Abstract
Heart failure (HF) is the leading cause of death around the world, the mortality caused by HF is growing rapidly, and has become a great threaten to both public health and economic growth. Dehydrocostus lactone (DHE) is the active constituent of Saussurea lappa and is widely used in traditional Chinese medicine for its multiple biological functions, including anti-inflammatory, antioxidant and anti-cancer. To the best of our knowledge, DHE's effect on HF has not been clarified. Thioredoxin-interacting protein (TXNIP) regulates the process of oxidative stress and inflammation and leads to an increase in oxidative stress via oxidization of thioredoxin, TXNIP promotes the activation of the immune response by its binding with the NOD-like receptor protein 3 inflammasome. An MTT assay revealed that the overexpression or inhibition of TXNIP markedly decreased or significantly increased the proliferation of H9c2 cells, respectively. Through reverse transcription-quantitative PCR (RT-qPCR) and western blotting, it was determined that the expression of proinflammatory cytokines was significantly decreased with the increased expression of anti-inflammatory cytokines in a TXNIP knockout model. Further study utilizing RT-qPCR and western blotting demonstrated that these effects may be mediated by the nuclear factor erythroid 2-related factor 2/heme oxygenase-1/NF-κB signaling pathway. In conclusion, TXNIP inhibition may promote the therapeutic effect of DHE on oxidative stress-induced damage.
Keywords: thioredoxin-interacting protein, dehydrocostus lactone, cardiomyocytes, doxorubicin, inflammation
Introduction
Heart failure (HF) is a cardiac disease induced by multiple factors, such as a high fat diet, lack of exercise and aging, that can be recognized by dysfunction in cardiac systolic and diastolic processes. The number of individuals affected by HF has increased from 5.7 million to 6.2 million in the US between 2013 and 2016, and HF-related deaths contributed to 13.4% of all deaths that occurred in the US in 2018(1). Previous studies reported that proinflammatory cytokines serve a critical role in promoting the development of HF (2) and that an increased concentration of cytokines and biomarkers are exhibited in patients with HF (3). Inflammation has been regarded as a possible therapeutic target for patients with HF (4). Thus, pathways regulating inflammation and oxidative responses may act as a treatment method for reducing cardiovascular disease risk. Dehydrocostus lactone (DHE) is a natural sesquiterpene lactone and the major compound in the roots of a well-known traditional Chinese herbal medicine, Saussurea lappa. DHE has presented anti-inflammatory and immunomodulatory effects in previous studies (5,6). A previous study also indicated that DHE protects HepG2 cells against oxidative stress via the induction of heme oxygenase-1 (HO-1) expression (7), and that this effect may be mediated by the inhibition of the NF-κB signaling pathway (8). Moreover, DHE has numerous biological functions, including anti-inflammatory properties, immunomodulatory capabilities and antitumor action (9). Previous studies found that DHE significantly inhibits the proliferation of cancer cells without affecting normal cells in breast and prostate cancer (10,11). Another study indicated that DHE reduces lipopolysaccharide-induced acute lung injury in a rat model via the inhibition of the inflammatory response (12).
Thioredoxin-interacting protein (TXNIP) is a protein that serves a critical role in multiple cellular processes, including metabolism, growth and antioxidative responses, by inhibiting the ability of cells to take up glucose (13). Previous research has indicated that activation of the inflammatory response is mediated by the activation of TXNIP (14), thus, we hypothesized that TXNIP inhibition could enhance the therapeutic effect of DHE in cardiomyocytes.
In the present study, a cell model of TXNIP overexpression and inhibition was established to study the proliferation of H9c2 cells and the expression of pro and anti-inflammatory genes. The involvement of the nuclear factor erythroid 2-related factor 2 (Nrf2)/HO-1/NF-κB signaling pathway was also investigated.
Materials and methods
Vector constructions
The TXNIP cDNA fragment was obtained using PCR with the following primers: Forward, 5'-AGTGATTGGCAGCAGGTC-3' and reverse, 5'-GGTGTCTGGGATGTTTAGG-3'. A blank pcDNA3.1 vector (cat. no. HG-VPH0683; HonorGene) and the product obtained from PCR were digested with KpnI (cat. no. R3142S; New England BioLabs, Inc.) and XhoI (cat. no. R0146S; New England BioLabs, Inc.) enzymes and the TXNIP fragment was linked with the digested vector using T4 DNA Ligase (cat. no. M0202S; New England BioLabs, Inc.). The pcDNA3.1-TXNIP overexpression vector (1.5 µg) was subsequently transfected into H9c2 cells using Lipofectamine™ 3000 Transfection Reagent (cat. no. L3000008; Thermo Fisher Scientific, Inc.) for 48 h at 37°C according to the manufacturer's protocol. Stable TXNIP-expressing H9c2 cells were screened using 1,000 µg/ml G418. After four weeks of screening, stable TXNIP overexpression cells were obtained and cells were used to perform western blotting and ELISA experiments.
The TXNIP knockdown vector was constructed as previously described (15). The knockout of the TXNIP gene was performed using the CRISPR-Cas9 system. A blank lentiCRISPR v2 vector (cat. no. 52961; Addgene) was digested using the BsmBI (cat. no. R0580S; New England BioLabs, Inc.) enzyme as recommended according to previous study (15) and oligos were constructed using the following gRNAs: Forward, 5'-CACCGGAGACAGACACCCGCCCATC-3' and reverse, 5'-CGATGGGCGGGTGTCTGTCTCCAAA-3', designed using an online CRISPR design tool (CRISPRdirect; https://crispr.dbcls.jp). The digested vector and oligos were ligated using quick ligase (cat. no. M0201S; New England BioLabs, Inc.) to construct the TXNIP knockdown vector [target gene sequence: Forward, 5'-GATGGGCGGGTGTCTGTCTC-3' and reverse, 5'-GAGACAGACACCCGCCCATC-3'; target exon, TXNIP-202 (Transcript ID: ENSRNOT00000028793.7; domain affected, IPR011022 (InterPro database)]. The vector was first transfected into 293T cells to assemble lentiviruses using Lipofiter transfect reagent (cat. no. KS-TRLF-200; Hanbio Biotechnology Co., Ltd.) at 37°C. The virus was collected at 72 h, after which it was subsequently transduced into H9c2 cells for 48 h at 37°C. Stable TXNIP-knockout cells were screened using 2 µg/ml puromycin. The blank pcDNA3.1 vector was used as a blank control for TXNIP overexpression detection and the blank lentiCRISPR v2 vector was used as a blank control for TXNIP knockout detection.
Cell culture, grouping and MTT assay
The rat cardiomyoblast H9c2 (cat. no. CRL-1446) and the human kidney epithelial 293T (cat. no. CRL-11268) cell lines were purchased from the American Type Culture Collection and cultured in high glucose DMEM (cat. no. 11965092; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (cat. no. 10091; Thermo Fisher Scientific, Inc.) in a 5% CO2 humid atmosphere at 37°C. Subsequently, the H9c2 cells were divided into four groups: i) The control group (NC); ii) the DHE treatment group (OT); iii) the DHE treatment combined with TXNIP inhibition group (OI); and iv) the DHE treatment combined with TXNIP overexpression group (OH). In all groups, H9c2 cells were first treated with 5 µM doxorubicin and incubated for 24 h at 37˚C (16) with or without 30 µM DHE for 24 h (17). The viability of cells was measured using an MTT assay. H9c2 cells were treated as described above, after which they were incubated with 5 mg/ml MTT reagent for 4 h at 37˚C. Cells were then dissolved in DMSO and absorbance was detected at 490 nm using a microplate reader (cat. no. PT-3502C; Potenov).
Ethics statement
This study was carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (18). The experimental protocols were approved by the Ethics Committee of Animal Experiments of Anqiu People's Hospital (Anqiu, China). All experiments were conducted in accordance with the Declaration of Helsinki.
Mouse model and grouping
A total of 32 C57BL/6 mice (age, 8-10 weeks; weight, 24-27 g) were purchased from the Xiamen University Laboratory Animal Center. Mice were kept in a 22-24˚C and 50-60% humidity atmosphere under a 12-h light/dark cycle, where food and water was freely available. Mice were randomly divided into the following four groups: i) The control group (NC); ii) the DHE treatment group (OT); iii) the DHE treatment combined with TXNIP inhibition group (OI); and iv) the DHE treatment combined with TXNIP overexpression group (OH). In order to construct the TXNIP overexpression and knockout mouse model, 200 µl of the TXNIP overexpression, knockout lentivirus vector or blank vector were injected in mice via the tail vein. The vector used for animal experiments was purchased from Hanbio Biotechnology Co., Ltd. A total of 32 mice were divided into two groups: The DHE-treated group (n=20) and the TXNIP expression detection group (n=12). Mice were treated with 15 mg/kg doxorubicin for 21 days to construct the HF model (19). In the DHE treatment groups, mice were treated with 5 mg/kg DHE for 21 days (12). For blood sample collection, 100 µl blood was collected through ophthalmic vein into a 1.5 ml tube, and incubated at room temperature for 1 h. After centrifugation at 1,500 x g at 4˚C for 10 min, serum was collected and subjected to ELISA. Then, mice in each group were anesthetized using sodium pentobarbital (100 mg/kg) via intraperitoneal injection and then sacrificed by cervical dislocation. For heart tissue assessment, the expression of TXNIP in full heart tissue of mice without DHE treatment was detected using western blotting analysis. The full heart tissue of mice treated with DHE was collected after mice were sacrificed. Heart tissues were stored at -80°C until further experimentation.
RNA extraction
RNA extraction was performed according to the protocol of the RNApure Tissue and Cell kit (cat. no. CW0560; CoWin Biosciences). Heart tissues and cells were first lysed with lysis buffer. After incubation at room temperature for 5 min, the mixture was centrifuged at 13,000 x g, 4°C for 5 min. Subsequently, the RNA samples were transferred into spin columns and centrifuged at 13,000 x g, 4°C for 1 min. After washing with washing buffer, RNA samples were eluted from the spin column using elution buffer after centrifugation at 13,000 x g, 4°C for 1 min. RNA sample concentration was detected using a spectrophotometer (UV5Nano; Mettler-Toledo International, Inc.). RNA samples were used for subsequent reverse transcription-quantitative PCR (RT-qPCR) experiments.
RT-qPCR
RT-qPCR experiments were performed using an UltraSYBR One Step RT-qPCR kit according to the manufacturer protocol (cat. no. CW2623; CoWin Biosciences). The reaction mixture was prepared as recommended by the manufacturer protocol and the reaction was performed with the following steps: Reverse transcription at 45˚C for 10 min; initial denaturation at 95˚C for 5 min; and 40 cycles of denaturation at 95˚C for 10 sec, annealing and elongation at 58˚C for 10 sec and final extension at 72˚C for 30 sec. The primers used for qPCR were as follows: Inducible nitric oxide synthase (iNOS) forward, 5'-CCCTTCAATGGTTGGTACATGG-3' and reverse, 5'-ACATTGATCTCCGTGACAGCC-3'; TNF-α forward, 5'-TTCTCATTCCTGCTTGTGG-3' and reverse, 5'-ACTTGGTGGTTTGCTACG-3'; IL-6 forward, 5'-CCACCAAGAACGATAGTCAA-3' and reverse, 5'-TTTCCACGATTTCCCAGA-3'; IL-1 forward, 5'-CCAGCTTCAAATCTCACAGCAG-3' and reverse, 5'-CTTCTTTGGGTATTGCTTGGGATC-3'; IL-10 forward, 5'-GGCAGATCTATGCTTGGCTCAGCACTG-3' and reverse, 5'-GCGATATCCCTGCAGTCCAGTAGACG-3'; cyclooxygenase-2 (COX-2) forward, 5'-TCACAGGCTTCCATTGACCAG-3' and reverse, 5'-CCGAGGCTTTTCTACCAGA-3'; NF-E2-Related Factor 2 (Nrf2) forward, 5'-GTCTTCACTGCCCCTCATC-3' and reverse, 5'-TCGGGAATGGAAAATAGCTCC-3'; HO-1 forward, 5'-AGAGTCCCTCACAGACAGAGTTT-3' and reverse, 5'-CCTGCAGAGAGAAGGCTACATGA-3'; NF-κB forward, 5'-ACGGGAGGGGAAGAAATCTATC-3' and reverse, 5'-AATGGCAAACTGTCTGTGAACA-3'; GAPDH forward, 5'-GATGCTGGTGCTGAGTATGTCG-3', reverse, 5'-GTGGTGCAGGATGCATTGCTCTGA-3'. GAPDH was used as an internal control. The expression of each target gene was calculated using 2-ΔΔCq method (20).
Western blotting
Heart tissues and cells from each group were lysed using RIPA lysis buffer (cat. no. CW2333; CoWin Biosciences) supplemented with a protease inhibitor cocktail (cat. no. CW2200; CWbio Biosciences). The concentration of proteins was determined using a BCA assay (cat. no. CW0014; CoWin Biosciences). Subsequently, 60 µg protein/lane was separated by SDS-PAGE on a 10% gel. After electrophoresis, the proteins were transferred onto PVDF membranes through Trans-Blot (cat. no. 1703940; Bio-Rad Laboratories, Inc.) transfer blotter. The membranes were first incubated with 5% skimmed milk for 1 h at room temperature, followed by incubation with the following primary antibodies at 4˚C overnight (all, Abcam; all, 1:1,000): Kelch like ECH associated protein 1 (Keap1; cat. no. ab226997; 1:1,000), Nrf2 (cat. no. ab31163), HO-1 (cat. no. ab68477), inhibitor of nuclear factor κB kinase (IKK; cat. no. ab178870), NF-κB (cat. no. ab32360), high mobility group box 1 (HMGB1; cat. no. ab18256), toll-like receptor (TLR)2 (cat. no. ab209217), TLR4 (cat. no. ab22048), IL-1β (cat. no. ab9722) and NLR family pyrin domain containing 3 (NLRP3; cat. no. ab263899), antibodies were purchased from Abcam). After washing with TBST (0.5% Tween-20) for three times, the membranes were subsequently incubated with goat-anti rabbit and goat-anti mouse HRP-labeled secondary antibodies at room temperature (1:1,000; cat. nos. ab6721 and ab6789; Abcam) for 1 h. The protein expression levels were detected using Immobilon Western HRP Substrate (cat. no. WBKLS0100; EMD Millpore) and were semi-quantified using Image Pro Plus 6.0 software (Media Cybernetics, Inc.). GAPDH was used as an internal control.
ELISA
ELISA was performed according to the protocol of each respective kit. The following kits were used and purchased from Abcam: iNOS (cat. no. ab285316), COX-2 (cat. no. ab210574), CCL9 (cat. no. ab240689), CXCL1 (cat. no. ab219044), CXCL9 (cat. no. ab203364), CXCL11 (cat. no. ab204519). Briefly, cultured medium and serum samples obtained from cells and mice were added to each well of a 96-well plate and incubated at room temperature for 3 h. Then, after washing four times with washing buffer, the samples were incubated with antibodies included in each ELISA kit according to the manufacturer's protocol at room temperature for 1 h, followed by incubation with HRP-streptavidin solution for 45 min at room temperature. After four incubations in washing buffer, the samples were incubated with one-step substrate reagent for 30 min at room temperature in the dark, then finally incubated with stop solution. The absorbance value was detected at 450 nm.
Statistical analysis
Data are presented as the mean ± SD. Experiments were repeated three times independently. The differences among groups were evaluated with one-way ANOVA and followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Detection of H9c2 cell viability and expression of TXNIP in cell and mice model of HF
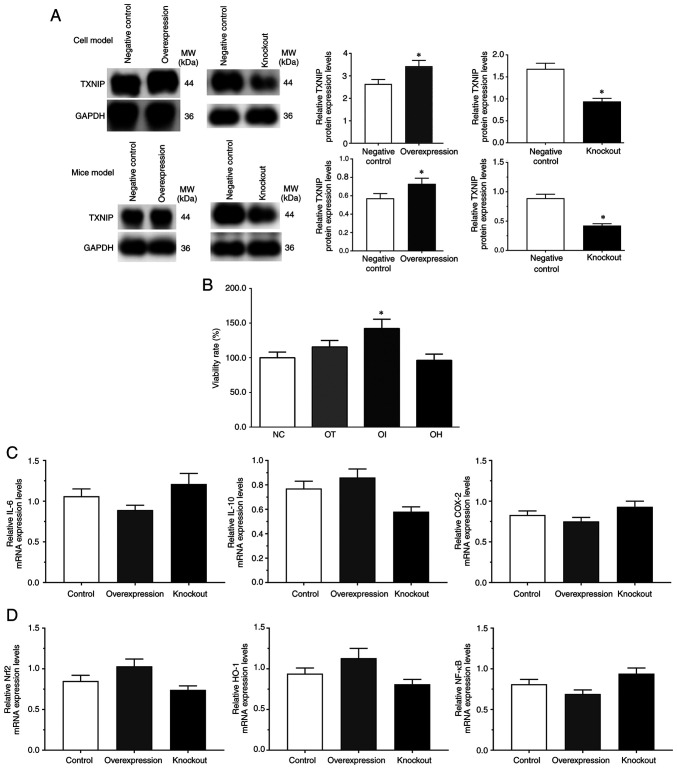
As presented in Fig. 1, the protein expression levels of TXNIP in the control (blank pcDNA3.1 vector), overexpression and control (blank lentiCRISPR v2) knockdown groups as well as the expression of TXNIP in the mice model of control, TXNIP overexpression and TXNIP knockout group, were detected using western blotting. The protein expression levels of TXNIP were significantly increased in the overexpression group and significantly decreased in the knockdown group compared with the controls (P<0.05; Fig. 1A). And the viability rates of H9c2 cells in the NC, OT, OI and OH groups were 100.0±8.8, 115.6±9.3, 142.2±13.3 and 96.2±9.1, respectively (Fig. 1B). The results indicated that the viability rate was significantly increased in the OI group but markedly reduced in the OH group compared with the NC group, which indicated that inhibition of TXNIP may serve a protective role in H9c2 cells. The mRNA expression levels of inflammatory-related genes and Nrf2/HO-1 signaling pathway genes, such as IL-6, IL-10, COX-2, Nrf2, HO-1 and NF-κB, in each group without DHE treatment was detected via RT-qPCR. Although the mRNA expression levels of these genes were slightly changed, no significant difference was observed (Fig. 1C and D).
Figure 1.
Evaluation of model construction using reverse transcription-quantitative PCR and western blotting, and cell viability rate assessment using an MTT assay. (A) TXNIP expression in overexpression and knockout groups without DHE treatment in H9c2 cells and mice. H9c2 cells and mice transfected with blank pcDNA3.1 vector were set as the negative control for the TXNIP overexpression group, and cells transfected with blank lantiCRISPR v2 vector were set as the negative control for the TXNIP knockout group. (B) DHE effect on H9c2 cell viability rate (%) compared with the control group. (C) Expression of inflammatory related genes without DHE treatment. (D) Expression of Nrf2/HO-1 signaling pathway genes without DHE treatment. *P<0.05 vs. control group. Data are presented as the mean ± SD. Each experiment was repeated three times independently. DHE, dehydrocostus lactone; TXNIP, thioredoxin-interacting protein; Nrf2, nuclear factor erythroid 2-related factor 2; HO-1, heme oxygenase-1; NC, negative control; OT, DHE treatment group; OI, DHE treatment combined with the TXNIP inhibition group; OH, DHE treatment combined with the TXNIP overexpression group; COX-2, cyclooxygenase 2.
mRNA expression levels of inflammation-related genes in mouse heart tissues and H9c2 cells
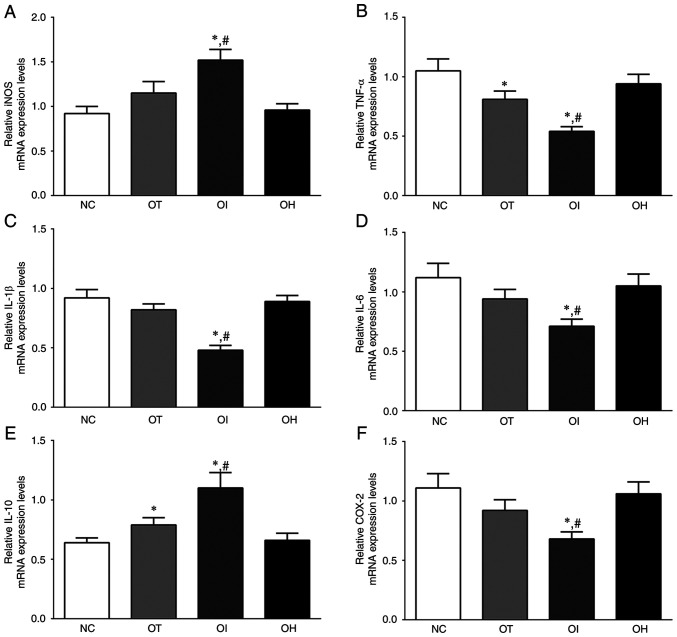
As presented in Fig. 2, the expression levels of iNOS, TNF-α, IL-1β, IL-6, IL-10 and COX-2 mRNA in H9c2 cells of the NC, OT, OI and OH groups were detected using RT-qPCR. Briefly, the expression of iNOS mRNA was significantly increased in OI group compared with the NC and OT groups (P<0.05). The expression of TNF-α in these groups was significantly decreased in OT and OI group compared with NC group (P<0.05), and was significantly decreased in the OI group compared with the OT group (P<0.05). The expression of IL-1β in these groups was significantly decreased in the OI group compared with the NC and OT groups (P<0.05). IL-10 expression levels were significantly increased in the OT and OI groups compared with the NC group (P<0.05), and were significantly increased in the OI group compared with the OT group (P<0.05). The expression of COX-2 in these groups was also significantly decreased in the OI group compared with NC and OT groups (P<0.05).
Figure 2.
Expression levels of inflammation and oxidative-reduction related mRNA in H9c2 cells using reverse transcription-quantitative PCR. (A) iNOS, (B) TNF-α, (C) IL-1, (D) IL-6, (E) IL-10 and (F) COX-2 in H9c2 cells. Data are presented as the mean ± SD. Each experiment was repeated three times independently. *P<0.05 vs. NC group; #P<0.05 vs. OT group. DHE, dehydrocostus lactone; TXNIP, thioredoxin-interacting protein; NC, negative control; OT, DHE treatment group; OI, DHE treatment combined with the TXNIP inhibition group; OH, DHE treatment combined the with TXNIP overexpression group; iNOS, inducible nitric oxide synthase; COX-2, cyclooxygenase-2.
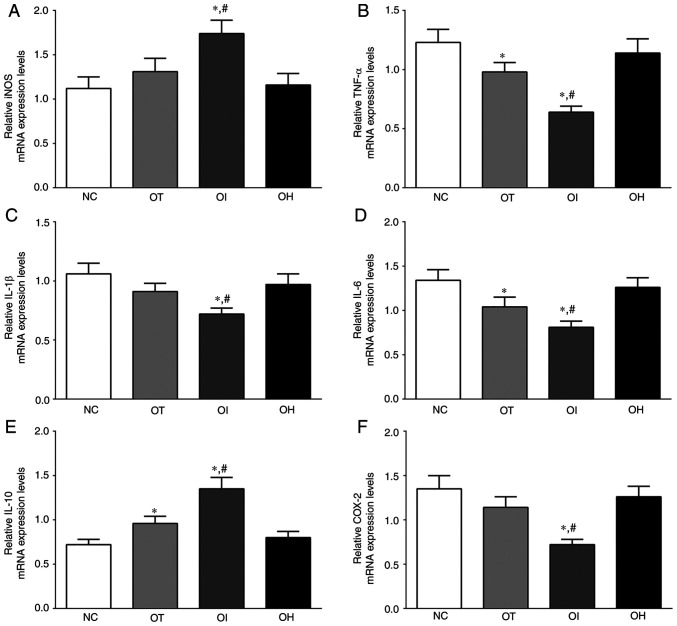
As presented in Fig. 3, the expression of iNOS, TNF-α, IL-1β, IL-6, IL-10 and COX-2 mRNA in mice heart tissue of NC, OT, OI and OH group was detected using RT-qPCR. The expression of iNOS in these groups was significantly increased in the OI group compared with the NC and OT group (P<0.05). The expression of TNF-α in these groups was significantly decreased in the OT and OI groups compared with the NC group (P<0.05), and was significantly decreased in the OI group compared with the OT group (P<0.05). IL-1β expression levels were significantly decreased in the OI group compared with the NC and OT groups (P<0.05). Additionally, the expression of IL-6 in these groups was significantly decreased in the OT and OI groups compared with the NC group (P<0.05), and was significantly decreased in the OI group compared with the OT group (P<0.05). The expression of IL-10 in these groups was significantly increased in OT and OI group compared with the NC group (P<0.05), and was significantly increased in the OI group compared with the OT group (P<0.05). The expression of COX-2 in these groups was significantly decreased in OI group compared with the NC and OT groups (P<0.05). iNOS and IL-10 mRNA expression levels were significantly increased after TXNIP knockout compared with the NC and OT groups, whereas the expression of proinflammatory factors, such as TNF-α, IL-1β, IL-6 and COX-2, was decreased following TXNIP knockout compared with the NC and OT groups. The results indicated that inhibition of TXNIP reduced the inflammatory response in H9c2 cells and in mouse heart tissues.
Figure 3.
Expression levels of inflammation and oxidative-reduction related mRNA in heart tissue of mice using reverse transcription-quantitative PCR. (A) iNOS, (B) TNF-α, (C) IL-1, (D) IL-6, (E) IL-10 and (F) COX-2 in mouse heart tissues. Data are presented as the mean ± SD. Each experiment was repeated three times independently. *P<0.05 vs. NC group; #P<0.05 vs. OT group. DHE, dehydrocostus lactone; TXNIP, thioredoxin-interacting protein; NC, negative control; OT, DHE treatment group; OI, DHE treatment combined with TXNIP inhibition group; OH, DHE treatment combined with TXNIP overexpression group; iNOS, inducible nitric oxide synthase; COX-2, cyclooxygenase-2.
Protein expression levels of the HO-1/NF-κB signaling pathway in mice heart tissues and H9c2 cells
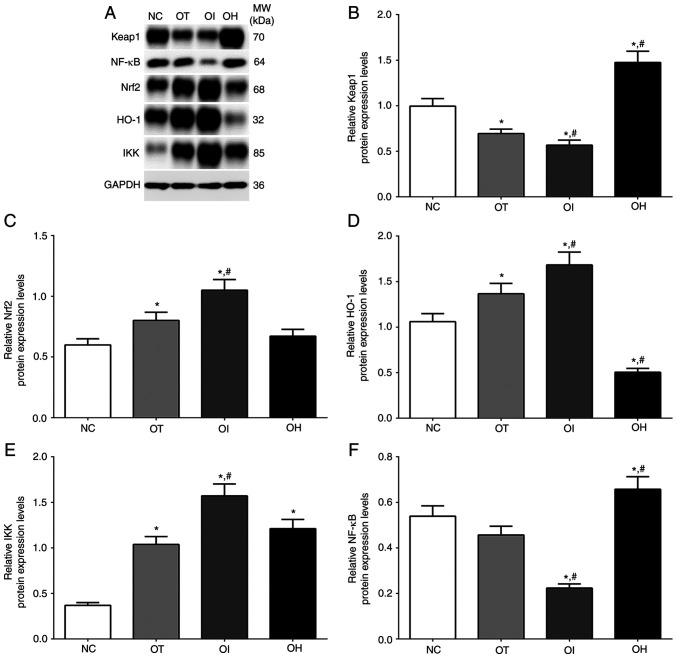
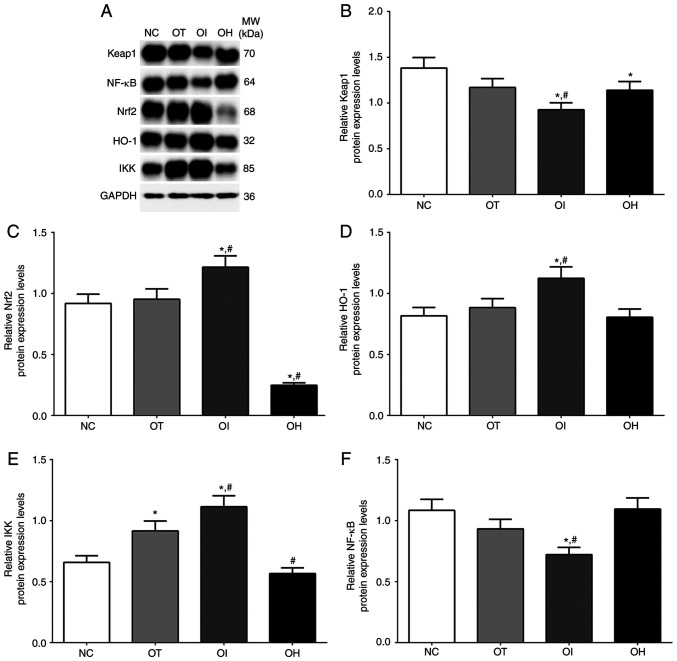
As presented in Fig. 4, the expression of the HO-1/NF-κB signaling pathway in NC, OT, OI and OH groups of H9c2 cells was detected using western blotting. The expression of Keap1 in these groups was significantly decreased in OT and OI groups, while significantly increased in the OH group compared with the NC group (P<0.05). Levels were also significantly decreased in the OI group and significantly increased in the OH group compared with the OT group (P<0.05). The expression of Nrf2 was significantly increased in OT and OI groups compared with the NC group (P<0.05), and significantly increased in the OI group compared with OT group (P<0.05). Additionally, the expression of HO-1 was significantly increased in OT and OI groups compared with the NC group (P<0.05), and significantly increased in the OI group and significantly decreased in the OH group compared with the OT group (P<0.05). The expression of IKK in these groups was significantly increased in all treatment groups compared with NC group (P<0.05), and was significantly increased in OI group compared with OT group (P<0.05). The expression of NF-κB in these groups was significantly decreased in OI group compared with NC and OT group (P<0.05), and was significantly increased in OH group compared with NC and OT group (P<0.05).
Figure 4.
Protein expression levels of HO-1 regulator proteins in H9c2 cells using western blot method. (A) Western blotting analysis and quantitative analysis of (B) Keap1, (C) Nrf2, (D) HO-1, (E) IKK and (F) NF-κB in H9c2 cells. Data are presented as the mean ± SD. Each experiment was repeated three times independently. *P<0.05 vs. NC group. #P<0.05 vs. OT group. DHE, dehydrocostus lactone; TXNIP, thioredoxin-interacting protein; NC, negative control; OT, DHE treatment group; OI, DHE treatment combined with TXNIP inhibition group; OH, DHE treatment combined with TXNIP overexpression group; Nrf2, nuclear factor erythroid 2-related factor 2; HO-1, heme oxygenase-1; Keap1, Kelch-like ECH-associated protein 1; MW, molecular weight.
As presented in Fig. 5, the expression of HO-1/NF-κB signaling pathway in NC, OT, OI and OH groups of heart tissues was detected using western blotting. The expression of Keap1 in these groups was significantly decreased in the OI group compared with NC and OT groups, and significantly decreased in the OH group compared with the NC group. The expression of Nrf2 in these groups was significantly increased in the OI group compared with NC and OT groups, while significantly decreased in OH group compared with the NC and OT groups. The expression of HO-1 in these groups was significantly increased in the OI group compared with the NC and the OT group. The expression of IKK in these groups was significantly increased in OT and OI group compared with the NC group, and was significantly increased in the OI group and significantly decreased in the OH group compared with the OT group. The expression of NF-κB in these groups was significantly decreased in the OI group compared with NC and OT groups.
Figure 5.
Protein expression levels of proinflammation proteins in heart tissue of mice using western blot method. (A) Western blotting analysis and quantitative analysis of (B) Keap1, (C) Nrf2, (D) HO-1, (E) IKK and (F) NF-κB. Data are presented as the mean ± SD. Each experiment was repeated three times independently. *P<0.05 vs. NC group. #P<0.05 vs. OT group. DHE, dehydrocostus lactone; TXNIP, thioredoxin-interacting protein; NC, negative control; OT, DHE treatment group; OI, DHE treatment combined with TXNIP inhibition group; OH, DHE treatment combined with TXNIP overexpression group; TLR, toll-like receptor; HMGB1, high mobility group protein B1; NLRP3, NOD-, LRR- and pyrin domain-containing protein 3; MW, molecular weight.
These results indicated that, in both cells and mice, the HO-1/NF-κB signaling pathway was activated after TXNIP knockout, as the expression of HO-1 was significantly increased. It was therefore hypothesized that TXNIP knockout may have a protective role in H9c2 cells and mouse heart tissues.
Protein expression levels of inflammation response-related molecules in mice heart tissues and H9c2 cells
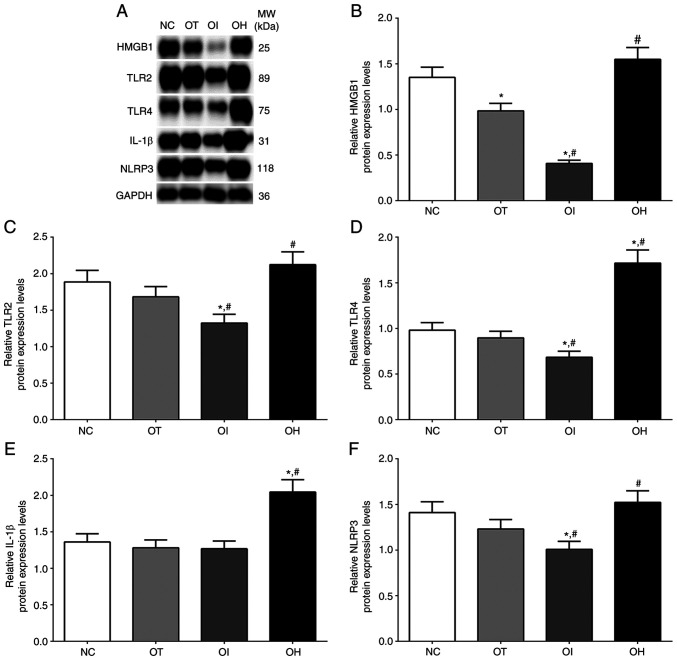
As presented in Fig. 6, the expression of HMGB1, TLR2, TLR4, IL-1β and NLRP3 in NC, OT, OI and OH group of H9c2 cells were detected using western blotting. The expression of HMGB1 in these groups was significantly decreased in OT and OI group compared with NC group (P<0.05), and was significantly decreased in OI group and significantly increased in OH group compared with OT group (P<0.05). The expression of TLR2 in these groups was significantly decreased in OI group compared with NC and OT group (P<0.05), and was significantly increased in OH group compared with OT group (P<0.05). The expression of TLR4 in these groups was significantly decreased in OI group compared with NC and OT group (P<0.05), and was significantly increased in OH group compared with NC and OT group (P<0.05). The expression of IL-1β in these groups was significantly increased in OH group compared with NC and OT group (P<0.05). The expression of NLRP3 in these groups was significantly decreased in OI group compared with NC and OT group (P<0.05), and significantly increased in OH group compared with OT group (P<0.05).
Figure 6.
Protein expression levels of HO-1 regulator proteins in H9c2 cells using western blot method. (A) Western blotting analysis and quantitative analysis of (B) HMGB1, (C) TLR2, (D) TLR4, (E) IL-1β and (F) NLRP3 in H9c2 cells. Data are presented as the mean ± SD. Each experiment was repeated three times independently. *P<0.05 vs. NC group. #P<0.05 vs. OT group. DHE, dehydrocostus lactone; TXNIP, thioredoxin-interacting protein; NC, negative control; OT, DHE treatment group; OI, DHE treatment combined with TXNIP inhibition group; OH, DHE treatment combined with TXNIP overexpression group; Nrf2, nuclear factor erythroid 2-related factor 2; HO-1, heme oxygenase-1; Keap1, Kelch-like ECH-associated protein 1; MW, molecular weight.
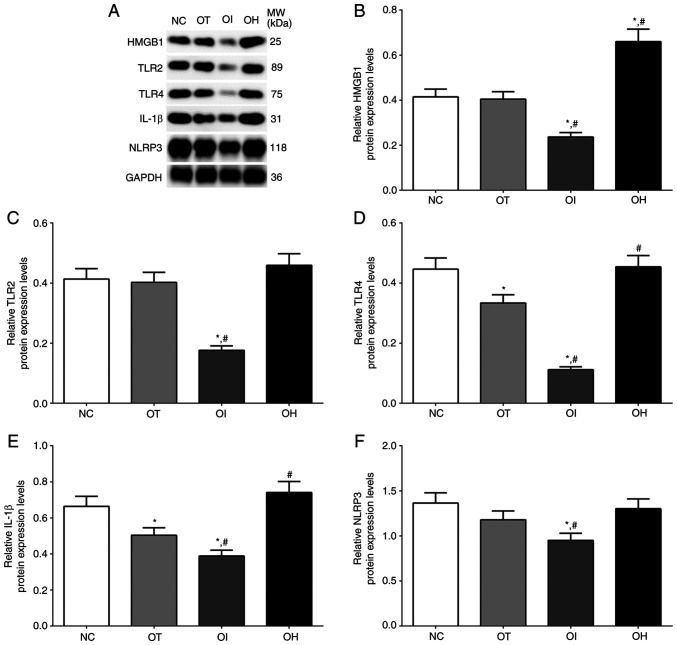
As for mouse heart tissues (Fig. 7), the expression of HMGB1, TLR2, TLR4, IL-1β and NLRP3 in NC, OT, OI and OH group of heart tissues were detected using western blotting. The expression of HMGB1 in these groups was significantly deceased in OI group compared with NC and OT group (P<0.05), and significantly increased in OH group compared with NC and OT group (P<0.05). The expression of TLR2 in these groups was significantly decreased in OI group compared with NC and OT group (P<0.05). The expression of TLR4 in these groups was significantly decreased in OT and OI group compared with NC group (P<0.05), and was significantly decreased in OI group compared with OT group and significantly increased in OH group compared with OT group (P<0.05). The expression of IL-1β in these groups was significantly decreased in OT and OI group compared with NC group (P<0.05), and was significantly decreased in OI group and significantly increased in OH group compared with OT group (P<0.05). The expression of NLRP3 in these groups was significantly decreased in OI group compared with NC and OT group (P<0.05).
Figure 7.
Expression of pro-inflammation proteins in mouse heart tissues using western blot method. (A) Western blotting analysis and quantitative analysis of (B) HMGB1, (C) TLR2, (D) TLR4, (E) IL-1β and (F) NLRP3 in H9c2 cells. Data are presented as the mean ± SD. Each experiment was repeated three times independently. *P<0.05 vs. NC group. #P<0.05 vs. OT group. DHE, dehydrocostus lactone; TXNIP, thioredoxin-interacting protein; NC, negative control; OT, DHE treatment group; OI, DHE treatment combined with TXNIP inhibition group; OH, DHE treatment combined with TXNIP overexpression group; TLR, toll-like receptor; HMGB1, high mobility group protein B1; NLRP3, NOD-, LRR- and pyrin domain-containing protein 3; MW, molecular weight.
These results indicated that TXNIP inhibition reduced the protein expression levels of inflammation-related molecules.
Concentration of inflammation-related and oxidation-reduction related factors in serum samples of mice and cultured medium of H9c2 cells
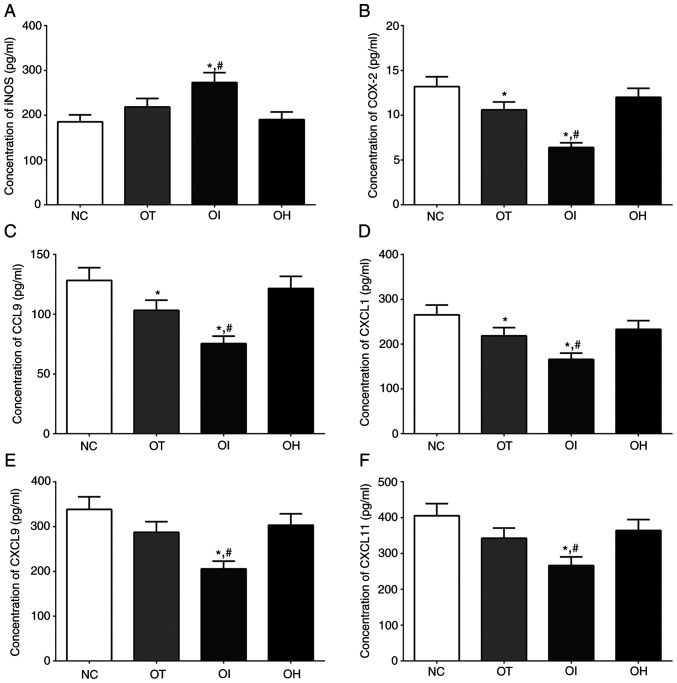
The concentrations of iNOS, which is related to the oxidative-reduction process, and COX-2, CCL9, CXCL1, CXCL9 and CXCL11 which were critical for the activation of inflammation process, in culture medium of H9c2 cells of NC, OT, OI and OH group was quantified using ELISA method and the results are presented in Fig. 8. The concentration of iNOS in NC, OT, OI and OH group of H9c2 cells was significantly increased in OI and OT group compared with NC group (P<0.05), and was significantly increased in OI group and significantly decreased in OH group compared with OT group (P<0.05). The concentration of COX-2 in these groups was significantly decreased in OT and OI group compared with NC group (P<0.05), and was significantly decreased in OI group compared with OT group (P<0.05). The concentration of CCL9 in these groups was significantly decreased in OT and OI group compared with NC group (P<0.05), and was significantly decreased in OI group compared with OT group (P<0.05). The changing in concentration of CXCL1 and CXCL9 in these groups presented a similar trend with CCL9 (P<0.05). And the concentration of CXCL11 in these groups was significantly decreased in OI group compared with NC and OT group (P<0.05).
Figure 8.
Detection of inflammation and oxidative-reduction related factors in H9c2 cells using ELISA method. (A) iNOS, (B) COX-2, (C) CCL9, (D) CXCL1, (E) CXCL9 and (F) CXCL11 in the culture medium of H9c2 cells. Data are presented as the mean ± SD. Each experiment was repeated three times independently. *P<0.05 vs. NC group. #P<0.05 vs. OT group. DHE, dehydrocostus lactone; TXNIP, thioredoxin-interacting protein; NC, negative control; OT, DHE treatment group; OI, DHE treatment combined with TXNIP inhibition group; OH, DHE treatment combined with TXNIP overexpression group; iNOS, inducible nitric oxide synthase; COX-2, cyclooxygenase-2; CCL9, C-C motif ligand 9; CXCL, chemokine C-X-C motif ligand.
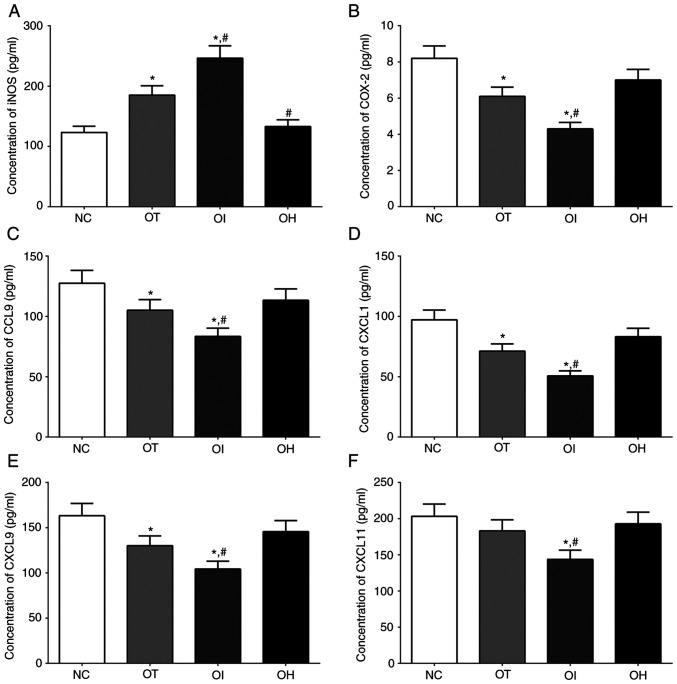
As reported in Fig. 9, The concentration of iNOS in NC, OT, OI and OH group of heart tissues was significantly increased in OI group compared with NC and OT group (P<0.05). The concentration of COX-2 in these groups was significantly decreased in OT and OI group compared with NC group (P<0.05), and was significantly decreased in OI group compared with OT group (P<0.05). The concentration of CCL9 and CXCL1 in these groups presented a similar trend with COX-2 (P<0.05). The concentration of CXCL9 and CXCL11 in these groups was significantly decreased in OI group compared with NC and OT group (P<0.05).
Figure 9.
Detection of inflammation and oxidative-reduction related factors in heart tissue of mice using ELISA method (A) iNOS, (B) COX-2, (C) CCL9, (D) CXCL1, (E) CXCL9 and (F) CXCL11 in mouse heart tissues. Data are presented as the mean ± SD. Each experiment was repeated three times independently. *P<0.05 vs. NC group. #P<0.05 vs. OT group. DHE, dehydrocostus lactone; TXNIP, thioredoxin-interacting protein; NC, negative control; OT, DHE treatment group; OI, DHE treatment combined with TXNIP inhibition group; OH, DHE treatment combined with TXNIP overexpression group; iNOS, inducible nitric oxide synthase; COX-2, cyclooxygenase-2; CCL9, C-C motif ligand 9; CXCL, chemokine C-X-C motif ligand.
These results indicated that DHE treatment decreased the secretion of inflammatory response-related factors and the knockout of TXNIP significantly enhanced this trend compared with control group.
Discussion
HF affects more than 5.7 million people in the US, according to previous data (21). In 2013, nearly 500,000 new cases occurred in people >55 years (22). DHE is obtained from Chinese herbs and can be used in the treatment of various diseases, such as liver cancer, colorectal cancer and leukemia (23,24). TXNIP, an endogenous inhibitor of thioredoxin, serves an important role in the resistance to oxidative stress induced by extracellular stimulations, including reactive oxygen species and hypoxia (25).
Under inflammatory conditions, TNF-α and the expression of other inflammation-related cytokines are induced by coagulation factors, such as factor VII and XI, further inducing the formation of complexes with factor VII and resulting in the activation of thrombin (26). Moreover, factor Xa, thrombin and the tissue factor-factor VII complex can increase the expression of TNF-α and other proinflammatory cytokines (27), promoting the blood coagulation process. Furthermore, factor Xa and thrombin directly affect the function of vascular cells (27) via protease-activated receptors and the nitric oxide (NO)/cGMP signaling pathway (28). The NO/cGMP signaling pathway has numerous effects on the vasculature and dysfunction of this signaling pathway leads to a reduction in cGMP production, which results in the occurrence of cardiovascular diseases (29). A previous study also indicated that the synthesis of NO inhibited the activity of TF (30) and prevented the formation of thrombi in circulation via its vasodilator and anti-aggregatory ability (31). The present study demonstrated that DHE significantly inhibited the expression of inflammation-related factors, including IL-1, IL-6, IL-10, COX-2 and TNF-α and had an inhibitory effect on the activation of the inflammatory response. Furthermore, the present study reported that DHE increased the expression levels of iNOS, which contributes to the protective effect of DHE.
TLRs are divided into two groups according to their cellular distribution: The cell membrane group (TLR1, 2, 4, 5 and 6) and the endosome membrane within the cell group (TLR3, 7, 8 and 9) (32). The transduction of cellular signals via TLRs to the nucleus leads to the activation of innate and adaptive immune responses via the myeloid differentiation primary response 88-dependent pathway and the Toll/IL-1 receptor domain-containing adapter-inducing IFN-β-dependent pathway, leading to the activation of the NF-κB signaling pathway (33). Under extracellular stimulation, the NF-κB signaling pathway is activated via TLRs, nod-like receptors or other cytokine receptors. Activation of the NF-κB signaling pathway further leads to the activation of NLRP3, initiating inflammasome activation (34). The NLRP3 inflammasome can recognize numerous stimuli and responds to endogenous factors that can lead to cellular damage, including uric acid crystals, drusen and extracellular ATP (35,36). TLR activation also induces the expression of pro-IL-1β (37). In the present study, the activation of these factors upon HF was inhibited after DHE treatment and was even lower after TNXIP inhibition, indicating that the activation of the inflammatory response in H9c2 cardiomyocytes and heart tissues was inhibited, and presented a protective role.
HO-1 is an enzyme presenting cytoprotective and antioxidant effects (38). The expression of HO-1 is regulated at the transcriptional level via the regulation of Nrf2(39). Under normal conditions, Nrf2 binds to Keap1(40) and when oxidated Nrf2 is released from Keap1 and translocated into the nucleus, further binding with antioxidant response element sequences of the HO-1 promoter region (41). The activation of HO-1 further leads to the activation of NF-κB, which inhibits the expression of IL-6, TNF-α and IL-1β and subsequently the inflammatory response process (42). In the present study, the protein expression levels of HO-1 were significantly increased after treatment with DHE combined with TXNIP konckout, presenting a protective effect (Fig. 10).
Figure 10.
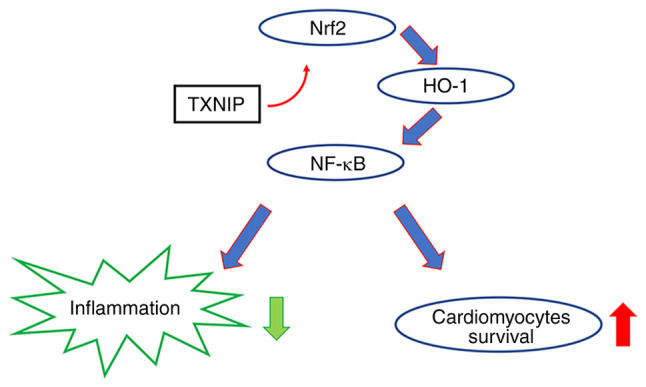
Schematic diagram of the experimental hypothesis. Knockout of TXNIP promotes the activation of HO-1 through the Nrf2 pathway, leading to a reduction in inflammation activation. TXNIP knockout therefore exerted a protective role in cardiomyocytes. Nrf2, nuclear factor erythroid 2-related factor 2; HO-1, heme oxygenase-1; TXNIP, thioredoxin-interacting protein.
Although the results indicated that TXNIP inhibition contributed to the therapeutic effect of DHE, the present study has numerous limitations. The detailed mechanism of how TXNIP inhibition promoted the effect of DHE is not fully understood and it should be explored in future experiments. The Nrf2/HO-1 signaling pathway expression in TXNIP overexpression and knockout groups without DHE treatment was detected using RT-qPCR only; in further experiments, the protein levels should also be detected. Other methods could contribute to explore the aforementioned mechanisms, such as mass spectrometry, RNA sequencing and chromatin immunoprecipitation sequencing. Clinical samples would also help understand the association between TXNIP and DHE, and the mechanism of TXNIP knockout promoting the function of DHE.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XZ and CC performed the cell, mice, protein and nucleic acid experiments, and wrote the manuscript. YH designed the experiment and revised the manuscript. XZ and YH confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (17). The present protocol and experiments were approved by the Ethics Committee of Animal Experiments of Anqiu People's Hospital (Anqiu, China; approval no. IACUC-20200920).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Delling FN, et al. Heart disease and stroke statistics-2020 Update: A report from the American Heart association. Circulation. 2020;141:e139–e596. doi: 10.1161/CIR.0000000000000757. [DOI] [PubMed] [Google Scholar]
- 2.Hofmann U, Frantz S. How can we cure a heart ‘in flame’? A translational view on inflammation in heart failure. Basic Res Cardiol. 2013;108(356) doi: 10.1007/s00395-013-0356-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ammirati E, Cannistraci CV, Cristell NA, Vecchio V, Palini AG, Tornvall P, Paganoni AM, Miendlarzewska EA, Sangalli LM, Monello A, et al. Identification and predictive value of interleukin-61 interleukin-101 and interleukin-62 interleukin-101 cytokine patterns in ST-elevation acute myocardial infarction. Circ Res. 2012;111:1336–1348. doi: 10.1161/CIRCRESAHA.111.262477. [DOI] [PubMed] [Google Scholar]
- 4.Bozkurt B, Torre-Amione G, Warren MS, Whitmore J, Soran OZ, Feldman AM, Mann DL. Results of targeted anti-tumor necrosis factor therapy with etanercept (ENBREL) in patients with advanced heart failure. Circulation. 2001;103:1044–1047. doi: 10.1161/01.cir.103.8.1044. [DOI] [PubMed] [Google Scholar]
- 5.Taniguchi M, Kataoka T, Suzuki H, Uramoto M, Ando M, Arao K, Magae J, Nishimura T, Otake N, Nagai K. Costunolide and dehydrocostus lactone as inhibitors of killing function of cytotoxic T lymphocytes. Biosci Biotechnol Biochem. 1995;59:2064–2067. doi: 10.1271/bbb.59.2064. [DOI] [PubMed] [Google Scholar]
- 6.Yuuya S, Hagiwara H, Suzuki T, Ando M, Yamada A, Suda K, Kataoka T, Nagai K. Guaianolides as immunomodulators. Synthesis and biological activities of dehydrocostus lactone, mokko lactone, eremanthin, and their derivatives. J Nat Prod. 1999;62:22–30. doi: 10.1021/np980092u. [DOI] [PubMed] [Google Scholar]
- 7.Jeong GS, Pae HO, Jeong SO, Kim YC, Kwon TO, Lee HS, Kim NS, Park SD, Chung HT. The alpha-methylenegamma-butyrolactone moiety in dehydrocostus lactone is responsible for cytoprotective heme oxygenase-1 expression through activation of the nuclear factor E2-related factor 2 in HepG2 cells. Eur J Pharmacol. 2007;565:37–44. doi: 10.1016/j.ejphar.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 8.Zhang HW, Liu YM, Fang X, Gu L, Luo C, Chen L, Wang Q. Vitamin D3 protects mice from Diquat-induced oxidative stress through the NF-κB/Nrf2/HO-1 signaling pathway. Oxid Med Cell Longev. 2021;2021(6776956) doi: 10.1155/2021/6776956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu YX, Jiang FJ, Liu G, Wang YY, Gao ZQ, Jin SH, Nie YJ, Chen D, Chen JL, Pang QF. Dehydrocostus lactone attenuates methicillin-resistant staphylococcus Aureus-induced inflammation and acute lung injury via modulating macrophage polarization. Int J Mol Sci. 2021;22(9754) doi: 10.3390/ijms22189754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HR, Kim JM, Kim MS, Hwang JK, Park YJ, Yang SH, Kim HJ, Ryu DG, Lee DS, Oh H, et al. Saussurea lappa extract suppresses TPA-induced cell invasion via inhibition of NF-κB-dependent MMP-9 expression in MCF-7 breast cancer cells. BMC Complement Altern Med. 2014;14(170) doi: 10.1186/1472-6882-14-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim EJ, Lim SS, Park SY, Shin HK, Kim JS, Park JH. Apoptosis of DU145 human prostate cancer cells induced by dehydrocostus lactone isolated from the root of Saussurea lappa. Food Chem Toxicol. 2008;46:3651–3658. doi: 10.1016/j.fct.2008.08.038. [DOI] [PubMed] [Google Scholar]
- 12.Nie Y, Wang Z, Chai G, Xiong Y, Li B, Zhang H, Xin R, Qian X, Tang Z, Wu J, Zhao P. Dehydrocostus lactone suppresses LPS-induced acute lung injury and macrophage activation through NF-κB signaling pathway mediated by p38 MAPK and Akt. Molecules. 2019;24(1510) doi: 10.3390/molecules24081510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masutani H, Yoshihara E, Masaki S, Chen Z, Yodoi J. Thioredoxin binding protein (TBP)-2/Txnip and alpha-arrestin proteins in cancer and diabetes mellitus. J Clin Biochem Nutr. 2012;50:23–34. doi: 10.3164/jcbn.11-36SR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai X, Liao R, Liu C, Liu S, Huang H, Liu J, Jin T, Guo H, Zheng Z, Xia M, et al. Epigenetic regulation of TXNIP-mediated oxidative stress and NLRP3 inflammasome activation contributes to SAHH inhibition-aggravated diabetic nephropathy. Redox Biol. 2021;45(102033) doi: 10.1016/j.redox.2021.102033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neville ES, Ophir S, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen JX, Zhang L, Liu H, Wang J, Li J, Yang Y, Wang Y, Cai H, Li R, Zhao Y. Salsolinol attenuates doxorubicin-induced chronic heart failure in rats and improves mitochondrial function in H9c2 cardiomyocytes. Front Pharmacol. 2019;10(1135) doi: 10.3389/fphar.2019.01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K, Zhou A, Ruan M, Jin Z, Lu J, Wang Q, Lu C. Dehydrocostus lactone suppresses ox-LDL-induced attachment of monocytes to endothelial cells. Am J Transl Res. 2019;11:6159–6169. [PMC free article] [PubMed] [Google Scholar]
- 18. National Research Council (US) Institute for Laboratory Animal Research. Guide for the Care and Use of Laboratory Animals. Washington (DC), National Academies Press (US), 1996. [PubMed] [Google Scholar]
- 19.Wang X, Gao Y, Tian Y, Liu X, Zhang G, Wang Q, Xie W, Liu K, Qian Q, Wang Q. Integrative serum metabolomics and network analysis on mechanisms exploration of Ling-Gui-Zhu-Gan Decoction on doxorubicin-induced heart failure mice. J Ethnopharmacol. 2020;250(112397) doi: 10.1016/j.jep.2019.112397. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. doi: 10.1006/meth.2001.1262. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402-408. [DOI] [PubMed] [Google Scholar]
- 21.Freddy AS, David G. Cardiac contractility modulation: A novel approach for the treatment of heart failure. Heart Fail Rev. 2016;21:645–660. doi: 10.1007/s10741-016-9571-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C, et al. Heart disease and stroke statistics-2017 update: A report from the American Heart Association. Circulation. 2017;135:e146–e603. doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng Z, Wang Y, Fan J, Lin X, Liu C, Xu Y, Ji W, Yan C, Su C. Costunolide and dehydrocostuslactone combination treatment inhibit breast cancer by inducing cell cycle arrest and apoptosis through c-Myc/p53 and AKT/14-3-3 pathway. Sci Rep. 2017;7(41254) doi: 10.1038/srep41254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun X, Kang H, Yao Y, Chen H, Sun L, An W, Jiang E, Wang S, Hu X. Dehydrocostus lactone suppressed the proliferation, migration, and invasion of colorectal carcinoma through the downregulation of eIF4E expression. Anticancer Drugs. 2015;26:641–648. doi: 10.1097/CAD.0000000000000229. [DOI] [PubMed] [Google Scholar]
- 25.Han X, Wu YC, Meng M, Sun QS, Gao SM, Sun H. Linarin prevents LPS-induced acute lung injury by suppressing oxidative stress and inflammation via inhibition of TXNIP/NLRP3 and NF-κB pathways. Int J Mol Med. 2018;42:1460–1472. doi: 10.3892/ijmm.2018.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iannucci J, Renehan W, Grammas P. Thrombin, a mediator of coagulation, inflammation, and neurotoxicity at the neurovascular interface: Implications for Alzheimer's disease. Front Neurosci. 2020;14(762) doi: 10.3389/fnins.2020.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pawlinski R, Pedersen B, Kehrle B, Aird WC, Frank RD, Guha M, Mackman N. Regulation of tissue factor and inflammatory mediators by Egr-1 in a mouse endotoxemia model. Blood. 2003;10:3940–3947. doi: 10.1182/blood-2002-07-2303. [DOI] [PubMed] [Google Scholar]
- 28.Scotton CJ, Krupiczojc MA, Königshoff M, Mercer PF, Lee YC, Kaminski N, Morser J, Post JM, Maher TM, Nicholson AG, et al. Increased local expression of coagulation factor X contributes to the fibrotic response in human and murine lung injury. J Clin Invest. 2009;1199:2550–2563. doi: 10.1172/JCI33288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motley ED, Eguchi K, Patterson MM, Palmer PD, Suzuki H, Eguchi S. Mechanism of endothelial nitric oxide synthase phosphorylation and activation by thrombin. Hypertension. 2007;49:577–583. doi: 10.1161/01.HYP.0000255954.80025.34. [DOI] [PubMed] [Google Scholar]
- 30.Stasch JP, Hobbs AJ. doi: 10.1007/978-3-540-68964-5_13. NO-independent, haem-dependent soluble guanylate cyclase stimlators. Handb Exp Pharmacol: 277-308, 2009 doi: 10.1007/978-3-540-68964-5_13. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y, Loscalzo J. Regulation of tissue factor expression in human microvascular endothelial cells by nitric oxide. Circulation. 2000;101:2144–2148. doi: 10.1161/01.cir.101.18.2144. [DOI] [PubMed] [Google Scholar]
- 32.Tschudi MR, Luscher TF. Nitric oxide: The endogenous nitrate in the cardiovascular system. Herz. 1996;21:50–60. [PubMed] [Google Scholar]
- 33.Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nat Rev Cancer. 2009;9:57–63. doi: 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- 34.Piras V, Selvarajoo K. Beyond MyD88 and TRIF pathways in Toll-like receptor signaling. Front Immunol. 2014;5(70) doi: 10.3389/fimmu.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, Fernandes-Alnemri T, Wu J, Monks BG, Fitzgerald KA, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franchi L, Muñoz-Planillo R, Núñez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol. 2012;13:325–332. doi: 10.1038/ni.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 38.Franchi L, Eigenbrod T, Núñez G. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J Immunol. 2009;183:792–796. doi: 10.4049/jimmunol.0900173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapitulnik J. Bilirubin: An endogenous product of heme degradation with both cytotoxic and cytoprotective properties. Mol Pharmacol. 2004;66:773–779. doi: 10.1124/mol.104.002832. [DOI] [PubMed] [Google Scholar]
- 40.Srisook K, Kim C, Cha YN. Molecular mechanisms involved in enhancing HO-1 expression: De-repression by heme and activation by Nrf2, the ‘one-two’ punch. Antioxid Redox Signa. 2005;l7:1674–1687. doi: 10.1089/ars.2005.7.1674. [DOI] [PubMed] [Google Scholar]
- 41.Farombi EO, Surh YJ. Heme oxygenase-1 as a potential therapeutic target for hepatoprotection. J Biochem Mol Biol. 2006;39:479–491. doi: 10.5483/bmbrep.2006.39.5.479. [DOI] [PubMed] [Google Scholar]
- 42.Owuor ED, Kong AN. Antioxidants and oxidants regulated signal transduction pathways. Biochem Pharmacol. 2002;64:765–770. doi: 10.1016/s0006-2952(02)01137-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.