuypers L., Seve A., Hocqueloux L., Prazuck T., Loriere E.S., REY F., Bruel T., Mouquet H., Andre E., Schwartz O. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 22.Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O’Connell A.-M., Simons D., Blomquist P.B., Zaidi A., Nash S., Aziz N.I.B.A., Thelwall S., Dabrera G., Myers R., Amirthalingam G., Gharbia S., Barrett J.C., Elson R., Ladhani S.N., Ferguson N., Zambon M., Campbell C.N., Brown K., Hopkins S., Chand M., Ramsay M., Bernal J.L. Effectiveness of COVID-19 vaccines against the Omicron (B.1.1.529) variant of concern. medRxiv. 2021 [Preprint]. doi: 10.1101/2021.12.14.21267615. [Google Scholar]
- 23.Yu X., Wei D., Xu W., Li Y., Li X., Zhang X., Qu J., Yang Z., Chen E. Reduced sensitivity of SARS-CoV-2 Omicron variant to booster-enhanced neutralization. Cell Discovery. 2022;8:4. doi: 10.1038/s41421-022-00375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cele S., Jackson L., Khan K., Khoury D.S., Moyo-Gwete T., Tegally H., Scheepers C., Amoako D., Karim F., Bernstein M., Lustig G., Archary D., Smith M., Ganga Y., Jule Z., Reedoy K., Cromer D., San J.E., Hwa S.-H., Giandhari J., Blackburn J.M., Gosnell B.I., Karim S.S.A., Hanekom W., von Gottberg A., Bhiman J., Lessells R.J., Moosa M.-Y.S., Davenport M.P., de Oliveira T., Moore P.L., Sigal A., NGS-SA, Team C-K SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv. 2021 [Preprint]. doi: 10.1101/2021.12.08.21267417. [Google Scholar]
- 25.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., Wang J., Wang Y., Niu X., Yang S., Liang H., Sun H., Li T., Yu Y., Cui Q., Liu S., Yang X., Du S., Zhang Z., Hao X., Shao F., Jin R., Wang X., Xiao J., Wang Y., Xie X.S. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen C.H., Schelde A.B., Moustsen-Helms I.R., Emborg H.-D., Krause T.G., Moelbak K., Valentiner-Branth P., Institut TIDPGaSS Vaccine effectiveness against SARS-CoV-2 infection with the Omicron or Delta variants following a two-dose or booster BNT162b2 or mRNA-1273 vaccination series: a Danish cohort study. medRxiv. 2021 [Preprint]. doi: 10.1101/2021.12.20.21267966. [Google Scholar]
- 27.Syed A.M., Ciling A., Khalid M.M., Sreekumar B., Kumar G.R., Silva I., Milbes B., Kojima N., Hess V., Shacreaw M., Lopez L., Brobeck M., Turner F., Spraggon L., Taha T.Y., Tabata T., Chen I.P., Ott M., Doudna J.A. Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles. medRxiv. 2021 doi: 10.1073/pnas.2200592119. [Preprint]. doi: 10.1101/2021.12.20.21268048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheward D.J., Kim C., Ehling R.A., Pankow A., Castro Dopico X., Martin D.P., Reddy S.T., Dillner J., Karlsson Hedestam G.B., Albert J., Murrell B. Variable loss of antibody potency against SARS-CoV-2 B.1.1.529 (Omicron) bioRxiv. 2021 [Preprint]. doi: 10.1101/2021.12.19.473354. [Google Scholar]
- 29.Haveri A., Solastie A., Ekström N., Österlund P., Nohynek H., Nieminen T., Palmu A.A., Melin M. Neutralizing antibodies to SARS-CoV-2 Omicron variant after 3rd mRNA vaccination in health care workers and elderly subjects and response to a single dose in previously infected adults. medRxiv. 2021 doi: 10.1002/eji.202149785. [Preprint]. doi: 10.1101/2021.12.22.21268273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arien K.K., Heyndrickx L., Michiels J., Vereecken K., Van Lent K., Coppens S., Pannus P., Martens G.A., Van Esbroeck M., Goossens M.E., Marchant A., Bartholomeeusen K., Desombere I. Three doses of the BNT162b2 vaccine confer neutralising antibody capacity against the SARS-CoV-2 B.1.1.529 (Omicron) variant of concern. medRxiv. 2021 doi: 10.1038/s41541-022-00459-z. [Preprint]. doi: 10.1101/2021.12.23.21268316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willett B.J., Grove J., MacLean O., Wilkie C., Logan N., De Lorenzo G., et al. The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism. medRxiv. 2022 [Preprint]. doi: 10.1101/2022.01.03.21268111. [Google Scholar]
- 32.Boschi C., Colson P., Bancod A., Moal V., La Scola B. Omicron variant escapes therapeutic mAbs contrary to eight prior main VOC. bioRxiv. 2022 doi: 10.1093/cid/ciac143. [Preprint]. doi: 10.1101/2022.01.03.474769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., et al. Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. bioRxiv. 2021 doi: 10.1016/j.cell.2021.12.046. [Preprint]. doi: 10.1101/2021.12.03.471045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banerjee A., Lew J., Kroeker A., Baid K., Aftanas P., Nirmalarajah K., Maguire F., Kozak R., McDonald R., Lang A., Gerdts V., Straus S.E., Gilbert L., Li A.X., Mozafarihasjin M., Walmsley S., Gingras A.-C., Wrana J.L., Mazzulli T., Colwill K., McGeer A.J., Mubareka S., Falzarano D. Immunogenicity of convalescent and vaccinated sera against clinical isolates of ancestral SARS-CoV-2, beta, delta, and omicron variants. bioRxiv. 2022 doi: 10.1016/j.medj.2022.04.002. [Preprint]. doi: 10.1101/2022.01.13.475409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulloa A.C., Buchan S.A., Daneman N., Brown K.A. Early estimates of SARS-CoV-2 Omicron variant severity based on a matched cohort study, Ontario, Canada. medRxiv. 2021 [Preprint]. doi: 10.1101/2021.12.24.21268382. [Google Scholar]
- 36.Wolter N., Jassat W., Walaza S., Welch R., Moultrie H., Groome M., Amoako D.G., Everatt J., Bhiman J.N., Scheepers C., Tebeila N., Chiwandire N., du Plessis M., Govender N., Ismail A., Glass A., Mlisana K., Stevens W., Treurnicht F.K., Makatini Z., Hsiao N-y, Parboosing R., Wadula J., Hussey H., Davies M.-A., Boulle A., von Gottberg A., Cohen C. Early assessment of the clinical severity of the SARS-CoV-2 Omicron variant in South Africa. medRxiv. 2021 doi: 10.1016/S0140-6736(22)00017-4. [Preprint]. doi: 10.1101/2021.12.21.21268116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.SARS-CoV-2 Variants of Concern and Variants Under Investigation in England. Technical Briefing: Update on Hospitalisation and Vaccine Effectiveness for Omicron VOC-21NOV-01 (B.1.1.529) UK Health Security Agency; London, United Kingdom: 2021. [Google Scholar]
- 38.Davies M.-A., Kassanjee R., Rousseau P., Morden E., Johnson L., Solomon W., et al. Outcomes of laboratory-confirmed SARS-CoV-2 infection in the Omicron-driven fourth wave compared with previous waves in the Western Cape Province, South Africa. medRxiv. 2022 doi: 10.1111/tmi.13752. [Preprint]. doi: 10.1101/2022.01.12.22269148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bentley E.G., Kirby A., Sharma P., Kipar A., Mega D.F., Bramwell C., Penrice-Randal R., Prince T., Brown J.C., Zhou J., Screaton G.R., Barclay W.S., Owen A., Hiscox J.A., Stewart J.P. SARS-CoV-2 Omicron-B.1.1.529 variant leads to less severe disease than Pango B and Delta variants strains in a mouse model of severe COVID-19. bioRxiv. 2021 [Preprint]. doi: 10.1101/2021.12.26.474085. [Google Scholar]
- 40.Abdelnabi R., Foo C.S., Zhang X., Lemmens V., Maes P., Slechten B., Raymenants J., André E., Weynand B., Dallemier K., Neyts J. The Omicron (B.1.1.529) SARS-CoV-2 variant of concern does not readily infect Syrian hamsters. Antiviral Res. 2022;198 doi: 10.1016/j.antiviral.2022.105253. 105253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diamond M., Halfmann P., Maemura T., Iwatsuki-Horimoto K., Iida S., Kiso M., et al. The SARS-CoV-2 B.1.1.529 Omicron virus causes attenuated infection and disease in mice and hamsters. Res Sq. 2021 [Preprint]. doi: 10.21203/rs.3.rs-1211792/v1. [Google Scholar]
- 42.McMahan K., Giffin V., Tostanoski L., Chung B., Siamatu M., Suthar M., Halfmann P., Kawaoka Y., Piedra-Mora C., Martinot A., Kar S., Andersen H., Lewis M.G., Barouch D.H. Reduced pathogenicity of the SARS-CoV-2 Omicron variant in hamsters. bioRxiv. 2022 doi: 10.1016/j.medj.2022.03.004. [Preprint]. doi: 10.1101/2022.01.02.474743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan S., Ye Z.-W., Liang R., Tang K., Zhang A.J., Lu G., Ong C.P., Poon V.K.-M., Chan C.C.-S., Mok B.W.Y., Qin Z., Xie Y., Sun H., Tsang J.O.-L., Yuen T.T.-T., Chik K.K.-H., Chan C.C.-Y., Cai J.-P., Luo C., Lu L., Yip C.C.-Y., Chu H., To K.K.-W., Chen H., Jin D.-Y., Yuen K.-Y., Chan J.F.W. The SARS-CoV-2 Omicron (B.1.1.529) variant exhibits altered pathogenicity, transmissibility, and fitness in the golden Syrian hamster model. bioRxiv. 2022 [Preprint]. doi: 10.1101/2022.01.12.476031. [Google Scholar]
- 44.SARS-CoV-2 Variants of Concern and Variants Under Investigation in England. Technical Briefing 34. UK Health Security Agency; London, United Kingdom: 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.