Abstract
Hydroxyl radical protein footprinting (HRPF) is a powerful and flexible technique for probing changes in protein topography. With the development of Fast Photochemical Oxidation of Proteins (FPOP), it became possible for researchers to perform HRPF in their laboratory on a very short timescale. While FPOP has grown significantly in popularity since its inception, adoption remains limited due to technical and safety issues involved in the operation of a hazardous Class IV UV laser, and irreproducibility often caused by improper laser operation and/or differential radical scavenging by various sample components. Here, we present a new integrated FOX (Flash OXidation) Protein Footprinting System. This platform delivers sample via flow injection to a facile and safe-to-use high pressure flash lamp with a flash duration of 10 μs FWHM. Integrated optics collect the radiant light and focus it into the lumen of a capillary flow cell. An inline radical dosimeter measures the hydroxyl radical dose delivered and allows for real-time compensation for differential radical scavenging. A programmable fraction collector collects and quenches only the sample that received the desired effective hydroxyl radical dose, diverting the carrier liquid and improperly oxidized sample to waste. We demonstrate the utility of the FOX Protein Footprinting System by determining the epitope of TNFα recognized by adalimumab. We successfully identify the surface of the protein that serves as the epitope for adalimumab, identifying four of the five regions previously noted by X-ray crystallography while seeing no changes in peptides not involved in the epitope interface. The FOX Protein Footprinting System allows for FPOP-like experiments with real-time dosimetry in a safe, compact and integrated benchtop platform.
Graphical Abstract
Introduction
Hydroxyl radical protein footprinting (HRPF) is a mass spectrometry-based structural covalent labeling technology that uses hydroxyl radicals generated in situ to probe the surface of proteins in diffuse aqueous solution [1–3]. While HRPF was initially reported using high-intensity X-rays from an X-ray synchrotron facility, methods for HRPF that could be carried out in a benchtop format using Fenton chemistry [4] or UV photolysis of hydrogen peroxide [5, 6] were reported soon after. However, these benchtop methods had the drawback of taking minutes to label proteins, which resulted in high background oxidation and could easily result in oxidation-induced conformational changes if not carefully controlled [5, 7]. In 2005, Hambly and Gross reported the development of Fast Photochemical Oxidation of Proteins, a method combining laser-induced UV photolysis of hydrogen peroxide (independently reported by Sze and Aye, [8]) with a continuous sample flow cell to allow for HRPF of a flexible volume of sample by using repeated laser pulses to label each volume with a single laser shot [9]. Unlike the previous continuous wave UV photolysis method, FPOP, when properly performed, completed the initial hydroxyl radical attack on a very short time scale, allowing for thorough labeling of protein structure in a researcher’s laboratory without evidence of oxidation-induced conformational changes [10]. FPOP has proven to be a powerful technique that quickly spread to other labs, with a powerful capability to perform thorough HRPF labeling under very flexible conditions [11].
While FPOP has proven to be a powerful technology, it has not yet been met with broad adoption, especially by the biopharmaceutical industry. There are several reasons for this lack of widespread adoption. FPOP most commonly uses a KrF excimer laser as an illumination source for photolysis of hydrogen peroxide. The KrF excimer laser requires frequent refills with fluorine gas. Fluorine gas is highly toxic at very low levels, requiring storage of gas cylinders in dedicated chemical fume hoods. The Class IV laser is itself a serious hazard. Exposure to either the direct beam or to specular reflections can cause immediate and permanent blindness to unprotected eyes. This requires specialized training and dedicated engineering controls to protect both the user and the people around them.
In addition to safety hazards, FPOP offers many experimental pitfalls to new investigators. As the UV laser pulse is invisible, alignment and adjustment of the optical bench can be very challenging and tedious. The broad reactivity of hydroxyl radicals, which is the key to their utility in labeling many amino acids in a single experiment, also causes difficulties. Almost any organic additive to the protein sample can react with the hydroxyl radicals [12], resulting in widely different radical scavenging backgrounds leading to uninterpretable and/or irreproducible results.
Here, we describe a new instrument, the Flash OXidation (FOX) Protein Footprinting System. Sample is introduced into the system via manual injection into an automated flow carrier. A fully enclosed high pressure flash lamp UV illumination source generates hydroxyl radicals in a single 10 μs FWHM flash, with integrated and fully enclosed optics that focuses the radiant light into the lumen of the capillary flow cell. A UV photometric absorbance detector, which measures the change in photometric absorbance of an internal standard radical dosimeter, [13] is used to perform real-time in-line radical dosimetry [14], with the detector positioned immediately downstream from the FOX system photolysis flow cell.
Experimental Section
Materials
Adenine and catalase from bovine liver were purchased from Sigma-Aldrich (St. Louis, MO). Acetonitrile, formic acid, hydrogen peroxide, LCMS-grade water, phosphate buffer solution (PBS), and tris base were purchased from Fisher Scientific (Fair Lawn, NJ). L-methionine amide was purchased from Chem-Impex (Wood Dale, IL). Mass spectrometry grade modified trypsin was obtained from Promega (Madison, WI). Recombinant human TNFα (Val77-Leu233) expressed in HEK293 cells was purchased from R&D Systems (Minneapolis, MN). Recombinant adalimumab was acquired from GlycoScientific (Athens, GA).
FOX Protein Footprinting System
The FOX™ Protein Footprinting System (GenNext Technologies, Inc., Half Moon Bay, CA) was used for the experiments described here (Figure 1). This system consists of four modules (flow injection module, FOX photolysis module, dosimetry module and sample collector) controlled by the FOX system control software (Figure S1, Supporting Information). Sample flows vertically from the bottom module to the top at a user-defined flow rate through a 250 μm ID fused silica capillary with UV-transparent coating (Molex-Polymicro Technologies, Phoenix, AZ; TSU 250350 deep UV transparent capillary) to help eject any bubbles formed during photolysis. The FOX Photolysis Module contains a custom power supply that delivers a very short pulse to drive the custom high-pressure Xe flash lamp, along with integrated optics to collect and focus the radiant light from the flash lamp into the fused silica capillary flow cell (Figure S2, Supporting Information). The power supply pulses are triggered by the FOX control software to deliver a photolysis flash at a user-defined frequency (up to 2 Hz). The FOX control software includes a calculator that allows the user to set flow rate and photolysis frequency to achieve a desired FOX exclusion volume (analogous to FPOP exclusion volume [9, 10]) using a simple plug flow model; more complex laminar flow modeling is not handled by the software [15]. The dosimetry module performs in-line real time dosimetry as previously described [14]. The dosimetry results are collected and reported by the FOX control software. Sample is collected by a computer-controlled sample collector that directs carrier fluid and/or improperly oxidized sample (as determined by real-time dosimetry) into waste, while collecting properly oxidized sample in a collection tube containing quench solution as described below.
Figure 1. FOX Protein Footprinting System.

Sample mixed with hydrogen peroxide and adenine internal standard radical dosimeter is introduced via flow injection through an injection port in the Fluidics Module (bottom). Sample is then pushed by a syringe pump upwards through a fused silica capillary through the FOX flash cell in the Photolysis Module (middle), where the sample is oxidized by photolysis of hydrogen peroxide. Sample is immediately carried to the Dosimetry Module (top), where the UV absorbance of the adenine dosimeter is measured at 265 nm. Finally, sample is deposited into quench solution in the computer-controlled sample collector (atop instrument), with carrier buffer diverted to waste.
Sample Flash Oxidation
TNFα and adalimumab were reconstituted to 29.4 μM and 40 μM, respectively, using PBS. TNFα was labeled with and without the presence of adalimumab. Samples were mixed with adenine, and immediately before labeling, 200 mM H2O2 was spiked 1:1 v/v into a single 10 μL sample for a final concentration of 100 mM H2O2, 1 mM adenine, 3 μM TNFα ± 1.5 μM adalimumab. For each condition, 20 μL was injected to overload the FOX Photolysis System’s 12 μL injection loop. Once triggered, the FOX Photolysis System’s Fluidics Module pumps the 12 μL sample at 7.5 μL/min using PBS as the carrier fluid. For each condition, the high-pressure Xe flash lamp either remained off (no lamp control) or flashed at a frequency of 1 Hz to photolyze H2O2 (FPOP sample). The lamp voltage was adjusted so that an equivalent concentration of effective hydroxyl radicals was generated (detected using in-line real time dosimetry) to label TNFα with and without adalimumab. Finally, as the bolus of sample reaches the end of the capillary, FOX Photolysis System’s product collector automatically rotates from a waste vial to a product vial containing 20 μL of quench solution (0.3 mg/mL of catalase to break down the remaining hydrogen peroxide and 35 mM methionine to scavenge any secondary oxidants). The unilluminated controls and FPOP samples were acquired in triplicates for TNFα with and without adalimumab.
Angiotensin II was prepared at a final concentration of 1 μM peptide, 200 mM hydrogen peroxide prior to direct injection via syringe pump into the flow path for oxidation using an early prototype version of the FOX Photolysis Module, with a flash fluence in the wavelength range of 200 – 280 nm of 6 mJ/mm2 and without inline dosimetry. Sample was quenched post-oxidation as described above for TNFα.
Sample Processing and LC-MS/MS
To reduce and denature the protein, TNFα samples were incubated in the presence of 100 mM Tris, pH 8.0, 1 mM CaCl2 and 10 mM DTT at 95 °C for 15 min. The samples were immediately cooled on ice for 3 min. To digest the samples, 1:20 ratio of trypsin: protein was added and the samples were incubated at 37 °C with rotation for 15 hr. The trypsin digestion reaction was stopped by heating the samples at 90 °C for 10 min.
A final concentration of 0.1% formic acid was added to the samples and injected via autosampler on a Dionex Ultimate 3000 nano-LC system coupled to an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific, San Jose, CA). The samples were first trapped on a 300 μm ID × 5 mm PepMap with 5 μm particle size and 100 Å pore size C18 trapping cartridge (Thermo Scientific), then back-eluted onto a 0.75 mm × 150 mm Acclaim PepMap with 2 μm particle size and 100 Å pore size C18 nanocolumn (Thermo Scientific). Peptides were separated using a gradient of solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile) at a flow rate of 0.30 μL/min. The gradient consisted of 2 to 35% solvent B over 28 min, ramped to 95% B over 4 min, held for 3 min, and then returned to 2% B over 2 min and held for 8 min. The spray voltage was 2450 volts, and the temperature of the heated capillary was 300 °C. The MS1 data were collected in positive ion mode in the Orbitrap with resolution of 60000 with the scan range from 250–2000 m/z. For TNFα, MS1 data collection was followed by eight subsequent MS2 scans on the top eight most abundant peptide ions by CID and EThcd in parallel to offer more diverse product ion spectra for peptide identification. These TNFα data have been deposited to the ProteomeXchange Consortium via the PRIDE [16] partner repository with the dataset identifier PXD023169. After peptides with significantly reduced oxidation in the presence of adalimumab were identified, samples were re-run to perform sub-peptide localization and quantitation of oxidation for those peptides. Targeted fragmentation was used for peptides showing significant changes in oxidation by using a mass inclusion list containing the m/z of the oxidized versions of the peptides. For CID, the isolation window was 3 m/z units and the data were collected in the ion trap mode. For EThcd, the supplemental activation collision energy was 6% and the data were collected in the Orbitrap with resolution of 30000. Calibrated charge-dependent ETD parameters were used.
FPOP Data Analysis
Unoxidized peptides were identified using Byonic version v3.9.4 (Protein Metrics) to determine identified peptide sequences, charge state(s) and retention time. The Byonic search parameters included all possible major oxidation modifications as variable modifications and the enzyme specificity was set to cleave the protein after lysine and arginine. The identified peptides’ monoisotopic mass, charge, score, and retention time were imported into FoxWare™ Protein Footprinting Software for the detection and quantification of FPOP modifications. FoxWare Protein Footprinting Software automatically generated a list of all possible oxidation product masses with a maximum number of three oxidation events per peptide. The XIC of all unmodified and modified MS1 ions were generated across the full retention time, and FoxWare Protein Footprinting Software automatically evaluated and filtered each XIC by its retention time, peak area, and MS1 spectra. All modified peptides’ XICs must be within a seven-minute retention time window determined by the unmodified peptide (i.e. four minutes before to three minutes after the unmodified peak apex; this window is fully definable by the user). Also, the modified peptides’ XICs must have a minimum peak area of 100,000 and an MS1 isotopic pattern with the C13 ion at least 0.15 to 3 times as intense as the C12 ion. All XICs that were automatically selected using FoxWare Protein Footprinting Software were used to quantify the extent of oxidation following equation 1:
The extent of oxidation between three replicates for each condition were automatically calculated and averaged in FoxWare Protein Footprinting Software. Finally, the background oxidation detected in the no lamp controls was subtracted from the oxidation in the FPOP samples to determine the extent of FPOP oxidation. Two-tailed unpaired t-tests were performed by FoxWare Protein Footprinting Software.
After identification of the peptides that showed significant changes in oxidation, samples were re-analyzed using data-independent acquisition, with CID MS/MS scans scheduled to sample peptide oxidation products throughout their elution time in a manual version of the method of Gross and coworkers [17]. We were unable to achieve definitive CID MS/MS identification of resolved sites of oxidation due to insufficient chromatographic resolution of peptide oxidation products combined with low MS/MS product ion signal, requiring spectral averaging to bring product ion signal to noise ratios to acceptable levels. However, we were able to restrict the regions of the peptide that were oxidized based on the manual identification of oxidized and unoxidized product ions. These sub-peptide assignments were used for analysis of the TNFα epitope of adalimumab.
Results and Discussion
High-pressure flash lamp output
Fast and stable illumination is key for the FOX system to properly emulate laser-based FPOP experiments. The custom high-pressure Xe flash lamp provides broad emission lines, with high UV intensity at short wavelengths where hydrogen peroxide absorbs photons strongly [18]. Flash lamp electrode design coupled with a custom power supply allows for very short light pulses (10 μs FWHM) combined with stable shot-to-shot light output at a set drive voltage (typical lamp t1/2 ≈ 171,000 pulses by adenine dosimetry, Figure 2). The duration of the illumination is significantly longer than the lifetime of the hydroxyl radical under most FPOP conditions [9, 19], making the duration of the pulse the rate-limiting step on labeling lifetime. While these light outputs are not as brief as those found with KrF excimer lasers (~20 ns), they are far shorter than typical exposures used for X-ray synchrotron radiolysis HRPF studies [3] and should be sufficient for thorough labeling of most proteins without oxidation-induced denaturation artifacts in the HRPF data [18].
Figure 2. Performance characteristics of FOX flash photolysis module.
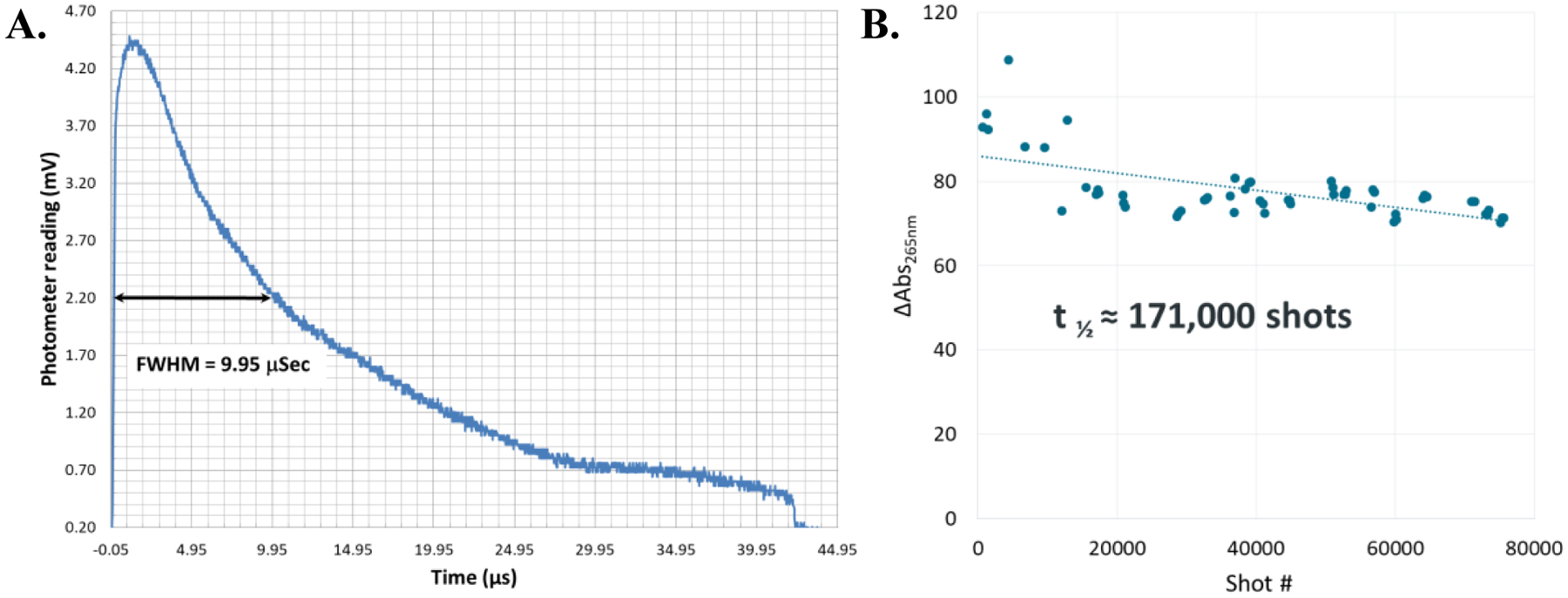
A. Light output of the flash lamp. The flash lamp reaches 90% of maximum output after 230 ns and has a flash width of 9.95 μs FWHM. The light output drops below detectable levels by 43 μs. B. Stability of the flash lamp at 750 V drive voltage, as measured by hydroxyl radical production via adenine dosimetry. Based on linear regression, the half-life of the lamp is approximately 171,000 shots. The fused silica capillary was replaced after 28,600 shots; this caused no apparent change in results.
FOX system oxidation of a model peptide
Oxidation of a model peptide, angiotensin II (DRVYIHPF) was carried out using an early prototype version of the FOX Photolysis Module to determine if the profile of peptide oxidation products generated were typical of other HRPF methods. Oxidation using the prototype FOX Photolysis Module generated standard additions of one oxygen atom (+16 Da), formation of a carbonyl (+14 Da), and multiple oxidation products that were combinations of the two (+30 Da and +32 Da). Analysis of the extracted ion chromatogram of the +16 Da and +32 Da oxidation products revealed that each mass represented a mixture of isomeric oxidation products, representing addition of the oxygen at different sites in the peptide sequence (Figure 3). No abundant products were observed that were not typically found with FPOP oxidation of the same peptide (data not shown). Overall, the FOX Photolysis Module oxidation profile was typical of those generated by FPOP and other HRPF methods.
Figure 3. FOX system oxidation products of angiotensin II.
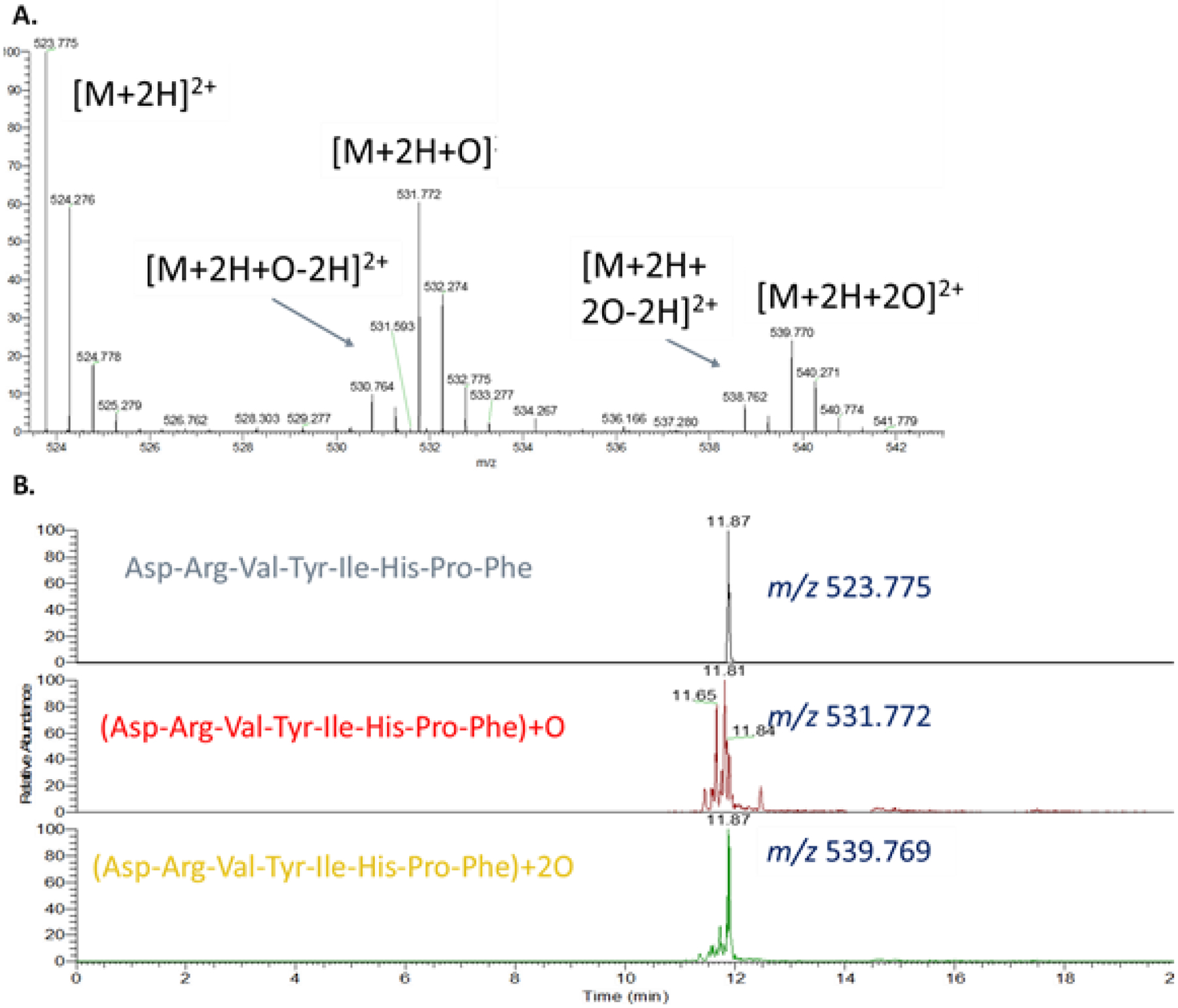
A. Average MS spectrum of doubly protonated angiotensin II and its oxidation products. B. Extracted ion chromatogram of doubly protonated angiotensin II and the two most abundant oxidation products. Each peak represents an oxidation product modified at a different location on the peptide. The large number of peaks indicates broad reactivity, as expected for an unstructured peptide without a single highly reactive residue.
Epitope mapping by HRPF with the FOX system
To test the accuracy of HRPF using the FOX system, we determined the HRPF footprint of tumor necrosis factor α (TNFα) alone, or in the presence of the biosimilar IgG therapeutic adalimumab. ELISA was performed to verify that our TNFα sample properly bound to our adalimumab biosimilar; binding response was more robust than anti-TNFα antisera from immunized rabbit, and comparable to binding to other TNFα samples to adalimumab (data not shown). TNFα alone or in a 2:1 TNFα to adalimumab molar ratio were introduced to the FOX system by flow injection, with each sample injected in triplicate and each injection collected and quenched separately to serve as an independent replicate. FOX system oxidation of TNFα with a flash lamp drive voltage of 715 V resulted in a change in adenine absorbance at 265 nm of 39.7±1.95 mAU. Addition of adalimumab scavenged significant amounts of hydroxyl radical, reducing the dosimeter response. FOX system flash lamp drive voltage was adjusted to 1000 V to compensate for the scavenging of adalimumab as previously reported for FPOP [14, 20], resulting in an adenine dosimetry ΔAbs265nm of 39.03±1.51 mAU (Figure 4).
Figure 4. Compensation for adalimumab radical scavenging in TNFα footprinting.
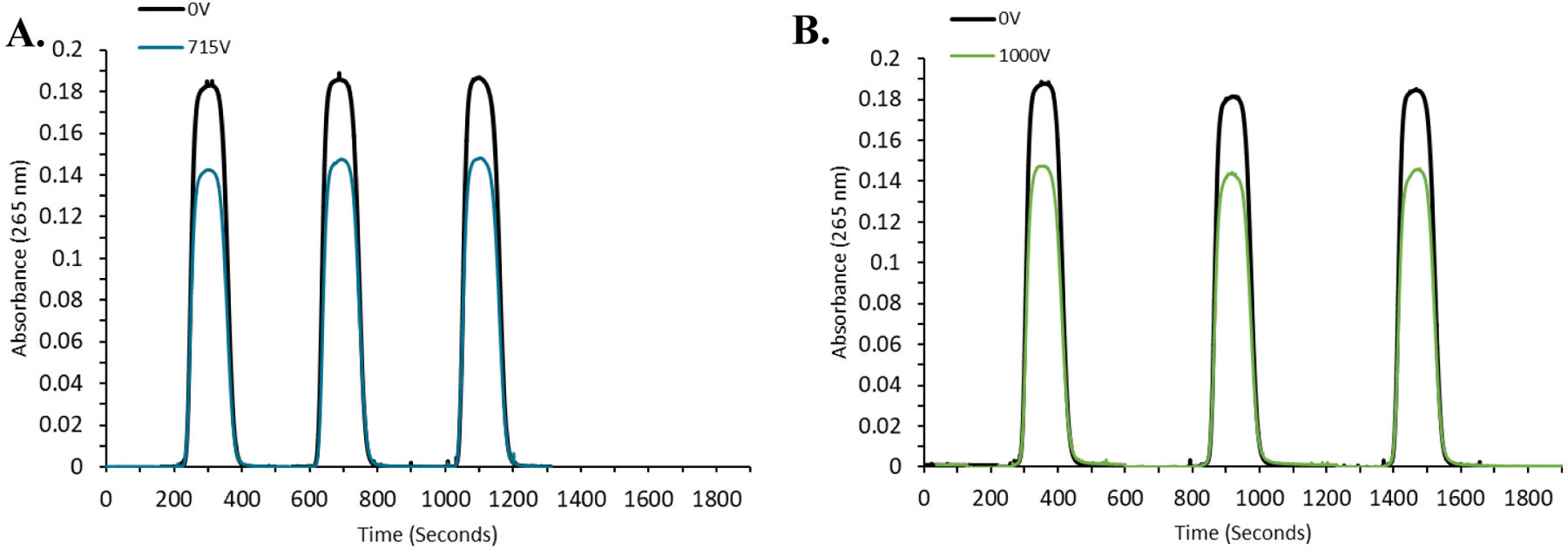
A. Flow injection real-time adenine dosimetry results for triplicate flow injections of TNFα at (black) 0V or (blue) 715V flash lamp drive voltage. Hydroxyl radicals generated at 715V drive voltage reduce the adenine absorbance at 265 nm by 39.7±1.95 mAU. B. Flow injection real-time adenine dosimetry results for triplicate flow injections of a 1:1 mixture of TNFα:adalimumab at (black) 0 V or (blue) 1000 V flash lamp drive voltage, giving an adenine ΔAbs265nm of 39.03±1.51 mAU.
Quenched samples were processed and analyzed as described in the Methods section. After identification of unoxidized peptides was performed using Byonic (Protein Metrics), oxidation products were identified and quantified using FoxWare Protein Footprinting Software (GenNext Technologies) with default settings. Analysis of the peptide oxidation products for the three replicates makes the reproducibility of FOX system oxidation apparent (Figures S3–S5, Supporting Information). Based on retention time profiles, the FOX system generated reproducible peptide oxidation products. Analysis of the oxidation events per peptide similarly showed highly reproducible oxidation by the FOX system (Figure 5). Just like FPOP, reproducibility of peptide oxidation measurements in FOX scales with the ion intensity of the peptide in LC-MS (Figure S6). The median coefficient of variation for peptides used to achieve maximum sequence coverage was 0.16, which compares very favorably to high quality FPOP data [21]. Protein oxidation was also very thorough, with every detected peptide used for maximum sequence coverage having quantifiable oxidation and the most reactive peptide (66–82, containing the TNFα’s only covered cysteine) reaching >1 oxidation event per peptide.
Figure 5. FOX system HRPF of TNFα in the presence or absence of adalimumab.
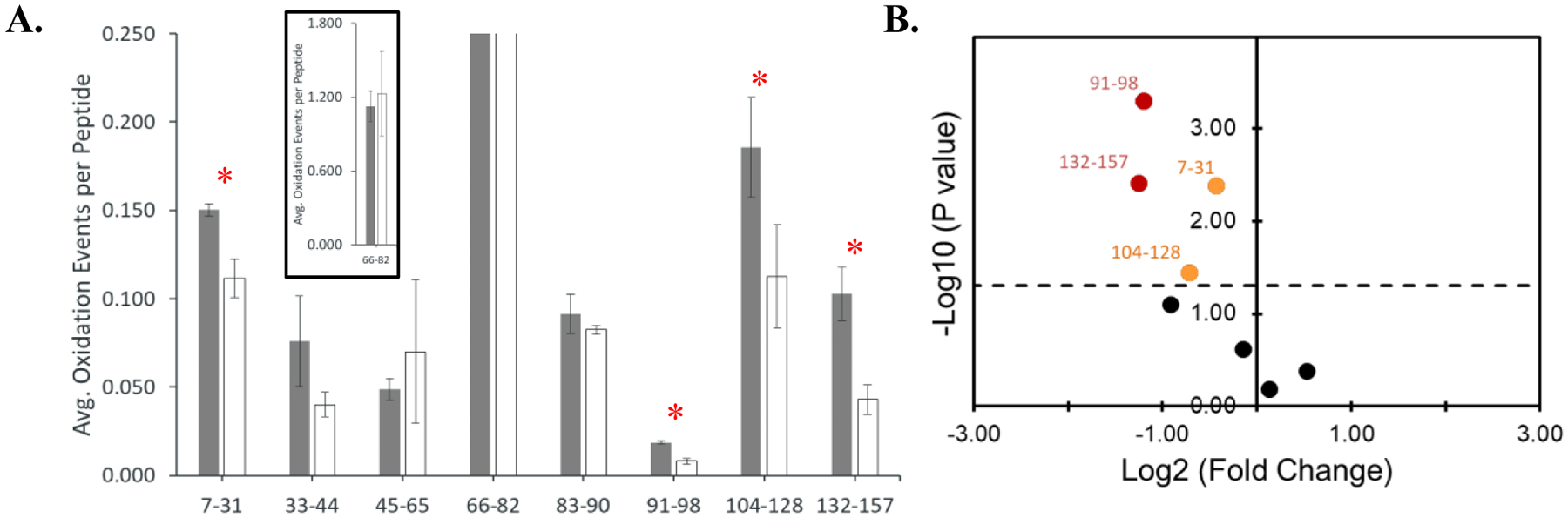
A. Histogram of FOX system HRPF peptide level results for TNFα alone (filled) or in the presence of adalimumab, compensated for scavenging (open) as reported by FoxWare Protein Footprinting software. (Inset) Peptide 66–82 results rescaled. Error bars represent one standard deviation from a triplicate measurement. Peptides that showed statistically significant protection upon adalimumab binding are marked with a red asterisk (p ≤ 0.05). B. Volcano plot of changes in TNFα FOX system HRPF results upon binding of adalimumab. Peptides that showed a statistically significant change that was at least a two-fold reduction in oxidation are colored red, while peptides that showed less than a two-fold reduction in oxidation are colored orange.
After peptide level oxidation was quantified by the FoxWare Protein Footprinting Software, sub-peptide assignment was attempted for the four peptides that showed significant changes in oxidation upon adalimumab addition (Figure 5). Samples were re-analyzed by LC-MS/MS, and the sub-peptide region was assigned using a manual version of the approach of Gross and co-workers [17]. While we were unable to separate and quantify peptide oxidation isomers chromatographically with sufficient signal and resolution for residue-level quantification, we were able to narrow down the sites of oxidation based on the product ion spectra of oxidized peptides. The identified oxidized regions, along with a comparison of FOX identified residues with interacting residues identified previously [22] are shown in Table 1.
Table 1.
Sub-peptide analysis of protected FOX oxidized TNFα peptides
| Adalimumab-protected TNFα peptide | Sub-peptide identified oxidized region | Interacting residues in the peptide |
|---|---|---|
| 7-TPSDKPVAHVVANPQAEGQLQWLNR-31 | 17-VANPQAEGQLQWL-29 | P19, E20, E23 |
| 91-VNLLSAIK-98 | 94-LSA-96 | V91, N92 |
| 104-ETPEGAEAKPWYEPIYLGGVFQLEK-128 | 104-ETPEGAEAKPWYE-116 | E110, A111, K112, P113 |
| 132-LSAEINRPDYLDFAESGQVYFGIIAL-157 | ND | E135, I136, N137, Y141, A145, E146 |
Structural analysis of the regions protected upon adalimumab binding are consistent with the previously published X-ray structure of the hexameric TNFα-adalimumab Fab complex (asymmetric unit: PDB ID 3WD5) [22] as reconstructed using the TNFα trimer (PDB ID 1TNF) as an alignment template [23]. Four peptides were found to have statistically significant protection upon oxidation, all of which contain at least one amino acid previously identified to be directly involved in the TNFα-adalimumab interface (Figure 6). Sub-peptide analysis of these peptides includes the directly interacting residues or, in the case of 91–98, amino acids immediately adjacent due to lack of oxidation of the directly interacting residues. FOX system HRPF missed oxidation of one significant part of the epitope, consisting of peptide 66–82. Interestingly, the amino acids in the loop contacting adalimumab are almost all inherently poorly reactive (K65, G66, Q67, T71, H72, T77, T79, and S81), while oxidation of this peptide is dominated by the highly reactive C69 (data not shown) which is not involved in the interface with adalimumab [22]. The sole amino acid in the interface of this peptide with relatively high inherent reactivity, H72, only interacts with adalimumab through the β-carbon [22] while the oxidation probes the imidazole ring [24]. Overall, the FOX system HRPF of the adalimumab epitope on TNFα is consistent with previously published X-ray crystallography data and known HRPF reactivity, capturing the antibody interaction spanning TNFα monomers as previously observed.
Figure 6. FOX system HRPF protection of TNFα plotted on the X-ray crystal structure of the TNFα-adalimumab Fab hexameric complex.
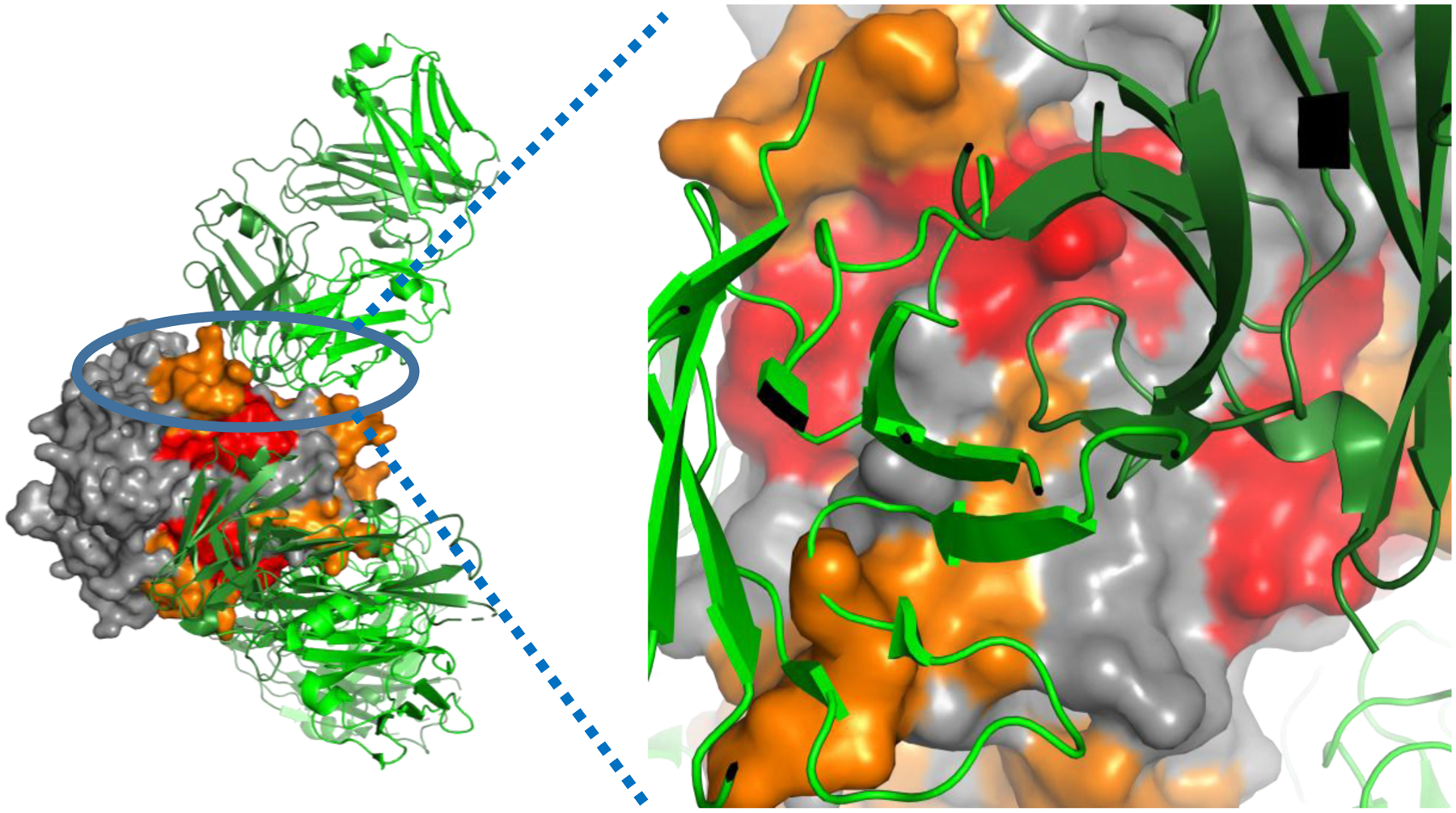
Left: Full view of the hexameric complex, with TNFα surface depicted and adalimumab Fab shown as a cartoon. TNFα regions that show no change in oxidation upon adalimumab binding are colored grey; TNFα regions that show <2x reduction in oxidation upon adalimumab binding are shown in orange; TNFα regions that show a >2x reduction in oxidation upon adalimumab binding are shown in red. The adalimumab Fab heavy chain is colored dark green, while the adalimumab light chain is colored light green. Right: Zoomed in view of the interface of one adalimumab Fab with the TNFα trimer surface, looking down the axis of the Fab. Each Fab interacts with two TNFα monomers in the trimer.
Conclusions
Previously published work has shown that the FOX system compares favorably to traditional laser-based FPOP in hydroxyl radical production as measured by adenine dosimetry and the ability to heavily modify proteins without labeling oxidatively altered conformations [18]. Here, we both investigate important figures of merit of the FOX system, as well as test its ability to generate reliable and useful structural results by mapping the adalimumab epitope on TNFα. Laser-based FPOP is generally considered to have a primary labeling lifetime of ~1 μs [9], although secondary radicals almost certainly persist much longer [25] and may result in slower oxidation of sulfur-containing residues [26–29]. The pulse width of the FOX system flash lamp means that primary radical reactions by flash oxidation are going to persist over 10–20 μs (Figure 2). While this duration of primary HRPF labeling is >10x longer than laser-based FPOP, it is still on a μs timescale, much shorter than X-ray synchrotron HRPF. It has also been shown to be fast enough to label proteins without oxidation-induced conformational change based on previously published measurement of dose-dependent labeling [18].
The flow injection sample introduction system is a significant departure from the syringe pump delivery used by traditional FPOP. The flow injection method allows for more reproducible sample delivery, faster sample-to-sample transition and ensures thorough capillary cleaning between samples to minimize carry-over. The use of a flow-injection system allows the FOX system to keep the flexibility of a flow system to handle widely varying sample volumes without alterations in optical path length of the photolytic light found in a static system [30], while maintaining the ability to perform real-time dosimetry and compensation [14, 19]. The FOX system flash lamps are able to fire ~171,000 shots before losing half of its hydroxyl radical generating capacity as measured by adenine dosimetry. As the inline dosimetry can be used to compensate for lamp output decay, this longevity allows for hundreds of samples to be modified per lamp, comparing quite favorably with KrF excimer laser gas replacement lifetimes. Combined with the efficiencies generated by flow injection, we have found the FOX system to be considerably more time-efficient compared to traditional laser-based FPOP. The use of the programmable sample collector ensures that, even with the relatively large volume of carrier buffer, only the volume containing the properly oxidized sample of interest is collected and quenched for analysis and the sample collection is highly reproducible. While improper operation of such a flow injection system could conceivably introduce additional error if the sample is improperly collected, the results generated here show that properly performed flow injection generates results with reproducibility very similar to the reproducibility reached by high quality traditional FPOP [21].
Analysis of the peptide oxidation products generated by the FOX system (Figure 3 and Figures S3–S5, Supporting Information) reveals oxidation product profiles that are readily familiar to any researcher acquainted with HRPF by FPOP or other established means. Peptide level analysis of the adalimumab epitope of TNFα as performed by automated software analysis using the FoxWare Protein Footprinting Software package gave accurate results for the antibody binding interface. The FOX system is able to generate FPOP-like results with integrated in-line real time radical dosimetry in a semi-automated fashion. The enclosure of the illumination source eliminates the severe eye hazard of the UV laser, and the elimination of fluorine gas from the workflow obviates the need for dedicated gas ventilation controls. With this new approach to the flash oxidation workflow, the major experimental obstacles preventing the adoption of FPOP by a wider audience due to difficulties and expense of setup and operation, hazards of KrF excimer laser operation, and irreproducibility due to inconsistent radical generation and/or scavenging have been overcome. The FOX system maintains many of the limitations common to FPOP and HRPF in general, however: the need for hydrogen peroxide to be added to the analyte protein; difficulties in thoroughly labeling samples at high concentration [31]; and secondary oxidation of sulfur-containing residues causing deviation from solvent accessibility-driven oxidation rates [27, 28].
While the FOX system was designed primarily for HRPF experiments, it is not limited in principle to these experiments. Viewed more broadly, the FOX system may offer benefits to a wide variety of structural biology-focused photochemistry. Hydroxyl radical-initiated free radical chemistry such as trifluoromethylation as recently reported [32, 33] could be performed using the FOX system, with many of the same benefits as described for HRPF experiments. Additionally, the broadband UV output and high intensity of the custom high-pressure Xe flash lamp can be used to enable flash photochemistry other than FPOP. For example, photo-initiated carbene-based footprinting [34–37] could be initiated using the FOX system, with highly controllable reaction times. Similarly, the FOX system could be used for photoaffinity labeling or photocrosslinking, offering broad UV irradiation potential and controllable irradiation parameters. With the development of appropriate internal controls to perform internal standardization analogous to hydroxyl radical dosimetry, the FOX system could provide real-time feedback on photoactivation success. While the broad-spectrum UV output increases the photochemistry accessible by the FOX system, researchers examining photosensitive biological systems may need to use filters to prevent inadvertent photoactivation by the photolysis source.
Supplementary Material
Synopsis:
A schematic representation of the FOX labeling process in the novel flash photolysis cell of the FOX Protein Footprinting System
Acknowledgments:
Development of the FOX laser-free FPOP system was supported by NIH SBIR 1 R43 GM125420-01. Development of FoxWare Protein Footprinting Software was supported by 1 R43 GM126617-01.
Financial Conflict of Interest Disclosure
J.S.S., E.E.C., R.O., R.W.E., D.H, and S.R.W. disclose a significant financial interest in GenNext Technologies, Inc., an early-stage company seeking to commercialize technologies for protein higher-order structure analysis. This manuscript and all data were reviewed by a knowledgeable scientist who has no financial conflict of interest, in accordance with the University of Mississippi FCOI management practices.
Footnotes
Supporting Information includes five figures showing the FOX photolysis system instrument control screen, a schematic of the FOX photolysis cell optical bench, and various data display screens from FoxWare protein footprinting software.
References
- 1.Chance MR Unfolding of apomyoglobin examined by synchrotron footprinting. Biochem Biophys Res Commun. 287(3), 614–621 (2001) [DOI] [PubMed] [Google Scholar]
- 2.Kiselar JG, Maleknia SD, Sullivan M, Downard KM, Chance MR Hydroxyl radical probe of protein surfaces using synchrotron X-ray radiolysis and mass spectrometry. International Journal of Radiation Biology. 78(2), 101–114 (2002) [DOI] [PubMed] [Google Scholar]
- 3.Chance MR, Farquhar ER, Yang S, Lodowski DT, Kiselar J Protein Footprinting: Auxiliary Engine to Power the Structural Biology Revolution. J Mol Biol. 432(9), 2973–2984 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharp JS, Becker JM, Hettich RL Protein surface mapping by chemical oxidation: structural analysis by mass spectrometry. Anal Biochem. 313(2), 216–225 (2003) [DOI] [PubMed] [Google Scholar]
- 5.Sharp JS, Becker JM, Hettich RL Analysis of protein solvent accessible surfaces by photochemical oxidation and mass spectrometry. Anal Chem. 76(3), 672–683 (2004) [DOI] [PubMed] [Google Scholar]
- 6.Sharp JS, Guo JT, Uchiki T, Xu Y, Dealwis C, Hettich RL Photochemical surface mapping of C14S-Sml1p for constrained computational modeling of protein structure. Anal Biochem. 340(2), 201–212 (2005) [DOI] [PubMed] [Google Scholar]
- 7.Venkatesh S, Tomer KB, Sharp JS Rapid identification of oxidation-induced conformational changes by kinetic analysis. Rapid Commun Mass Spectrom. 21(23), 3927–3936 (2007) [DOI] [PubMed] [Google Scholar]
- 8.Aye TT, Low TY, Sze SK Nanosecond laser-induced photochemical oxidation method for protein surface mapping with mass spectrometry. Anal Chem. 77(18), 5814–5822 (2005) [DOI] [PubMed] [Google Scholar]
- 9.Hambly DM, Gross ML Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J Am Soc Mass Spectrom. 16(12), 2057–2063 (2005) [DOI] [PubMed] [Google Scholar]
- 10.Gau BC, Sharp JS, Rempel DL, Gross ML Fast photochemical oxidation of protein footprints faster than protein unfolding. Anal Chem. 81(16), 6563–6571 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu XR, Rempel DL, Gross ML Protein higher-order-structure determination by fast photochemical oxidation of proteins and mass spectrometry analysis. Nat Protoc. 15(12), 3942–3970 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buxton GV, Greenstock CL, Helman WP, Ross AB Critical-Review of Rate Constants for Reactions of Hydrated Electrons, Hydrogen-Atoms and Hydroxyl Radicals (.OH/.O-) in Aqueous-Solution. Journal of Physical and Chemical Reference Data. 17(2), 513–886 (1988) [Google Scholar]
- 13.Xie B, Sharp JS Hydroxyl Radical Dosimetry for High Flux Hydroxyl Radical Protein Footprinting Applications Using a Simple Optical Detection Method. Anal Chem. 87(21), 10719–10723 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharp JS, Misra SK, Persoff JJ, Egan RW, Weinberger SR Real Time Normalization of Fast Photochemical Oxidation of Proteins Experiments by Inline Adenine Radical Dosimetry. Anal Chem. 90(21), 12625–12630 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konermann L, Stocks BB, Czarny T Laminar Flow Effects During Laser-Induced Oxidative Labeling for Protein Structural Studies by Mass Spectrometry. Analytical Chemistry. 82(15), 6667–6674 (2010) [DOI] [PubMed] [Google Scholar]
- 16.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Perez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaino JA The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47(D1), D442–D450 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niu B, Appleby TC, Wang R, Morar M, Voight J, Villasenor AG, Clancy S, Wise S, Belzile JP, Papalia G, Wong M, Brendza KM, Lad L, Gross ML Protein Footprinting and X-ray Crystallography Reveal the Interaction of PD-L1 and a Macrocyclic Peptide. Biochemistry. 59(4), 541–551 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinberger SR, Chea EE, Sharp JS, Misra SK Laser-free Hydroxyl Radical Protein Footprinting to Perform Higher Order Structural Analysis of Proteins. Journal of Visualized Experiments. e61861 (In-press (2020)) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Misra SK, Sharp JS Enabling Real-Time Compensation in Fast Photochemical Oxidations of Proteins for the Determination of Protein Topography Changes. J Vis Exp. 163), (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misra SK, Orlando R, Weinberger SR, Sharp JS Compensated Hydroxyl Radical Protein Footprinting Measures Buffer and Excipient Effects on Conformation and Aggregation in an Adalimumab Biosimilar. AAPS J. 21(5), 87 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abolhasani Khaje N, Mobley CK, Misra SK, Miller L, Li Z, Nudler E, Sharp JS Variation in FPOP Measurements Is Primarily Caused by Poor Peptide Signal Intensity. J Am Soc Mass Spectrom. 29(9), 1901–1907 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu S, Liang S, Guo H, Zhang D, Li H, Wang X, Yang W, Qian W, Hou S, Wang H, Guo Y, Lou Z Comparison of the inhibition mechanisms of adalimumab and infliximab in treating tumor necrosis factor alpha-associated diseases from a molecular view. J Biol Chem. 288(38), 27059–27067 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eck MJ, Sprang SR The structure of tumor necrosis factor-alpha at 2.6 A resolution. Implications for receptor binding. J Biol Chem. 264(29), 17595–17605 (1989) [DOI] [PubMed] [Google Scholar]
- 24.Garrison WM Reaction Mechanisms in the Radiolysis of Peptides, Polypeptides, and Proteins. Chemical Reviews. 87(2), 381–398 (1987) [Google Scholar]
- 25.Vahidi S, Konermann L Probing the Time Scale of FPOP (Fast Photochemical Oxidation of Proteins): Radical Reactions Extend Over Tens of Milliseconds. J Am Soc Mass Spectrom. 27(7), 1156–1164 (2016) [DOI] [PubMed] [Google Scholar]
- 26.Saladino J, Liu M, Live D, Sharp JS Aliphatic peptidyl hydroperoxides as a source of secondary oxidation in hydroxyl radical protein footprinting. J Am Soc Mass Spectrom. 20(6), 1123–1126 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie B, Sood A, Woods RJ, Sharp JS Quantitative Protein Topography Measurements by High Resolution Hydroxyl Radical Protein Footprinting Enable Accurate Molecular Model Selection. Sci Rep. 7(1), 4552 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaur P, Kiselar J, Yang S, Chance MR Quantitative protein topography analysis and high-resolution structure prediction using hydroxyl radical labeling and tandem-ion mass spectrometry (MS). Mol Cell Proteomics. 14(4), 1159–1168 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang W, Ravikumar KM, Chance MR, Yang S Quantitative mapping of protein structure by hydroxyl radical footprinting-mediated structural mass spectrometry: a protection factor analysis. Biophys J. 108(1), 107–115 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riaz M, Misra SK, Sharp JS Towards high-throughput fast photochemical oxidation of proteins: Quantifying exposure in high fluence microtiter plate photolysis. Anal Biochem. 561–562(32–36 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong X, Wren JC, Konermann L Effects of protein concentration on the extent of gamma-ray-mediated oxidative labeling studied by electrospray mass spectrometry. Anal Chem. 79(16), 6376–6382 (2007) [DOI] [PubMed] [Google Scholar]
- 32.Cheng M, Asuru A, Kiselar J, Mathai G, Chance MR, Gross ML Fast Protein Footprinting by X-ray Mediated Radical Trifluoromethylation. J Am Soc Mass Spectrom. 31(5), 1019–1024 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng M, Zhang B, Cui W, Gross ML Laser-Initiated Radical Trifluoromethylation of Peptides and Proteins: Application to Mass-Spectrometry-Based Protein Footprinting. Angew Chem Int Ed Engl. 56(45), 14007–14010 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jumper CC, Bomgarden R, Rogers J, Etienne C, Schriemer DC High-resolution mapping of carbene-based protein footprints. Anal Chem. 84(10), 4411–4418 (2012) [DOI] [PubMed] [Google Scholar]
- 35.Gomez GE, Monti JL, Mundo MR, Delfino JM Solvent mimicry with methylene carbene to probe protein topography. Anal Chem. 87(19), 10080–10087 (2015) [DOI] [PubMed] [Google Scholar]
- 36.Manzi L, Barrow AS, Scott D, Layfield R, Wright TG, Moses JE, Oldham NJ Carbene footprinting accurately maps binding sites in protein-ligand and protein-protein interactions. Nat Commun. 7(13288 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang B, Rempel DL, Gross ML Protein Footprinting by Carbenes on a Fast Photochemical Oxidation of Proteins (FPOP) Platform. J Am Soc Mass Spectrom. 27(3), 552–555 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


