Supplemental Digital Content is available in the text.
Keywords: cardiomyopathy, dilated; circadian clocks; diet, high-fat; heart failure; lipid metabolism; obesity
Background:
The nuclear receptor Rev-erbα/β, a key component of the circadian clock, emerges as a drug target for heart diseases, but the function of cardiac Rev-erb has not been studied in vivo. Circadian disruption is implicated in heart diseases, but it is unknown whether cardiac molecular clock dysfunction is associated with the progression of any naturally occurring human heart diseases. Obesity paradox refers to the seemingly protective role of obesity for heart failure, but the mechanism is unclear.
Methods:
We generated mouse lines with cardiac-specific Rev-erbα/β knockout (KO), characterized cardiac phenotype, conducted multi-omics (RNA-sequencing, chromatin immunoprecipitation sequencing, proteomics, and metabolomics) analyses, and performed dietary and pharmacological rescue experiments to assess the time-of-the-day effects. We compared the temporal pattern of cardiac clock gene expression with the cardiac dilation severity in failing human hearts.
Results:
KO mice display progressive dilated cardiomyopathy and lethal heart failure. Inducible ablation of Rev-erbα/β in adult hearts causes similar phenotypes. Impaired fatty acid oxidation in the KO myocardium, in particular, in the light cycle, precedes contractile dysfunctions with a reciprocal overreliance on carbohydrate utilization, in particular, in the dark cycle. Increasing dietary lipid or sugar supply in the dark cycle does not affect cardiac dysfunctions in KO mice. However, obesity coupled with systemic insulin resistance paradoxically ameliorates cardiac dysfunctions in KO mice, associated with rescued expression of lipid oxidation genes only in the light cycle in phase with increased fatty acid availability from adipose lipolysis. Inhibition of glycolysis in the light cycle and lipid oxidation in the dark cycle, but not vice versa, ameliorate cardiac dysfunctions in KO mice. Altered temporal patterns of cardiac Rev-erb gene expression correlate with the cardiac dilation severity in human hearts with dilated cardiomyopathy.
Conclusions:
The study delineates temporal coordination between clock-mediated anticipation and nutrient-induced response in myocardial metabolism at multi-omics levels. The obesity paradox is attributable to increased cardiac lipid supply from adipose lipolysis in the fasting cycle due to systemic insulin resistance and adiposity. Cardiac molecular chronotypes may be involved in human dilated cardiomyopathy. Myocardial bioenergetics downstream of Rev-erb may be a chronotherapy target in treating heart failure and dilated cardiomyopathy.
Clinical Perspective.
What Is New?
We characterized the in vivo cardiac functions of Rev-erbα/β, druggable components of the circadian clock whose agonists and antagonists can benefit the heart in preclinical studies.
Obesity coupled with systemic insulin resistance paradoxically ameliorates cardiac dysfunctions in a mouse model with a disrupted cardiac circadian clock and rescues lipid oxidation genes only in the sleep cycle in phase with increased fatty acid availability from adipose lipolysis.
The temporal coordination between cardiac clock–mediated anticipation and nutrient-induced response in myocardial metabolism may contribute to the obesity paradox.
What Are the Clinical Implications?
Alterations of cardiac molecular circadian clock correlate with severities of cardiac dilation in patients who have heart failure with dilated cardiomyopathy.
Systemic insulin resistance, under certain conditions, might be cardioprotective by promoting adipose lipolysis and the availability of free fatty acids to the heart
Dietary factors or pharmacological reagents that alter myocardial metabolism can have distinct cardiac outcomes depending on the time of the day.
Editorial, see p 465
The time-of-the-day difference is known for the incidence of many adverse cardiovascular events, including myocardial infarction, sudden cardiac death, ischemic stroke, and ventricular arrhythmia.1 Robust diurnal rhythms are also observed for heart-related physiological factors such as sleep, consummatory behaviors, hormones, blood clotting or fibrinolysis factors, the renin-angiotensin-aldosterone system, the autonomic nervous system, and the heart itself.1–4 These diurnal rhythms are driven conjointly by the response to and the anticipation of environmental changes. The anticipation is mediated by the molecular circadian clock machinery composed of multiple transcription factors forming positive and negative feedback loops.5–7 The molecular clock machinery is ubiquitously expressed in numerous tissues and organs. It is unclear whether cardiac molecular clock dysfunction in humans is associated with the progression of any naturally occurring heart diseases.
The key clock components Rev-erbα and Rev-erbβ are nuclear receptors with ligand-binding domains, making them more druggable than general transcription factors.8 Small-molecule Rev-erb agonists and antagonists are available and show promising therapeutic efficacy in sleep, mood, or metabolic disorders in animal models.8,9 Both Rev-erb agonists and antagonists show beneficial effects to the heart. Rev-erb agonists can mitigate pressure overload–induced cardiac hypertrophy, myocardial infarction, or myocardial ischemia-reperfusion in mice.10–13 Rev-erb antagonist increases myocardial ischemia-reperfusion tolerance ex vivo at the sleep-to-wake transition.14 The paradoxical beneficial effects of both agonists and antagonists may be related to the temporal oscillation and phase-specific effect of Rev-erb or the timing of the drug administration. It is unclear whether these small molecules work directly on the heart or through other peripheral tissues, because how cardiac Rev-erb regulates heart function was not studied in vivo.
Obesity paradox refers to the clinical observation that, although obesity is a risk factor for heart failure, patients with heart failure who have higher body mass index have a better prognosis than those with lower body mass index.15,16 It was unknown whether obesity merely reflects overall better health or is cardioprotective. In animal models, diet-induced obesity can ameliorate heart failure under certain conditions,17 but diet-induced obesity can impair cardiac functions, and prolonged high-fat diets can model diabetic cardiomyopathy.18 In studying a genetic mouse model with cardiomyocyte-specific disruption of the molecular circadian clock, we accidentally found that the temporal coordination between clock-mediated anticipation and nutrient-induced response in myocardial metabolism may contribute to the obesity paradox.
Methods
Mice and Regular Laboratory Studies
Rev-erbαloxP (Nr1d1tm1.2Rev, MGI ID 5426700), Rev-erbβloxP (Nr1d2tm1.1Rev, MGI ID 5426699), αMHC-Cre (RRID:IMSR_JAX:011038), and αMHC-MerCreMer (RRID:IMSR_JAX:005657) mouse lines have been previously described.19–22 All the animal procedures followed the guidelines of the Institutional Animal Care and Use Committee at Baylor College of Medicine. The isolation and culture of adult mouse cardiomyocytes follow the established procedures.23,24 The pcDNA3.1-E4bp4 plasmid (Addgene 34572)25 was cotransfected with a green fluorescent protein plasmid. Radioactive isotope tracer studies were as described previously.26,27 Detailed procedures are described in the Expanded Methods section of Supplemental Material, including Tables S1 and S2.
RNA Sequencing and Chromatin Immunoprecipitation Sequencing
RNA-sequencing results of Bmal1-CKO versus wild-type (WT) hearts28 were downloaded from GSE43073. Bmal1 chromatin immunoprecipitation sequencing data29 were downloaded from GSE110602. Other detailed procedures are described in the Supplemental Material.
Proteomics, Lipidomics, and Metabolomics
Proteomics analysis was described previously.30,31 Lipids were extracted and detected as described.32–34 Metabolites were extracted using described methods.35,36 Detailed procedures are described in the Supplemental Material.
Human Tissues and Clinical Data
All human hearts were obtained from the Heart Transplant Center of Fuwai Hospital, Beijing, China. The study was approved by the Ethics Committee of Fuwai Hospital. Patients were diagnosed with idiopathic dilated cardiomyopathy (DCM) according to the ESC guideline.37 Organ procurement organizations coordinate the donor consent and the assessment of the potential donor organ.38 Detailed procedures are described in the Supplemental Material, including Tables S3 through S5.
Statistics
Statistical parameters and results for each panel are described in Table S6.
Data Availability
RNA sequencing (GSE152372) and chromatin immunoprecipitation sequencing (GSE153150) data are available in GEO. Proteomics data are available in the MassIVE repository (MSV MSV000088075). Metabolomics and lipidomics data are available in National Metabolomics Data Repository, Project ID: PR001208, Project DOI: 10.21228/M8JX2X, Study ID: ST001915, ST001916, and ST001917. The data are available from the corresponding author on reasonable request.
Results
Rev-CKO Mice Develop Progressive Contractile Dysfunction and Lethal Heart Failure
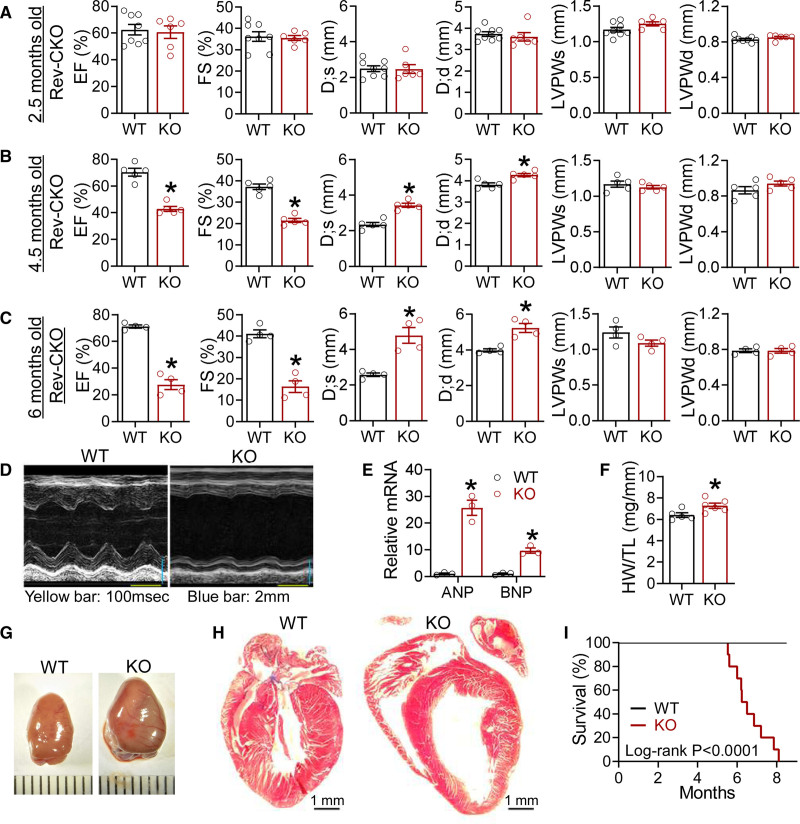
Rev-erbα and Rev-erbβ share high sequence homology with redundant roles in circadian rhythm.39 Therefore, we generated cardiomyocyte-specific Rev-erbα/β double knockout mice (referred to as Rev-CKO or simply KO) to avoid functional compensation by crossbreeding mice bearing Rev-erbαloxP and Rev-erbβloxP alleles19,20 with the αMHC-Cre line.21 Littermates bearing Rev-erbα/βloxP alleles without Cre were used as the WT control. KO mice were born at the expected Mendelian ratio. Western blot and quantitative reverse transcription polymerase chain reaction analysis confirmed the efficient depletion of Rev-erb in the heart but not in the skeletal muscle or liver (Figure S1A and S1B). On Cre recombination, the floxed Rev-erbα allele generates a truncated Rev-erbα protein without the DNA-binding domain (DBD).20 The high expression of the truncated Rev-erbα protein (DBDm) in the KO heart (Figure S1A) is attributable to disrupted proteolysis40–42 or negative transcriptional autoregulation43 (Figure S1B). KO mice did not show obvious abnormalities in the diurnal rhythm of locomotor activity, energy expenditure, respiratory exchange ratio, or food intake (Figure S1C–S1H). KO mice showed normal left ventricle size, wall thickness, and contractile function at 2.5 months of age (Figure 1A and Figure S2A). However, at 4.5 months of age, KO mice displayed impaired contractile function with an enlarged left ventricular diameter but no obvious thickening of the ventricular wall (Figure 1B and Figure S2B). By 6 months of age, the ejection fraction fell to 20% to 30%, and the left ventricle chamber got further dilated (Figure 1C and Figure S2C). Female KO mice at 6 months also developed similar contractile dysfunctions and cardiac dilation (Figure 1D and Figure S2C). αMHC-Cre mice showed normal contractile functions at this age (Figure S2D). The contractile dysfunction in KO mice was associated with elevated expression of atrial natriuretic peptide and B-type natriuretic peptide; Figure 1E) and obviously enlarged hearts (Figure 1F and 1G and Figure S2C). The left ventricle of the KO heart was dilated but without obvious fibrosis (Figure 1G). Inflammatory genes in KO hearts were not upregulated at 2 months of age but were increased at 4.5 months of age compared with WT (Figure S2E). There were no obvious changes in cardiomyocyte cell size or cell death at a young age (Figure S2F–S2H). Most KO mice died at between 6 and 8 months of age (Figure 1I). Postmortem inspection of KO mice revealed normal gross morphology of multiple organs, except for pulmonary congestion and enlarged heart (Figure S2I). In summary, cardiomyocyte-specific KO of Rev-erbα/β caused progressive contractile dysfunction, leading to DCM and lethal heart failure.
Figure 1.
Cardiomyocyte-specific ablation of Rev-erb causes progressive and lethal heart failure. A, Echocardiography analysis in WT and KO male mice at 2.5 months of age, n=8 for WT and n=6 for KO. B, Echocardiography analysis in WT and KO male mice at 4.5 months of age, n=5 mice. C, Echocardiography analysis in WT and KO male mice at 6 months of age, n=4 mice. D, Representative M mode images at 6 months of age. E, Quantitative reverse transcription polymerase chain reaction analysis of the heart at 5 months of age, n=mice. F, Heart weight (HW) to tibia length (TL) ratio at 6 months of age, n=5 for WT and n=6 for KO. G, Representative gross picture of hearts at 6 months of age. H, Trichrome stain at 6 months of age. I, Survival of WT and KO female mice, n=13 for WT and n=10 for KO mice, the P value is based on the log-rank Mantel-Cox test. Data are mean±SEM. Each dot represents an individual mouse. *P<0.05 between groups by 2-sided t test. See Table S6 for statistical details. ANP indicates atrial natriuretic peptide; BNP, B-type natriuretic peptide; D;d, left ventricular dimension at end-diastole; D;s, left ventricular dimension at end-systole; EF, ejection fraction; FS, fractional shortening; KO, knockout; LVPWd, left ventricular posterior wall thickness at end-diastole; LVPWs, left ventricular posterior wall thickness at end-systole; and WT, wild type.
Inducible Deletion of Rev-erb in Adult Hearts Causes Heart Failure and Cardiac Dilation
To address the potential developmental confounding effects, we crossbred Rev-erbα/β double-floxed mice with the inducible cardiomyocyte-specific Cre line αMHC-MerCreMer.22 The resultant inducible cardiomyocyte-specific KO mice (referred to as “MCM”) were born at the expected Mendelian ratio with no obvious abnormalities. quantitative reverse transcription polymerase chain reaction confirmed efficient deletion of Rev-erbα/β after tamoxifen injection at 2 months of age (Figure S3A and S3B). At 4.5 months after Cre activation, MCM mice displayed contractile dysfunctions comparable to those in KO mice at 4.5 months of age (Figure S3C and S3D), suggesting that it takes a similar amount of time for the phenotype to develop in both mouse lines. The MCM line also showed elevated expression of atrial natriuretic peptide/B-type natriuretic peptide and heart weight at 4.8 months after Cre activation (Figure S3E and S3F). These results exclude the developmental disruption as a major contributing factor in the cardiac dysfunction of KO mice.
Rev-erb Regulates the Oscillatory Expression of Metabolic Genes in the Heart
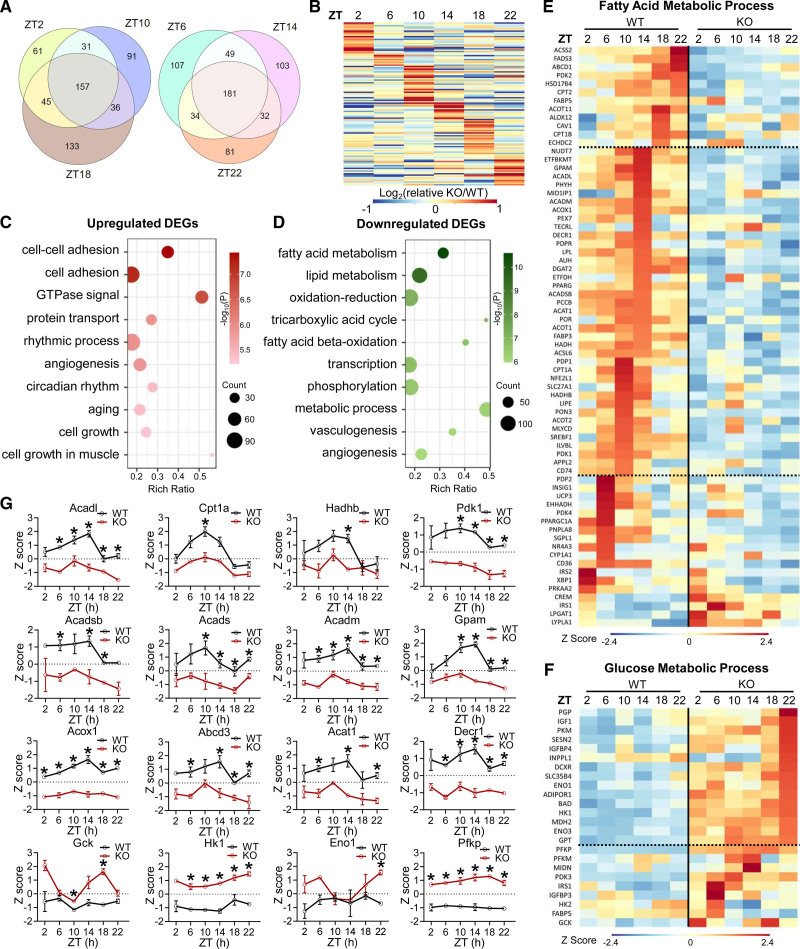
We analyzed gene expression in KO mice at a young age to exclude secondary effects of contractile dysfunctions. Quantitative reverse transcription polymerase chain reaction analyses of total RNA extracted from ventricular heart tissues and purified adult mouse cardiomyocytes (Figure S4A and S4B) showed similar fold changes between KO and WT, in line with that, up to 85% of the heart volume is from cardiomyocytes.44 The MCM heart also showed similar changes (Figure S4C). We collected ventricular heart tissues every 4 hours at Zeitgeber time (ZT) 2, 6, 10, 14, 18, and 22 at 2 months of age and identified differentially expressed genes (DEGs) between KO and WT at each ZT (Figure S4D). Common DEGs shared by all ZTs constitute <50% of the DEGs, and ≈25% to 35% of DEGs at each ZT were unique to that ZT (Figure 2A). The fold change for common DEGs at all 6 ZT times also showed robust ZT-specific differences (Figure 2B). Upregulated DEGs (KO versus WT) were enriched in cell adhesion and circadian rhythm, whereas downregulated DEGs were enriched in metabolic processes, especially fatty acid oxidation (FAO; Figure 2C and 2D and Figure S4E and S4F), supporting a central role of Rev-erb in myocardial energy metabolism.
Figure 2.
Cardiac Rev-erb regulates the transcriptomic diurnal rhythm in the heart. A, Overlap of differentially expressed genes (DEGs) in KO vs WT at the indicated ZTs. DEGs cutoff: q<0.05 and |log2Fold-Change| >1. B, Relative fold change (KO/WT) at each ZT (after normalization to the average fold change in 6 ZTs) for common DEGs among all time points. C, Top enriched biological process (BP) from Gene Ontology analysis for pooled upregulated DEGs (KO vs WT, q<0.05) at multiple ZTs. D, Top enriched biological process (BP) from Gene Ontology analysis for pooled downregulated DEGs (KO vs WT, q<0.05) at multiple ZTs. E, Heat map of DEGs involved in fatty acid metabolic process. F, Heat map of DEGs involved in glucose metabolic process. G, Relative gene expression levels, n=3 mice at each ZT for each genotype. Data are represented as mean±SEM. All z scores are based on the average and standard deviation of all samples at all time points. *adjusted P<0.05 by 2-way ANOVA with Bonferroni multiple comparisons test. See Table S6 for statistical details. KO indicates knockout; WT, wild type; and ZT, Zeitgeber time.
Among DEGs involved in fatty acid metabolism, many displayed diurnal oscillation in WT hearts, with the peak expression around the light-to-dark transition, a temporal pattern disrupted in KO hearts that showed constant low expression levels (Figure 2E and 2G). In contrast, among DEGs involved in glucose metabolism, many gained a more robust oscillatory pattern in KO hearts than in WT hearts, with the peak expression in the dark cycle (Figure 2F and 2G). These results suggest that KO hearts may have reduced FAO, in particular, at the late light cycle, and enhanced glucose utilization, in particular, at the late dark cycle, compared with WT. Thus, a physiological role of cardiac Rev-erb is to enhance FAO genes in the light cycle but to counteract diet-mediated activation of glucose metabolic genes in the dark cycle.
Rev-erb Regulates Fatty Acid Metabolic Genes Through Multiple Modes
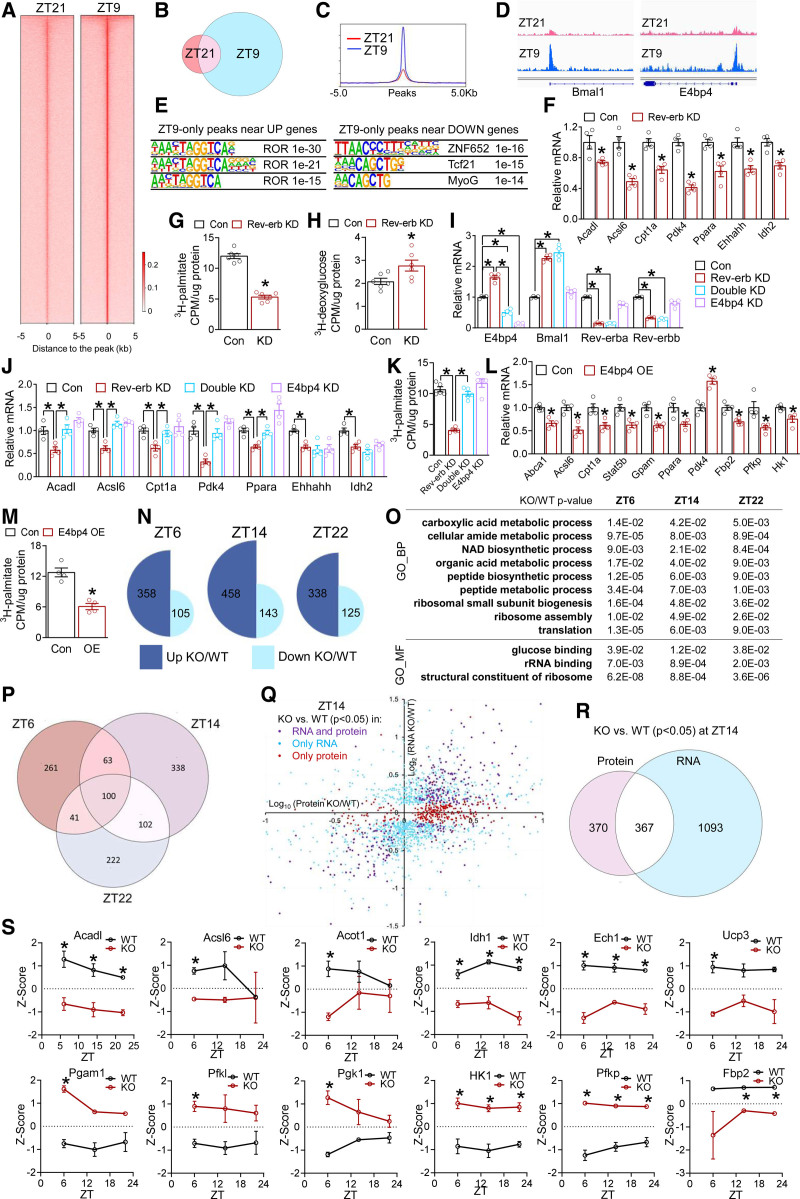
Cardiac Rev-erb mRNA peaks at ZT6–9 and reaches the nadir at ZT18–21 (Figure S5A). Rev-erbα protein follows shortly after, with the peak expression at around ZT9–12 and the nadir expression at ZT21–24 (Figure S1A). We used the anti–PCM-1 (pericentriolar material 1) antibody to isolate cardiomyocyte nuclei at ZT9 and ZT21 at 2.5 months of age and performed Rev-erbα chromatin immunoprecipitation sequencing. The genome-wide binding peaks of Rev-erbα in cardiomyocytes at ZT9 are 5 times greater than at ZT21 (Figure 3A–3C). Classic Rev-erb target genes Bmal1 and E4bp4 carried strong Rev-erb peaks in the promoters (Figure 3D). The canonical Rev-erb motif ROR-response element was the top enriched motif in high-stringency ZT9-only Rev-erbα peaks near DEGs that were upregulated in KO versus WT (Figure 3E), which validates the specificity of the chromatin immunoprecipitation assay and is in line with the role of Rev-erb as a transcription repressor.45 The ROR-response element was not among the top enriched motifs near downregulated DEGs (Figure 3E). Many downregulated metabolic genes did not have Rev-erb peaks nearby (Figure S5B). These results suggest that Rev-erb may regulate the upregulated DEGs directly, but may regulate the downregulated DEGs indirectly through another transcription repressor.
Figure 3.
Rev-erb uses multiple modes to regulate cardiac transcriptome and proteome. A, Heat map of Rev-erbα chromatin immunoprecipitation sequencing signals in isolated cardiomyocyte nuclei at all peaks. B, Overlap of Rev-erbα binding peaks at ZT9 and ZT21. C, The average profile of Rev-erbα binding peaks at ZT21 and ZT9. D, Browser tracks of Rev-erbα peaks at Bmal1 and E4bp4. E, Top enriched motif and P values in high-stringency Rev-erbα peaks within 10 kb of the transcription start sites of differentially expressed genes that were upregulated (UP) or downregulated (DOWN) in KO vs WT hearts, respectively. F, RT-qPCR analysis in AC16 cells transfected with small interfering RNA to knockdown Rev-erbα/β (Rev-erb KD) or small interfering RNA control (Con). Each dot represents an independent well of cells (F–M). *P<0.05 between 2 groups by 2-sided t test for F through M. G, Fatty acid oxidation (FAO) rate in AC16 cells after knockdown of Rev-erb (KD, or Rev-erb KD), n=6 wells. H, Glucose uptake assay in AC16 cells after knockdown of Rev-erb (KD, or Rev-erb KD), n=6 wells. I and J, RT-qPCR analysis in AC16 cells after knockdown of Rev-erb (Rev-erb KD) or E4bp4 (E4bp4 KD) or both (Double KD), n=4 wells. K, FAO rate in AC16 cells, n=5 wells. L, RT-qPCR analysis in AC16 cells overexpressing E4bp4 (E4bp4 OE) or green fluorescent protein as a control (Con), n=4 wells. M, FAO rate in AC16 cells overexpressing E4bp4 (OE), n=4 wells. N, Differentially expressed proteins (DEPs) between KO and WT at the indicated ZTs (|Log2FC| >0.58 and P<0.05 by t test). O, Shared Gene Ontology terms of DEPs between KO and WT at all ZTs, as analyzed by iPathwayGuide. P, Overlap of DEPs in KO vs WT at the indicated ZTs. DEPs cutoff: |Log2FC| >0.58 and P<0.05 by t test. Q and R, Correlation and overlap of the differentially expressed genes and DEPs (KO vs WT) at ZT14 using P<0.05 as the cutoff for both RNA and protein. S, Relative protein levels based on the average and standard deviation of all samples at all time points, n=3 mice. *adjusted P<0.05 by 2-way ANOVA with Bonferroni multiple comparisons test. Data are represented as mean±SEM. *P<0.05 between groups by 2-sided t test, unless otherwise stated. See Table S6 for statistical details. BP indicates biological process; KD, knockdown; KO, knockout; MF, molecular function; ROR, retinoid-related orphan receptor; RT-qPCR, quantitative reverse transcription polymerase chain reaction; WT, wild type; and ZT, Zeitgeber time.
E4bp4 is a transcription factor of high binding similarity with altered histone acetylation markers in Rev-erb–depleted cardiomyocytes.46 E4bp4 is a known transcription repressor and a target of Rev-erb,47 with a robust Rev-erbα peak at its transcription start site at ZT9 (Figure 3D). E4bp4 expression was upregulated in the KO versus WT hearts (Figure S5C). In addition, E4bp4 was known to regulate lipid metabolism in the intestine and liver.48,49 These observations prompted us to test whether the upregulated E4bp4 accounts for the downregulated lipid metabolic genes in KO cardiomyocytes. Knockdown of Rev-erbα/β in AC16 cardiomyocytes downregulated FAO genes and FAO flux rate (Figure 3F and 3G) and increased glucose uptake (Figure 3H). The combined knockdown of both Rev-erbα/β and E4bp4 rescued the expression of FAO genes and the FAO flux compared with Rev-erb knockdown alone (Figure 3I–3K), without affecting glucose uptake (Figure S5D). Similar results were obtained in adult mouse cardiomyocytes (Figure S5E–S5G). Overexpression of E4bp4 in AC16 cells (Figure S5H) downregulated FAO genes (Figure 3L), reduced FAO rate (Figure 3M), and did not alter glucose uptake (Figure S5I). Although other clock components, such as Bmal1, may also play a role in the gene expression perturbation in KO hearts (Figure S6), the maximum KO/WT difference in E4bp4 in the late light cycle support that E4bp4 contributes to impaired FAO in KO hearts at this time.
Cardiac Rev-erb Regulates the Myocardial Proteome
To address how cardiac Rev-erb regulates the myocardial protein content, we performed proteomic profiling at ZT6, ZT14, and ZT22 in both WT and KO hearts at 10 weeks of age. More than 4000 proteins were detected and quantified in each sample. Approximately 3-fold more proteins were upregulated than downregulated in KO versus WT at each ZT (Figure 3N). Differentially expressed proteins (KO versus WT) were highly dependent on ZT, with only 17% to 22% of the differentially expressed proteins at each ZT shared by the other ZTs (Figure 3P). Although differentially expressed proteins at different ZTs had limited overlap at the level of individual proteins, they shared common functionality enrichment in metabolic processes (Figure 3O). These findings further support that cardiac Rev-erb regulates the diurnal rhythm of energy metabolism. DEGs and differentially expressed proteins correlate with each other at variable degrees (Figure 3Q and Figure S7A–S7G). Approximately 40% to 50% of proteins showed differential expression at the protein level but not at the RNA level. Conversely, 70% to 80% of protein-coding genes showed differential expression at the RNA level but not at the protein level (Figure 3R, Figure S7D and S7G). Despite such discrepancy, metabolic enzymes show consistent differential expression at both RNA and protein levels (Figure 3S). FAO enzymes were more prominently downregulated in KO versus WT hearts in the light cycle than in the dark cycle, whereas enzymes involved in carbohydrate metabolism were upregulated (Figure 3S). These results support that cardiac Rev-erb promotes FAO while repressing glucose metabolism.
Cardiac Rev-erb Regulates the Diurnal Rhythm of Myocardial Metabolism
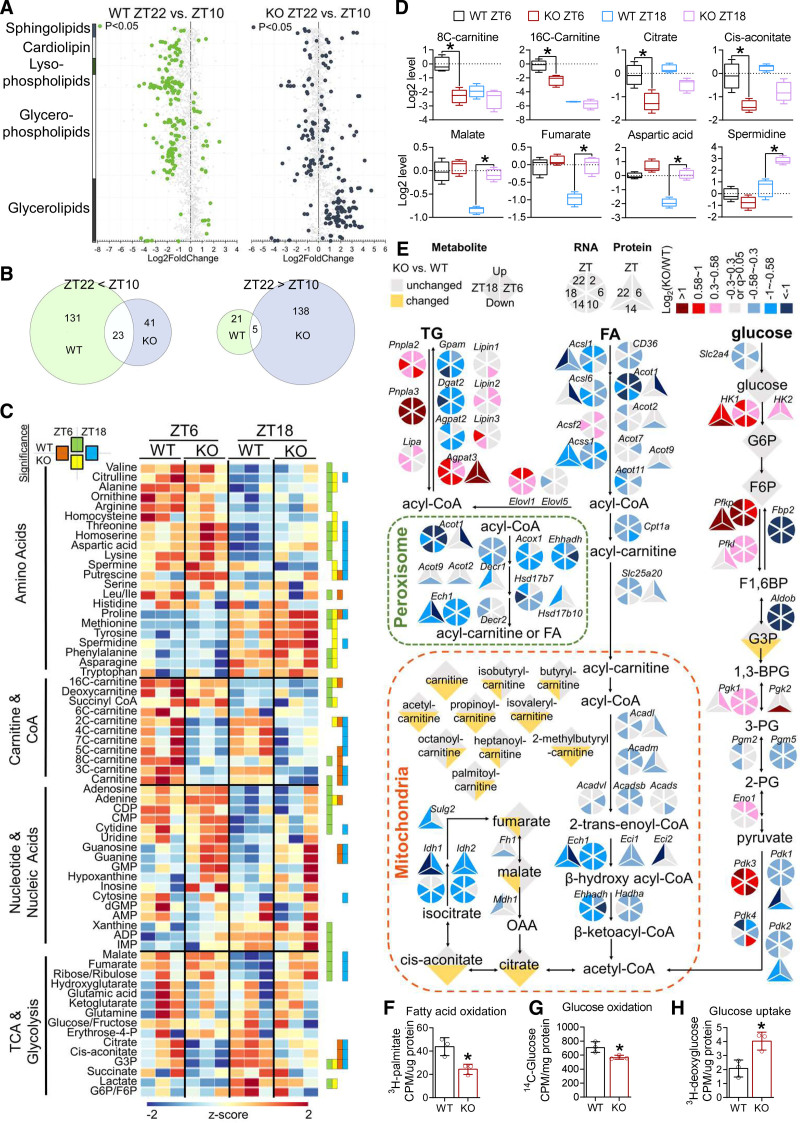
Lipidomic analysis at 3 months of age identified that 180 lipid species, mostly cardiolipin and glycerophospholipids, displayed robust diurnal rhythm in WT hearts (Figure 4A). Only 28 lipid species retained the same rhythmicity in the KO heart (Figure 4A and 4B). In addition to the loss of rhythmicity, the KO heart also showed a remarkable gain of rhythmicity for >100 lipid species, mostly glycerolipids (Figure 4A and 4B). This led to an overall flip of the rhythmicity in KO versus WT hearts. Most oscillatory lipids in WT hearts had higher content at ZT10 than at ZT22, whereas the majority of oscillatory lipids in KO hearts were higher at ZT22 than at ZT10 (Figure 4B). As a result, several lipid species, in particular, cardiolipin and glycerophospholipids, were higher in KO versus WT hearts at ZT22 (Figure S7H). These results suggest that cardiolipin and glycerophospholipid metabolism in the WT heart is more active in the light cycle than in the dark cycle, and such a diurnal rhythm was dependent on cardiac Rev-erb. Rev-erb ablation caused a gain of rhythmicity for glycerolipid metabolism that peaked in the dark cycle. Because glycerolipids constitute a major class of dietary lipids, these changes may be an outcome of substrate competition attributable to overreliance on carbohydrate metabolism.
Figure 4.
Cardiac Rev-erb regulates the diurnal rhythm of myocardial oxidative metabolism. A, Cardiac lipid species with differential levels for the indicated comparisons, n=3 mice for each genotype at indicated ZT. B, The number of lipid species with a significant difference for the indicated comparisons. C, The relative abundance of metabolites in the WT and KO heart. Significant differences for the indicated comparisons (adjusted P values for t test at an false discovery rate (Benjamini Hochberg method) threshold of <0.25) were indicated by green (WT_ZT6 vs WT_ZT18), yellow (KO_ZT6 vs KO_ZT18), orange (WT_ZT6 vs KO_ZT6), and blue (WT_ZT18 vs KO_ZT18), n=3 mice for each genotype at indicated ZT. D, The relative abundance of metabolites in the WT and KO hearts. y axis shows the log2 conversion of the relative abundance after normalization to the average relative abundance of WT at ZT6. Box plots center lines, limits, and whiskers represent the median, quartile, and minimum/maximum values, respectively; n=3 mice for each group. *Adjusted P values for t test at a false discovery rate (Benjamini Hochberg method) threshold of <0.25 between 2 groups. E, Metabolic pathways with highlighted genes, proteins, and metabolites that were altered in the KO vs WT hearts. F, Fatty acid oxidation rate in adult mouse cardiomyocytes isolated from 2-month-old male mice, n=3 mice. G, Glucose oxidation rate in adult mouse cardiomyocytes isolated from 2-month-old male mice, n=3 mice. H, Glucose uptake rate in adult mouse cardiomyocytes isolated from 2-month-old male mice, n=3 mice. Data are mean±SEM. *P<0.05 between groups by 2-sided t test unless otherwise stated. See Table S6 for statistical details. CoA indicates coenzyme A; 1,3BPG, 1,3-bisphospoglyceric acid; F1,6BP, fructose 1,6-bisphosphate; F6P, fructose 6-phosphate; FA, fatty acids; G3P, glyceraldehyde 3-phosphate; G6P, glucose 6-phosphate; KO, knockout; OAA, oxaloacetate; 2PG, 2-phosphoglyceric acid; 3PG, 3-phosphoglyceric acid; TCA, tricarboxylic acid cycle; TG, triglycerides; WT, wild type; and ZT, Zeitgeber time.
Metabolomics analysis at 10 weeks of age showed that WT hearts did not display much oscillation for short-chain acyl-carnitines and tricarboxylic acid cycle intermediates (Figure 4C). In contrast, many amino acids and nucleotides show oscillation in WT hearts with higher levels in the light/fasting cycle. About half of the detected metabolites were changed in KO versus WT hearts at either ZT6 or ZT18. The KO/WT differences were more prominent at ZT6 than ZT18 for long-chain acyl-carnitines and tricarboxylic acid cycle metabolites immediately downstream of acetyl coenzyme A, such as citrate and aconitate (Figure 4C and 4D), which is consistent with the more prominent impairment of FAO in the light cycle than in the dark cycle. The KO/WT differences were more prominent at ZT18 than ZT6 for tricarboxylic acid cycle intermediates closely related to carbohydrate metabolism, such as malate and fumarate, and some amino acids and polyamines, as well (Figure 4C and 4D), in line with the increased carbohydrate metabolism in KO versus WT hearts in the dark cycle. Integrated analysis of the multi-omics data suggests an overall reduction in oxidative lipid metabolism in the KO versus WT hearts, in particular, in the light cycle, and a reciprocal increase in glycolysis, in particular, in the dark cycle (Figure 4E). Mitochondrial DNA copy numbers and protein content of oxidative phosphorylation complexes remained largely unchanged between KO and WT heart at 10 weeks of age (Figure S8A and S8B), whereas the respiratory enzyme activity of succinate dehydrogenase showed a trend of reduction in KO hearts (Figure S8C), suggesting that Rev-erb regulates more transient changes in mitochondrial metabolic pathways, although mitochondrial content or biogenesis might be secondarily affected. Primary cardiomyocytes isolated from 2-month-old mice (Figure S8D) showed a reduction in FAO and glucose oxidation (Figure 4F and 4G) and an increase in glucose uptake (Figure 4H). The oxygen consumption capacity of the AC16 cardiomyocytes was consistently reduced after Rev-erb knockdown (Figure S8E). These results demonstrate that Rev-erb depletion impaired oxidative metabolism and caused metabolic inflexibility before contractile dysfunctions.
Dietary Lipid Alone Does Not Affect Cardiac Dysfunctions in KO Mice
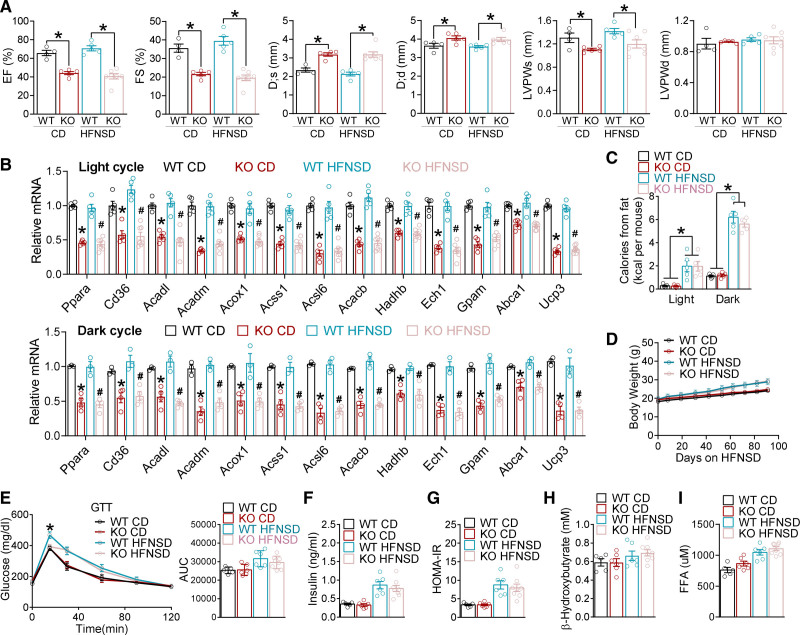
If the impaired FAO is the cause of the distorted fuel preference toward carbohydrates and the subsequent heart failure in KO mice, it might be possible to alleviate the contractile dysfunction by boosting FAO capacity to relieve its overreliance on glycolysis. Because high-fat diets can upregulate PPARα (peroxisome proliferator-activated receptor alpha)-associated FAO genes in the heart and uncouple cardiac glycolysis from glucose oxidation,50,51 we sought to test the hypothesis by feeding mice a high-fat no-sucrose diet (HFNSD; Table S2) starting at 8 weeks of age. Contrary to what we expected, echocardiography at 5 months of age did not reveal improvement in cardiac functions (Figure 5A and Figure S9A). HFNSD did not alter the expression of lipid metabolic genes or proteins in KO hearts (Figure 5B and Figure S9B), although it increased dietary lipid intake in the dark cycle compared with the chow diet (Figure 5C). In addition to dietary lipids in the dark cycle, adipose lipolysis releases free fatty acids (FFAs) as another major source of lipids for the heart, especially in the light cycle when insulin is low. HFNSD caused only a mild increase in body weight (Figure 5D), glucose intolerance (Figure 5E), baseline insulin levels (Figure 5F), and the Homeostatic Model Assessment index of insulin resistance (Figure 5G) compared with chow diet, without affecting the serum ketone (Figure 5H) or FFA levels (Figure 5I) in both WT and KO mice. The slight improvement of glucose tolerance in KO versus WT mice on HFNSD is consistent with the overreliance on glucose utilization. These data demonstrate that increasing dietary lipid supply in the dark cycle by itself does not rescue the cardiac dysfunctions in KO mice.
Figure 5.
HFNSD does not affect cardiac dysfunctions in KO mice. A, Echocardiography analysis in 5-month-old female mice on chow diet (CD) or high-fat no-sucrose diet (HFNSD), n≥4 mice. B, quantitative reverse transcription polymerase chain reaction analysis in the hearts of WT and KO female mice on CD or HFNSD, n≥5 mice. Mice were harvested at light cycle (ZT6–9) or dark cycle (ZT18–21). *P<0.05 between WT_CD and KO_CD by 2-sided t test. #P<0.05 between WT_HFNSD and KO_HFNSD 2 two-sided t test. C, Calories intake from dietary lipids during light and dark cycles in female mice fed with CD or HFNSD, n=5 mice. *P<0.05 between CD and HFNSD by 2-sided t test for both genotypes. D, Body weight gain in female mice fed with CD or HFNSD, n≥6 mice. HFNSD started at 8 weeks of age. E, Glucose tolerance test (GTT) and the area under the curve (AUC) for GTT at 5 months of age, n=5 mice for WT_CD, n=6 mice for KO_CD and WT_HFNSD, n=8 for KO_HFNSD. *P<0.05 between WT_HFNSD and KO_HFNSD by 2-sided t test. F and G, Basal insulin levels for insulin and the relative Homeostatic Model Assessment for Insulin Resistance (HOMA-IR) at 5 months of age, n≥5 mice. H and I, Blood ketone or free fatty acids (FFA) levels at 5 months of age in the light cycle, n≥5 mice. Each dot represents an individual mouse. Data are mean±SEM. *P<0.05 between groups by 2-sided t test unless otherwise stated. See Table S6 for statistical details. D;d indicates left ventricular dimension at end-diastole; D;s, left ventricular dimension at end-systole; EF, ejection fraction; FS, fractional shortening; KO, knockout; LVPWd, left ventricular posterior wall thickness at end-diastole; LVPWs, left ventricular posterior wall thickness at end-systole; and WT, wild type.
Obesity and Insulin Resistance Delay Cardiac Dysfunctions in KO Mice
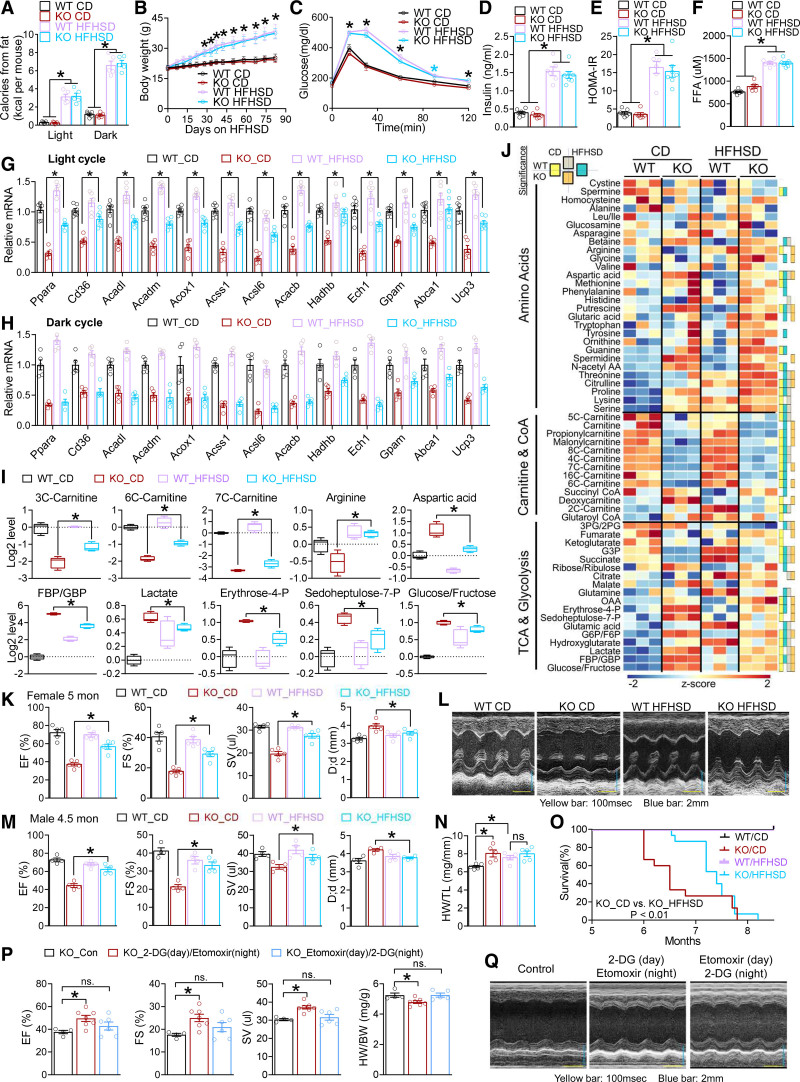
We sought to increase lipid availability in the light cycle by promoting robust adiposity and systemic insulin resistance so that FFA availability from adipose lipolysis in the light cycle can be potentiated. A high-fat high-sucrose diet (HFHSD; Table S2) not only increased dietary lipids in the dark cycle (Figure 6A and Figure S10A), but also caused >10 g body weight gain after 3 months compared with HFNSD (Figures 6B and 5D). HFHSD also led to more severe glucose intolerance (Figure 6C and Figure S10B), higher baseline insulin levels (Figure 6D), higher Homeostatic Model Assessment index of insulin resistance (Figure 6E), and higher FFAs in the light cycle (Figure 6F) in both WT and KO mice compared with HFNSD (Figure 5E–5I), without altering blood ketone levels (Figure S10C). It is remarkable that the downregulated fatty acid metabolic genes (KO versus WT) and proteins were rescued in part by HFHSD in the light cycle (Figure 6G and Figure S10D), but not in the dark cycle (Figure 6H). These results suggest that increased FFA availability in the light cycle upregulates the expression of lipid metabolic genes, probably through lipid-sensing signaling pathways. Metabolomics analysis at 4.5 months of age showed lower carnitine and coenzyme A species in KO versus WT hearts and a reciprocal higher glycolytic intermediates and amino acids on chow diet (Figure 6I), in line with the impaired FAO of KO hearts and overreliance on glucose and amino acids for bioenergetics. All these KO/WT differences on chow diet were ameliorated by HFHSD (Figure 6I and 6J). These data suggest that providing more FFAs for myocardial oxidative metabolism in the light cycle can in part rescue the impaired oxidative metabolism in the KO heart and relieve the overreliance on carbohydrate and amino acids for bioenergetics.
Figure 6.
HFHSD rescues cardiac dysfunctions in Rev-CKO mice. A, Calorie intake from dietary lipids during light and dark cycles in female mice fed with CD or high-fat high-sucrose diet (HFHSD), n=5 mice. B, Body weight of female mice. HFHSD started at 8 weeks of age, n≥5 mice. C, Glucose tolerance test (GTT) at 5 months of age, n=8 mice for WT_CD, n= mice for KO_CD and WT_HFHSD, n=8 for KO_HFHSD. *(blue) P<0.05 between CD and HFHSD in KO mice. D and E, Basal insulin levels and the relative Homeostatic Model Assessment for Insulin Resistance (HOMA-IR) in female mice at 5 months of age, n≥6 mice. F, Blood free fatty acid (FFA) levels in female mice at 5 months of age in the light cycle, n ≥ 6 mice. *P<0.05 between CD and HFHSD by 2-sided t test for both genotypes for A through F. G and H, quantitative reverse transcription polymerase chain reaction analysis of the hearts harvested at ZT6–9 (light cycle) or ZT18–21 (dark cycle) from WT and KO female mice on HFHSD at 5 months of age, n≥6 mice. HFHSD started at 8 weeks of age. I and J, Metabolomics analysis in the cardiac ventricles of WT and KO male mice at 4.5 months of age, n=3 mice. Mice were harvested at ZT6. HFHSD started at 8 weeks of age. Color boxes on the right represent significant differences (adjusted P values for t test at a false discovery rate [Benjamini Hochberg method] threshold of <0.25) for any 2-group comparisons as indicated. Box plots center lines, limits, and whiskers represent the median, quartile, and minimum/maximum values, respectively. *Adjusted P values for t test at an false discovery rate (Benjamini Hochberg method) threshold of <0.25 between 2 indicated groups. K and L, Echocardiography analysis (n=5 mice) and representative M mode images in female mice at 5 months of age. HFHSD started at 8 weeks of age. M, Echocardiography analysis of male mice at 4.5 months of age on HFHSD, n=4 mice. HFHSD started at 8 weeks of age. N, Heart weight (HW) to tibia length (TL) ratio in male mice at 5 months of age, n=5 mice. O, Survival curve, female mice, n=15, analyzed with Kolmogorov-Smirnov test. P and Q, Echocardiography analysis (n=4 mice for KO_Con, n=7 mice for KO_2-DG (day)/etomoxir (night), n= 6 mice for KO_Etomoxir (day)/2-DG (night), the HW/TL ratio, and representative M mode images of male KO mice at 5 months of age after chronic treatment of 2-DG and etomoxir at the indicated time of the day. The sample size is indicated in the figure unless otherwise stated. Each dot represents an individual mouse. Data are mean±SEM. *P<0.05 between groups by 2-sided t test. See Table S6 for statistical details. BW indicates body weight; CD, chow diet; CoA, coenzyme A; D;d, left ventricular dimension at end-diastole; 2-DG, 2-deoxyglucose; EF, ejection fraction; F6P, fructose 6-phosphate; FBP, d-fructose 1,6-bisphosphate; FS, fractional shortening; GBP, d-glucitol 1,6-bisphosphate; G3P, glyceraldehyde 3-phosphate; G6P, glucose 6-phosphate; KO, knockout; OAA, oxaloacetate; 2PG, 2-phosphoglyceric acid; 3PG, 3-phosphoglyceric acid; SV, stroke volume; TCA, tricarboxylic acid cycle; and WT, wild type.
The ejection fraction and ventricle dilation of KO mice was alleviated by HFHSD at 5 months of age (Figure 6K and 6L and Figure S10E). Similar results were obtained in both sexes (Figure 6M and Figure S10F). HFHSD also masked the heart weight differences between WT and KO hearts (Figure 6N). The HFHSD-mediated alleviation was temporary. By 7 months of age, hearts from HFHSD-fed KO mice still showed enlarged hearts with atrial mural thrombus (Figure S10G and S10H). The rapid precipitation of cardiac dysfunction may be attributable to obesity-induced hemodynamic alterations or systemic inflammation as mice continue to grow more obese. Thus, adverse effects of HFHSD counteract and prevail over its beneficial effects over time. As a result, HFHSD did not alter the maximum lifespan of KO mice but significantly prolonged the median survival age of KO mice by >1 month (Figure 6O). To further address the potential role of dietary sugar, we supplemented drinking water with glucose, sucrose, or fructose. These high-sugar diets did not cause as prominent obesity as HFHSD and did not alleviate KO cardiac dysfunctions (Figure S11A–S11C), supporting the notion that dietary macronutrient itself is insufficient to rescue KO cardiac dysfunctions. The HFHSD data collectively support the insights from the multi-omics profiling and suggest that metabolic disruption contributes to heart failure in KO mice.
Time-Dependent Effects of Metabolism-Targeting Drugs
We sought to use small molecules targeting metabolism to treat heart failure in KO mice. Because many FAO enzymes were downregulated in the KO heart compared with the WT heart, direct activation of a specific step of the FAO pathway might do more harm than good because it may cause accumulation of specific lipid species at some steps of the FAO pathway. We took advantage of the reciprocal fuel competition between fatty acids and glucose, because blocking glucose utilization could activate FAO and vice versa. We chose 2-deoxyglucose, an inhibitor of glycolysis, and etomoxir, an FAO inhibitor that was tested for treating heart failure.52 We treated mice with 1 dose every 2 days with 2 different timing regimens: 2-deoxyglucose at ZT2 (day) + etomoxir at ZT14 (night) and etomoxir at ZT2 (day) + 2-deoxyglucose at ZT14 (night). After continuous treatment from 3 to 5 months of age, the first regimen ameliorated contractile dysfunctions in KO mice, whereas the second regimen did not (Figure 6P and 6Q). This result suggests that aligning the metabolic pathway activity with the substrate availability rhythm can ameliorate heart failure. Therefore, timing matters for drug therapies targeting cardiac metabolism.
Molecular Clock Chronotypes in Human Hearts With Dilated Cardiomyopathy
The cardiac dilation in Rev-CKO mice prompted us to explore whether cardiac molecular clock function is associated with the severity of cardiac dilation in humans. We performed a retrospective analysis of the clock gene expression in failing hearts from patients with DCM who received heart transplants (Table S3). Rev-erbα and Bmal1 genes are among those with the most robust diurnal expression rhythm in human hearts.14,53 The expression level of Rev-erbα in the afternoon (14:00 pm to 18:00 pm) is ≈50% of that in the morning (6:00 am to10:00 am), whereas Bmal1 has the opposite phase distribution in normal hearts (Figure S12A). Thus, the Rev-erbα/Bmal1 gene expression ratio in the heart can serve as a marker for assessing the functionality of the cardiac molecular clock. We selected failing hearts collected in the morning or afternoon for gene expression analysis, using the average of normal hearts to set the baseline. After calculating the Rev-erbα/Bmal1 expression ratio, we defined the morning samples (collected from 6:00 to 10:00 am) above the average of normal hearts (3 samples collected from 6:00 to 10:00 am, 3 samples collected from 14:00 to 18:00 pm) as a molecular chronotype (type A), whereas those below the baseline as another molecular chronotype (type B), with the reversed criteria for the afternoon samples (collected from 14:00 to 18:00 pm; Figure S12B and S12C). We have 20 patients in type A and 16 patients in type B, with type A more similar to the temporal pattern in normal hearts.
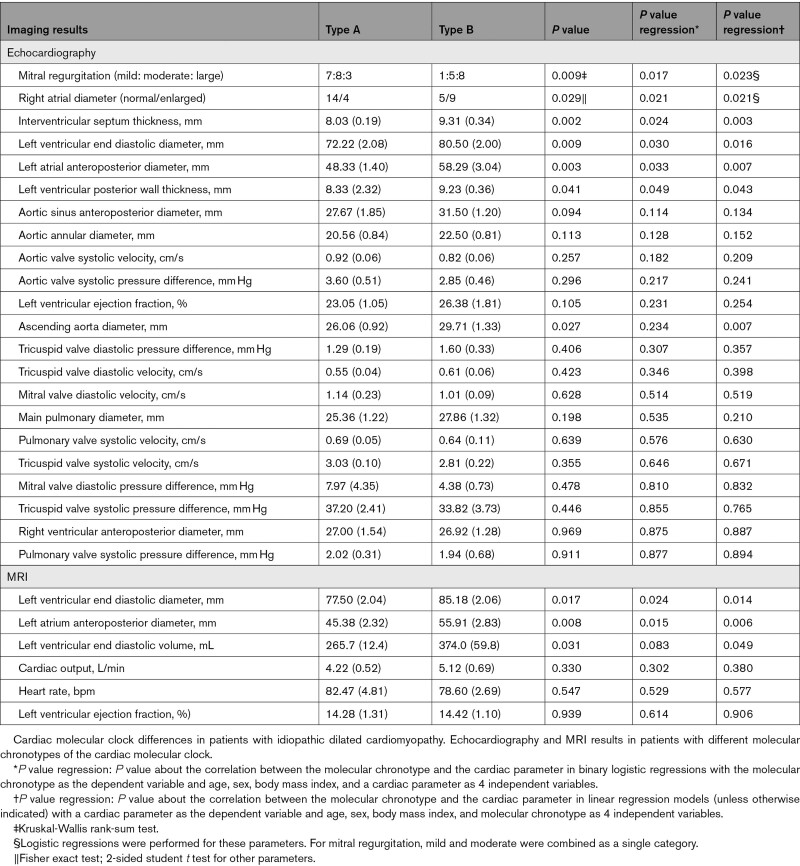
There were no significant differences between the 2 molecular chronotypes in age, sex, body mass index, or preoperative medication (Table S4). The cardiac contractile functions and most hemodynamic parameters did not show a difference (Table), which is expected from patients who received heart transplants. However, echocardiography showed that hearts with molecular chronotype B had increased left ventricular end-diastolic diameter compared with molecular chronotype A (Table). Mitral regurgitation was also more represented in the type B group, in line with the more severe ventricular dilation. These results were consistent with the MRI results, and the correlation remains significant after correcting for age, sex, and body mass index (Table) or in male patients only (Table S5). These data suggest that the cardiac molecular chronotype correlates with the severity of cardiac dilation in human DCM.
Table 1.
Cardiac Functions in Patients With Dilated Cardiomyopathy
Discussion
Our results revealed that the diurnal rhythm of many metabolites and lipid species in the heart is regulated by cardiac Rev-erb without affecting the behavioral or feeding diurnal rhythm. Compared with the WT heart, the Rev-CKO heart shows impaired FAO in the light cycle and overreliance on carbohydrates in the dark cycle. Disruption of the temporal organization of myocardial bioenergetics in the KO heart is likely a cause of progressive contractile dysfunction and lethal heart failure. This notion is supported by several lines of evidence. (1) Inducible depletion of Rev-erb in adult hearts caused similar heart failure and cardiac dilation as embryonic depletion of Rev-erb, excluding developmental defects as confounding factors for cardiac dysfunction in KO mice. (2) Nonbiased omics analysis pinpoints the metabolic process as a top altered functional pathway in KO versus WT hearts. (3) The metabolic disruption precedes contractile dysfunctions. (4) The HFHSD ameliorates contractile dysfunction and cardiac dilation in KO mice. (5) Metabolism-targeting drugs ameliorate contractile dysfunctions in KO mice in a time-dependent manner. In addition to metabolic changes, other processes could also play a role in the cardiac dysfunctions of KO hearts, including cell death, cell adhesion, sarcomere organization, or calcium signaling. Because metabolic changes and lipid species accumulation precedes systolic dysfunction, we speculate that metabolic disturbance impairs systolic function attributable to mismatched fuel preference and fuel availability. The associated lipotoxicity and perturbed protein homeostasis might contribute to inflammation, cardiac remodeling, and cardiac dilation.
In the normal physiological condition in mice, cardiac FAO gene expression is higher in the late light cycle than in the late dark cycle, which aligns with a higher baseline FFA availability in the late light cycle from elevated adipose lipolysis. The rise of the cardiac Rev-erb in the light cycle represses E4BP4 that further represses FAO genes, which accounts for the rise of FAO genes in the late light cycle. This is supported by a dampened oscillation with a constant low expression of FAO genes in the KO heart. Conversely, the glucose metabolic genes showed a more oscillatory pattern in the KO heart than in the WT heart, suggesting that the rise of Rev-erb expression in the late light cycle serves as an anticipatory mechanism to avoid overactivation of glycolytic genes in response to dietary carbohydrates. This mechanism may help maintain the lipids as the dominant myocardial fuel source throughout the day despite the robust diurnal rhythm of feeding behaviors under normal physiological conditions. Things take an interesting turn under the pathological condition of obesity and insulin resistance. Although the WT heart showed only moderate upregulation of FAO genes in the light cycle on chronic HFHSD feeding, the KO heart showed a greater degree of upregulation, suggesting that Rev-erb prevents the oscillation from being hijacked and overstimulated by HFHSD, probably by counteracting the sensitivity of lipid-sensing signaling pathways. This regulatory mechanism can be cardioprotective because the overactivation of lipid-sensing pathways can cause an imbalance between lipid uptake and lipid oxidation, which leads to heart failure.54
The alleviation of the cardiac dysfunction in the obesity condition is reminiscent of the obesity paradox. The cardioprotective effects of HFHSD are temporary, likely because prolonged HFHSD generates adverse effects as mice continue to get more obese, which is different from humans. The HFNSD provided the same surplus of dietary lipid as HFHSD but did not rescue heart failure in KO mice, suggesting that dietary lipid surplus in the feeding cycle is not cardioprotective. The 2 diets are not ketogenic and do not differ in the unsaturated fatty acid levels, suggesting that these factors do not play a role. The obvious difference between HFNSD and HFHSD is that HFHSD causes more prominent obesity and systemic insulin resistance, which promotes adipose lipolysis and the availability of FFAs to the heart. Both adiposity and insulin resistance seem important for the phenotypic rescue because the former provides a big FFA reservoir, whereas the latter promotes the lipolysis flux rate. Further supporting this notion is the temporal alignment of the FFAs availability and FAO gene expression rescue. FFAs from lipolysis are abundant in the light cycle when the insulin level is low, consistent with that HFHSD rescues FAO gene expression in the KO heart in the light cycle but not in the dark cycle. By comparison, HFNSD does not elevate FFAs drastically in the light cycle and does not rescue FAO gene expression. Considering that circadian disruption is a risk factor for heart failure, the data from Rev-erb KO mice suggest that the obesity paradox, especially in the presence of circadian disruption, could be contributed by increased cardiac FFA supply from adipose lipolysis in the fasting cycle because of insulin resistance and adiposity. Further study is warranted to determine whether this concept applies to other heart failure models.
The contractile and metabolic changes of the KO heart bear similarities and distinctions with the Bmal1 null mice55,56 or cardiomyocyte-specific Bmal1 KO (CBK) mice. Most CBK mice died at 7 to 12 months of age, with cardiac hypertrophy and ventricular fibrosis at 7 to 9 months of age.28,57 Inducible ablation of Bmal1 in cardiomyocytes caused arrhythmia.58 In comparison, most Rev-CKO mice died at 6 to 8 months of age with cardiac dilation but without obvious ventricular wall thickening or fibrosis. The CBK heart showed increased FAO and reduced glucose oxidation,28,59 whereas the Rev-CKO cardiomyocytes showed reduced oxidation of both fatty acids and glucose but increased glucose uptake. The residual function of Rev-erbα DBDm does not seem significant because the Rev-CKO mouse line displays a phenotype very similar to the double KO line without truncated DBDm.46 The reduced FAO in Rev-CKO hearts is in line with the metabolic changes in skeletal muscles of Rev-erbα whole-body KO mice,60 although Rev-CKO hearts do not show a reduction in mitochondrial DNA copy number or mitochondrial oxidative phosphorylation protein complex levels, suggesting that Rev-erb may have different functions in cardiac muscles than in skeletal muscles.
Circadian disruption is a risk factor for heart diseases. Shift work is associated with increased cardiac events. Human cardiac molecular chronotype is correlated with the severity of cardiomyopathy. The Rev-CKO mouse model further demonstrates that a disrupted cardiac clock can play a causal role in DCM and heart failure. We do not suggest that Rev-erb is the only pathway through which clock disruption can affect metabolism. However, because the Rev-CKO mice can mimic some aspects of the cause in naturally occurring human heart diseases, the amelioration of heart failure in Rev-KO mice with dietary or pharmacological interventions supports myocardial bioenergetics as a chronotherapy target in treating heart failure. The dietary intervention result suggests that it might be cardioprotective to boost FAO in the sleep cycle by supplying FFAs. However, we do not advocate using etomoxir or 2-deoxyglucose directly in humans. These prototype compounds served as a proof of concept that a chronotherapy strategy to inhibit FAO in the wake cycle and glycolysis in the sleep cycle might be beneficial for patients who have heart failure with circadian disruption.
Article Information
Acknowledgments
We thank Dr Lin at the Neuropathology Core (supported by U54HD083092) for the histology study; Drs Liu and Li at Baylor College of Medicine (BCM) for technical assistance; Drs Ait-Oudhia and Vaidya at the University of Florida for providing the AC16 cell line; Drs Wan and Liu at BCM for some of the RNA-sequencing analyses; and Dr Xiao at the University of Texas Health Science Center, Dr Wang at BCM, Dr Shi at Wenzhou Medical University, and Dr Yao at the Northern Illinois University for statistical advice. Drs Sun and Zhang conceived the study. Dr S. Song collected most data from mice and cell culture. Dr Tien performed chromatin immunoprecipitation sequencing (ChIP-seq). Dr Cui collected data from human patients. Dr J. Song supervised the human study. Dr Basil analyzed ChIP-seq and RNA-sequencing data. Dr Zhu established the cell culture system. Drs Gong, Zhu, and Cao and V. Mundra bred and maintained the mouse lines. Dr Li constructed plasmids. H. Li helped with echocardiography. Drs Fan and Xie provided consultation and assistance with cardiomyocyte isolation. Drs Choi and Jung performed proteomics analyses. Drs Luo and Chang provided consultation and training on echocardiography. Dr Xue performed some quantitative reverse transcription polymerase chain reaction validations. Dr Zhou assisted with histology analyses. A.R. Ortiz, B. Stork, and Dr York provided consultation on glucose oxidation assay. Dr Putluri supervised metabolomics and lipidomics analyses. Dr Chu provided fellowship for Dr S. Song. Drs S. Song, Tien, Cui, Basil, Choi, Jung, Zhang, and Sun analyzed the data. Dr S. Song, Cui, Zhang, and Sun interpreted the data. Drs J. Song, Zhang, and Sun obtained funding. Drs S. Song and Sun wrote the manuscript with input from the other authors.
Sources of Funding
The Proteomics Core and Metabolomics Core were supported by the Cancer Prevention and Research Institute of Texas (CPRIT) Core Facility Support Award RP210227 “Proteomic and Metabolomic Core Facility” and intramural funds from the Dan L. Duncan Comprehensive Cancer Center (DLDCCC) at Baylor College of Medicine (BCM; P30CA125123). Chromatin immunoprecipitation sequencing was done at the Genomics Core that was supported by P30DK56338, P30CA125123, and S10OD02346901. The echocardiography was done at the BCM Mouse Metabolism and Phenotyping Core that was supported by R01DK114356 and UM1HG006348. The cell sorting was done at the BCM Cytometry and Cell Sorting Core (CPRIT-RP180672, National Institutes of Health (NIH) P30CA125123, and S10RR024574). The laboratories of the authors were supported by the Shanghai Municipal Science and Technology Major Project (2017SHZDZX01; to Dr J. Song), American Diabetes Association ADA1-19-PDF-012 (to Dr Zhou), NIH grants U01CA214263 (to Dr Putluri), HL123551, HL143067 (to Dr Zhang), American Heart Association (AHA30970064), and HL153320, DK111436, AG069966, AG070687, and ES027544 (to Dr Sun). We also thank John S. Dunn Foundation, Mrs. Clifford Elder White Graham Endowed Research Fund, Cardiovascular Research Institute at BCM, the DLDCCC, the Specialized Programs of Research Excellence (SPORE) program (P50CA126752), the Gulf Coast Center for Precision Environmental Health (P30ES030285), and the Texas Medical Center Digestive Diseases Center (P30 DK056338).
Disclosures
None.
Supplemental Material
Expanded Methods
Figures S1–S12
Tables S1–S6
Supplementary Material
Nonstandard Abbreviations and Acronyms
- DCM
- dilated cardiomyopathy
- DEGs
- differentially expressed genes
- FAO
- fatty acid oxidation
- FFAs
- free fatty acids
- HFHSD
- high-fat high-sucrose diet
- HFNSD
- high-fat no-sucrose diet
- ZT
- Zeitgeber time
S. Song, C.-L. Tien, and H. Cui contributed equally.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCULATIONAHA.121.056076.
For Sources of Funding and Disclosures, see page 463.
Contributor Information
Shiyang Song, Email: fwsongjiangping@126.com.
Chih-Liang Tien, Email: iamonster@hotmail.com.
Hao Cui, Email: cuihao0087@sina.com.
Paul Basil, Email: basil@connect.hku.hk.
Ningxia Zhu, Email: znx103@163.com.
Yingyun Gong, Email: gongyingyun@126.com.
Wenbo Li, Email: huili.hl@outlook.com.
Hui Li, Email: huili.hl@outlook.com.
Qiying Fan, Email: Qiyingfan87@gmail.com.
Jong Min Choi, Email: jongmin.choi@bcm.edu.
Weijia Luo, Email: weijia@tamu.edu.
Yanfeng Xue, Email: xueyanfeng1990@163.com.
Rui Cao, Email: 13572537171@yeah.net.
Wenjun Zhou, Email: wenjunz@bcm.edu.
Andrea R. Ortiz, Email: andrea.ortiz@bcm.edu.
Brittany Stork, Email: brittany.stork@bcm.edu.
Vatsala Mundra, Email: vm18@rice.edu.
Nagireddy Putluri, Email: putluri@bcm.edu.
Brian York, Email: york@bcm.edu.
Maoping Chu, Email: chmping@126.com.
Jiang Chang, Email: jchang@ibt.tamhsc.edu.
Sung Yun Jung, Email: syjung@bcm.edu.
Liang Xie, Email: liangx@bcm.edu.
References
- 1.Thosar SS, Butler MP, Shea SA. Role of the circadian system in cardiovascular disease. J Clin Invest. 2018;128:2157–2167. doi: 10.1172/JCI80590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chellappa SL, Vujovic N, Williams JS, Scheer FAJL. Impact of circadian disruption on cardiovascular function and disease. Trends Endocrinol Metab. 2019;30:767–779. doi: 10.1016/j.tem.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peliciari-Garcia RA, Darley-Usmar V, Young ME. An overview of the emerging interface between cardiac metabolism, redox biology and the circadian clock. Free Radic Biol Med. 2018;119:75–84. doi: 10.1016/j.freeradbiomed.2018.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crnko S, Du Pré BC, Sluijter JPG, Van Laake LW. Circadian rhythms and the molecular clock in cardiovascular biology and disease. Nat Rev Cardiol. 2019;16:437–447. doi: 10.1038/s41569-019-0167-4 [DOI] [PubMed] [Google Scholar]
- 5.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006 [DOI] [PubMed] [Google Scholar]
- 7.Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science. 2016;354:994–999. doi: 10.1126/science.aah4965 [DOI] [PubMed] [Google Scholar]
- 8.Kojetin DJ, Burris TP. REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov. 2014;13:197–216. doi: 10.1038/nrd4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gloston GF, Yoo SH, Chen ZJ. Clock-Enhancing Small Molecules and Potential Applications in Chronic Diseases and Aging. Front Neurol. 2017;8:100. doi: 10.3389/fneur.2017.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alibhai FJ, LaMarre J, Reitz CJ, Tsimakouridze EV, Kroetsch JT, Bolz SS, Shulman A, Steinberg S, Burris TP, Oudit GY, et al. Disrupting the key circadian regulator CLOCK leads to age-dependent cardiovascular disease. J Mol Cell Cardiol. 2017;105:24–37. doi: 10.1016/j.yjmcc.2017.01.008 [DOI] [PubMed] [Google Scholar]
- 11.Stujanna EN, Murakoshi N, Tajiri K, Xu D, Kimura T, Qin R, Feng D, Yonebayashi S, Ogura Y, Yamagami F, et al. Rev-erb agonist improves adverse cardiac remodeling and survival in myocardial infarction through an anti-inflammatory mechanism. PLoS One. 2017;12:e0189330. doi: 10.1371/journal.pone.0189330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Zhang R, Tien CL, Chan RE, Sugi K, Fu C, Griffin AC, Shen Y, Burris TP, Liao X, et al. REV-ERBα ameliorates heart failure through transcription repression. JCI Insight. 2017;2:95177. doi: 10.1172/jci.insight.95177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reitz CJ, Alibhai FJ, Khatua TN, Rasouli M, Bridle BW, Burris TP, Martino TA. SR9009 administered for one day after myocardial ischemia-reperfusion prevents heart failure in mice by targeting the cardiac inflammasome. Commun Biol. 2019;2:353. doi: 10.1038/s42003-019-0595-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montaigne D, Marechal X, Modine T, Coisne A, Mouton S, Fayad G, Ninni S, Klein C, Ortmans S, Seunes C, et al. Daytime variation of perioperative myocardial injury in cardiac surgery and its prevention by Rev-Erbα antagonism: a single-centre propensity-matched cohort study and a randomised study. Lancet. 2018;391:59–69. doi: 10.1016/S0140-6736(17)32132-3 [DOI] [PubMed] [Google Scholar]
- 15.Abel ED, Litwin SE, Sweeney G. Cardiac remodeling in obesity. Physiol Rev. 2008;88:389–419. doi: 10.1152/physrev.00017.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horwich TB, Fonarow GC, Clark AL. Obesity and the obesity paradox in heart failure. Prog Cardiovasc Dis. 2018;61:151–156. doi: 10.1016/j.pcad.2018.05.005 [DOI] [PubMed] [Google Scholar]
- 17.Stanley WC, Dabkowski ER, Ribeiro RF, Jr, O’Connell KA. Dietary fat and heart failure: moving from lipotoxicity to lipoprotection. Circ Res. 2012;110:764–776. doi: 10.1161/CIRCRESAHA.111.253104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riehle C, Bauersachs J. Of mice and men: models and mechanisms of diabetic cardiomyopathy. Basic Res Cardiol. 2018;114:2. doi: 10.1007/s00395-018-0711-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE, et al. Gene regulation. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science. 2015;348:1488–1492. doi: 10.1126/science.aab3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agah R, Frenkel PA, French BA, Michael LH, Overbeek PA, Schneider MD. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100:169–179. doi: 10.1172/JCI119509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, Penninger JM, Molkentin JD. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89:20–25. doi: 10.1161/hh1301.092687 [DOI] [PubMed] [Google Scholar]
- 23.Judd J, Lovas J, Huang GN. Isolation, culture and transduction of adult mouse cardiomyocytes. J Vis Exp. 2016;114:54012. doi: 10.3791/54012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alam P, Maliken BD, Ivey MJ, Jones SM, Kanisicak O. Isolation, transfection, and long-term culture of adult mouse and rat cardiomyocytes. J Vis Exp. 2020;164:10.3791/61073. doi: 10.3791/61073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith AM, Qualls JE, O’Brien K, Balouzian L, Johnson PF, Schultz-Cherry S, Smale ST, Murray PJ. A distal enhancer in Il12b is the target of transcriptional repression by the STAT3 pathway and requires the basic leucine zipper (B-ZIP) protein NFIL3. J Biol Chem. 2011;286:23582–23590. doi: 10.1074/jbc.M111.249235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong S, Zhou W, Fang B, Lu W, Loro E, Damle M, Ding G, Jager J, Zhang S, Zhang Y, et al. Dissociation of muscle insulin sensitivity from exercise endurance in mice by HDAC3 depletion. Nat Med. 2017;23:223–234. doi: 10.1038/nm.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akie TE, Cooper MP. Determination of fatty acid oxidation and lipogenesis in mouse primary hepatocytes. J Vis Exp. 2015:e52982. doi: 10.3791/52982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young ME, Brewer RA, Peliciari-Garcia RA, Collins HE, He L, Birky TL, Peden BW, Thompson EG, Ammons BJ, Bray MS, et al. Cardiomyocyte-specific BMAL1 plays critical roles in metabolism, signaling, and maintenance of contractile function of the heart. J Biol Rhythms. 2014;29:257–276. doi: 10.1177/0748730414543141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beytebiere JR, Trott AJ, Greenwell BJ, Osborne CA, Vitet H, Spence J, Yoo SH, Chen Z, Takahashi JS, Ghaffari N, et al. Tissue-specific BMAL1 cistromes reveal that rhythmic transcription is associated with rhythmic enhancer-enhancer interactions. Genes Dev. 2019;33:294–309. doi: 10.1101/gad.322198.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung SY, Choi JM, Rousseaux MW, Malovannaya A, Kim JJ, Kutzera J, Wang Y, Huang Y, Zhu W, Maity S, et al. An anatomically resolved mouse brain proteome reveals Parkinson disease-relevant pathways. Mol Cell Proteomics. 2017;16:581–593. doi: 10.1074/mcp.M116.061440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saltzman AB, Leng M, Bhatt B, Singh P, Chan DW, Dobrolecki L, Chandrasekaran H, Choi JM, Jain A, Jung SY, et al. gpGrouper: a peptide grouping algorithm for gene-centric inference and quantitation of bottom-up proteomics data. Mol Cell Proteomics. 2018;17:2270–2283. doi: 10.1074/mcp.TIR118.000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piyarathna DWB, Rajendiran TM, Putluri V, Vantaku V, Soni T, von Rundstedt FC, Donepudi SR, Jin F, Maity S, Ambati CR, et al. Distinct lipidomic landscapes associated with clinical stages of urothelial cancer of the bladder. Eur Urol Focus. 2018;4:907–915. doi: 10.1016/j.euf.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purwaha P, Gu F, Piyarathna DWB, Rajendiran T, Ravindran A, Omilian AR, Jiralerspong S, Das G, Morrison C, Ambrosone C, et al. Unbiased lipidomic profiling of triple-negative breast cancer tissues reveals the association of sphingomyelin levels with patient disease-free survival. Metabolites. 2018;8:E41. doi: 10.3390/metabo8030041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vantaku V, Dong J, Ambati CR, Perera D, Donepudi SR, Amara CS, Putluri V, Ravi SS, Robertson MJ, Piyarathna DWB, et al. Multi-omics integration analysis robustly predicts high-grade patient survival and identifies CPT1B effect on fatty acid metabolism in bladder cancer. Clin Cancer Res. 2019;25:3689–3701. doi: 10.1158/1078-0432.CCR-18-1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kornberg MD, Bhargava P, Kim PM, Putluri V, Snowman AM, Putluri N, Calabresi PA, Snyder SH. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science. 2018;360:449–453. doi: 10.1126/science.aan4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vantaku V, Putluri V, Bader DA, Maity S, Ma J, Arnold JM, Rajapakshe K, Donepudi SR, von Rundstedt FC, Devarakonda V, et al. Epigenetic loss of AOX1 expression via EZH2 leads to metabolic deregulations and promotes bladder cancer progression. Oncogene. 2020;39:6265–6285. doi: 10.1038/s41388-019-0902-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mestroni L, Maisch B, McKenna WJ, Schwartz K, Charron P, Rocco C, Tesson F, Richter A, Wilke A, Komajda M. Guidelines for the study of familial dilated cardiomyopathies. Collaborative Research Group of the European Human and Capital Mobility Project on Familial Dilated Cardiomyopathy. Eur Heart J. 1999;20:93–102. doi: 10.1053/euhj.1998.1145 [DOI] [PubMed] [Google Scholar]
- 38.Branch of Organ Transplantation of Chinese Medical Association. Branch of Organ Transplantation of Chinese Medical Association Process and specification of Chinese donation after citizen′s death (2019 edition). ORGAN TRANSPLANTATION. 2019;10:122–127. doi: 10.3969/j.issn.1674-7445.2019.02.003 [Google Scholar]
- 39.Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin L, Joshi S, Wu N, Tong X, Lazar MA. E3 ligases Arf-bp1 and Pam mediate lithium-stimulated degradation of the circadian heme receptor Rev-erb alpha. Proc Natl Acad Sci U S A. 2010;107:11614–11619. doi: 10.1073/pnas.1000438107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chini CC, Escande C, Nin V, Chini EN. DBC1 (Deleted in Breast Cancer 1) modulates the stability and function of the nuclear receptor Rev-erbα. Biochem J. 2013;451:453–461. doi: 10.1042/BJ20121085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao X, Hirota T, Han X, Cho H, Chong LW, Lamia K, Liu S, Atkins AR, Banayo E, Liddle C, et al. Circadian amplitude regulation via FBXW7-targeted REV-ERBα degradation. Cell. 2016;165:1644–1657. doi: 10.1016/j.cell.2016.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adelmant G, Bègue A, Stéhelin D, Laudet V. A functional Rev-erb alpha responsive element located in the human Rev-erb alpha promoter mediates a repressing activity. Proc Natl Acad Sci U S A. 1996;93:3553–3558. doi: 10.1073/pnas.93.8.3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou P, Pu WT. Recounting cardiac cellular composition. Circ Res. 2016;118:368–370. doi: 10.1161/CIRCRESAHA.116.308139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harding HP, Lazar MA. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol Cell Biol. 1995;15:4791–4802. doi: 10.1128/MCB.15.9.4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dierickx P, Zhu K, Carpenter BJ, Jiang C, Vermunt MW, Xiao Y, Luongo T, Yamamoto T, Martí-Pàmies I, Mia S.Circadian REV-ERBs repress E4bp4 to activate NAMPT-dependent NAD+ biosynthesis and sustain cardiac function. Nat Cardiovasc Res. Published online December 23, 2021. doi: 10.1038/s44161-021-00001-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duez H, van der Veen JN, Duhem C, Pourcet B, Touvier T, Fontaine C, Derudas B, Baugé E, Havinga R, Bloks VW, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology. 2008;135:689–698. doi: 10.1053/j.gastro.2008.05.035 [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Kuang Z, Yu X, Ruhn KA, Kubo M, Hooper LV. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science. 2017;357:912–916. doi: 10.1126/science.aan0677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang M, Zhang D, Zhao Z, Sit J, Saint-Sume M, Shabandri O, Zhang K, Yin L, Tong X. Hepatic E4BP4 induction promotes lipid accumulation by suppressing AMPK signaling in response to chemical or diet-induced ER stress. FASEB J. 2020;34:13533–13547. doi: 10.1096/fj.201903292RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chess DJ, Lei B, Hoit BD, Azimzadeh AM, Stanley WC. Effects of a high saturated fat diet on cardiac hypertrophy and dysfunction in response to pressure overload. J Card Fail. 2008;14:82–88. doi: 10.1016/j.cardfail.2007.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inserte J, Aluja D, Barba I, Ruiz-Meana M, Miró E, Poncelas M, Vilardosa Ú, Castellano J, Garcia-Dorado D. High-fat diet improves tolerance to myocardial ischemia by delaying normalization of intracellular PH at reperfusion. J Mol Cell Cardiol. 2019;133:164–173. doi: 10.1016/j.yjmcc.2019.06.001 [DOI] [PubMed] [Google Scholar]
- 52.Holubarsch CJ, Rohrbach M, Karrasch M, Boehm E, Polonski L, Ponikowski P, Rhein S. A double-blind randomized multicentre clinical trial to evaluate the efficacy and safety of two doses of etomoxir in comparison with placebo in patients with moderate congestive heart failure: the ERGO (etomoxir for the recovery of glucose oxidation) study. Clin Sci (Lond). 2007;113:205–212. doi: 10.1042/CS20060307 [DOI] [PubMed] [Google Scholar]
- 53.McTiernan CF, Lemster BH, Bedi KC, Margulies KB, Moravec CS, Hsieh PN, Shusterman V, Saba S. Circadian pattern of ion channel gene expression in failing human hearts. Circ Arrhythm Electrophysiol. 2021;14:e009254. doi: 10.1161/CIRCEP.120.009254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finck BN, Han X, Courtois M, Aimond F, Nerbonne JM, Kovacs A, Gross RW, Kelly DP. A critical role for PPARalpha-mediated lipotoxicity in the pathogenesis of diabetic cardiomyopathy: modulation by dietary fat content. Proc Natl Acad Sci U S A. 2003;100:1226–1231. doi: 10.1073/pnas.0336724100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lefta M, Campbell KS, Feng HZ, Jin JP, Esser KA. Development of dilated cardiomyopathy in Bmal1-deficient mice. Am J Physiol Heart Circ Physiol. 2012;303:H475–H485. doi: 10.1152/ajpheart.00238.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kohsaka A, Das P, Hashimoto I, Nakao T, Deguchi Y, Gouraud SS, Waki H, Muragaki Y, Maeda M. The circadian clock maintains cardiac function by regulating mitochondrial metabolism in mice. PLoS One. 2014;9:e112811. doi: 10.1371/journal.pone.0112811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ingle KA, Kain V, Goel M, Prabhu SD, Young ME, Halade GV. Cardiomyocyte-specific Bmal1 deletion in mice triggers diastolic dysfunction, extracellular matrix response, and impaired resolution of inflammation. Am J Physiol Heart Circ Physiol. 2015;309:H1827–H1836. doi: 10.1152/ajpheart.00608.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schroder EA, Lefta M, Zhang X, Bartos DC, Feng HZ, Zhao Y, Patwardhan A, Jin JP, Esser KA, Delisle BP. The cardiomyocyte molecular clock, regulation of Scn5a, and arrhythmia susceptibility. Am J Physiol Cell Physiol. 2013;304:C954–C965. doi: 10.1152/ajpcell.00383.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mia S, Kane MS, Latimer MN, Reitz CJ, Sonkar R, Benavides GA, Smith SR, Frank SJ, Martino TA, Zhang J, et al. Differential effects of REV-ERBα/β agonism on cardiac gene expression, metabolism, and contractile function in a mouse model of circadian disruption. Am J Physiol Heart Circ Physiol. 2020;318:H1487–H1508. doi: 10.1152/ajpheart.00709.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woldt E, Sebti Y, Solt LA, Duhem C, Lancel S, Eeckhoute J, Hesselink MK, Paquet C, Delhaye S, Shin Y, et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med. 2013;19:1039–1046. doi: 10.1038/nm.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing (GSE152372) and chromatin immunoprecipitation sequencing (GSE153150) data are available in GEO. Proteomics data are available in the MassIVE repository (MSV MSV000088075). Metabolomics and lipidomics data are available in National Metabolomics Data Repository, Project ID: PR001208, Project DOI: 10.21228/M8JX2X, Study ID: ST001915, ST001916, and ST001917. The data are available from the corresponding author on reasonable request.