Abstract
Objectives:
Determine whether preoperative dietary pre-habilitation with a low-fat, high-fiber diet reverses the impact of western diet (WD) on the intestinal microbiota and improves postoperative survival
Background:
The majority of surgical patients consume a WD. We have previously demonstrated that WD fed mice subjected to an otherwise recoverable surgical injury (30% hepatectomy), antibiotics, and a short period of starvation developed reduced survival (29%) compared to mice fed a low-fat, high-fiber standard chow (SD) (100%).
Methods:
Mice were subjected to 6 weeks of a WD and underwent dietary pre-habilitation with SD prior to exposure to antibiotics, starvation, and surgery. 16S rRNA gene sequencing was utilized to determine microbiota composition. Mass spectrometry measured short chain fatty acids and functional prediction from 16S gene amplicons were utilized to determine microbiota function.
Results:
As soon as at 24 hours, dietary pre-habilitation of WD mice resulted in restoration of bacterial composition of the stool microbiota, transitioning from Firmicutes dominant to Bacteroidetes dominant. However, during this early pre-habilitation, stool butyrate per microbial biomass remained low and postoperative mortality remained unchanged from WD. Microbiota function demonstrated reduced butyrate contributing taxa as potentially responsible for failed recovery. However, after 7 days of prehabilitation (7DP), there was greater restoration of butyrate producing taxa and survival improved (29% vs 79% vs 100%: 7DP vs WD vs SD, p <0.001).
Conclusion:
The deleterious effects of WD on the gut microbiota can be restored after 7 days of dietary pre-habilitation. Moreover, stool markers may define the readiness of the microbiome to withstand the process of surgery and may be predictive of enhanced survival.
Mini-Abstract:
Pre-operative dietary pre-habilitation with a high-fiber, low-fat diet reverses the negative impact of a low-fiber, high-fat western diet on the intestinal microbiome and improves postoperative survival. Temporal measurements of stool microbiota composition and function offer potential non-invasive markers to determine the microbiome readiness for surgery after dietary pre-habilitation.
Introduction:
Despite bundled care and meticulous attention to asepsis, postoperative infection rates remain at unacceptable levels and are among the most costly complications1,2. Following elective surgery, evidence suggest that the pathogens causing postoperative infection originate from the gastrointestinal tract microbiota3–5. When operating within the abdomen, antibiotics that empirically target the gut microbiota have reduced postoperative infection rates to an all-time low6,7. Yet this approach carries the unwanted consequences of also eliminating many of the health-promoting microbiota that may guide postoperative recovery8. The response of the microbiota to antibiotics is in part determined by diet9 as the variability of the gut microbiome at the individual level is closely correlated to diet10. However, diet and its consequences on the gut microbiota over the course of surgical intervention has received little attention11. Although preoperative dietary interventions such as the use of immunonutrition and synbiotics have been proposed, their benefit in elective surgery remains unclear12–14. The extent to which diet can be manipulated to maintain the resilience of the microbiota to antibiotic exposure, starvation, and surgical injury is unknown.
The consumption of highly processed low fiber, high fat foods 15, has been demonstrated to have major implications on the health-promoting effects of the normal microbiota. Processed diets result in a decrease in microbial populations involved in the production of short chain fatty acids (SCFAs)16–18, key metabolites that drive a recovery-directed immune response following surgical injury19. As a result of adherence to a western diet, as many as 30% of surgical patients undergoing major surgery can be considered obese20 which in turn is a major risk factor for the development of septic complications 21. Therefore, a more complete understanding of how diet alters the microbiome in the context of outcome from surgical injury may inform novel approaches to reduce morbidity and mortality. We recently demonstrated that WD fed mice become sensitive to the overall process of surgery that includes antibiotics, pre-operative starvation, and a sterile injury (30% hepatectomy) (ASH)5. WD- fed mice subjected to ASH displayed high mortality rates associated with the presence of virulent and resistant pathogens originating from the gut microbiota5. Importantly, when mice were fed a plant-based, low-fat, high-fiber standard chow diet (SD) under the conditions of ASH, they recovered uneventfully and did not develop infection-related mortality22. Similarly, we previously demonstrated that WD-induced alterations in the gut microbiota adversely affected anastomotic healing allowing collagenase producing microbiota to bloom 23. Remarkably, this effect could be reversed by short term dietary pre-habilitation that involved the feeding with SD to mice just prior to surgery12. Taken together, these studies suggest that the effects of a WD can be reversed by short-term dietary pre-habilitation and may mitigate the collateral damage on the microbiota that invariably occurs when surgical patients are subjected to short term starvation, antibiotics and surgical injury.
In the present study, we hypothesized that short-term dietary prehabilitation (DietPreHab) prior to a major surgical stress (i.e 30% hepatectomy) can improve post-operative outcome in mice chronically fed a WD. The aims of this study were to determine the time-dependence of the DietPreHab to alter surgical outcomes in mice, its associated changes in the microbiome and to identify potential stool-based microbiota-related markers that can predict readiness for surgery when mice are subjected to antibiotics, short-term starvation, and a sterile surgical injury.
Materials and methods:
Mouse Husbandry:
Six week old C57BL/6 mice (Charles River Laboratory) were housed in a temperature-controlled 12h light/dark cycled rooms at University of Chicago. Mice were fed on either standard rodent chow (Envigo) or a Western diet (Bio-Serv, catalog no. S3282) for 6 weeks. Following 6 weeks of WD, mice subjected to DietPreHab were transitioned to SD. All experiments were in accordance with National Institute of Health (NIH) guidelines and approval was obtained from University of Chicago Institutional Animal Care and Use Committee (IACUC protocol 71744).
Surgical Procedure:
Our previously described model of antibiotics, starvation, and hepatectomy was utilized5. After 6 weeks on their diets and/or completion of DietPreHab, mice were subjected to antibiotics twice per day for 5 days consisting of cefoxitin (30mg/kg i.p., Hikma Pharmaceuticals, Eatontown, NJ) and clindamycin (70mg/kg p.o.; Clindrops; Hebry Schein, Dublin, OH). Prior to surgery, mice were subjected to 12 hours of starvation, but allowed water ad libitum. Mice were anesthetized using 80mg/kg ketamine and 5mg/kg xylazine administered i.p. A midline laparotomy was performed, the left lobe of the liver was exteriorized, and 30% of the liver was resected. Following surgery, the mice were resuscitated with 1cc of warmed NS s.c., given 0.05mg/kg buprenorphine, and allowed to recover on a warming pad. Mice were assessed every 6 hours for signs of sepsis and scored using an established sepsis scores: score 1- ambulatory, active, normal coat, normal feces; 2- ambulatory, active, ruffled fur (mild sepsis); 3- ruffled fur, hunched poster, increased respirations (moderate sepsis); 4- ruffled fur, hunched posture, increased respirations, slow and staggering gait (severe sepsis); and 5- animal on side, minimally responsive, rapid and shallow respirations, gasping (moribund). Mice were sacrificed when they reached a sepsis score of 4.
16S rRNA sequencing and sequence data analysis.
Microbial DNA extraction from cecal contents and stool was performed using a Power Fecal DNA isolation kit (Qiagen, Carlsbad, CA). For library preparation, DNA was amplified using the barcoded 12-bp Golay primer set designed for the Earth Microbiome Project (EMP). PCR was performed according to the manufacturer’s protocol using the EMP primers, mPNA, AccuStart II PCR ToughMix, and the extracted DNA (Quntabio). After amplification, the PCR products were quantified by using a PicoGreen dsDNA quantitation assay (Invitrogen). The results of the quantification were used to normalize the amount of DNA from the PCR product used for sequencing and ensure that each amplicon was represented evenly during sequencing. Finally, an aliquot of the final pool was taken, and the DNA was purified by using an Agencourt AMPure XP PCR* purification system (Beckman-Coulter). The samples were then run on an Illumina MiSeq at Argonne National Laboratory (150 bp × 2).
Qiime224 was utilized for16S rRNA gene sequence analysis and demux emp-paired-end command was used to demultiplex and join the paired-end reads. Quality filtering was performed using Deblur. Taxonomy and OTUs were assigned using a Greengenes classifier. Sequences were further analyzed utilizing the Phyloseq25 package within R. Samples were rarefied to a depth of 10,000 reads per sample. For the alpha diversity, the Shannon index was used, and the beta diversity was analyzed using nonmetric multidimensional scaling (NMDS) plots that were generated based on a weighted UniFrac dissimilarity matrix. To determine significantly different OTUs between groups of interest, the DESeq226 package within R was utilized and OTUs were compared between days of dietary pre-habilitation. Significant OTUs were determined by a P value (false discovery rate [FDR]) cutoff of <0.05).
PICRUSt27 analysis was performed to predict microbiota function. The picrust2_out_pipeline function was utilized using a nearest sequence taxon index cutoff of 2. Contributions of samples and individual taxa to enzymes, designated by enzyme commission numbers, and pathways were determined utilizing KEGG designations. Taxon relative functional abundance was determined by the relative abundance of a given taxa multiplied by the number of gene copies present within the given taxa. The taxon relative functional abundance was compared across all significant taxa. In order to focus on butyrate producing pathways, enzymes and pathways specific to butyrate production were selected. STAMP28 pipeline was used to compare the microbiota function between diets and dietary prehab days using Benjamin Hochberg correction. Taxa and functional shift scores were calculated within FishTaco29 as previously described to determine taxa contribution to functional pathways. Briefly, the shift scores represent Wilcoxon rank sum statistics. Contribution between taxa attenuating and enhancing individual pathways were displayed for both case and control.
qPCR analysis
Real-time PCRs were carried out in duplicate on an ABI StepOne plus real-time PCR system sequence detector with 2x FastStart SYBR green mix. All qPCR mixtures contained 10ul of 2xFast-Start SYBR green with dye1, 0.5ul of each forward and reverse primer (final concentration, 0.4mM), and 9ul of the DNA (equilibrated 25ng DNA). PCR primers(Integrative DNA Technologies) were utilized for Firmicutes, Bacteroidetes, and universal 16S (Table S1)30. Relative abundance as determined by PCR was estimated as:
Bacterial load was determined by using a standard curve of 16S amplification from E. coli DNA to allow for estimating bacterial DNA/mg stool.
Gas Chromotography-mass spectrometry
SCFA were extracted from mouse feces, utilizing approximately 50mg of mouse feces, using diethyl ether (Fisher Scientific), derivatized using N-tert-Butyldimethylsilyl methyltrifluoroacetamide with 1% tert Butyldimethylchlorosilane (Sigma) and run on an Agilent Single Quad GC–MS(5977A Single Quad and 7890B GC). 4-methylvalonic acid was spiked into each sample as an internal control and used to measure extraction efficiencies. Standard curves were generated utilizing butyric acid, propionic acid, and acetic acids. All values are normalized to sample mass and amounts of SCFA was calculated based off of generated standard curves for each individual SCFA.
Results:
DietPreHab attenuates WD associated weight changes and improves post-surgical outcome
In attempts to reverse the postoperative mortality seen after WD feeding, WD fed mice were transitioned to 3 or 7 days of DietPreHab (3DP versus 7DP) (Figure 1A). Following DietPreHab, the mice were administered 5 days of antibiotics, underwent 12 hours of starvation, and a 30% hepatectomy (ASH). DietPreHab led to a reduction in weight gain in WD-fed mice with the greatest attenuation of weight gain noted in the 7DP group (49.17 vs 46.7 vs 44.3 vs 16.0%: WD vs 3DR vs 7DR vs SD, p <0.01) (Figure 1B,C). The 5 days of antibiotics led to ~ 15% weight loss with a non-significant attenuation of antibiotic induced weight loss in the SD and 7DP mice (Figure 1D). Survival after exposure to the ASH was dependent on diet, and dramatically improved with 7DP (29% vs 38% vs 79% vs 100%: WD vs 3DR vs 7DR vs SD, Figure 1D P< 0.05). Standard culture techniques of the blood, liver, and spleen demonstrated no growth among moribund mice. However, Serratia marcescens, a pathogen previously identified to be selected for by ASH treatment in this model, was detected in the spleen using DNA and varied among treatment groups. Levels of S. marcescens DNA were decreased in DietPreHab survivors, but not in moribund DietPreHab and WD mice (Supplemental Figure S1), although not statistically significant.
Figure 1:
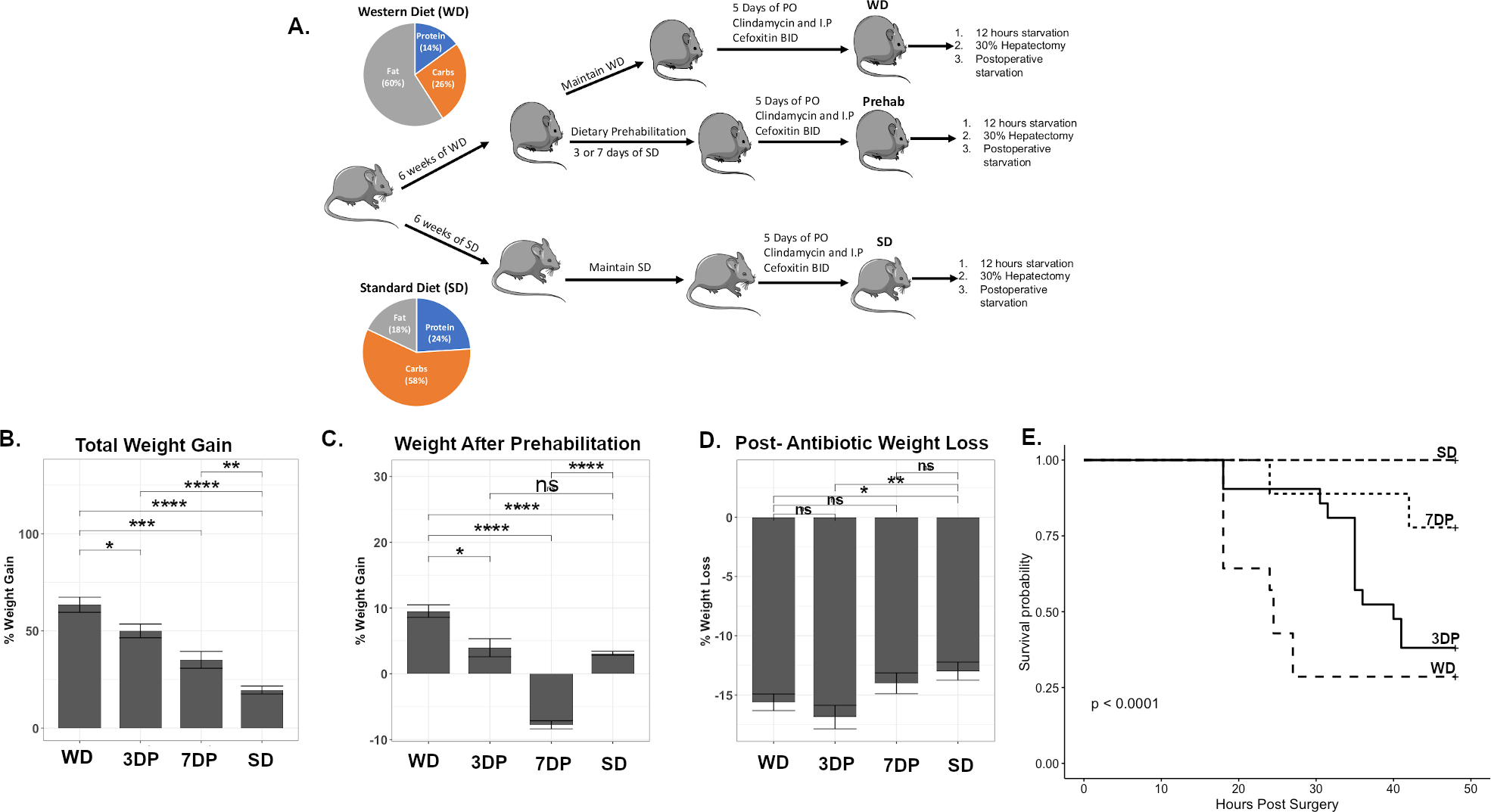
Short course of preoperative dietary prehabilitation (DietPreHab) results in improved response to antibiotic exposure, starvation, and surgical injury. The experimental design (A) demonstrates how the mice are subjected to their dietary intervention along with the diet composition of SD and WD represented within the pie chart. The total percent weight gain at the end of the dietary interventions (B),after prehabilitation (C),and the amount of weight loss following 5 days of antibiotic exposure (D) are displayed. The survival curves (E) demonstrate the survival differences between WD (n = 14), SD (n = 14), 3DP (n = 21), and 7DP (n = 9) * p< 0.05, ** p< 0.01, *** p< 0.001, **** p <0.0001
DietPreHab causes significant compositional alterations in the gut microbiota.
The preoperative stool microbiota was assessed to determine whether a non-invasive biomarker could be used to reflect the adequacy of DietPreHab to reverse the impact of WD. After DietPreHab, there was a significant reduction in alpha diversity in both 3DP and 7DP (Figure 2A). Beta diversity based on weighted unifrac demonstrated a distinct clustering of SD separate from WD, 7DP, and 3DP (Figure 2B). When assessed at the phyla level, the stool microbiota of both 3DP and 7DP was similar to that of SD (Figure 2C). There was a significant increase in the abundance of Bacteroidetes and a significant reduction in Proteobacteria and Firmicutes after both 3 and 7 days of pre-habilitation (i.e., 3DP, 7DP) (Figure 2D–F).
Figure 2:
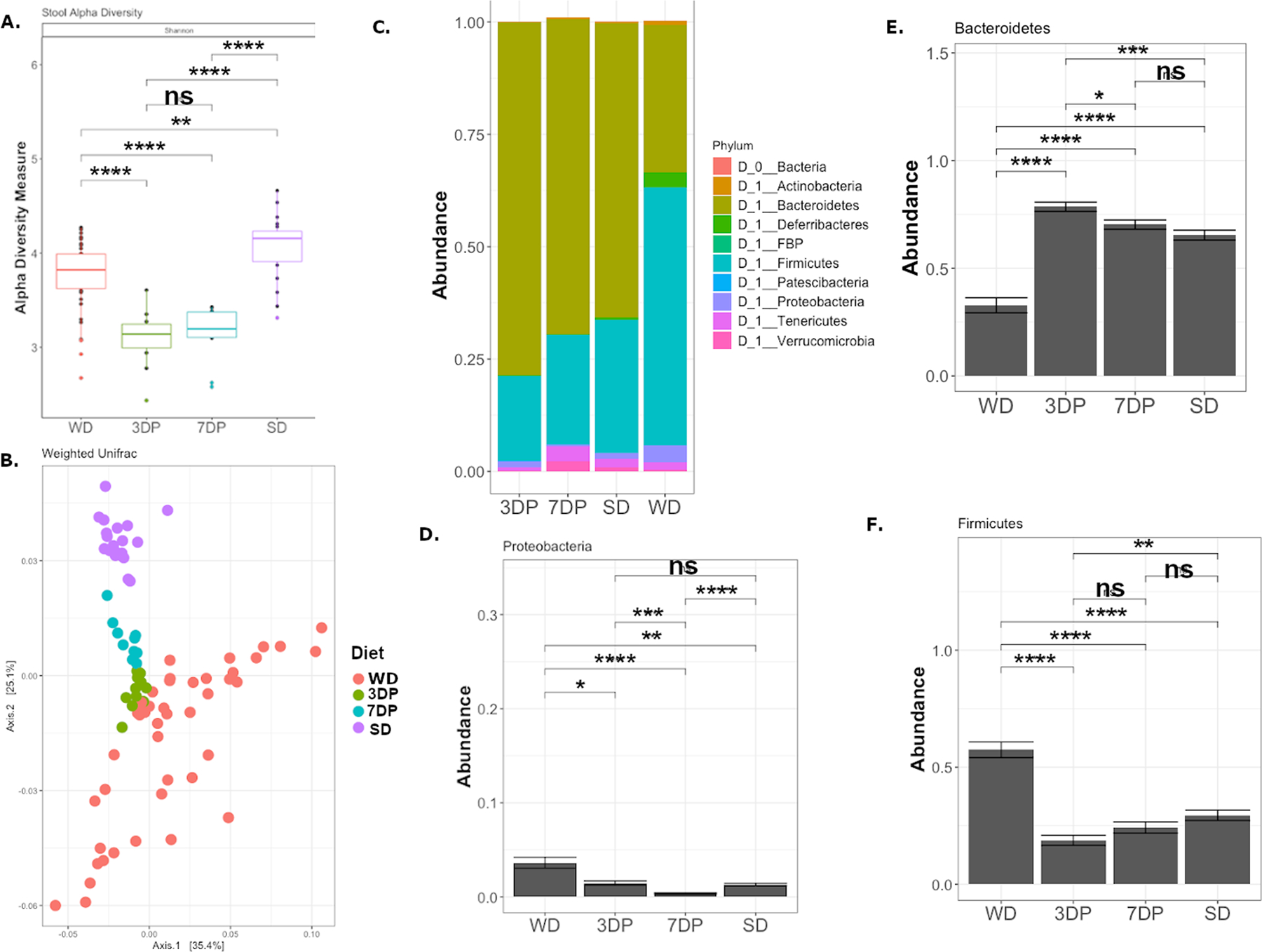
Dietary prehabilitation results in restoration of SD stool microbiota composition after 3 days of prehabilitation. 16S rRNA sequencing was performed on stool microbiota for WD (n = 44), SD (n = 20), 3DP (n = 11), and 7DP (n = 10) mice prior to antibiotic exposure. (A.)The Shannon alpha diversity is reduced after dietary prehabilitation, and (B.) the beta diversity as measured by weighted unifrac demonstrates distinct clustering of the WD, SD, and prehabilitation (3DP and 7DP) stool microbiota. The composition of the stool microbitoa at the phyla level demonstrates similarities between SD, 7DP, and 3DP (C). (D) The abundance of Proteobacteria, Bacteroidetes (E), and Firmicutes (F) were compared across SD, WD, 3DP, and 7DP. * p< 0.05, ** p< 0.01, *** p< 0.001, **** p <0.0001
The functional sequelae of DietPreHab on the microbiota were investigated by measuring the fecal levels of SCFAs (butyrate, propionate, and acetate) using GC-MS. SCFA levels were measured in the cecum of WD and SD fed mice as the cecum harbors the densest biomass of bacteria in the gut. WD resulted in a significant reduction in butyrate and acetate within the cecum (Supplemental Figure S2). When stool SCFAs were assessed as nmol per mg of expelled stool, there was a similar trend as seen in the cecum with SD fed mice having higher levels of butyrate and acetate as compared to WD fed mice (Figure 3A–C). Interestingly, butyrate levels dropped and remained low in 3DP mice. In contrast, butyrate levels after 7 days of pre-habilitation trended toward levels observed with a SD, not statistically significant.
Figure 3:
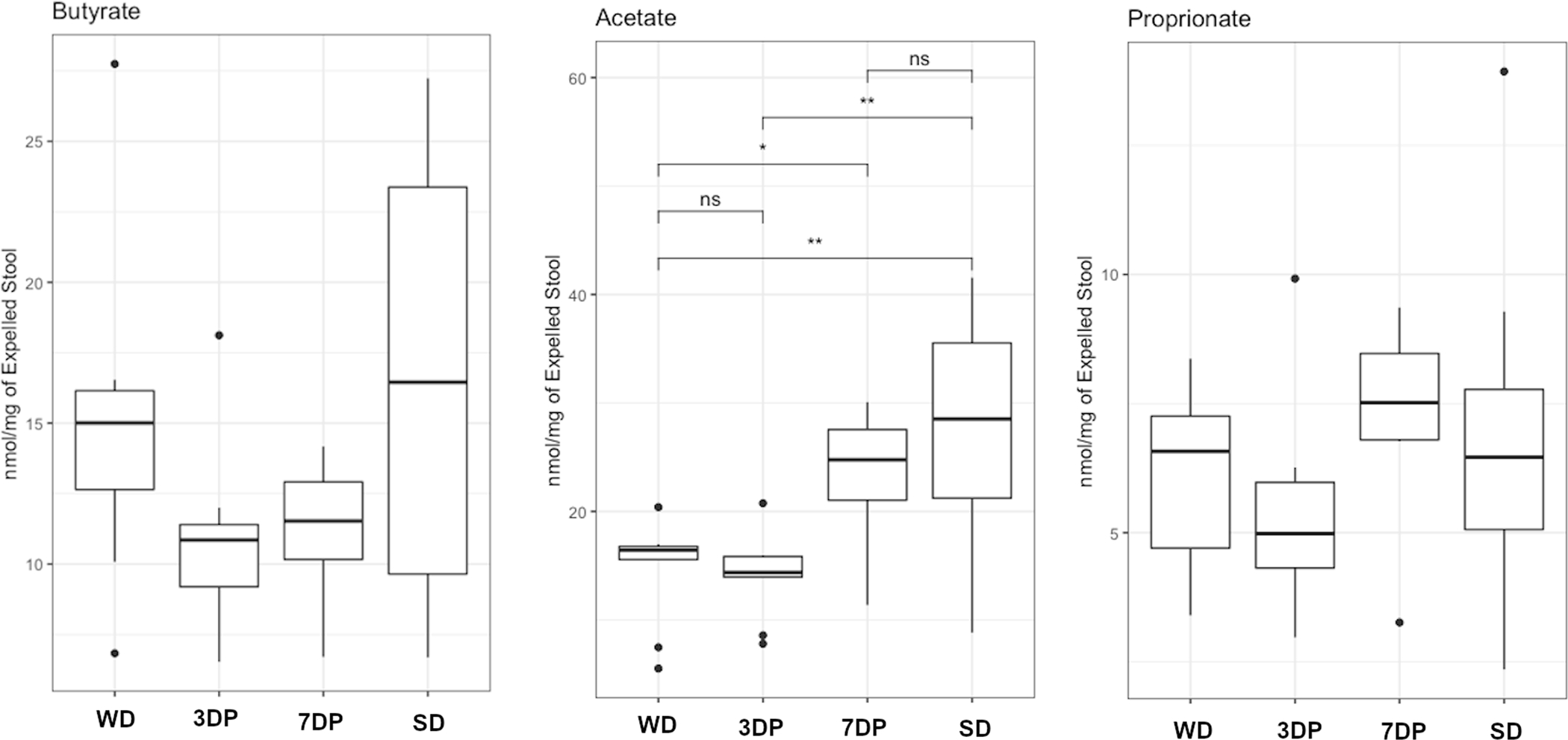
Stool SCFA are altered during DietPreHab. Stool butyrate levels are decreased in 3DP (n = 5) and 7DP (n = 5) compared to both SD (n = 5) and WD (n=5). Stool acetate levels are reduced in WD and 3DP mice compared to 7DP and SD mice. Propioinate levels are similar across all groups.
Rapid bloom of Bacteroidetes by DietPreHab fails to improve postoperative survival
We have seen a transition from a Firmicutes dominance in WD to a Bacteroidetes dominance seen in SD, 3DP, and 7DP. In order to determine if DietPreHab induced alterations in the abundance of Bacteroidetes and Firmicutes was sufficient to alter outcome, mice began the regimen of DietPreHab and stool levels of Bacteroidetes and Firmicutes were measured daily by qPCR (Figure 4A). Once the stool microbiota composition switched from Firmicutes to Bacteroidetes dominance, mice received antibiotics, starvation and underwent a 30% hepatectomy (ASH), referred to as the prediction group (Figure 4A). Prior to starting DietPreHab, all mice displayed a Firmicutes dominance microbiome over Bacteroidetes with an average Firmicutes abundance of 65.2% and Bacteroidetes abundance of 31.9%. After 24 hours of pre-habilitation, 75% of the prediction group mice had a reversal to Bacteroidetes dominance and all mice had a reversal to Bacteroidetes dominance after 48 hours. However, the prediction group had outcomes similar to WD indicating that single time measurements of Bacteroidetes and Firmicutes was not sufficient to improved survival from ASH (Figure 4B).
Figure 4:
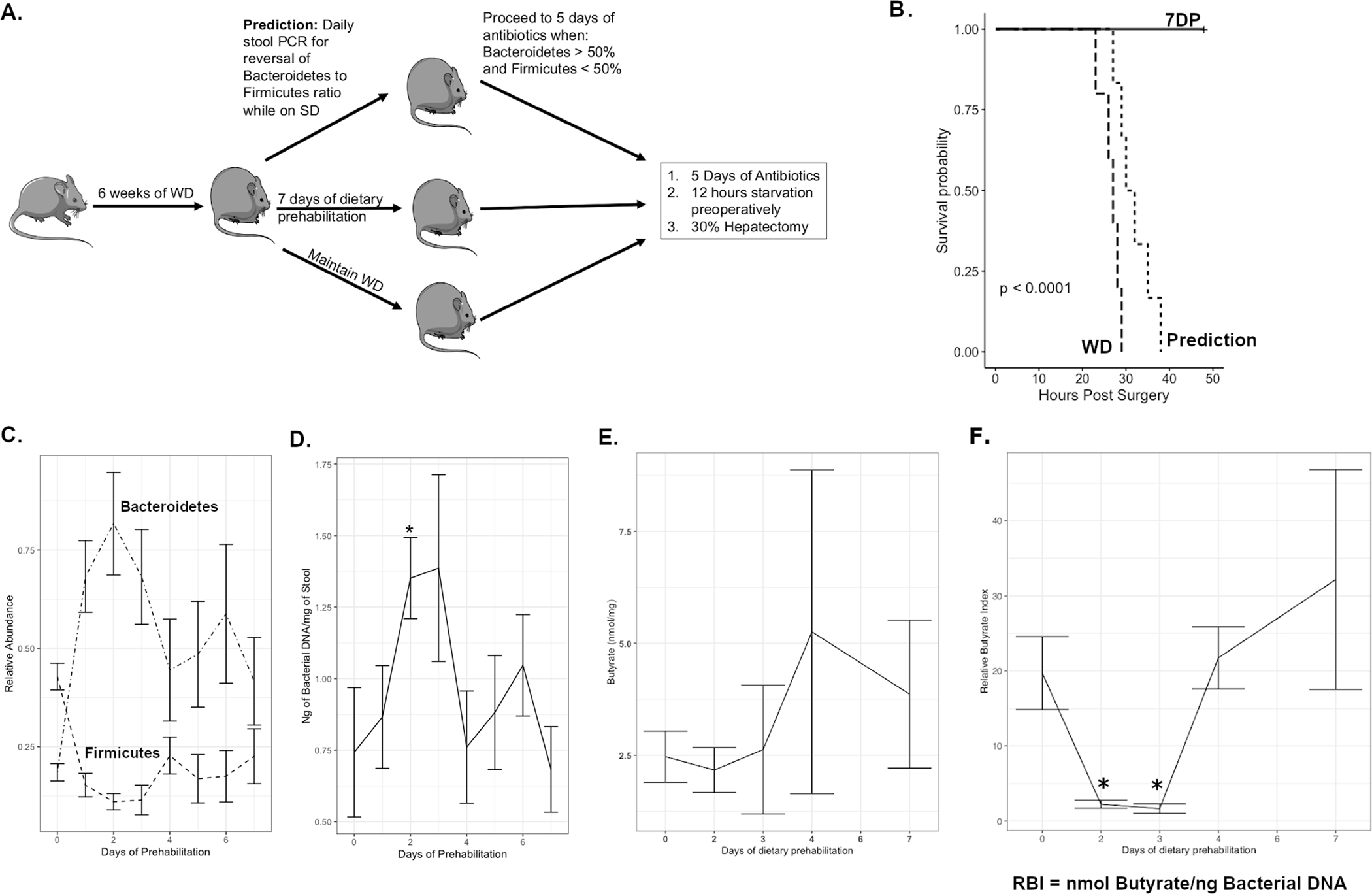
Microbiota compositional changes are not sufficient to predict survival during dietary prehabilitation. (A.)The experimental design outlining the prediction group which underwent daily PCR of the stool to determine when the mice would proceed to antibiotics, starvation and surgery. The survival differences between 7DP (n = 5), Prediction (n = 8), and WD (n = 5) are outlined in B. The abundance of Bacteroidetes and Firmicutes as measured by PCR during the course of dietary pre-habilitation (D, p < 0.001 Kruskal-Wallis) and bacterial load within the stool (E, Kruskal-Wallis p = 0.07, wilcox pairwise comparison p < 0.05 on day 2 vs day 0 and day 1). The stool butyrate levels (F) and relative butyrate index(RBI) which is the butyrate normalized to the bacterial load (G) demonstrate the alteration during DietPreHab (Kruskal-wallis p < 0.05, pairwise day 0 compared to day 2 (p = 0.06) and day 3 (p = 0.01))
In order to clarify the changes in the microbiota over the course of DietPreHab, alterations in Bacteroidetes and Firmicutes were measured daily during DietPreHab. There was a significant spike in levels of Bacteroidetes (Kruskal-Wallis, p < 0.001) and a drop in Firmicutes (Kruskal-Wallis, p < 0.001) after 24–48 hours of DietPreHab, followed by a gradual decrease of Bacteroidetes and increase of Firmicutes reaching stable levels after 1 week (Figure 4C). In addition to the spike in abundance of Bacteroidetes at the early DietPreHab period, there was an increase in bacterial load (ng bacterial DNA/mg of stool, Kruskall-wallis p = 0.07) in the first two days that then decreased reaching a stable level between 4 and 7 days of DietPreHab (Figure 4D).
Given the failure of fecal composition to predict outcomes, we focused on fecal SCFAs levels during DietPreHab. Over the course of DietPreHab, there was an initial reduction in fecal butyrate and acetate followed by a steady increase; however, these levels were never completely restored to the levels of SD fed mice. The initial reduction in stool SCFA may represent an increase in utilization by the host or other bacteria after a deprivation of SCFAs associated with WD (Figure 4E), suggesting that spot concentrations of SCFA in feces may not be an adequate indication of the amount of SCFA present. To better understand the relationship between bacterial alterations and their contribution to butyrate levels, a relative butyrate index was developed. The relative butyrate index (RBI) was calculated by normalizing the butyrate levels to the bacterial load. When the RBI was calculated, a significant decrease in RBI on day 2 and 3 (p <0.05) was observed with DietPreHab followed by an increase at day 7. Taken together, these data indicate that neither stool microbiota compositional changes alone nor SCFA concentrations alone are sufficient to predict survival. Rather, the functional output of the microbiota relative to bacterial biomass present may provide a more accurate index for tracking how the microbiota respond to dietary manipulation.
WD and SD mice display distinct butyrate producing taxa
To better understand the functional changes occurring within the stool microbiota, the functional profile of WD and SD fecal microbiota were predicted from 16S rRNA gene amplicon sequencing using PICRUSt27. This allows for the determination of baseline functional changes prior to DietPreHab. WD and SD stool microbiota had distinct functional profiles (Figure 5A) with over 200 significantly altered pathways.
Figure 5:
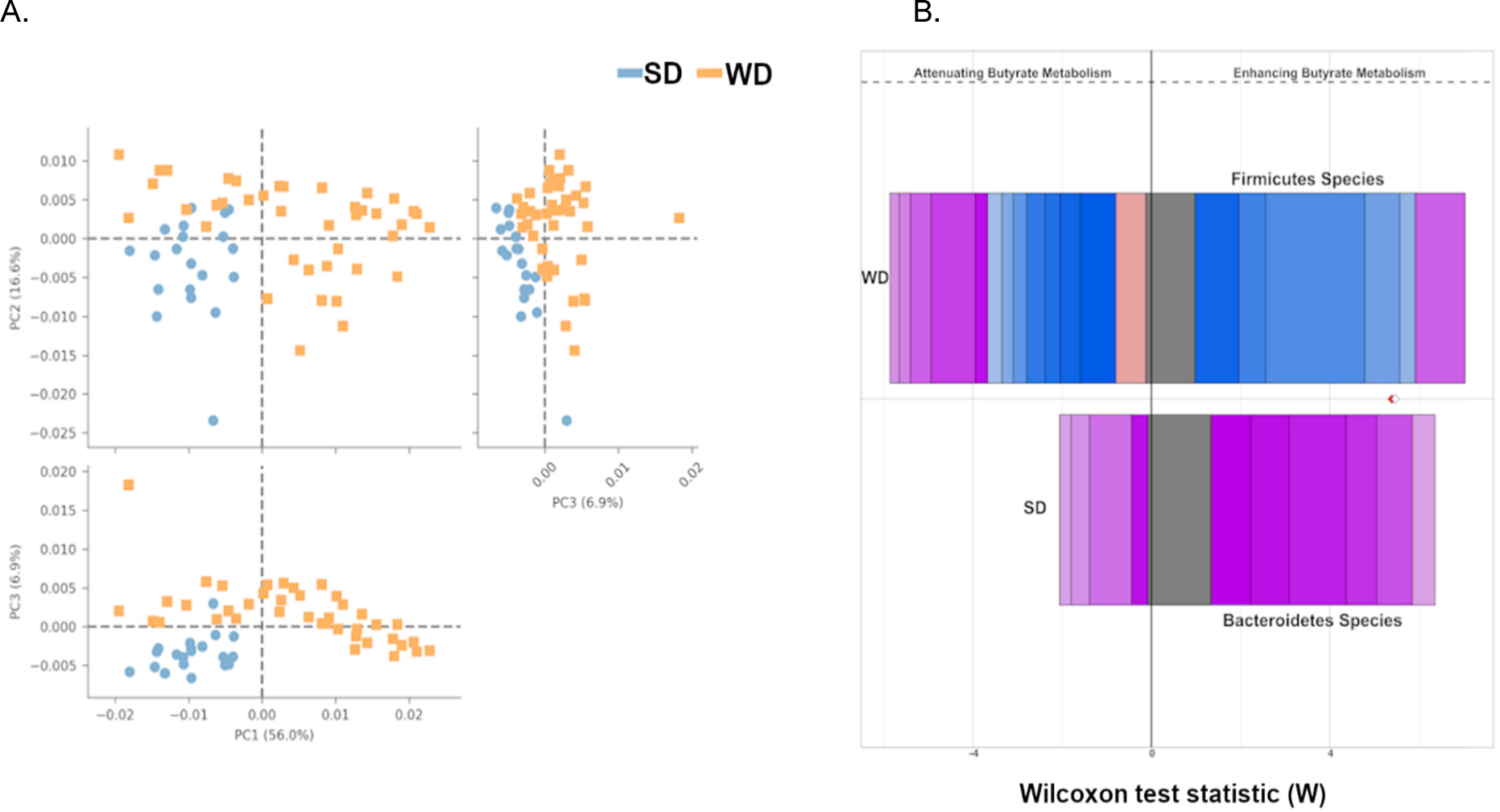
WD and SD stool microbiota have distinct functional profiles. The WD and SD microbiota profiles demonstrate clustering by diet after analysis with STAMP (A). FishTaco was used to assess the contributions of differentially abundant taxa to significantly altered pathways between SD and WD. The Wilcoxon rank sum score represents the degree of alterations contributed by the taxa for a given pathway. The top bar represents WD taxa contributing to the pathway and the bottom bar represents SD taxa contributing to the pathway. The bars to the left of center represent taxa that attenuate the pathway and bars to the right of center represent taxa that enhance the pathway. The different colors on the bar represent different taxa. Comparison of butyrate metabolism between WD and SD (B) demonstrates significantly different taxa contributing to butyrate metabolism between SD (Bacteroidetes – Pink/Purple) and WD (Firmicutes - Blue).
When assessed from a compositional standpoint, there were approximately 600 unique taxa that were significantly, differentially abundant between SD and WD fed mice. These taxa were predominantly of the order Bacteroidales in SD fed mice and Clostridiales in WD fed mice. Using FishTaco29, individual taxa contributions to the microbiota functional profile were determined (Supplemental Figure S3). When specifically focusing on butyrate metabolism, there was a significant difference in the taxa associated with butyrate production in WD and SD stool microbiota (Figure 5B). When comparing the taxa contributing to butyrate production, SD mice tended to have more taxa from the order Bacteroidales compared to WD fed mice having taxa from the order Clostridiales. However, when the abundance of butyrate producing enzymes were assessed across the entire stool microbiota, there were no major differences in abundance of butyrate pathways between WD and SD stool microbiota suggesting that the reduced butyrate production measured in the cecum was likely a consequence of elimination of dietary fiber rather than complete elimination of butyrate producers (Supplemental Figure S4).
DietPreHab results in early reduction of butyrate contributing taxa followed by its rapid restoration
Given the significant alteration in the functional profile of WD and SD stool microbiota, the functional alterations during DietPreHab were determined using PICRUSt. The alteration seen in the RBI during DietPreHab and its correlation with survival led to a focus on butyrate production which can be produced by four general pathways from glucose, succinate, lactate, and acetate31 (Figure 6A). Traditionally butyrate producing taxa are quanitifed by the terminal enzymes butyrate kinase (bukenz) and butyrate:acetyl-coA transferase (butenz)32. In order to determine how the alterations in the stool microbiota during DietPreHab contributed to butyrate production, the predicted presence of butyrate related enzymes within the significantly altered taxa during DietPreHab were analyzed to determine their contribution to butyrate production over the course of DietPreHab.
Figure 6:
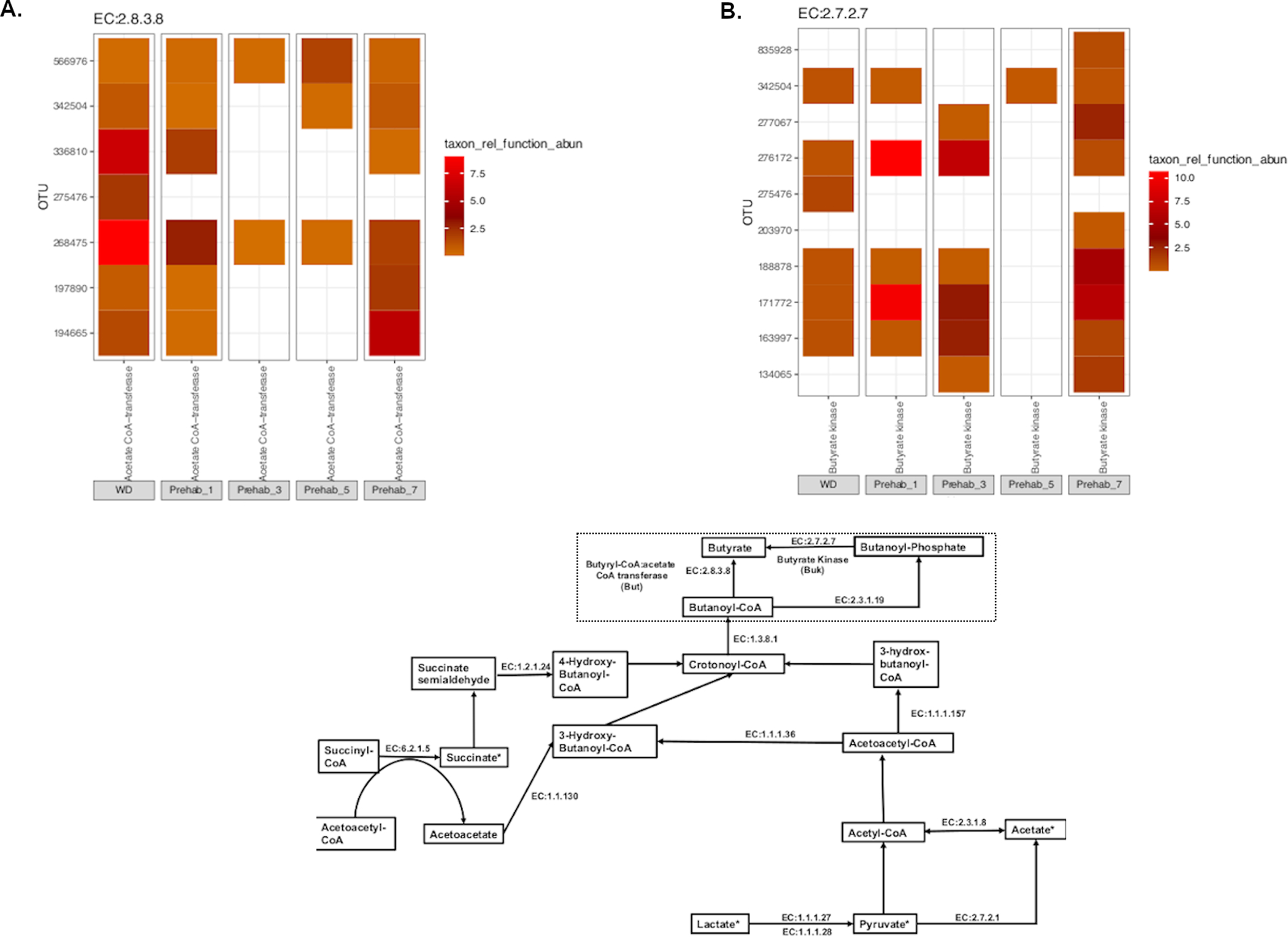
Butyrate contributing taxa are initially reduced during DietPreHab and restored at day 7. When assessing the contributions of butyrate to butyrate metabolism, individual enzymes that have been shown to contribute to butyrate metabolism were queried amongst all significantly differentially abundant taxa during DietPreHab as determined by DESeq (p < 0.05, FDR). Each individual enzyme corresponds to KEGG enzyme call number and can be related to butyrate prediction as indicated by the pathway in A (modified from KEGG pathway 00650). The heatmaps for each individual enzyme represent the relative taxon contribution abundance to a given enzyme. The end enzymes But (EC:2.8.2.8) and Buk (EC:2.7.2.7) are demonstrated in B and C respectively.
When focusing on the terminal enzymes bukenz and butenz, there is an initial reduction in bukenz and butenz containing taxa during early DietPreHab that is restored by day 7 (Figure 6B,C). This pattern of initial reduction and then restoration at the end of DietPrehab was consistent for the majority of butyrate related taxa (Supplement Figure S5). The new taxa present in 7DP containing the enzyme bukenz included 4 additional members of the order Clostridiales. Butyrate producing bacteria were initially decreased during DietPreHab and then were replaced by a new community of butyrate producing bacteria by day 7 of prehabilitation corresponding with the trend in RBI. This initial loss of butyrate producers may explain why DietPreHab was ineffective at improving surgical outcomes prior to 7 days. Additionally, when the functional profile of the stool microbiota on the early, ineffective days of DietPreHab (days 1 and 3) was assessed, the functional profile focused on cell wall synthesis and division corresponding with the rapid increase in bacterial load (Supplemental Figure S6) which correlates with increased bacterial load.
Discussion:
In the line with our previous findings5, the current work confirmed the adverse influence of a high-fat, low-fiber WD on the intestinal microbiome and its association with a poor outcome when the host is exposed to antibiotics, short-term starvation and operative trauma. The outcome of surgically operated mice exposed to WD was able to be reversed with as little as 7 days of dietary pre-habilitation using SD. Results from this model also revealed stool-based biomarkers may provide insight into how to assess microbiota readiness for surgery in the context of dietary pre-habilitation. We identified butyrate, a major secreted product of anaerobic bacteria that has known beneficial effects on the immune system, as a potential candidate biomarker 19,33–35.
An interesting finding in the present study was that when mice were fed a WD alone, it did not lead to complete loss of butyrate and butyrate-producing taxa. The attenuation of butyrate in this model, most striking in the cecum, may have been related to the absence of dietary fiber in WD as others have shown18,36. These findings, when combined with our previous data demonstrating that survival of mice following a 30% hepatectomy and exposure to healthcare associated pathogens can be improved when butyrate producing taxa are restored with a fecal transplant, suggest a central role for butyrate on mortality in this model19, and are reaffirmed with the alterations in butyrate-taxa seen in DietPreHab.
Data from this study also suggests that the utility of single time point microbiota measurements in the stool may not accurately reflect the dynamic changes to the intestinal microbiota that occur during dietary interventions. During dietary pre-habilitation, there is clearly an immediate response of the microbiota to the dietary intervention (Figure 4C) followed by a steady progression toward homeostasis after 1 week. Failure to account for both functional (i.e., SCFA) and compositional changes of the intestinal microbiota over time may limit the ability to accurately determine the readiness of the intestinal microbiota, and hence the host immune system, for a major surgical stress.
There are several limitation to this study. We performed our experiments utilizing a single strain of mice from a single vendor. This allowed for controlling the background genetics of the mice and allowed for maintaining consistency within the microbiome; however, the results of dietary pre-habilitation may be dependent on the baseline microbiota and genetic background of the mice. Additionally, a western diet has many negative metabolic alterations of the host that are known to increase the risk for surgical complications21,37 which we did not account for or directly assay. It is known that a western diet associated microbiome has a major impact on the host immune system and response to infection16,38. No cytokines or serum markers of inflammation were assessed in this study. Lastly, the precise cause of mortality among mice fed a WD and subjected to the combined exposure of ASH remains unknown. Despite extensive culturing of organs, a distinct pattern of bacterial dissemination of pathobiota from among the gut microbiota was not established as causal to death in this model as we have observed in many similar studies with this model5. Capturing the microbes responsible for mortality may require higher resolution next generation sequencing technology applied across multiple organs beyond the spleen using qPCR as performed here.
Supplementary Material
Supplemental Table S1. PCR primers
Figure S1: Total number of Serratia marcescens reads per mg of splenic tissue. Prehab non-survivors (n = 5) represents mice that underwent DietPreHab and did not survive ASH. Prehab-Survivors (n = 5) represent mice that underwent DietPreHab and survive ASH. SD (n = 5) mice underwent ASH after SD feeding and all survived. WD (n= 5) mice underwent ASH after WD and none survived.
Figure S2: GC-MS was performed on total cecal content for SD (n=5) and WD (n =5 ) to measure total amount of butyrate, acetate, and cecum
Figure S3: FishTaco analysis of 16S rRNA amplicons for SD and WD mice to determine functional profile of the microbiota. The top 20 most significant pathways are displayed on the y axis. Each color represents different taxa that either attenuate or enhance the referenced pathway. Top bar represents WD and bottom bar represents SD.
Figure S4: PICRUSt analysis was performed on 16S rRNA amplicons and the relative abundance of pathways that contribute to butyrate synthesis was compared between SD and WD mice.
Figure S5: Significant taxa over the course of DietPreHab determined by DESeq2 were compared for their contribution to butyrate pathway (K). Each panel (A-J) represents an enzyme within the butyrate synthesis pathway (K). The taxon relative abundance takes into account the presence of the given gene within the genome of the taxa multiplied by its relative abundance.
Figure S6: Using PICRUSt and STAMP, the functional profile of the microbiota during dietary prehab are compared on PCoA plot (A). FishTaco analysis (B) demonstrates a comparison between Day 3 and WD for the top 20 most significantly different pathways. Top bar for each pathway represents 3 days of DietPreHab and the bottom bar represents Day 0 (WD). Each color bar represents a unique taxa. The bars to the left represent taxa that attenuate function and those to the right represent taxa that enhance the given pathway.
Acknowledgments
Funding source: National Institutes of Health, NIGMS 5R01GMO62344-19
REFERENCES:
- 1.Waltz PK & Zuckerbraun BS Surgical Site Infections and Associated Operative Characteristics. Surg. Infect. 18, 447–450 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Zimlichman E et al. Health Care–Associated Infections: A Meta-analysis of Costs and Financial Impact on the US Health Care System. JAMA Intern. Med. 173, 2039 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Krezalek MA et al. Can Methicillin-resistant Staphylococcus aureus Silently Travel From the Gut to the Wound and Cause Postoperative Infection? Modeling the “Trojan Horse Hypothesis”: Ann. Surg. 267, 749–758 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Pickard JM & Núñez G Pathogen Colonization Resistance in the Gut and Its Manipulation for Improved Health. Am. J. Pathol. 189, 1300–1310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyoju SK et al. Mice Fed an Obesogenic Western Diet, Administered Antibiotics, and Subjected to a Sterile Surgical Procedure Develop Lethal Septicemia with Multidrug-Resistant Pathobionts. mBio 10, e00903–19, /mbio/10/4/mBio.00903–19.atom (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rollins KE, Javanmard-Emamghissi H, Acheson AG & Lobo DN The Role of Oral Antibiotic Preparation in Elective Colorectal Surgery: A Meta-analysis. Ann. Surg. 270, 43–58 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scarborough JE, Mantyh CR, Sun Z & Migaly J Combined Mechanical and Oral Antibiotic Bowel Preparation Reduces Incisional Surgical Site Infection and Anastomotic Leak Rates After Elective Colorectal Resection: An Analysis of Colectomy-Targeted ACS NSQIP. Ann. Surg. 262, 331–337 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Alverdy JC & Shogan BD Preparing the bowel for surgery: rethinking the strategy. Nat. Rev. Gastroenterol. Hepatol. 16, 708–709 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng KM et al. Recovery of the Gut Microbiota after Antibiotics Depends on Host Diet, Community Context, and Environmental Reservoirs. Cell Host Microbe 26, 650–665.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolodziejczyk AA, Zheng D & Elinav E Diet–microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 17, 742–753 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Zmora N, Suez J & Elinav E You are what you eat: diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 16, 35–56 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Challine A et al. Impact of Oral Immunonutrition on Postoperative Morbidity in Digestive Oncologic Surgery: A Nation-Wide Cohort Study. Ann. Surg. Publish Ahead of Print, (2019). [DOI] [PubMed] [Google Scholar]
- 13.Adiamah A, Skořepa P, Weimann A & Lobo DN The Impact of Preoperative Immune Modulating Nutrition on Outcomes in Patients Undergoing Surgery for Gastrointestinal Cancer: A Systematic Review and Meta-analysis. Ann. Surg. 270, 247–256 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Chowdhury AH et al. Perioperative Probiotics or Synbiotics in Adults Undergoing Elective Abdominal Surgery: A Systematic Review and Meta-analysis of Randomized Controlled Trials. Ann. Surg. 1 (2019) doi: 10.1097/SLA.0000000000003581. [DOI] [PubMed] [Google Scholar]
- 15.Sonnenburg ED & Sonnenburg JL The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol. 17, 383–390 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Napier BA et al. Western diet regulates immune status and the response to LPS-driven sepsis independent of diet-associated microbiome. Proc. Natl. Acad. Sci. 116, 3688–3694 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Las Heras V et al. Short-term consumption of a high-fat diet increases host susceptibility to Listeria monocytogenes infection. Microbiome 7, 7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh A, De Vadder F, Kovatcheva-Datchary P & Bäckhed F From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 165, 1332–1345 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Kim SM et al. Fecal microbiota transplant rescues mice from human pathogen mediated sepsis by restoring systemic immunity. Nat. Commun. 11(1):2354. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wahl TS et al. The Obese Colorectal Surgery Patient: Surgical Site Infection and Outcomes. Dis. Colon Rectum 61, 938–945 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurunathan U et al. Association of Obesity With Septic Complications After Major Abdominal Surgery: A Secondary Analysis of the RELIEF Randomized Clinical Trial. JAMA Netw. Open 2, e1916345 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaborin A et al. Phosphate-Containing Polyethylene Glycol Polymers Prevent Lethal Sepsis by Multidrug-Resistant Pathogens. Antimicrob. Agents Chemother. 58, 966–977 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyoju SK et al. Low-fat/high-fibre diet prehabilitation improves anastomotic healing via the microbiome: an experimental model: Diet and anastomotic healing after colonic surgery. Br. J. Surg. 107, 743–755 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolyen E et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMurdie PJ & Holmes S phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Douglas GM, Beiko RG & Langille MGI Predicting the Functional Potential of the Microbiome from Marker Genes Using PICRUSt. in Microbiome Analysis (eds. Beiko RG, Hsiao W & Parkinson J) vol. 1849 169–177 (Springer New York, 2018). [DOI] [PubMed] [Google Scholar]
- 28.Parks DH, Tyson GW, Hugenholtz P & Beiko RG STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manor O & Borenstein E Systematic Characterization and Analysis of the Taxonomic Drivers of Functional Shifts in the Human Microbiome. Cell Host Microbe 21, 254–267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y-W et al. Use of 16S rRNA Gene-Targeted Group-Specific Primers for Real-Time PCR Analysis of Predominant Bacteria in Mouse Feces. Appl. Environ. Microbiol. 81, 6749–6756 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esquivel-Elizondo S, Ilhan ZE, Garcia-Peña EI & Krajmalnik-Brown R Insights into Butyrate Production in a Controlled Fermentation System via Gene Predictions. mSystems 2, mSystems.00051–17, e00051–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vital M, Howe AC & Tiedje JM Revealing the Bacterial Butyrate Synthesis Pathways by Analyzing (Meta)genomic Data. mBio 5, e00889–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang PV, Hao L, Offermanns S & Medzhitov R The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. 111, 2247–2252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulthess J et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity 50, 432–445.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji J et al. Microbial metabolite butyrate facilitates M2 macrophage polarization and function. Sci. Rep. 6, 24838 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalby MJ, Ross AW, Walker AW & Morgan PJ Dietary Uncoupling of Gut Microbiota and Energy Harvesting from Obesity and Glucose Tolerance in Mice. Cell Rep. 21, 1521–1533 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shariq OA et al. Does Metabolic Syndrome Increase the Risk of Postoperative Complications in Patients Undergoing Colorectal Cancer Surgery?: Dis. Colon Rectum 62, 849–858 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Christ A, Lauterbach M & Latz E Western Diet and the Immune System: An Inflammatory Connection. Immunity 51, 794–811 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1. PCR primers
Figure S1: Total number of Serratia marcescens reads per mg of splenic tissue. Prehab non-survivors (n = 5) represents mice that underwent DietPreHab and did not survive ASH. Prehab-Survivors (n = 5) represent mice that underwent DietPreHab and survive ASH. SD (n = 5) mice underwent ASH after SD feeding and all survived. WD (n= 5) mice underwent ASH after WD and none survived.
Figure S2: GC-MS was performed on total cecal content for SD (n=5) and WD (n =5 ) to measure total amount of butyrate, acetate, and cecum
Figure S3: FishTaco analysis of 16S rRNA amplicons for SD and WD mice to determine functional profile of the microbiota. The top 20 most significant pathways are displayed on the y axis. Each color represents different taxa that either attenuate or enhance the referenced pathway. Top bar represents WD and bottom bar represents SD.
Figure S4: PICRUSt analysis was performed on 16S rRNA amplicons and the relative abundance of pathways that contribute to butyrate synthesis was compared between SD and WD mice.
Figure S5: Significant taxa over the course of DietPreHab determined by DESeq2 were compared for their contribution to butyrate pathway (K). Each panel (A-J) represents an enzyme within the butyrate synthesis pathway (K). The taxon relative abundance takes into account the presence of the given gene within the genome of the taxa multiplied by its relative abundance.
Figure S6: Using PICRUSt and STAMP, the functional profile of the microbiota during dietary prehab are compared on PCoA plot (A). FishTaco analysis (B) demonstrates a comparison between Day 3 and WD for the top 20 most significantly different pathways. Top bar for each pathway represents 3 days of DietPreHab and the bottom bar represents Day 0 (WD). Each color bar represents a unique taxa. The bars to the left represent taxa that attenuate function and those to the right represent taxa that enhance the given pathway.


