Abstract
Preimplantation genetic testing (PGT) for monogenic disorders and assisted reproductive technology have evolved and progressed in tandem. PGT started with single-cell polymerase chain reaction (PCR) followed by fluorescent in situ hybridisation for a limited number of chromosomes, later called ‘preimplantation genetic diagnosis (PGD) version 1’. This review highlights the various molecular genetic techniques that have evolved to detect specific inherited monogenic disorders in the preimplantation embryo. Literature review in English was performed in PubMed from 1990 to 2021, using the term ‘preimplantation genetic diagnosis’. With whole-genome amplification, multiple copies of embryonic DNA were created. This helped in avoiding misdiagnosis caused by allele dropout. Multiplex fluorescent PCR analysed informative short tandem repeats (STR) and detected mutations simultaneously on automated capillary electrophoresis sequencers by mini-sequencing. Comparative genomic hybridisation (CGH) and array CGH were used for 24 chromosome aneuploidy screening. Subsequently, aneuploidies were detected by next-generation sequencing using single-nucleotide polymorphism arrays, while STR markers were used for haplotyping. ‘PGD version 2’ included accurate marker-based diagnosis of most monogenic disorders and detection of aneuploidy of all chromosomes. Human leukocyte antigen matching of embryos has important implications in diagnosis and cure of haemoglobinopathies and immunodeficiencies in children by means of matched related haematopoietic stem cell transplantation from an unaffected ‘saviour sibling’ obtained by PGT.
KEYWORDS: Haplotyping, human leucocyte antigen matching, haematopoietic stem cell transplantation, preimplantation genetic diagnosis, preimplantation genetic testing, preimplantation genetic testing for monogenic disorders PGT-M (evolving), technologies, saviour sibling
INTRODUCTION
Preimplantation genetic testing (PGT) has evolved dramatically over the past 30 years, ever since Handyside et al. reported the first twin live births, following in vitro fertilisation (IVF) and preimplantation diagnosis (PID) in two couples at risk of transmitting the X-linked recessive disorders, adrenoleukodystrophy and X-linked mental retardation.[1,2] At that time, it was only possible to identify the sex of the embryo by amplifying a Y-linked repeat sequence in single cells biopsied at cleavage stages and avoid transferring male embryos due to a 50% chance of being affected, an option preferred to prenatal diagnosis, wherein an affected pregnancy would be terminated.
The terminology preimplantation genetic diagnosis (PGD) was used for many years for the diagnosis of both-single gene disorders by molecular methods[3,4,5,6,7] and restricted number of chromosome aneuploidies or translocations by fluorescence in situ hybridisation (FISH).[8,9,10,11] With the advent of 24 chromosome aneuploidy screening by array comparative genomic hybridisation (aCGH), the terms PGD for aneuploidies,[12,13] preimplantation genetic screening (PGS),[14,15] or comprehensive chromosome screening (CCS) were interchangeably used.[16,17,18] Subsequently, the uniform terminology PGT was formulated by the International Committee for Monitoring Assisted Reproductive Technologies, in partnership with the American Society for Reproductive Medicine, European Society of Human Reproduction and Embryology, International Federation of Fertility Societies, March of Dimes, Asian Pacific Initiative on Reproduction, International Federation of Gynaecology and Obstetrics, and others.[19] PGT was further categorised into PGT for aneuploidy (PGT-A), PGT for structural rearrangements (PGT-SR) and PGT for monogenic disorders (PGT-M)[20,21,22] [Table 1].
Table 1.
Comparison of earlier and present terminologies for preimplantation genetics
| Earlier nomenclature | Present nomenclature |
|---|---|
| PGD for single-gene disorders | PGT-M |
| PGD-A/PGS/CCS | PGT-A |
| PGD for chromosome rearrangements | PGT-SR |
PGD=Preimplantation genetic diagnosis, PGD-A=Preimplantation genetic diagnosis for aneuploidies, PGS=Preimplantation genetic screening, CCS=Comprehensive chromosome screening, PGT-M=Preimplantation genetic testing for monogenic disorders, PGT-A=Preimplantation genetic testing for aneuploidies, PGT-SR=Preimplantation genetic testing for chromosomal structural rearrangements
Couples primarily opting for PGT-M are now advised to simultaneously perform PGT-A as unaffected embryos selected by PGT-M could have chromosome aneuploidies. Hence, using both PGT-M and PGT-A, only euploid embryos free of disease are selected to improve live birth rates.[23]
Objective
The objective of this review is to describe the evolution and utility of various molecular techniques used in PGT-M and to understand the novel advances over the past three decades.
SEARCH METHODS
Literature review in English was primarily performed in PubMed, from January 1999 to August 2021, using the term ‘preimplantation genetic diagnosis'. Titles and relevant abstracts were manually evaluated to eliminate papers not pertaining to PGD or PGT-M and evolution of methodologies [Figure 1]. Review articles were checked for cross-references. Full texts were obtained through Clinical Key, Academia and Google. Informative chapters from a recent textbook on PGT were searched for as well.[24,25,26]
Figure 1.
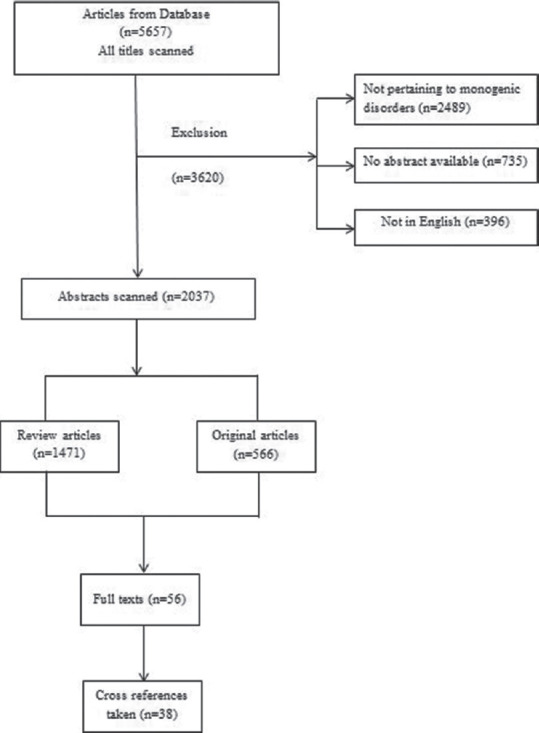
Literature review strategy
INDICATIONS FOR PREIMPLANTATION GENETIC TESTING FOR MONOGENIC DISORDERS
PGT-M can be offered for all monogenic disorders, to identify pathogenic or likely pathogenic loci.[27] Indication could be for common or rare diseases and for specific nuclear or mitochondrial genes.[28] The nuclear loci may have an autosomal dominant inheritance pattern with a 50% risk of transmitting the gene mutation to the progeny, e.g., Huntington disease or hereditary cancers due to pathogenic variations in genes such as breast cancer 1 (BRCA1), BRCA2, adenomatous polyposis coli, ataxia telangiectasia mutated, MutL homolog 1 (MLH1) and MutS homolog 2 (MLH2). Autosomal recessive disorders have a 25% risk of recurrence in each pregnancy as in beta-thalassaemia and sickle cell anaemia, while X-linked disorders such as Duchenne muscular dystrophy (DMD), haemophilia, adrenoleukodystrophy and fragile-X have a 50% chance of males being affected and a 50% chance of females being carriers. Other disorders where PGT-M can be used include achondroplasia, cystic fibrosis, epidermolysis bullosa, Tay-Sachs disease, ichthyosis, long QT syndrome, mucopolysaccharidosis, osteogenesis imperfecta, phenylketonuria, polycystic kidney disease, retinoblastoma, retinitis pigmentosa, Rett syndrome, spinal muscular atrophy, spinocerebellar ataxia, tuberous sclerosis and Usher syndrome. PGT-M is also useful when a couple is unable to find human leucocyte antigen (HLA)-matched donors from registries for a haematopoietic stem cell transplantation (HSCT) to cure their child affected with a haematologic or autoimmune disorder,[29,30] by enabling selection of HLA-matched unaffected embryos for transfer. PGT for sex selection and family balancing is not permitted in India.
EVOLUTION OF TECHNOLOGIES
The single-cell simplex polymerase chain reaction (PCR), performed by Handyside et al. in 1990,[1] subsequently showed unreliable amplification of a 680 base pair fragment while studying the sickle cell containing region of the beta globin gene in the DNA of single blastomeres from intact embryos. The same technique applied to a cluster of about 10 embryonic cells, however, showed successful amplification more frequently.[31] Amplification is necessary to increase the limited amount of genomic DNA available from embryo biopsies. Allele dropout (ADO) caused by amplification failure of one of the two parental alleles was first described in 1991 as a ‘partial amplification failure', causing a potential source of misdiagnosis for both dominant and recessive diseases.[32]
Multiplex PCR subsequently became the choice for monogenic disorders. The gold standard method of PCR was replaced by genome-wide technologies, such as fluorescent PCR and fragment analysis on automated sequencers. There was a shift in biopsy time from a single blastomere biopsy at cleavage stage or utilisation of polar body biopsy to utilising five to eight trophectoderm cells obtained at the blastocyst stage.[33,34] The most important measure to control and detect ADO as well as account for contamination was the inclusion of closely linked informative short tandem repeat (STR) markers in the multiplex PCR reaction. STRs, also known as microsatellites, are a sequence of two or more base pairs, repeated a number of times in such a way that they lie adjacent to each other on the chromosome. These determine an individual's inherited traits and parentage. The co-amplification of STR markers with or without the pathogenic variant amplicon(s) at the level of a single or few cells yields more accurate results and this approach known as haplotyping has been very useful.[35] Haplotypes are combinations of several phase-determined polymorphic markers and are valuable for studies of disease association.
Nested PCR, a modification of PCR intended to reduce non-specific binding in products due to the amplification of unexpected primer binding sites, had to be carried out. The tedious process of multiplexing was facilitated by the availability of commercial PCR multiplex kits. The development of fluorescent-labelled primers led to the method of multiplex fluorescent PCR, a sensitive method for allele detection.
Some of the single-gene disorders where PGD was applied in the 1990s include cystic fibrosis,[36,37] fragile X,[38] DMD,[39] Tay-Sachs disease,[40] haemophilia,[41] RhD blood typing,[42] Marfan syndrome,[43] sickle cell anaemia, ornithine transcarbamylase deficiency,[44] beta-thalassaemia and myotonic dystrophy.[45] Aneuploidy testing of a limited number of chromosomes by FISH was more common. FISH was also used for the detection of unbalanced translocations.[46,47] Cell recycling was attempted, although it was observed that the ADO rate was higher. Due to the low amount of input DNA from embryo biopsies in PGT-M, sensitive DNA amplification techniques were needed.
Whole-genome amplification (WGA) overcame the problem of the minute quantities of genomic DNA obtained, by amplifying the entire genome of a single or few cells up to several micrograms and providing sufficient template for many standard downstream applications such as PCR, single-nucleotide polymorphism (SNP) arrays and next-generation sequencing (NGS). A SNP is a variation at a single position in a DNA sequence among individuals. If a SNP occurs within a gene, then there is more than one allele. SNPs are used to identify disease causing genes, by sequencing.
In 1992, the first WGA methods of degenerate oligonucleotide primed PCR[48] and primer extension pre-amplification[49] were published. These methods allowed WGA from a single cell. This was a major technical improvement which together with blastocyst-stage biopsy and the introduction of vitrification, revolutionised PGT.
HAPLOTYPING
Haplotyping determines the group of alleles within a genetic segment on a single chromosome, which are inherited together. The haplotype which is common in the family members with the familial pathogenic variant is referred to as the high-risk haplotype (or mutant), whereas the haplotype without the familial pathogenic variant is known as the low-risk or wild-type haplotype. Therefore, genetic markers located close to the gene of interest are genotyped in DNA samples from the couple and relevant family members with known genetic status during the pre-clinical workup. Informative genetic markers which flank the locus of interest and allow differentiation of the parental haplotypes are selected for use in the clinical test. The clinical test can be either direct, when the pathogenic variant plus linked genetic markers are assessed, or indirect, when testing is only based on haplotyping. If the high-risk haplotype is determined during workup, an indirect testing method can be applied. Alternatively, a direct method is chosen where the detection of the pathogenic variant is combined with the genetic markers for haplotype confirmation. This is also known as preimplantation genetic haplotyping.
HUMAN LEUCOCYTE ANTIGEN (HLA) HAPLOTYPE MATCHING
HLA haplotype matching of embryos is carried out to select unaffected embryos that are HLA compatible with the couple's affected child, for future HSCT or umbilical stem cell transplant. HLA typing is usually combined with PGT for a specific monogenic disorder, mainly a haemoglobinopathy, solid tumour, immunodeficiency or immune disorder.[50,51,52]
HAEMOGLOBINOPATHIES
Haemoglobinopathies, caused by inherent mutations in genes coding for globin synthesis, comprise all genetic disorders of haemoglobin formation and utilisation. Symptomatic haemoglobinopathy is the most important monogenic disease in the world.[53] Haemoglobinopathies result in substantial mortality and morbidity. They commonly occur in populations of Africa, the Mediterranean area and Southeast Asia regions. In India, β-thalassaemia is the most commonly encountered single-gene disorder. About 10,000–20,000 babies with thalassaemia major are born every year in India.[54] About 3%–4% of the Indian population carries the β-thalassaemia gene mutation. Its incidence is much higher in North-eastern India and in the Lohana, Marwadi, Sindhi and Aggarwal communities. Certain individuals of the Islamic and Sikh religions are more susceptible. Individuals with β-thalassaemia major need blood transfusions, chelations, and multiple hospitalisation cycles, putting pressure on parents, siblings, extended family, medical services and society. The probability of finding an allele match for the Indian population in multinational HLA registries is 16% and only about 0.008% in Indian registries.[55,56,57] Occasionally, it may not be possible to determine the high-risk haplotype during pre-PGT workup if there is a de novo pathogenic variant, or if no relevant family DNA samples are obtained. In these cases, it may be determined during the clinical cycle based on the results from the biopsied blastocysts, through an affected embryo.[22] However, this is a complex procedure requiring multidisciplinary collaboration.
ALLOGENIC HAEMATOPOIETIC STEM CELL TRANSPLANTATION
Allogenic HSCT was pioneered in 1957 by Thomas et al.[58] The first birth of a child who was HLA matched by PGD to save an elder sibling suffering from a haematological disorder called Fanconi anaemia was reported by Verlinsky et al. in 2001.[59] This success was achieved after five IVF cycles and testing 41 embryos over 4 years. Kuliev et al. in 2005 reported their experience of PGT-M for haemoglobin disorders with HLA matching.[60] Following this, several groups all over the world reported success. In 2014, Kuliev and Rechitsky described the chance of obtaining unaffected euploid embryos in different inheritance patterns. The chances of getting a thalassaemia unaffected 100% HLA-matched embryo to an affected sibling were 18.75% and thalassaemia unaffected fully HLA-matched euploid embryo were 9.4%.[61] Among the numerous disease-linked STRs present, most families have at least two or three informative ones which help provide a diagnosis.[62]
We reported recently our first successful ‘Saviour Sibling’ birth in Maharashtra, India, for an older sibling with beta-thalassaemia major.[63]
THE PROCEDURE OF HAPLOTYPING
After WGA, STR-based haplotyping by targeted PCR can be carried out. A combination of STRs in proximity to the mutation site, having different alleles on all four parental chromosomes, is selected, to track the inheritance of each chromosome from the parent to the embryo. The probability of multiple loci on the same chromosome being affected in a cell is very low; hence, this strategy of using flanking STR markers, combined with mutation detection, is commonly used for PGT-M. The comparatively short length of the STRs, together with the sensitivity of fluorescent PCR analysed by capillary electrophoresis with automated sequencers, helps in multiplex PCR, which is the amplification of multiple target sequences from a single, or a few cells.[2] Mutation detection can also be done simultaneously using single fluorescent base extension reactions, or mini-sequencing for single-nucleotide substitutions, with specially designed primers. An indirect single-cell HLA typing protocol based on a multiplex fluorescent PCR of STR markers scattered throughout the HLA complex was reported in 2005 for preimplantation HLA matching.[64]
Accurate high-resolution typing for the HLA Class I and Class II loci using NGS has improved HLA matching accuracy and reduced rejection rates for both related and unrelated donor transplants.[58] HSCT shows superior outcomes with fewer complications and higher survival rate when a matched sibling donor is available. NGS has the advantage of potentially providing nucleotide-resolution data for genetic analysis, thus overcoming the difficulties associated with the absence of STR informativity and the requirement of a workup on relatives. If the disease-causing variant (s) in a family can be directly detected in the embryo-biopsy samples, an affected embryo can also be used for haplotyping.
HUMAN LEUCOCYTE ANTIGEN MATCHING FOR BETA-THALASSAEMIA
Currently, PGT-M for HLA matching of embryos is done by combining direct and indirect genetic studies of the HLA region using fluorescent PCR. Fragment analysis of PCR products by capillary electrophoresis is done using AB 3130. The direct study involves PCR amplification of the haemoglobin subunit beta (HBB) gene sequence, while the indirect study involves PCR amplification of polymorphic markers on loci such as D11S988, D11S1338, D11S1760, D11S1871, D11S2351, D11S4181, D11S4891 and D11S4957 linked to the HBB gene. PCR amplification of polymorphic markers linked to the HLA region includes D6S1560, D6S1583, D6S1629, D6S1683, D6S2924, MOG3 and TNFa. PGT-M is subsequently carried out on the embryo biopsies after WGA, using only the informative STR and/or SNP markers previously defined in the pre-PGT-M study by fluorescent PCR.
Subsequently, PGT-A for all chromosomes is carried out by NGS, using the PGS kit followed by data analysis on software which aligns the reads using the human genome build ‘hg19'. This PGT-A test cannot detect chromosome gains and losses below 10 Mb, balanced structural rearrangements, low levels of mosaicism, uniparental disomy (UPD), haploidy, triploidy and single-gene variants. The chance of identifying matched, unaffected embryos for recessive conditions is 18.8%.[7]
A geneticist experienced in pedigree and linkage analysis should determine which familial DNA samples are needed for a reliable and accurate diagnosis. Samples should be collected from prospective parents and close relatives with known disease status. When using commercially available NGS-based validated protocols, it is still recommended to carry out an implementation validation of the complete wet and dry laboratory workflow before clinical use.
SINGLE-NUCLEOTIDE POLYMORPHISM ARRAYS
SNP arrays are high-density oligo-arrays made up of several million probes, allowing genotyping of innumerable selected SNPs across all chromosomes in a single reaction. SNPs consist of two alleles A and B, which could be homozygous (AA or BB) or heterozygous (AB), irrespective of the actual nucleotides A, T, G or C which may be present.[2,6] The analysis and interpretation are based on the amount of fluorescence and the ratio of hybridisation intensities for A and B (allele frequencies). The same principle of linkage-based testing applies to targeted multiplex PCR and SNP array, though a locus specific pre-clinical workup is not required for SNP arrays, reducing the waiting time for the couples. Only a short workup of DNA of the couple, and an affected or unaffected child or grandparent, is required to establish phasing, as biallelic SNP markers, by definition, can only distinguish two out of the four parental chromosomes. The four different sets of genome-wide SNP markers can be identified by determining the genotypes of the parents and working out which of the AB alleles at each position is present on individual chromosomes using, for example, an existing child of known disease status. The SNP array platform can be used for double indications, such as two monogenic disorders or a monogenic disorder with HLA matching.[65] It can also concurrently analyse PGT-M and PGT-A, as both SNP genotype and chromosome copy number information are available from the raw data set. SNP arrays can detect aneuploidies, polyploidies and UPD. The two measures providing evidence of the copy number state are the log R ratio and the B-allele frequency.[66] Another application of haplotyping by SNP array is to detect balanced translocations or inversions and differentiate between normal and balanced translocation carriers, if the aberrations are inherited and relevant samples of the family are available for testing.
KARYOMAPPING
Karyomapping was developed by Handyside et al. and is based on SNP analysis using a bead array to genotype 300K SNPs genome-wide, Mendelian analysis and an algorithm incorporated to avoid errors caused by ADO.[67] Karyomapping can also be used for accurate selection of HLA-matched embryos, along with aneuploidy screening. However, STRs assist in identification in cases of loss of heterozygosity (LOH) in the target region.[68] Further, since there are markers for each parental chromosome, meiotic trisomies, monosomies and subchromosomal deletions can be identified.[2]
SEQUENCING
Sequencing by NGS involves DNA fragmentation and library preparation of templates with adapters which contain barcodes for a cost-effective analysis with many samples in the same run. The single-molecule templates are subjected to parallel sequencing directly, called third generation, or after clonal amplification known as second generation. The sequence reads are then mapped to a reference genome. The genomic coverage or read depth refers to the number of reads found at a given position.[24] A low coverage is sufficient for PGT-A. For monogenic disorders, sequencing at high coverage is required, hence only targeted sequencing is carried out for PGT-M, together with PGT-A by NGS or PGT-SR by aCGH as a separate test,[69] though recent integrated analysis tools allow a combination of both.
TARGETED SEQUENCING
Targeted sequencing helps in detecting known and novel variants in selected sets of genes or genomic regions in a cost-effective, rapid way. Gene sequencing can be accomplished using several different DNA sequencing methods, depending on the scale, such as increasing the read depth across the mutation site. The mutation loci can be captured in a pre-amplification reaction with mutation-specific primers. MARSALA (mutated allele revealed by sequencing with aneuploidy and linkage analysis) is one such method.[70] The targeted amplification is coupled with locus-specific pre-clinical workup.
TEST DEVELOPMENT/PRECLINICAL PREIMPLANTATION GENETIC TESTING FOR MONOGENIC DISORDERS WORKUP
Before starting a PGT cycle, the pre-examination process should be completed successfully. The laboratory test development includes informativity testing or segregation analysis and validation.[34,71]
TRANSPORT PREIMPLANTATION GENETIC TESTING
The biopsied trophectoderm cells from the IVF laboratory are transported to a genetics unit specialising in cellular molecular diagnostics. Transport PGT services have expanded substantially, despite the challenges related with transport to different cities.[27]
EXCLUSION TESTING
In families with a history of late-onset neurodegenerative disorders such as Huntington disease, individuals at risk who want to avoid pre-symptomatic testing but wish for their own biological unaffected children may opt for PGT with exclusion testing.[72] This is possible by indirect testing with selection of embryos carrying the haplotype of the unaffected prospective grandparent for transfer. Pre-clinical informativity testing is done for DNA samples of the couple and the parents of the partner at risk only. Exclusion testing recognises the right of the parent to not know whether they are themselves affected while enabling them to have children not affected by the disease. Genetic implication counselling is an essential part of this procedure.[73] Some couples will have an unnecessary IVF procedure even if the partner in question does not have the deleterious variant, due to the indirect testing needed for non-disclosure. In case direct testing is carried out, but the carrier status is not disclosed to the couple, besides maintaining extreme confidentiality, fake embryo transfers may be performed to keep the couple unaware of the carrier status.[20]
PGT as a diagnostic tool is empowered by genome-wide sequencing, allowing a generic protocol for monogenic and chromosomal disorders, including detection of de novo mutations and repeat expansions.[74] Reduction of the cost could be achieved by reducing the complexity of the libraries and decreasing the number of reads.
COMPREHENSIVE PREIMPLANTATION GENETIC TESTING
Initially, comprehensive PGT-M and PGT-A were carried out with two biopsies. The polar body was biopsied for chromosome screening by aCGH and a day 3 blastomere was biopsied for genetic analysis of a single-gene disorder by multiplex PCR.[75,76] In 2011, a WGA product was used to carry out SNP microarray for aneuploidy together with PCR-based linkage analysis for GM1 gangliosidosis from a single biopsy.[22,77] Rechitsky et al. in 2015 reported the first systematic experience demonstrating improved pregnancy rates and reduced spontaneous miscarriage rates, especially in women with advanced age.[78] A large-scale study in 2017 also showed an increase in clinical pregnancy rates per transfer from 33.6% to 49% by avoiding transfer of embryos with a low developmental potential.[79] The recent high throughout single-cell genotyping technologies enable studying hundreds of thousands of informative SNPs for genome-wide haplotyping by the methods of karyomapping and haplarithmisis which is integrated into single-cell haplotyping and imputation of linked disease variants computational workflow.[80] The linkage disequilibrium principle is used to detect the disease-causing mutations and assign them to parental haplotype blocks on all chromosomes by genotyping and genome-wide SNP analysis. The advantage of this method is that it can be used for any familial single-gene disorder, without the need for developing family specific protocols. Screening of multiple genes, aneuploidies, UPD, LOH and balanced translocations is also possible simultaneously. It has been shown that 0PN and 1PN zygotes which are normally discarded can develop into balanced diploid embryos.[81] Inclusion of such zygotes in comprehensive PGT can increase the number of available embryos for transfer per IVF cycle by over 20%.[82]
PreimplantationCCS using ChromInst, a single-cell overnight sequencing protocol, was reported by Gao et al. in 2021 where single-cell WGA and NGS library construction were integrated into a two-step PCR procedure of about 2.5 h reaction time. New strategies are under way for genome-wide haplotyping, such as long fragment read technology, and a microfluidic device to separate and amplify homologue chromosomes of singe metaphase cells.
ETHICAL ISSUES
PGT with exclusion testing is prohibited in some countries. In autosomal dominant conditions, embryos which have inherited the haplotype of the affected grandparent will have a 50% chance of being affected and will be discarded, though they also have a 50% chance of being healthy. Psychological evaluation of the parents should be carried out to make sure that they do not consider the saviour sibling only as an instrument to save their older child. This is one of the major issues of ethical debates. The fate of unaffected non-HLA–matched embryos should also be discussed.
UTILITY OF PREIMPLANTATION GENETIC TESTING FOR MONOGENIC DISORDERS
PGT-M can be used for any inherited single gene disorder, to select unaffected embryos. This includes cancer predisposition genes, late onset disorders such as Huntington disease and non-life–threatening conditions such as non-syndromic sensorineural hearing loss. Currently, globally, PGT-M is used for many conditions such as thalassaemia, haemophilia, cystic fibrosis, fragile X syndrome, adrenoleukodystrophy, polycystic kidney disease, endocrine diseases, mitochondrial disorders and many more. We have carried out PGT-M for a variety of monogenic disorders such as inherited cancer genes, neurofibromatosis, sickle cell anaemia, DMD, Leigh syndrome, Norrie's Disease, retinoblastoma, cardiac disorders, Tay-Sach disease, propionic academia, hereditary inclusion body myopathy, Huntington chorea and G6PD deficiency,[83,84,85] including for BRCA1[86] and haplotyping and saviour sibling for thalassaemia major.[87] PGT gives more confidence for single embryo transfer, reducing pregnancy complications of multiple gestation. It decreases the miscarriage rates as aneuploidy is avoided. However, testing does not guarantee that a suitable embryo will be available for transfer and couples need to be aware of this before embarking on a PGT cycle.
GLIMPSES INTO THE FUTURE OF PREIMPLANTATION GENETIC TESTING
Variant haplophasing around the target by long read sequencing is a recent novel approach to PGT, using third-generation sequencing (TGS) as part of a general workup for PGT-SR and PGT-M.[88] Detailed parental phased SNP profiles around targets are used for selection of informative polymorphic markers to simplify and facilitate clinical PGT designs which allow discrimination between carrier and non-carrier embryos. This enables rapid selection of closely linked informative markers around the region of interest for patient-specific test design and has the ability to set the phase without additional blood samples from the extended family. This method can also distinguish between balanced and normal embryos in cases of translocations.[89]
Blastocentesis is an experimental alternative biopsy approach to retrieve cell-free DNA released into the inner cavity. Efforts are also on-going for non-invasive PGT from embryonic DNA in spent culture medium after IVF, thus bypassing embryo biopsy.[90] This is based on the current established methodology of non-invasive prenatal testing from cell-free foetal fraction DNA in maternal plasma for aneuploidies.[26]
Many genes responsible for cell lineage and differentiation during organogenesis, explaining lack of embryo development, will be known and integrated into PGT-M or cell-free DNA platforms and panels, with the aim of selecting embryos which will increase live births in ART.[26] PGT has been carried out for over 600 disorders, and this may even increase to 1000, as expanded carrier screening has led to an increase in the usage of PGT-M. This applies to adult-onset disorders as well, as carriers may desire that their children should not inherit the mutant allele. An emerging field now is PGT for polygenic disease risk.[91,92,93] Efforts are on linking the genetic constitution of the embryo to its morphology, implantation potential and transcriptome using single-cell RNA sequencing.[94]
SUMMARY
Technologies yielding data on genotyping as well as chromosome copy number with concurrent PGT-M and PGT-A have greatly improved the live birth rate, enabling single embryo transfer. Automated sequencing, mini-sequencing and real-time PCR have further refined diagnostic capability. WGA of single blastocyst biopsies ensures multifactor PGT without compromising embryo viability. A semi-nested direct and indirect testing system minimises embryo misdiagnosis risk due to ADO, non-specific amplification or contamination. The diagnostic PGT approach has greatly evolved over the years. HLA haplotyping by linkage analysis is the most common generic approach, though SNP arrays and NGS are also being used.[58] TGS with the portable nanopore is the latest technology that has revolutionised the area of genomics by bringing DNA sequencing from large laboratories to clinics using long read sequencing information. This shows promise for genomics point of care testing in the field of IVF and PGT.[36] The impact of genomic medicine has been the strongest in reproductive diagnostics, and the time has come to acknowledge the same.
CONCLUSION
The role of PGT is shifting from diagnostics to therapeutics, being used not only to avoid conception of affected children but also to give birth to healthy children who are potential saviours. The medical community and society at large should be aware of the availability of advanced techniques so that children and their families affected by these lethal disorders can enjoy a better quality of life.
There are certain ethical objections raised that the future child will be an instrument to cure another child. However, a broader view entails that this technology is an exception as it is lifesaving and enhances the family structure, improving the quality of life of not only the saviour sibling and the affected child but of the entire family and society at large.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Handyside AH, Kontogianni EH, Hardy K, Winston RM. Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification. Nature. 1990;344:768–70. doi: 10.1038/344768a0. [DOI] [PubMed] [Google Scholar]
- 2.Cimadomo D, Rienzi L, Capalbo A, Rubio C, Innocenti F, Garcia-Pascual CM, et al. The dawn of the future: 30 years from the first biopsy of a human embryo. The detailed history of an ongoing revolution. Hum Reprod Update. 2020;26:453–73. doi: 10.1093/humupd/dmaa019. [DOI] [PubMed] [Google Scholar]
- 3.Grifo JA, Boyle A, Tang YX, Ward DC. Preimplantation genetic diagnosis.In situ hybridization as a tool for analysis. Arch Pathol Lab Med. 1992;116:393–7. [PubMed] [Google Scholar]
- 4.Ao A. Preimplantation genetic diagnosis of inherited disease. Indian J Exp Biol. 1996;34:1177–82. [PubMed] [Google Scholar]
- 5.Wells D, Sherlock JK. Strategies for preimplantation genetic diagnosis of single gene disorders by DNA amplification. Prenat Diagn. 1998;13:1389–401. [PubMed] [Google Scholar]
- 6.Sermon K, Steirteghem AV, Liebaers I. Preimplantation genetic diagnosis. Lancet. 2004;363:1633–41. doi: 10.1016/S0140-6736(04)16209-0. [DOI] [PubMed] [Google Scholar]
- 7.Kuliev A, Verlinsky Y. Place of preimplantation genetic diagnosis in genetic practice. Am J Med Genet A. 2005;134A:105–10. doi: 10.1002/ajmg.a.30635. [DOI] [PubMed] [Google Scholar]
- 8.Munné S, Lee A, Rosenwaks Z, Grifo J, Cohen J. Diagnosis of major chromosome aneuploidies in human preimplantation embryos. Hum Reprod. 1993;8:2185–91. doi: 10.1093/oxfordjournals.humrep.a138001. [DOI] [PubMed] [Google Scholar]
- 9.Munné S, Weier HU. Simultaneous enumeration of chromosomes 13, 18, 21, X, and Y in interphase cells for preimplantation genetic diagnosis of aneuploidy. Cytogenet Cell Genet. 1996;75:263–70. doi: 10.1159/000134497. [DOI] [PubMed] [Google Scholar]
- 10.Munné S, Sandalinas M, Escudero T, Velilla E, Walmsley R, Sadowy S, et al. Improved implantation after preimplantation genetic diagnosis of aneuploidy. Reprod Biomed Online. 2003;7:91–7. doi: 10.1016/s1472-6483(10)61735-x. [DOI] [PubMed] [Google Scholar]
- 11.Colls P, Goodall N, Zheng X, Munné S. Increased efficiency of preimplantation genetic diagnosis for aneuploidy by testing 12 chromosomes. Reprod Biomed Online. 2009;19:532–8. doi: 10.1016/j.rbmo.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Capalbo A, Romanelli V, Cimadomo D, Girardi L, Stoppa M, Dovere L, et al. Implementing PGD/PGD-A in IVF clinics: Considerations for the best laboratory approach and management. J Assist Reprod Genet. 2016;33:1279–86. doi: 10.1007/s10815-016-0768-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubio C, Bellver J, Rodrigo L, Castillón G, Guillén A, Pellicer A, et al. PGD-A in advanced maternal age: Final results of a day- 3 RCT. Reprod Biomed Online. 2018;36:e8. [Google Scholar]
- 14.Blockeel C, Schutyser V, De Vos A, Verpoest W, De Vos M, Staessen C, et al. Prospectively randomized controlled trial of PGS in IVF/ICSI patients with poor implantation. Reprod Biomed Online. 2008;17:848–54. doi: 10.1016/s1472-6483(10)60414-2. [DOI] [PubMed] [Google Scholar]
- 15.Gleicher N, Kushnir VA, Barad DH. Preimplantation genetic screening (PGS) still in search of a clinical application: A systematic review. Reprod Biol Endocrinol. 2014;12:22. doi: 10.1186/1477-7827-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahdouh EM, Balayla J, García-Velasco JA. Impact of blastocyst biopsy and comprehensive chromosome screening technology on preimplantation genetic screening: A systematic review of randomized controlled trials. Reprod Biomed Online. 2015;30:281–9. doi: 10.1016/j.rbmo.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 17.Wells D, Alfarawati S, Fragouli E. Use of comprehensive chromosomal screening for embryo assessment: Microarrays and CGH. Mol Hum Reprod. 2008;14:703–10. doi: 10.1093/molehr/gan062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen M, Wei S, Hu J, Quan S. Can comprehensive chromosome screening technology improve IVF/ICSI outcomes. A meta-analysis? PLoS One. 2015;10:e0140779. doi: 10.1371/journal.pone.0140779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zegers-Hochschild F, Adamson GD, Dyer S, Racowsky C, Schmidt L, Sokol R, et al. The international glossary on infertility and fertility care.2017. Fertil Steril. 2017;108:393–406. doi: 10.1016/j.fertnstert.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Carvalho F, Coonen E, Goossens V, Kokkali G, Rubio C, Meijer-Hoogeveen M, et al. ESHRE PGT consortium good practice recommendations for the organisation of PGT. Hum Reprod Open. 2020;2020:hoaa021. doi: 10.1093/hropen/hoaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.ESHRE PGT-SR/PGT-A Working Group. Coonen E, Rubio C, Christopikou D, Dimitriadou E, Gontar J, Goossens V, et al. ESHRE PGT Consortium good practice recommendations for the detection of structural and numerical chromosomal aberrations. Hum Reprod Open. 2020;2020:hoaa017. doi: 10.1093/hropen/hoaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ESHRE PGT-M Working Group. Carvalho F, Moutou C, Dimitriadou E, Dreesen J, Giménez C, Goossens V, et al. ESHRE PGT consortium good practice recommendations for the detection of monogenic disorders. Hum Reprod Open. 2020;2020:hoaa018. doi: 10.1093/hropen/hoaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treff NR, Zimmerman R, Bechor E, Hsu J, Rana B, Jensen J, et al. Validation of concurrent preimplantation genetic testing for polygenic and monogenic disorders, structural rearrangements, and whole and segmental chromosome aneuploidy with a single universal platform. Eur J Med Genet. 2019;62:103647. doi: 10.1016/j.ejmg.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 24.De Rycke M, Verdyck . Preimlantation genetic testing for monogenic disorders. In: Griffin DK, Harton GL, editors. Preimplantation Genetic Testing Recent Advances in Reproductive Medicine. 1st ed. Boca Raton, London, New York: CRC Press; 2020. pp. 77–86. [Google Scholar]
- 25.Tsuiko O, Vermeesh JR. Novel methods in preimplantation genetic testing. In: Griffin DK, Harton GL, editors. Preimplantation Genetic Testing Recent Advances in Reproductive Medicine. 1st ed. Boca Raton, London, New York: CRC Press; 2020. pp. 87–95. [Google Scholar]
- 26.Simpson JL, Rechitsky S, Kuliev A. Preimplantation genetic testing in the future. In: Griffin DK, Harton GL, editors. Preimplantation Genetic Testing Recent advances in Reproductive Medicine. 1st ed. Boca Raton, London, New York: CRC Press; 2020. pp. 151–6. [Google Scholar]
- 27.Rycke MD, Berckmoes V. Preimplantation genetic testing for monogenic disorders. Genes (Basel) 2020;11:871–86. doi: 10.3390/genes11080871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smeets HJ, Sallevelt SC, Dreesen JC, de Die-Smulders CE, de Coo IF. Preventing the transmission of mitochondrial DNA disorders using prenatal or preimplantation genetic diagnosis. Ann N Y Acad Sci. 2015;1350:29–36. doi: 10.1111/nyas.12866. [DOI] [PubMed] [Google Scholar]
- 29.Duarte RF, Labopin M, Bader P, Basak GW, Bonini C, Chabannon C, et al. Indications for haematopoietic stem cell transplantation for haematological diseases, solid tumours and immune disorders: Current practice in Europe 2019. Bone Marrow Transplant. 2019;54:1525–52. doi: 10.1038/s41409-019-0516-2. [DOI] [PubMed] [Google Scholar]
- 30.De Rycke M, De Vos A, Belva F, Berckmoes V, Bonduelle M, Buysse A, Keymolen K, et al. Preimplantation genetic testing with HLA matching: From counseling to birth and beyond. J Hum Genet. 2020;65:445–54. doi: 10.1038/s10038-020-0732-z. [DOI] [PubMed] [Google Scholar]
- 31.Pickering SJ, McConnell JM, Johnson MH, Braude PR. Reliability of detection by polymerase chain reaction of the sickle cell containing region of the beta-globin gene in single human blastomeres. Hum Reprod. 1992;7:630–36. doi: 10.1093/oxfordjournals.humrep.a137710. [DOI] [PubMed] [Google Scholar]
- 32.Navidi W, Arnheim N. Using PCR in preimplantation genetic disease diagnosis. Hum Reprod. 1991;6:836–49. doi: 10.1093/oxfordjournals.humrep.a137438. [DOI] [PubMed] [Google Scholar]
- 33.Kokkali G, Coticchio G, Bronet F, Celebi C, Cimadomo D, Goossens V, Liss J, et al. ESHRE PGT Consortium and SIG Embryology good practice recommendations for polar body and embryo biopsy for PGT. Hum Reprod Open. 2020:1–12. doi: 10.1093/hropen/hoaa020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thornhill AR, de Die Smulders CE, Geraedts JP, Harper JC, Harton GL, Lavery SA, et al. ESHRE PGD Consortium ‘Best practice guidelines for clinical preimplantation genetic diagnosis (PGD) and preimplantation genetic screening (PGS) Hum Reprod. 2005;20:35–48. doi: 10.1093/humrep/deh579. [DOI] [PubMed] [Google Scholar]
- 35.Spits C, De Rycke M, Verpoest W, Lissens W, Van Steirteghem A, Liebaers I, et al. Preimplantation genetic diagnosis for Marfan syndrome. Fertil Steril. 2006;86:310–20. doi: 10.1016/j.fertnstert.2005.12.059. [DOI] [PubMed] [Google Scholar]
- 36.Ao A, Handyside A, Winston RM. Preimplantation genetic diagnosis of cystic fibrosis (delta F508) Eur J Obstet Gynecol Reprod Biol. 1996;65:7–10. doi: 10.1016/0028-2243(95)02294-3. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, Lissens W, Silber SJ, Devroey P, Liebaers I, Van Steirteghem A. Birth after preimplantation diagnosis of the cystic fibrosis delta F508 mutation by polymerase chain reaction in human embryos resulting from intracytoplasmic sperm injection with epididymal sperm. JAMA. 1994;272:1858–60. [PubMed] [Google Scholar]
- 38.Dreesen JC, Geraedts JP, Dumoulin JC, Evers JL, Pieters MH. RS46 (DXS548) genotyping of reproductive cells: Approaching preimplantation testing of the fragile-X syndrome. Hum Genet. 1995;96:323–29. doi: 10.1007/BF00210416. [DOI] [PubMed] [Google Scholar]
- 39.Liu J, Lissens W, Van Broeckhoven C, Lofgren A, Camus M, Liebaers I, et al. Normal pregnancy after preimplantation DNA diagnosis of a dystrophin gene deletion. Prenat Diagn. 1995;15:351–8. doi: 10.1002/pd.1970150409. [DOI] [PubMed] [Google Scholar]
- 40.Gibbons WE, Gitlin SA, Lanzendorf SE, Kaufmann RA, Slotnick RN, Hodgen GD. Preimplantation genetic diagnosis for Tay-Sachs disease: Successful pregnancy after pre-embryo biopsy and gene amplification by polymerase chain reaction. Fertil Steril. 1995;63:723–8. doi: 10.1016/s0015-0282(16)57472-x. [DOI] [PubMed] [Google Scholar]
- 41.Lissens W, Sermon KD. Preimplantation genetic diagnosis: Current status and new developments. Hum Reprod. 1997;12:1756–61. doi: 10.1093/humrep/12.8.1756. [DOI] [PubMed] [Google Scholar]
- 42.Avner R, Reubinoff BE, Simon A, Zentner BS, Friedmann A, Mitrani-Rosenbaum S, et al. Management of rhesus isoimmunization by preimplantation genetic diagnosis. Mol Hum Reprod. 1996;2:60–2. doi: 10.1093/molehr/2.1.60. [DOI] [PubMed] [Google Scholar]
- 43.Harton GL, Tsipouras P, Sisson ME, Starr KM, Mahoney BS, Fugger EF, et al. Preimplantation genetic testing for Marfan syndrome. Mol Hum Reprod. 1996;2:713–5. doi: 10.1093/molehr/2.9.713. [DOI] [PubMed] [Google Scholar]
- 44.Morsy M, Takeuchi K, Kaufmann R, Veeck L, Hodgen GD, Beebe SJ. Preclinical models for human pre-embryo biopsy and genetic diagnosis. II. Polymerase chain reaction amplification of deoxyribonucleic acid from single lymphoblasts and blastomeres with mutation detection. Fertil Steril. 1992;57:431–8. doi: 10.1016/s0015-0282(16)54859-6. [DOI] [PubMed] [Google Scholar]
- 45.Verlinsky Y, Munné S, Simpson JL, Kuliev A, Ao A, Ray P, et al. Current status of preimplantation diagnosis. J Asst Reprod Genet. 1997;14:72–5. doi: 10.1007/BF02765773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munné S, Morrison L, Fung J, Márquez C, Weier U, Bahçe M, et al. Spontaneous abortions are reduced after pre-conception diagnosis of translocations. J Assist Reprod Genet. 1998;15:290–6. doi: 10.1023/A:1022544511198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munne S. Preimplantation genetic diagnosis of numerical and structural chromosome abnormalities. Reprod Biomed Online. 2002;4:183–96. doi: 10.1016/s1472-6483(10)61938-4. [DOI] [PubMed] [Google Scholar]
- 48.Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A. Degenerate oligonucleotide-primed PCR: General amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–25. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- 49.Zhang L, Cui X, Schmitt K, Hubert, Navidi W, Arnheim N. Whole genome amplification from a single cell: Implications for genetic analysis. Proc Natl Acad Sci U S A. 1992;89:5847–51. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kakourou G, Kahraman S, Ekmekci GC, Tac HA, Kourlaba G, Kourkouni E, et al. The clinical utility of PGD with HLA matching: A collaborative multi-centre ESHRE study. Hum Reprod. 2018;33:520–30. doi: 10.1093/humrep/dex384. [DOI] [PubMed] [Google Scholar]
- 51.Kakourou G, Vrettou C, Moutafi M, Traeger-Synodinos J. Preimplantation HLA matching: The production of a saviour child. Best Pract Res Clin Obstet Gynaecol. 2017;44:76–89. doi: 10.1016/j.bpobgyn.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 52.Sureda A, Bader P, Cesaro S, Dreger P, Duarte RF, Dufour C, et al. Indications for allo- and auto-SCT for haematological diseases, solid tumours and immune disorders: Current practice in Europe. Bone Marrow Transplant. 2015;50:1037–56. doi: 10.1038/bmt.2015.6. [DOI] [PubMed] [Google Scholar]
- 53.Chandy M. Developing a National Thalassemia Control Programme for India. In: Ghosh K, Colah R, editors. Control and management of Thalassemia and other Hemoglobinopathies in the Indian Subcontinent. Synoptic Views. Mumbai: National Institute of Immunohaematology; 2008. pp. 46–9. [Google Scholar]
- 54.National Health Mission Guidelines on Hemoglobinopathies in India. Prevention and Control of Hemoglobinopathies in India. Thalassemia, Sickle Cell Disease and other Variant Hemoglobins. Ministry of Health and Family Welfare, Government of India. 2016 [Google Scholar]
- 55.Roy P. Beta thalassemia: An Indian perspective. Adv Biotechnol Microbiol. 2019;14:555893. [Google Scholar]
- 56.Angelucci E, Matthes-Martin S, Baronciani D, Bernaudin F, Bonanomi S, Cappellini MD, et al. Hematopoietic stem cell transplantation in thalassemia major and sickle cell disease: Indications and management recommendations from an international expert panel. Haematologica. 2014;99:811–20. doi: 10.3324/haematol.2013.099747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tiwari AK, Bhati-Kushwaha H, Kukreja P, Mishra VC, Tyagi N, Sharma A, et al. Probability of finding marrow unrelated donor (MUD) for an Indian patient in a multi-national human leukocyte antigen (HLA) registry. Indian J Hematol Blood Transfus. 2015;31:186–95. doi: 10.1007/s12288-014-0416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas D, Lochte L, Jr, Lu C, Ferrebee W. Intravenous infusion of bone marrow in patients receiving radiation and chemotherapy. N Engl J Med. 1957;257:491–6. doi: 10.1056/NEJM195709122571102. [DOI] [PubMed] [Google Scholar]
- 59.Verlinsky Y, Rechitsky S, Schoolcraft W, Strom C, Kuliev A. Preimplantation diagnosis for Fanconi anemia combined with HLA matching. JAMA. 2001;285:3130–3. doi: 10.1001/jama.285.24.3130. [DOI] [PubMed] [Google Scholar]
- 60.Kuliev A, Rechitsky S, Verlinsky O, Tur-Kaspa I, Kalakoutis G, Angastiniotis M, et al. Preimplantation diagnosis and HLA typing for haemoglobin disorders. Reprod Biomed Online. 2005;11:362–70. doi: 10.1016/s1472-6483(10)60845-0. [DOI] [PubMed] [Google Scholar]
- 61.Kuliev A, Verlinsky O, Rechitsky S. Preimplantation HLA typing for stem cell transplantation treatment of hemoglobinopathies. Thalassemia Rep. 2014;4:99–101. [Google Scholar]
- 62.Renwick PJ, Trussler J, Ostad-Saffari E, Fassihi H, Black C, Braude P, et al. Proof of principle and first cases using preimplantation genetic haplotyping a paradigm shift for embryo diagnosis. Reprod Biomed Online. 2006;13:110–9. doi: 10.1016/s1472-6483(10)62024-x. [DOI] [PubMed] [Google Scholar]
- 63.Parikh FR, Athalye AS, Naik DJ, Naik NJ, Sanap MV, Sanap RR, et al. Successful preimplantation genetic testing for beta Thalassemia and HLA haplotyping with live birth of a savior sibling and cryopreservation of umbilical cord blood stem cells for future use: A case report. Indian J Hematol Blood Transfus. 2020;36(Suppl 1):S203. [Google Scholar]
- 64.Fiorentino F, Kahraman S, Karadayi H, Biricik A, Sertyel S, Karlikaya G, Saglam Y, et al. Short tandem repeats haplotyping of the HLA region in preimplantation HLA matching. Eur J Hum Genet. 2005;13:953–8. doi: 10.1038/sj.ejhg.5201435. [DOI] [PubMed] [Google Scholar]
- 65.G, Lang K, Quenzel P, Bohme I, Sauter J, Hofmann JA, et al. 2.7 million samples genotyped for HLA by next generation sequencing: Lessons learned. BMC Genomics. 2017;18:161. doi: 10.1186/s12864-017-3575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, et al. QuantiSNP: An objective Bayes hidden-Markov model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–25. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Natesan SA, Bladon AJ, Coskun S, Qubbaj W, Prates R, Munne S, et al. Genome-wide karyomapping accurately identifies the inheritance of single-gene defects in human preimplantation embryos in vitro. Genet Med. 2014;16:838–45. doi: 10.1038/gim.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J, Bao-Min L, Li R, Guo J, Xu Y, Pan JF, et al. Karyomapping in preimplantation genetic testing for β-thalassemia combined with HLA matching: A systematic summary. J Asst Reprod Genet. 2019;36:2515–23. doi: 10.1007/s10815-019-01595-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Treff NR, Fedick AM, Tao X, Devkota B, Taylor D, Scott RT., Jr Evaluation of targeted next-generation sequencing-based preimplantation genetic diagnosis of monogenic disease. Fertil Steril. 2013;99:1377–84.e6. doi: 10.1016/j.fertnstert.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 70.Yan L, Huang L, Xu L, Huang J, Ma F, Zhu X, et al. Live births after simultaneous avoidance of monogenic diseases and chromosome abnormality by next-generation sequencing with linkage analyses. Proc Natl Acad Sci U S A. 2015;112:15964–9. doi: 10.1073/pnas.1523297113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thornhill AR, Snow K. Molecular diagnostics in preimplantation genetic diagnosis. J Mol Diagn. 2002;4:11–29. doi: 10.1016/S1525-1578(10)60676-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Van Rij MC, De Rademaeker M, Moutu C, Dressen JC, De Rycke M, Liebaers I, et al. Preimplantation genetic diagnosis (PGD) for Huntington's disease: The experience of three European centres. Eur J Hum Genet. 2012;20:368–75. doi: 10.1038/ejhg.2011.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shenfield F, Pennings G, Devroey P, Sureau C, Tarlatzis B, Cohen J. Taskforce 5: Preimplantation genetic diagnosis. Hum Reprod. 2003;18:649–51. doi: 10.1093/humrep/deg110. [DOI] [PubMed] [Google Scholar]
- 74.Long N, Qiao Y, Xu Z, Tu J, Lu Z. Recent advances and application in whole-genome multiple displacement amplification. Quant Biol. 2020;8:279–94. [Google Scholar]
- 75.Obradors A, Fernandez E, Oliver-Bonet M, Rius M, Wells D, Benet J, et al. Birth of a healthy boy after a double factor PGD in a couple carrying a genetic disease and at risk for aneuploidy: Case report. Hum Reprod. 2008;23:1949–56. doi: 10.1093/humrep/den201. [DOI] [PubMed] [Google Scholar]
- 76.Obradors A, Fernandez E, Oliver-Bonet M, Rius M, Benet J, Navarro J, et al. Outcome of twin babies free of Von Hippel-Lindau disease after a double-factor preimplantation genetic diagnosis: Monogenetic mutation analysis and comprehensive aneuploidy screening. Fertil Steril. 2009;91:933. e1–7. doi: 10.1016/j.fertnstert.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 77.Brezina PR, Benner A, Rechitsky S, Kuliev A, Pauling D, Kearns WG, et al. Single-gene testing combined with single nucleotide polymorphism microarray preimplantation genetic diagnosis for aneuploidy: A novel approach in optimizing pregnancy outcome. Fertil Steril. 2011;95:1786.e5–8. doi: 10.1016/j.fertnstert.2010.11.025. [DOI] [PubMed] [Google Scholar]
- 78.Rechitsky S, Kuliev A, Goodman A, Pakhalchuk T, Ramos GS, Zlatopolsky Z, et al. First systematic experience of preimplantation genetic diagnosis for single-gene disorders, and/or preimplantation human leukocyte antigen typing, combined with 24-chromosome aneuploidy testing. Fertil Steril. 2015:10503–12. doi: 10.1016/j.fertnstert.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 79.Minasi MG, Fiorentino F, Ruberti A, Biricik A, Cursio E, Greco E, et al. Genetic diseases and aneuploidies can be detected with a single blastocyst biopsy: A successful clinical approach. Hum Reprod. 2017;32:1770–7. doi: 10.1093/humrep/dex215. [DOI] [PubMed] [Google Scholar]
- 80.Esteki MZ, Dimitriadou E, Mateiu L, Melotte C, Kumar P, Das R, et al. Concurrent whole-genome haplotyping and copy-number profiling of single cells. Am J Hum Genet. 2015;96:894–912. doi: 10.1016/j.ajhg.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Destouni A, Dimitriadou E, Masset H, Debrock S, Melotte C, Bogaert K, et al. Genome-wide haplotyping embryos developing from 0PN and 1PN zygotes increases transferrable embryos in PGT-M. Hum Reprod. 2018;33:2302–11. doi: 10.1093/humrep/dey325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen S, Yin X, Zhang S, Xia J, Liu P, Xie P, et al. Comprehensive preimplantation genetic testing by massively parallel sequencing. Hum Reprod. 2021;36:236–47. doi: 10.1093/humrep/deaa269. [DOI] [PubMed] [Google Scholar]
- 83.Madon PF, Athalye AS, Parikh FR. Preimplantation genetic diagnosis. In: Sahetya R, Purandare H, editors. Principles and Practice of Fetal Medicine. 1st ed. New Delhi: Jaypee Brothers; 2016. pp. 210–7. [Google Scholar]
- 84.Parikh FR, Athalye AS, Naik NJ, Naik DJ, Sanap RR, Madon PF. Preimplantation genetic testing: Its evolution where are we today? J Hum Reprod Sci. 2018;11:306–14. doi: 10.4103/jhrs.JHRS_132_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nair SB, Athalye AS, Sanap RR, Warang DJ, Padyal PM, Madon PF, et al. Prenatal Diagnosis and preimplantation genetic diagnosis in hematological disorders a preliminary data from India. J Transl Sci. 2018;4:1–3. [Google Scholar]
- 86.Parikh FR, Naik DJ, Naik NJ, Sanap RR, Madon PF, Sanap MV, et al. First successful twin delivery in India after preimplantation genetic diagnosis for BRCA1 mutation. Indian J Appl Res. 2018;8:338–9. [Google Scholar]
- 87.Athalye AS, Naik DJ, Sanap RR, Naik NJ, Sanap MV, Warang DJ, et al. Preimplantation genetic testing for beta thalassemia and other hematological disorders. Indian J Haematol Blood Transfus. 2020;36(Suppl 1):S204. [Google Scholar]
- 88.Cheng Y, Yu Q, Ma M, Wang H, Tian S, Zhang W, et al. Variant haplophasing by long-read sequencing: A new approach to preimplantation genetic testing workups. Fertil Steril. 2021;116:774–83. doi: 10.1016/j.fertnstert.2021.04.012. [DOI] [PubMed] [Google Scholar]
- 89.Margolis C, Werner M, Jalas C. Variant haplophasing by long-read sequencing: Proof of concept in preimplantation genetic workup and an opportunity to distinguish balanced and normal embryos. Fertil Steril. 2021;116:668–9. doi: 10.1016/j.fertnstert.2021.06.055. [DOI] [PubMed] [Google Scholar]
- 90.Ben-Nagi J, Odia R, Gonzalez XV, Heath C, Babariya D, SenGupta S, et al. The first ongoing pregnancy following comprehensive aneuploidy assessment using a combined blastocenetesis, cell free DNA and trophectoderm biopsy strategy. J Reprod Infertil. 2019;20:57–62. [PMC free article] [PubMed] [Google Scholar]
- 91.Treff NR, Eccles J, Marin D, Messick E, Lello L, Gerber J, et al. Preimplantation genetic testing for polygenic disease relative risk reduction: Evaluation of genomic index performance in 11,883 adult sibling pairs. Genes. 2020;11:648. doi: 10.3390/genes11060648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rubio C, Simón C. Embryo genetics. Genes (Basel) 2021;12:118. doi: 10.3390/genes12010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Treff NR, Marin D, Lello L, Hsu S, Tellier LC. Preimplantation genetic testing: Preimplantation genetic testing for polygenic disease risk. Reproduction. 2020;160:A13–7. doi: 10.1530/REP-20-0071. [DOI] [PubMed] [Google Scholar]
- 94.Tšuiko O, Fernandez Gallardo E, Voet T, Vermeesch JR. Preimplantation genetic testing: Single-cell technologies at the forefront of PGT and embryo research. Reproduction. 2020;160:A19–31. doi: 10.1530/REP-20-0102. [DOI] [PubMed] [Google Scholar]


