Abstract
Type 2 diabetes (T2D) is characterized by pathophysiological alterations in lipid metabolism. One strategy to understand the molecular mechanisms behind these abnormalities is to identify cis-regulatory elements (CREs) located in chromatin-accessible regions of the genome that regulate key genes. In this study we integrated assay for transposase-accessible chromatin followed by sequencing (ATAC-seq) data, widely used to decode chromatin accessibility, with multi-omics data and publicly available CRE databases to identify candidate CREs associated with T2D for further experimental validations. We performed high-sensitive ATAC-seq in nine human liver samples from normal and T2D donors, and identified a set of differentially accessible regions (DARs). We identified seven DARs including a candidate enhancer for the ACOT1 gene that regulates the balance of acyl-CoA and free fatty acids (FFAs) in the cytoplasm. The relevance of ACOT1 regulation in T2D was supported by the analysis of transcriptomics and proteomics data in liver tissue. Long-chain acyl-CoA thioesterases (ACOTs) are a group of enzymes that hydrolyze acyl-CoA esters to FFAs and coenzyme A. ACOTs have been associated with regulation of triglyceride levels, fatty acid oxidation, mitochondrial function, and insulin signaling, linking their regulation to the pathogenesis of T2D. Our strategy integrating chromatin accessibility with DNA binding and other types of omics provides novel insights on the role of genetic regulation in T2D and is extendable to other complex multifactorial diseases.
Keywords: ATAC-seq, regulatory element, T2D, liver
Introduction
While insulin resistance and altered glucose metabolism are considered the main drivers of the pathogenesis of type 2 diabetes (T2D), the focus has moved to the study of anomalies in lipid metabolism to better understand the onset and progression of the disease. Increased level of triglycerides and low-density lipoproteins with reduction of high-density lipoprotein are the key features of dyslipidemia in T2D patients and are associated with increased risk for cardiovascular disease (Krauss, 2004). From a clinical standpoint, lipid-lowering drugs such as statins and fibrates are used alone or in combination to inhibit cholesterol synthesis or modulate the transcription of several genes participating in lipid metabolism.
Long-chain acyl-CoA thioesterases (ACOTs) are a group of enzymes that hydrolyze acyl-CoA esters to free fatty acids (FFAs) and coenzyme A (CoASH). ACOTs are grouped into type I monomeric ACOTs 1–6 and type II multimeric ACOTs 7–13 based on their molecular weights. Human genes encoding for type I ACOTs are clustered on chromosome 14q24.3 and have ubiquitous subcellular localization with specific isoforms present in cytosol (ACOT1 and ACOT6), mitochondria (ACOT2), and peroxisomes (ACOT4) (Hunt and Alexson, 2002; Kirkby et al., 2010).
Several cellular processes are activated by acyl-CoAs and inhibited by FFAs or vice versa, highlighting the importance of ACOTs in maintaining cellular homeostasis (Hunt et al., 2006). ACOTs have been associated with regulation of triglyceride levels (Kang et al., 2012), fatty acid oxidation (Moffat et al., 2014), mitochondrial function (Kawano et al., 2014), and insulin signaling (Yang et al., 2012) in brown adipose, liver, and heart tissues (Bakshi et al., 2018).
ACOT1 plays a central role in regulating hepatic fatty acid oxidation, oxidative stress, and inflammation during fasting (Franklin et al., 2017). These findings provide a link that might help explain the progression of nonalcoholic fatty liver disease and its close association with insulin resistance and T2D.
The hydrolysis of acyl-CoAs that is catalyzed by ACOTs directly affects the extent of mitochondrial β-oxidation preventing the transport of FFAs into the mitochondria. Uptake of FFAs that exceeds the oxidation capacity could lead to lipotoxicity, whereas increased FFAs oxidation could lead to elevated production of reactive oxygen species. By tuning the cytosolic balance of acyl-CoA and FFAs, ACOTs regulate oxidative capacity through substrate availability and signaling (Franklin et al., 2017). In the cytosol, ACOT1 is involved in the modulation of the pool of long chain (C12–C20) saturated and monounsaturated acyl-CoAs and FFAs that act also as regulators of ligand-activated transcription factors (TFs).
Nuclear receptors such as peroxisome proliferator-activated receptor alpha (PPARα) and hepatocyte nuclear factor 4 alpha (HNF4α) can be activated or inhibited by acyl-CoAs and, in turn, bind regulatory elements tuning the expression of ACOT1 in a loop manner (Dongol et al., 2007). The ability of ACOT1 to ultimately control the availability of ligands for PPARα and HNF4α makes it a relevant gene to better understand the molecular mechanism contributing toward impaired lipid metabolism in T2D and other types of the diabetic spectrum (e.g., gestational diabetes or HNF4α related maturity onset of the young diabetes).
DNA accessibility plays a key role on gene regulation in the human genome. cis-regulatory elements (CREs), such as enhancers, promoters, silencers, and insulators overlap open chromatin regions and are able to bind different TFs that activate or repress target genes (Gontarz et al., 2020). ATAC-seq (assay for transposase-accessible chromatin followed by sequencing) is widely used to decode chromatin accessibility with high sensitivity.
In this study we performed high-sensitive ATAC-seq in nine human liver samples from normal and T2D donors and identified a set of differentially accessible regions (DARs). DARs marked putative regulatory elements including a candidate enhancer for the ACOT1 gene that regulates the balance of acyl-CoA and FFAs in the cytoplasm. Our strategy that integrates chromatin accessibility with DNA binding and other omics data provides novel insights on the role of genetic regulation in complex multifactorial diseases such as T2D.
Materials and Methods
Liver biosamples and nuclei preparation
Frozen liver tissues samples (n = 11) were obtained from the human tissue laboratory (Supplementary Table S1), a biobank for multi-organ donors funded by the excellence of diabetes research in Sweden (EXODIAB), following the Uppsala Regional Ethics Committee approval (Dnr: 2014/391). Three samples were harvested from donors diagnosed with T2D (HbA1c ≥6.5%) and eight from control donors characterized by normoglycemia (CTRL). Each liver sample was processed using the gentleMACS dissociator (Miltenyi) program E to create a suspension of single nuclei. Approximately 5 mg of frozen liver tissue was transferred into a gentleMACS C-tube with 10 mL of 0.250 M sucrose solution. The homogenate was filtered using 100 μm MACS SmartStrainers and the volume adjusted to 15 mL with the sucrose solution.
After centrifugation at 600 g at 4°C in an Eppendorf 5810R Centrifuge the supernatant was discarded, the nuclei in the pellet were resuspended in 1 mL of phosphate-buffered saline (PBS) with 0.04% bovine serum albumin and filtered through a MACS SmartStrainers (30 μm). The purity and concentration of the nuclei suspension was evaluated using the Countess™ II Automated Cell Counter (Thermo Scientific, Waltham, MA) by staining with trypan blue.
Tn5 transposome assembly
Tn5 transposase was ordered from the Protein Science Facility at Karolinska Institutet. The Tn5 assembly was performed by following previous description (Chen et al., 2016). The adaptor oligonucleotides (Supplementary Table S2) used for Tn5 transposome assembly were synthesized by Integrated DNA Technology. Equimolar Tn5MErev and Tn5ME-A, Tn5ME-B and Tn5MErev were mixed in separate PCR tubes, respectively, and placed on a PCR machine to run the following program for annealing: 95°C for 5 min, ramping down to 25°C with −0.1°C/s.
For Tn5 transposome assembly, a 50 μL reaction containing 2 μL 50 μM Tn5MErev/Tn5ME-A, 2 μL 50 μM Tn5MErev/Tn5ME-B, 20 μL glycerol, 11.87 μL 2 × dialysis buffer (100 mM HEPES-KOH at pH 7.2, 0.2 M NaCl, 0.2 mM EDTA, 2 mM DTT, 0.2% Triton X-100, 20% glycerol), 8.61 μL H2O, and 5.52 μL 18.18 μM Tn5 were mixed gently with pipette and incubated for 1 h at 25°C. Activity assay was performed to confirm good quality of Tn5 transposome followed by storage at −20°C until use.
ATAC-seq library preparation and sequencing
ATAC-seq libraries were prepared following the previously described standard Omni-ATAC protocol (Corces et al., 2017). In brief, for each liver sample, two technical replicates containing 50,000 nuclei each were pelleted at 500 g for 5 min at 4°C. The pellets were individually resuspended in 50 μL lysis buffer (10 mM Tris-Cl, pH 7.4; 10 mM NaCl; 3 mM MgCl2; 0.1% Igepal CA-630; 0.1% Tween-20 and 0.01% Digitonin) and centrifuged at 500 g for 10 min. The supernatant was removed and each nuclei pellet was resuspended in a 50 μL tagmentation buffer (25 μL 2 × TD buffer, 16.5 μL PBS, 0.5 μL 10% Tween-20, 0.5 μL 1% Digitonin, 5 μL nuclease-free H2O, and 2.5 μL Tn5 transposome) and incubated at 37°C for 30 min. Next, MinElute PCR Purification Kit (Qiagen) was used for DNA purification.
The following library preparation was performed following the previously described standard protocol (Buenrostro et al., 2015). All libraries were sequenced on an Illumina Nova-seq platform at Novogene Europe according to standard procedure.
ATAC-seq data analysis and DARs definition
We performed pair-end sequencing on the 11 liver duplicate samples from the EXODIAB cohort. Trimmed adaptor sequences from the FastQ files (https://github.com/TheJacksonLaboratory/ATAC-seq/blob/master/auyar/pyadapter_trim.py) were mapped to the hg19 reference genome using bowtie2 using the parameter—very-sensitive (Langmead and Salzberg, 2012). Resulting BAM files were sorted using samtools v.1.9 (Li et al., 2009), PCR and optical duplicates were removed using Picard v.1.119 (http://broadinstitute.github.io/picard/), and reads were filtered for alignment quality of >Q10. Reads mapping to mitochondria were removed. Samples with <10,000 peaks in either replica or replicates with Pearson correlation coefficient <0.8 were discarded.
BAM files were lifted to the reference genome hg38, replicates of samples were merged using samtools and peaks were called using MACS2 (Gaspar, 2018) with −q = 0.1. Next, we used the program featureCounts from the package Subread (Liao et al., 2014) to summarize the number of sequence reads of each sample over merged genomic features that were calculated as the merged set of peaks called in all samples. Features with <75 reads in all samples, <500 reads on average across samples, and standard deviation >100 across samples were excluded from subsequent analysis. We used the R package limma (Ritchie et al., 2015) to perform a differential analysis on the log2-transformed matrix of counts while accounting for the confounding factors age, sex, and body mass index.
After correcting for multiple testing, we identified seven statistically significant DARs with p < 0.05 and log-fold change (logFC) >1.8 (Table 1). Significant DARs were then overlapped with several publicly available datasets (Boix et al., 2021; Meuleman et al., 2020; Moore et al., 2020; Vierstra et al., 2020) (Supplementary Table S3). We also downloaded the full set of TFs ChIP-seq and DNase-seq experiments related to liver from the ENCODE 3 project (Supplementary Table S4). We created a set of unique peaks by merging signals originating from the same ChIP-seq and DNase-seq regardless of liver localization. This set consisted of a set of narrowPeak files, one for each TF or DNase signal.
Table 1.
Differentially Accessible Regions Identified from Assay for Transposase-Accessible Chromatin Followed by Sequencing of Liver Tissue in Controls and Type 2 Diabetes Donors
| DAR coordinates (hg38) | logFCa | Adjusted p | Closest gene | Registry of cCREs from ENCODE | ||
|---|---|---|---|---|---|---|
| chr14:73536843-73537501 | 3.369 | 9.33692029E-11 | ACOT1, HEATR4 | EH38E1727203b | EH38E1727204c | |
| chr17:43360659-43361492 | 2.223 | 1.67375632E-03 | ARL4D, DHX8, TMEM106A | EH38E1864240b | EH38E1864241b | EH38E1864242c |
| chr1:120850829-120851349 | −2.132 | 3.32913025E-03 | NBPF26, PPIAL4A, AC241377.2 | EH38E1381833c | EH38E1381834b | |
| chr1:32760164-32760576 | 2.074 | 4.84295471E-03 | KIAA1522, YARS, S100PBP | EH38E1335310d | ||
| chr7:83162614-83163011 | 1.971 | 1.23236816E-02 | PCLO, SEMA3E, CACNA2D1 | EH38E2568071b | EH38E2568072c | |
| chr4:49514497-49514687 | 1.876 | 2.84249499E-02 | CWH43, OCIAD2, OCIAD1 | EH38E2296070e | ||
| chr8:10713891-10714177 | 1.851 | 3.15049009E-02 | SOX7, C8orf74, RP1L1 | EH38E2610261b | ||
Estimate of the log2-fold change.
Proximal enhancer like.
Promoter like.
Distal enhancer like.
Dnase-H3K4me3.
cCREs, candidate cis-regulatory elements; DAR, differentially accessible region; logFC, log-fold change.
This collection of unique peaks was provided as input to tfNet (Diamanti et al., 2016) that was executed with the following settings: -na—distance 50—lScore 2—neigh 20, 60—overlap 5—acc bed. tfNet resulted in a collection of putative regulatory regions that mark clusters of co-occurring TF binding sites.
Data availability
Raw sequencing data were deposited in the GEO database under the accession number: GSE173277 and are available from the corresponding author upon request.
Results
Definition of DARs in T2D liver
ATAC-seq was performed on 11 liver samples from the EXODIAB biobank. Results from two samples were discarded after QC (see Methods and Supplementary Table S1) and downstream analysis was performed on a cohort of nine samples that consisted of six controls and three T2D donors. Differential analysis revealed seven unique DARs between cases and controls. The seven DARs were intersected with the ENCODE three blacklisted regions to filter out elements that generate artifact signal in certain regions of the genome like centromeres, telomeres, and satellite repeats (Amemiya et al., 2019).
Subsequently, DARs were characterized using several publicly available datasets of liver-specific genomic annotations including EpiMap, DNaseI hypersensitive sites (DHSs), and candidate CREs from the ENCODE project. The recent release of ENCODE 3 (Moore et al., 2020) provided a new comprehensive set of high-quality DHS maps instrumental to annotate the human regulatory landscape (Meuleman et al., 2020). DHSs provide an excellent complementary source of information to annotate open chromatin regions defined by the ATAC-seq data.
We overlapped DHSs defined in liver biosamples by ENCODE with the seven significant DARs (Supplementary Table S5). The majority of DARs overlapped two or more DHSs reflecting the larger size of our identified ATAC-seq regions (average length of 471 bp, range 190 to 833) compared with the DHSs (average length of 204 bp). One of the major improvements in the DHSs datasets from ENCODE 3 is the annotation of DHSs with 16 biological components that reflect the biological context, that is, tissues or cell lines where a specific DHS is more active/present.
Two DARs were marked by the DHS-annotated component “digestive,” which includes liver biosamples from ENCODE, hence of higher relevance to our study. The remaining DARs were marked by DHSs annotated as “tissue invariant,” usually associated with the regulation of housekeeping genes, or with other biological components less relevant to T2D.
Next, we overlapped the identified DARs with a collection of high-resolution epigenomic annotations from the EpiMap repository. The EpiMap database (Boix et al., 2021) aggregates epigenetic datasets from the ENCODE, Roadmap, and GGR projects providing detailed chromatin state annotation of regulatory elements across more than 800 biosamples and 18 marks/assays. In our study we overlapped the seven DARs with chromatin states defined in liver, hepatic stellate cells, and hepatocyte biosamples. Overall, three of seven DARs were annotated as enhancers in at least one of the liver-related biosamples (Supplementary Table S5).
Characterization of the ACOT1 DAR
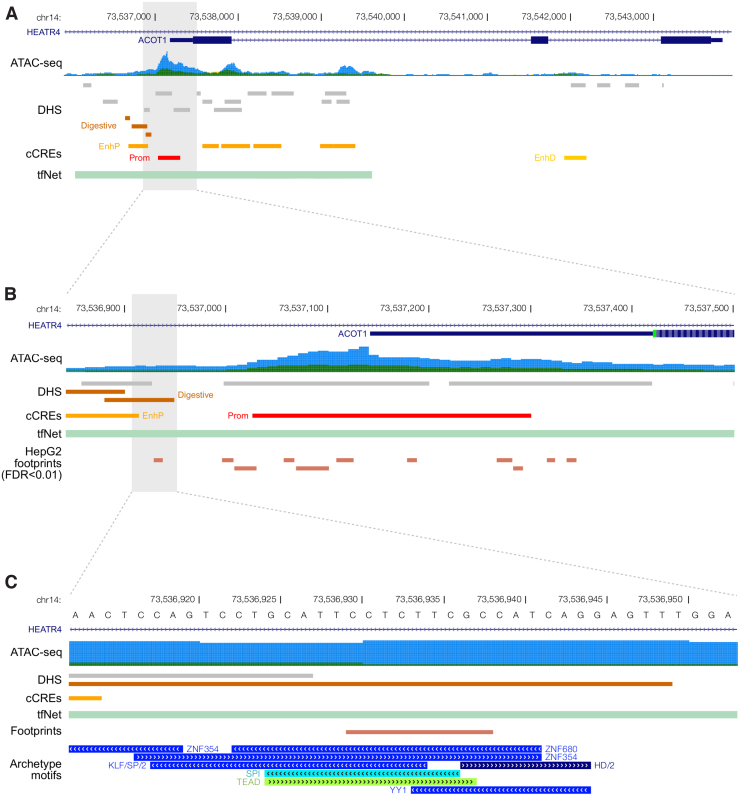
ACOT1 is expressed in liver, adipose tissue, skeletal muscle, and other metabolic tissues (Lonsdale et al., 2013). The ATAC-seq region with the highest differential accessibility between controls and T2D samples was identified in the promoter region of the ACOT1 gene (ACOT1-DAR). This region was significantly more open in T2D compared with controls suggesting higher gene expression in the disease state. The ACOT1-DAR span 658 bp (Fig. 1A) and overlap the promoter region, 5′UTR and part of the first exon of ACOT1. It also overlaps the first intron of HEATR4 encoding for the HEAT Repeat Containing four protein expressed mostly in the testis, brain, and nerves and only at low levels in the liver (Lonsdale et al., 2013) (Supplementary Fig. S1).
FIG. 1.
Open chromatin and regulatory elements landscape in the ACOT1 genomic region. (A) Open chromatin regions defined by ATAC-seq in CTRL (dark green) and T2D (blue) liver samples (ATAC-seq), publicly available datasets of liver-specific genomic annotations including DHSs and candidate CREs from the ENCODE project (cCREs) and regulatory regions defined using the tfNet tool (tfNet). (B) ACOT1-DAR. A track with footprints defined in HepG2 is shown at the bottom. (C) Base pair resolution of the putative regulatory element with archetype motif clusters. ACOT, acyl-CoA thioesterase; ATAC-seq, assay for transposase-accessible chromatin followed by sequencing; cCRE, candidate cis-regulatory element; DAR, differentially accessible region; DHS, DNaseI hypersensitive site; T2D, type 2 diabetes.
We first compared the ACOT1-DAR with the DHSs data from ENCODE (Fig. 1A). Six DHSs overlapped ACOT1-DAR of which three DHSs were active in the liver. One of them was annotated with a “digestive” component suggesting a tissue-specific accessibility. Next, we intersected the ACOT1-DAR with the Registry of candidate cis-regulatory elements (cCREs) derived from ENCODE. Two cCREs defined in liver overlapped the ACOT1-DAR namely EH38E1727203 with a proximal enhancer-like signature and EH38E1727204 with a promoter-like signature (Fig. 1B).
Motifs in DARs footprints and tfNet
ENCODE 3 provides 286 distinct motif clusters that take into account the redundancy and overlap between various TF motif model databases. Within each cluster motifs are aligned to each other to generate an archetypal consensus motif (Vierstra et al., 2020). We found 138 unique TFs overlapping the ACOT1-DAR and an overrepresentation (42.7%) of archetypal motif clusters belonging to the C2H2 zinc finger DNA binding domain. For a more focused analysis of TF motifs, we selected footprints defined in liver from ENCODE and intersect them with the DHS region annotated with a “digestive” component overlapping the ACOT1-DAR (highlighted region, Fig. 1B).
This region overlapped seven distinct archetypal motif clusters: ZNF354, ZNF680, SPI, TEAD, KLF/SP/2, YY1, and HD/2. The computational tool tfNet identifies putative regulatory regions and genomic signal interactions based on ChIP-seq data (Diamanti et al., 2016). tfNet identified a regulatory region defined by ChIP-seq and DNase-seq signals from several TFs belonging to the motif clusters annotated in the footprint region: SP1 and PATZ1 (KLF/SP/2 cluster), TEAD3 (TEAD), YY1 (YY1), HOXA5, and MIXL1 (HD/2). With the exception of HOXA5 and MIXL1, GTEx data confirmed the liver expression of these TFs that are candidate regulators of ACOT1 expression.
Transcriptomics
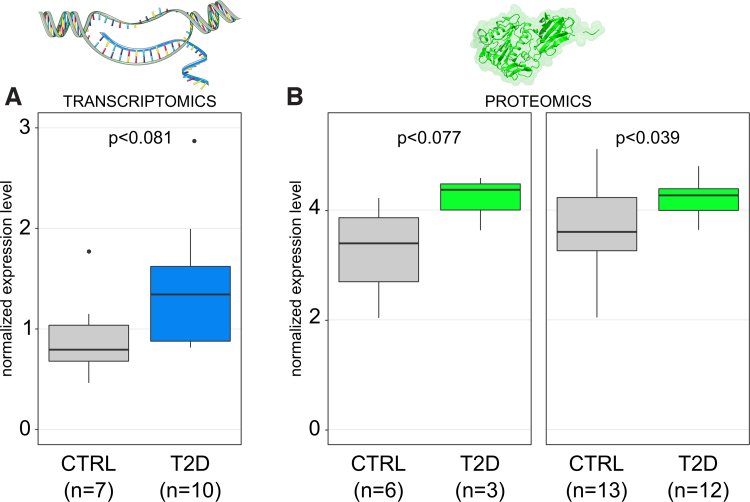
ACOT1-DAR is annotated in EpiMap as an active enhancer in hepatocytes (BSS00554). Of interest, ACOT1-DAR is annotated as a repressed or quiescent chromatin state in other liver and hepatic stellate cells in EpiMap suggesting a cell-type-specific regulatory activity. To further investigate the hypothesis of a hepatocyte-specific regulation of ACOT1 transcription we used in-house single nuclei transcriptomics (Cavalli et al., 2020) and publicly available single cell transcriptomics data (Aizarani et al., 2019; MacParland et al., 2018; Ramachandran et al., 2019). This confirmed a hepatocyte-specific expression of ACOT1. Because the region was more open in T2D we investigated the expression of the gene. An increasing trend in the expression of ACOT1 was observed in liver biopsies of T2D patients compared with nondiabetic controls analyzed by serial analysis of gene expression (Misu et al., 2010) (Fig. 2A).
FIG. 2.
Increased transcription and translation of ACOT1 in T2D. p Values were calculated from a t-test. (A) SAGE data for ACOT1 expression in liver CTRL and T2D samples. (B) ACOT1 levels in liver tissue from MS Proteomics performed in the EXODIAB cohort. Results are presented from the subcohort of nine samples analyzed in this study (3 T2D + 6 CTRL) and from the whole EXODIAB cohort (12 T2D + 13 CTRL). SAGE, serial analysis of gene expression.
Proteomics and metabolomics
We recently performed mass spectrometry-based proteomics in liver samples of a larger EXODIAB cohort including the nine samples considered in this study. To evaluate the downstream effects of an increased accessibility in T2D liver of the regulatory element at the promoter of ACOT1, we investigated the level of the ACOT1 protein. In the larger cohort (13 controls +12 T2D samples) ACOT1 was significantly higher in T2D than in controls (t-test, p = 0.04; Fig. 2B). A trend of higher expression of ACOT1 in T2D was seen in the nine samples in this study (p = 0.077; Fig. 2B) (Diamanti et al., 2021). This was in line with the finding of more open chromatin structure and the trend of higher gene expression in the liver of T2D.
Moreover, pathway enrichment analysis of the proteomics data highlighted proteins with altered levels in T2D and drivers of the overall deregulation of specific molecular pathways. The results showed that ACOT1 was one of the leading proteins altered in the “Metabolism of lipids” pathway in T2D (data not shown). Based on a former metabolomics study in the same EXODIAB cohort (Diamanti et al., 2019), we were able to observe the levels of metabolites that could be affected by a higher expression of ACOT1. The thioesterase enzyme catalyzes the hydrolysis of coenzyme A esters, specifically long-chain acyl-CoAs (C12 to C20), into FFAs and CoA, hence an increase in ACOT1 activity could result in an increase of long-chain FFAs.
The metabolomics analysis on controls and T2D liver samples revealed an overall increase of FFAs in liver with C16:0 and C18:0 significantly increased in T2D liver samples and a trend for elevated levels of C16:0-OH, C18:1, C20:4 (Diamanti et al., 2019). Thus, an increased level of ACOT1 in T2D liver is supported by signals in chromatin, by expression of gene and protein and by the effect on metabolites.
Discussion
One strategy to dissect gene regulation of complex diseases is to investigate genomic regions with significantly altered accessibility between cases and controls. ATAC-seq offers a high-resolution approach to identify open chromatin regions with putative gene regulatory elements. In this study we performed ATAC-seq on a cohort of liver samples from T2D donors and healthy controls, and identified seven differentially accessible genomic regions. They are candidate CREs that could be involved in the pathogenesis of T2D. ATAC-seq detects the average accessibility of gene regulatory elements in a population of cells. Although ChIP-seq gives a more qualitative definition of CREs, it is also more expensive, depends on antibodies, and bias can be introduced, for example, by crosslinking and sonication.
The integration of ATAC-seq with multi-omics data can help refine the definition of the CREs and build support to select promising candidate regulatory elements for further experimental validations. We presented a proof-of-concept approach aimed at identifying genomic regulatory elements in the context of T2D. Our results suggest that such a multi-omics data integration strategy could be applied to the study of other related diseases, for example, nonalcoholic fatty liver disease and dyslipidemias or T2D complications.
In this study, motif analysis within the ACOT1-DAR used the intersection of signals from (1) a DHS in liver biosamples and annotated as digestive and (2) a footprint defined in HepG2. A scan for motifs clusters revealed an overrepresentation of TFs with the C2H2 zinc finger DNA binding domain. Recently the Tn5 transposase preferential motif has been identified and shown to spike at C2H2 zinc finger TFs (Martins et al., 2018). However, by integrating ENCODE ChIP-seq data from liver, we could support the presence of a motif with the experimental evidence of a specific TF binding to the footprint. This supported that the TFs SP1, PATZ1, TEAD3, and YY1 are relevant for further experimental validation as putative mediator of ACOT1 expression.
In summary, our data suggest that ACOT1 is deregulated in T2D. The primary finding was by ATAC-seq showing increased open chromatin in T2D in key regulatory sequences of the gene. The relevance of ACOT1 regulation in T2D was supported by the analysis of multi-omics data in liver tissue. Microarray data confirmed higher expression of ACOT1 in T2D samples of a different cohort, whereas single cells and nuclei transcriptomics profiles in liver tissue showed a hepatocyte-specific expression pattern. Proteomics data also supported an increased level of ACOT1 in liver in T2D samples. The possible downstream effects of an increased activity of ACOT1 were supported by metabolomics with an increase of long-chain FFAs in T2D liver samples compared with controls.
ATAC-seq is a valuable tool to detect regulatory elements relevant for disease. However, some limitations must be addressed: sample availability might be a limiting step considering that the total number of cells defines library complexity, so too few cells would result in under-transposition and too many in over-transposition. The donor cohort size is also relevant because it can affect the length and resolution of the accessible regions. Because ATAC-seq relies on insertion to accessible DNA, rather than digestion of protected DNA, the technique is susceptible to sequencing contamination by mitochondrial DNA (Klein and Hainer, 2020). ATAC-seq also requires high sequencing coverage to achieve effective footprinting.
Finally, not all TFs have a corresponding motif and open chromatin information alone cannot be used to infer the binding profiles of such factors. Moreover, some motifs could potentially to be bound by multiple sequence-specific TFs that indicate the value of an integrative approach combining ATAC-seq with ChIP-seq and other -omics techniques (Ma and Zhang, 2020).
Conclusions
Studying chromatin accessibility by ATAC-seq in healthy and T2D liver tissue, we identified 7 DARs that mark putative regulatory elements including a candidate enhancer for the ACOT1 gene that regulates the balance of acyl-CoA and FFAs in the cytoplasm. Integration of transcriptomics and proteomics data in liver tissue highlighted the relevance of ACOT1 regulation in T2D. Our strategy that integrates chromatin accessibility with DNA binding and other types of omics provides novel insights toward the role of genetic regulation in complex multifactorial diseases such as T2D.
Supplementary Material
Abbreviations Used
- ACOT
acyl-CoA thioesterase
- ATAC-seq
assay for transposase-accessible chromatin followed by sequencing
- CRE
cis-regulatory element
- cCRE
candidate cis-regulatory element
- DAR
differentially accessible region
- DHS
DNaseI hypersensitive site
- FFA
free fatty acid
- HNF4α
hepatocyte nuclear factor 4 alpha
- logFC
log-fold change
- PBS
phosphate-buffered saline
- PPARα
peroxisome proliferator-activated receptor alpha
- T2D
type 2 diabetes
- TF
transcription factor
Author Disclosure Statement
The authors declare they have no financial conflicts of interest.
Funding Information
The study was supported by grants from SciLifeLab (C.W.), the Swedish Diabetes Foundation (DIA 2017-269) (C.W.), EXODIAB (C.W.), the family Ernfors Fund (C.W.), AstraZeneca (C.W.) and the Swedish Research Council (VR-2016-06794, VR-2017-02074 to X.C.).
Supplementary Material
References
- Aizarani N, Saviano A, Sagar, et al. (2019). A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya HM, Kundaje A, and Boyle AP. (2019). The ENCODE blacklist: Identification of problematic regions of the genome. Sci Rep 9, 9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi I, Brown SHJ, Brandon AE, et al. (2018). Increasing Acyl CoA thioesterase activity alters phospholipid profile without effect on insulin action in skeletal muscle of rats. Sci Rep 8, 13967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boix CA, James BT, Park YP, Meuleman W, and Kellis M. (2021). Regulatory genomic circuitry of human disease loci by integrative epigenomics. Nature 590, 300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ. (2015). ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109, 21.9.1–21.9.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli M, Diamanti K, Pan G, et al. (2020). A multi-omics approach to liver diseases: Integration of single nuclei transcriptomics with proteomics and HiCap bulk data in human liver. OMICS 24, 180–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shen Y, Draper W, et al. (2016). ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat Methods 13, 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, et al. (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14, 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamanti K, Cavalli M, Pan G, et al. (2019). Intra- and inter-individual metabolic profiling highlights carnitine and lysophosphatidylcholine pathways as key molecular defects in type 2 diabetes. Sci Rep 9, 9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamanti K, Cavalli M, Pereira MJ, et al. (2021). Organ-specific metabolic pathways distinguish prediabetes, type 2 diabetes and normal tissues. bioRxiv. DOI: 10.1101/2021.05.09.443296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamanti K, Umer HM, Kruczyk M, et al. (2016). Maps of context-dependent putative regulatory regions and genomic signal interactions. Nucleic Acids Res 44, 9110–9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongol B, Shah Y, Kim I, Gonzalez FJ, and Hunt MC. (2007). The acyl-CoA thioesterase I is regulated by PPARα and HNF4α via a distal response element in the promoter. J Lipid Res 48, 1781–1791. [DOI] [PubMed] [Google Scholar]
- Franklin MP, Sathyanarayan A, and Mashek DG. (2017). Acyl-CoA thioesterase 1 (ACOT1) regulates PPARα to couple fatty acid flux with oxidative capacity during fasting. Diabetes 66, 2112–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar JM. (2018). Improved peak-calling with MACS2. bioRxiv. DOI: 10.1101/496521. [DOI] [Google Scholar]
- Gontarz P, Fu S, Xing X, et al. (2020). Comparison of differential accessibility analysis strategies for ATAC-seq data. Sci Rep 10, 10150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt MC, and Alexson SEH. (2002). The role Acyl-CoA thioesterases play in mediating intracellular lipid metabolism. Prog Lipid Res 41, 99–130. [DOI] [PubMed] [Google Scholar]
- Hunt MC, Rautanen A, Westin MAK, Svensson LT, and Alexson SEH. (2006). Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional evolution results in a reduced number of human peroxisomal ACOTs1. FASEB J 20, 1855–1864. [DOI] [PubMed] [Google Scholar]
- Kang HW, Niepel MW, Han S, Kawano Y, and Cohen DE. (2012). Thioesterase superfamily member 2/acyl-CoA thioesterase 13 (Them2/Acot13) regulates hepatic lipid and glucose metabolism. FASEB J 26, 2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y, Ersoy BA, Li Y, Nishiumi S, Yoshida M, and Cohen DE. (2014). Thioesterase superfamily member 2 (Them2) and phosphatidylcholine transfer protein (PC-TP) interact to promote fatty acid oxidation and control glucose utilization. Mol Cell Biol 34, 2396–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkby B, Roman N, Kobe B, Kellie S, and Forwood JK. (2010). Functional and structural properties of mammalian acyl-coenzyme A thioesterases. Prog Lipid Res 49, 366–377. [DOI] [PubMed] [Google Scholar]
- Klein DC, and Hainer SJ. (2020). Genomic methods in profiling DNA accessibility and factor localization. Chromosome Res 28, 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss RM. (2004). Lipids and lipoproteins in patients with type 2 diabetes. Diabetes Care 27, 1496. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL. (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics (Oxford, England) 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W. (2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, et al. (2013). The genotype-tissue expression (GTEx) project. Nat Genet 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, and Zhang Y. (2020). Profiling chromatin regulatory landscape: Insights into the development of ChIP-seq and ATAC-seq. Mol Biomed 1, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacParland SA, Liu JC, Ma X-Z, et al. (2018). Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 9, 4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins AL, Walavalkar NM, Anderson WD, Zang C, and Guertin MJ. (2018). Universal correction of enzymatic sequence bias reveals molecular signatures of protein/DNA interactions. Nucleic Acids Res 46, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuleman W, Muratov A, Rynes E, et al. (2020). Index and biological spectrum of human DNase I hypersensitive sites. Nature 584, 244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misu H, Takamura T, Takayama H, et al. (2010). A liver-derived secretory protein, selenoprotein P, causes insulin resistance. Cell Metab 12, 483–495. [DOI] [PubMed] [Google Scholar]
- Moffat C, Bhatia L, Nguyen T, et al. (2014). Acyl-CoA thioesterase-2 facilitates mitochondrial fatty acid oxidation in the liver. J Lipid Res 55, 2458–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JE, Purcaro MJ, Pratt HE, et al. (2020). Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, Dobie R, Wilson-Kanamori JR, et al. (2019). Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, Lazar J, Sandstrom R, et al. (2020). Global reference mapping of human transcription factor footprints. Nature 583, 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Chen C, Wang H, et al. (2012). Protective effects of Acyl-coA thioesterase 1 on diabetic heart via PPARα/PGC1α signaling. PLoS One 7, e50376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data were deposited in the GEO database under the accession number: GSE173277 and are available from the corresponding author upon request.