Abstract
A collection of Campylobacter fetus strains, including both C. fetus subsp. fetus and C. fetus subsp. venerealis, were phenotypically identified to the subspecies level and genotypically typed by PCR and amplified fragment length polymorphism (AFLP) analysis. Phenotypic subspecies determination methods were unreliable. Genotyping of the strains by PCR and AFLP showed a clear discrimination between the two subspecies.
Campylobacter fetus can cause disease in both animals and humans. This species is divided into C. fetus subspecies fetus and C. fetus subspecies venerealis on the basis of biochemical differences (16). C. fetus subsp. venerealis appears to have a restricted host species specificity and may cause fertility problems in cows. C. fetus subsp. fetus, on the other hand, is commonly recovered from the intestinal tract of a number of animal species and may cause abortion and infertility in sheep and cattle. It can also cause serious systemic disease in humans (13). Testing for bovine C. fetus infection and subtyping of isolates are statutory requirements for the import and export of bovine semen and embryos (1). Subspecies differentiation of C. fetus is generally done on the basis of growth in the presence of 1% glycine (12). However, glycine tolerance can be mediated by phages (2), and differences in the glycine tolerance of a C. fetus strain have been described (7). Other tests, such as selenite reduction and cefoperazone resistance, are considered only indicative (9, 15). Therefore, the phenotypic assays, on which the discrimination between these two subspecies is based, are considered to be poorly robust.
Genotyping techniques have been successfully developed for the genus Campylobacter (17). Recently, the use of amplified fragment length polymorphism (AFLP) analysis for genotyping C. jejuni and C. coli has been described (5, 6). The discriminatory power of this technology is comparable to that of pulsed-field gel electrophoresis (3). The aim of this study was to determine the value of AFLP typing, biochemical typing, and typing by PCR (8) for subtyping C. fetus.
Bacteriology.
Sixty-nine bacterial strains from three geographical regions were grown at 37°C under microaerobic conditions (6% O2, 7% CO2, 7% H2, 80% N2). For discrimination between C. fetus subsp. fetus and C. fetus subsp. venerealis, growth in the presence of 1% (wt/vol) glycine was determined. Forty-seven of the strains were typed as C. fetus subsp. fetus, and 22 were typed as C. fetus subsp. venerealis (Table 1). However, for eight strains, the results of the glycine test were not consistently reproducible and were difficult to interpret. Difficulties associated with phenotypic tests for differentiating between C. fetus subsp. venerealis and C. fetus subsp. fetus are well recognized (9, 10). As the biochemical tests are unpredictable, alternative methods of subspecies differentiation have been investigated.
TABLE 1.
Campylobacter fetus strain information and typing results
| Code | Sourceg | Yr | Resultsh for:
|
|||
|---|---|---|---|---|---|---|
| Biochemical test | PCR | AFLP | ||||
| 10354 (ATCC 19438) | B | 1962 | V | V | V | |
| 10842 (ATCC 27374) | O | 1972 | F | F | F | |
| IZ 2149-90a | B | 1999 | F | F | F | |
| IZ 2149-80a | B | 1999 | F | F | F | |
| 497779a | O | 1998 | F | F | F | |
| 501340a | B | 1999 | F | F | F | |
| 930075a | B | 1999 | F | F | F | |
| SZ-10941b | O | 1999 | F | F | F | |
| SZ-1074b | O | 1999 | F | F | F | |
| 5.5.21b | ?i | ≤1993j | V | V | V | |
| 5.5.22b | ? | ≤1993 | V | V | V | |
| 5.5.41b | ? | 1992 | F | F | F | |
| 5.5.42b | O | 1994 | F | F | F | |
| 5.5.43b | O | 1994 | F | F | F | |
| 5.5.47b | O | 1996 | F | F | F | |
| 5.5.48b | O | 1996 | F | F | F | |
| 481b | O | 1999 | F | F | F | |
| SZ 599-2bb | O | 1999 | F | F | F | |
| 601b | O | 1999 | F | F | F | |
| 602b | O | 1999 | F | F | F | |
| 18156b | B | 1984 | V | V | V | |
| 44168b | B | 1984 | V | V | V | |
| 492116b | B | 1998 | F | F | F | |
| 98/v156c | B | 1998 | F | F | F | |
| 98/v444c | B | 1998 | F | F | F | |
| 98/v445c | B | 1998 | F | V | F | |
| 98/v635c | B | 1998 | F | F | F | |
| V51/99c | B | 1999 | F | F | F | |
| XV-98c | O | 1998 | F | F | F | |
| 2RV-98c | O | 1998 | F | F | F | |
| 8F-98c | O | 1998 | F | F | F | |
| BT 36-98c | B | 1998 | F | F | F | |
| 0194-98c | O | 1998 | F | F | F | |
| 64d | O | 1999 | F | F | F | |
| 68d | O | 1999 | F | F | F | |
| 74d | O | 1999 | F | F | F |
| 80d | O | 1999 | F | F | F |
| 122d | O | 1999 | F | F | F |
| 123d | O | 1999 | F | F | F |
| Cfve | ? | ? | V | F | F |
| 89/8/5396e | ? | ? | F | F | F |
| 8e | ? | ? | V | F | F |
| 10e | ? | ? | V | F | F |
| 44.1e | ? | ? | V | F | F |
| 135e | ? | ? | V | F | F |
| 136e | ? | ? | V | F | F |
| 154.4e | ? | ? | V | F | F |
| 248.1e | ? | ? | F | F | F |
| 5396/7e | ? | ? | F | F | F |
| Faf | B | 1998 | F | F | F |
| Maf | B | 1998 | F | F | F |
| Paf | B | 1998 | F | F | F |
| Qaf | B | 1998 | F | F | F |
| V1af | B | 1998 | F | F | F |
| V2af | B | 1998 | F | F | F |
| Yaf | B | 1998 | F | F | F |
| 23af | B | 1998 | F | F | F |
| 24af | B | 1998 | F | F | F |
| 26af | B | 1998 | F | F | F |
| 97/v549cf | B | 1997 | V | V | V |
| 97/v557cf | B | 1997 | V | V | V |
| 97/v561cf | B | 1997 | V | V | V |
| 97/v562cf | B | 1997 | V | V | V |
| 97/v566cf | B | 1997 | V | V | V |
| 97/v569cf | B | 1997 | V | V | V |
| 97/v570cf | B | 1997 | V | V | V |
| 97/v571cf | B | 1997 | V | V | V |
| 97/v572cf | B | 1997 | V | V | V |
| 97/v574cf | B | 1997 | V | V | V |
Obtained from ID-Lelystad, Lelystad, The Netherlands.
Obtained from Animal Health Service, Boxtel, Deventer, and Drachten, The Netherlands.
Obtained from Veterinary Laboratory Agency, Weybridge, United Kingdom.
Obtained from Veteriner Kontrol ve Arastirma Enstitusu, Konyn, Turkey.
Obtained from Onderstepoort Veterinary Institute, Onderstepoort, South Africa.
Outbreak isolate.
B, strain isolated from bovine; O, strain isolated from ovine.
F, C. fetus subsp. fetus; V, C. fetus subsp. venerealis.
?, data unknown.
≤1993, in or before 1993.
PCR subtyping.
A subspecies-specific PCR was performed as previously described (8). Of the PCR primers, one primer set was directed to both C. fetus subspecies (±750 bp), whereas another primer set only amplified a C. fetus subsp. venerealis-specific band (142 bp). All strains showed the C. fetus-specific band. In general the results are consistent with the phenotypic tests. However, 7 of the 54 C. fetus subsp. fetus strains identified by PCR were negative for growth on 1% glycine. In these cases the PCR results were supported by data from the AFLP, suggesting that strains were mistyped by the biochemical method. Interestingly, these seven strains were part of a group of nine strains from South Africa. These results may indicate some evolutionary distinction between C. fetus subsp. fetus strains from South Africa and those from other geographical regions. Unfortunately, appropriate clinical data were not available for these strains to determine whether this difference was host, disease, or epidemiology related. Conversely, 1 out of the 15 C. fetus subsp. venerealis strains (98/v445) identified by PCR was positive for growth on 1% glycine. For this strain, the biochemical typing and PCR results were inconsistent and the AFLP results did not correlate with the PCR. This strain was also tested in a routine immunofluorescence assay but did not react with C. fetus-specific antiserum. It appears that this strain shows aberrant behavior phenotypically as well as genotypically.
The molecular basis of this PCR test and the relevance of this DNA difference to disease presentation are currently unknown. Although the PCR subtyped only one C. fetus subsp. fetus strain as C. fetus subsp. venerealis, it should be clear that the consequences of such a mistyping may be serious for import and export of animals and the veterinary health status of a country. In the original study that described the PCR, two isolates typed as C. fetus subsp. venerealis were considered C. fetus subsp. fetus by a probabilistic identification score (8). Before this PCR assay can be recommended as a stand-alone test for statutory test purposes, more strains should be typed.
AFLP subtyping.
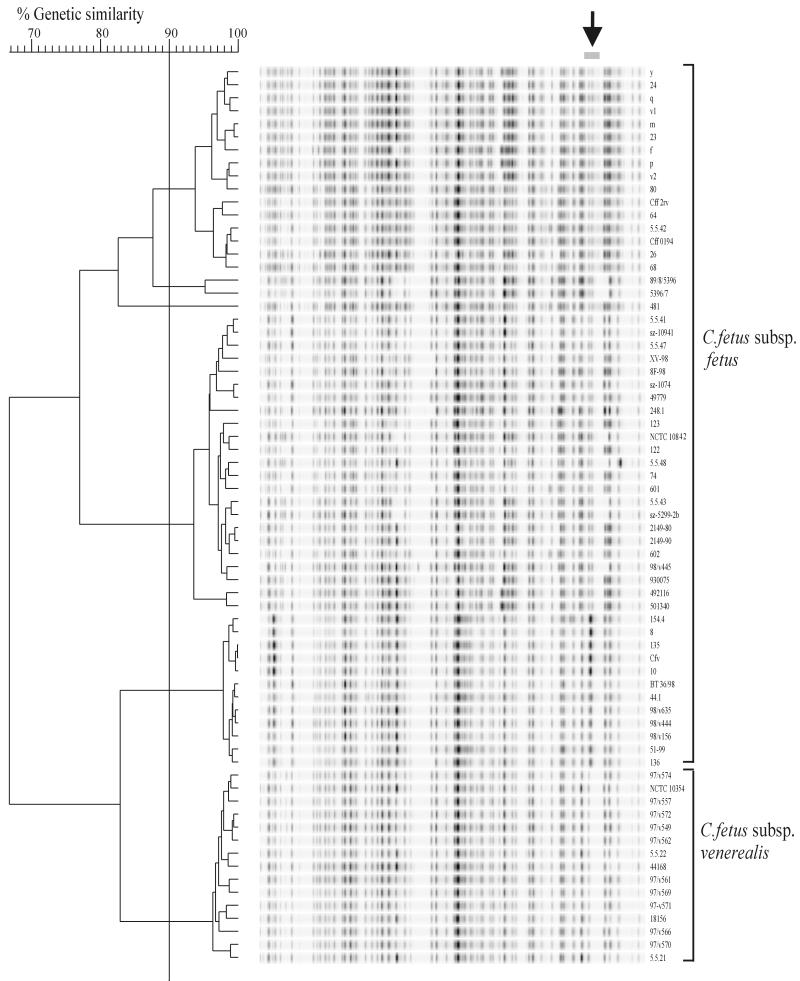
AFLP typing was performed according to the adapted PE Applied Biosystems protocol previously described (5). The obtained AFLP pattern consisted of approximately 55 to 60 bands in all strains (Fig. 1). Differentiation was initially based on the whole profile that clustered all the C. fetus subsp. venerealis strains together and divided the C. fetus subsp. fetus strains into several clusters. Using cluster analysis on only a small region of the pattern (Fig. 1) improved the discrimination between C. fetus subsp. venerealis strains and the C. fetus subsp. fetus clusters. The patterns showed extensive homology, which may therefore restrict the use of this technique for typing individual strains. This was partly shown by fingerprinting isolates obtained from two outbreaks. For C. fetus subsp. venerealis, isolates from the outbreaks were not clustered separately from unrelated strains, whereas strains from a C. fetus subsp. fetus outbreak could be separated, but only when the whole pattern was analyzed (data not shown).
FIG. 1.
Dendrogram showing the AFLP banding patterns of 69 C. fetus strains. Cluster analysis was based on the similarity levels among bands in region 841 to 879 of the banding patterns (arrow). The different clusters of C. fetus subsp. fetus and C. fetus subsp. venerealis are indicated. The percentages of genetic similarity among banding patterns are shown.
On the basis of previous DNA-DNA hybridization and taxonomic studies, the division of C. fetus into two subspecies has been questioned (4, 14, 18). Nevertheless these subspecies have clear differences in clinical presentation. The AFLP, which samples the whole bacterial genome, appears to support the subspecies differentiation. The evidence from the PCR and AFLP for DNA differences between these two subspecies is now compelling. Whether these differences are relevant to the pathophysiology of this group of organisms has yet to be determined. Our results confirm previous attempts to subtype C. fetus strains using molecular techniques. Pulsed field gel electrophoresis also showed relatively homologous patterns among different C. fetus strains although some evidence of subspecies differentiation has been described (10, 11).
In conclusion the results of this investigation indicate that biochemical assays currently used for differentiation of C. fetus subspecies are unreliable. In contrast, a recently developed PCR technique may have considerable value in routine diagnosis. However, more isolates have to be tested to have an indication about the incidence of aberrant strains. AFLP analysis was shown to be a suitable method for subspecies differentiation.
Acknowledgments
We thank Alan Rigter, Ellard Kruijt, and Ank van Zijderveld from ID-Lelystad for technical assistance and M. Henton (South Africa), L. Guler (Turkey), E. G. Hartman and I. J. Visser (The Netherlands), and J. Shreeve (United Kingdom) for supplying strains.
We also thank the Ministry of Agriculture Fisheries and Foods, GB, for partly funding this study.
REFERENCES
- 1.Anonymous. OIE manual of standards for diagnostic tests and vaccines 3rd ed. Paris, France: Office International des Epizooties; 1996. pp. 256–266. [Google Scholar]
- 2.Chang W, Ogg J. Transduction and mutation to glycine tolerance in Vibrio fetus. Am J Vet Res. 1971;32:649–653. [PubMed] [Google Scholar]
- 3.De Boer P, Duim B, Rigter A, van der Plas J, Jacobs-Reitsma W F, Wagenaar J A. Computer-assisted analysis and epidemiological value of genotyping methods for Campylobacter jejuni and Campylobacter coli. J Clin Microbiol. 2000;38:1940–1946. doi: 10.1128/jcm.38.5.1940-1946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denes A S, Lutze-Wallace C L, Cormier M L, Garcia M M. DNA fingerprinting of Campylobacter fetus using cloned constructs of ribosomal RNA and surface array protein genes. Vet Microbiol. 1997;54:185–193. doi: 10.1016/s0378-1135(96)01273-4. [DOI] [PubMed] [Google Scholar]
- 5.Duim B, Wassenaar T M, Rigter A, Wagenaar J. High-resolution genotyping of Campylobacter strains isolated from poultry and humans with amplified fragment length polymorphism fingerprinting. Appl Environ Microbiol. 1999;65:2369–2375. doi: 10.1128/aem.65.6.2369-2375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duim B, Ang C W, van Belkum A, Rigter A, van Leeuwen W J, Endtz H P, Wagenaar J A. Amplified fragment length polymorphism analysis of Campylobacter jejuni strains isolated from chickens and from patients with gastroenteritis or Guillain-Barré or Miller Fisher syndrome. Appl Environ Microbiol. 2000;66:3917–3923. doi: 10.1128/aem.66.9.3917-3923.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey S M, Greenwood J R. Relationships among catalase-positive campylobacters determined by deoxyribonucleic acid-deoxyribonucleic acid hybridization. Int J Syst Bacteriol. 1983;33:275–284. [Google Scholar]
- 8.Hum S, Quinn K, Brunner J, On S L. Evaluation of a PCR assay for identification and differentiation of Campylobacter fetus subspecies. Aust Vet J. 1997;75:827–831. doi: 10.1111/j.1751-0813.1997.tb15665.x. [DOI] [PubMed] [Google Scholar]
- 9.On S L W. Identification methods for campylobacters, helicobacters, and related organisms. Clin Microbiol Rev. 1996;9:405–422. doi: 10.1128/cmr.9.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.On S L W, Harrington C. Evaluation of numerical analysis of PFGE-DNA profiles for differentiating of Campylobacter fetus subspecies by comparison with phenotypic, PCR, and 16S rDNA sequencing methods. J Appl Microbiol. 2001;90:285–293. doi: 10.1046/j.1365-2672.2001.01247.x. [DOI] [PubMed] [Google Scholar]
- 11.Salama S M, Garcia M M, Taylor D E. Differentiation of the subspecies of Campylobacter fetus by genomic sizing. Int J Syst Bacteriol. 1992;42:446–450. doi: 10.1099/00207713-42-3-446. [DOI] [PubMed] [Google Scholar]
- 12.Smibert R M. Genus Campylobacter Sebald and Véron 1963, 907AL. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. pp. 111–118. [Google Scholar]
- 13.Thompson S A, Blaser M J. Pathogenesis of Campylobacter fetus infections. In: Nachamkin I, Blaser M J, editors. Campylobacter. 2nd ed. Washington, D.C.: American Society for Microbiology; 2000. pp. 321–347. [Google Scholar]
- 14.Vandamme P, Falsen E, Rossau R, Hoste B, Segers P, Tytgat R, de Ley J. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int J Syst Bacteriol. 1991;41:88–103. doi: 10.1099/00207713-41-1-88. [DOI] [PubMed] [Google Scholar]
- 15.Vandamme P. Taxonomy of the family Campylobacteriaceae. In: Nachamkin I, Blaser M J, editors. Campylobacter. 2nd ed. Washington, D.C.: American Society for Microbiology; 2000. pp. 3–26. [Google Scholar]
- 16.Véron M, Chatelain R. Taxonomic study of the genus Campylobacter Sebald and Véron and designation of the neotype strain for the type species. Campylobacter fetus (Smith and Taylor) Sebald and Véron. Int J Syst Bacteriol. 1973;23:122–134. [Google Scholar]
- 17.Wassenaar T M, Newell D G. Genotyping of Campylobacter spp. Appl Environ Microbiol. 2000;66:1–9. doi: 10.1128/aem.66.1.1-9.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wesley I V, Wesley R D, Cardella M, Dewhirst F E, Paster B J. Oligodeoxynucleotide probes for Campylobacter fetus and Campylobacter hyointestinalis based on 16S rRNA sequences. J Clin Microbiol. 1991;29:1812–1817. doi: 10.1128/jcm.29.9.1812-1817.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]