Significance
One cannot help but marvel at the precise organization of microtubule polymers in cellular structures such as the axoneme and the spindle. However, our understanding of the biochemical mechanisms that sculpt these arrays comes largely from in vitro experiments with a small number (one or two) of microtubules. This is somewhat akin to studying the architecture of multilane highways by studying one-lane streets. Here, we directly visualize depolymerizing microtubule arrays at individual microtubule and protofilament resolution using atomic force microscopy. Our results reveal differences in microtubule depolymerase activity and provide insights into how these differences in enzymatic activity on the nanometer scale can result in the differential remodeling of multimicrotubule arrays on the micron-length scale.
Keywords: atomic force microscopy, cytoskeleton, microtubule arrays, axoneme, kinesin
Abstract
The dynamic reorganization of microtubule-based cellular structures, such as the spindle and the axoneme, fundamentally depends on the dynamics of individual polymers within multimicrotubule arrays. A major class of enzymes implicated in both the complete demolition and fine size control of microtubule-based arrays are depolymerizing kinesins. How different depolymerases differently remodel microtubule arrays is poorly understood. A major technical challenge in addressing this question is that existing optical or electron-microscopy methods lack the spatial-temporal resolution to observe the dynamics of individual microtubules within larger arrays. Here, we use atomic force microscopy (AFM) to image depolymerizing arrays at single-microtubule and protofilament resolution. We discover previously unseen modes of microtubule array destabilization by conserved depolymerases. We find that the kinesin-13 MCAK mediates asynchronous protofilament depolymerization and lattice-defect propagation, whereas the kinesin-8 Kip3p promotes synchronous protofilament depolymerization. Unexpectedly, MCAK can depolymerize the highly stable axonemal doublets, but Kip3p cannot. We propose that distinct protofilament-level activities underlie the functional dichotomy of depolymerases, resulting in either large-scale destabilization or length regulation of microtubule arrays. Our work establishes AFM as a powerful strategy to visualize microtubule dynamics within arrays and reveals how nanometer-scale substrate specificity leads to differential remodeling of micron-scale cytoskeletal structures.
The dynamic formation and dismantling of protein arrays underlie a broad range of cellular functions in both prokaryotes and eukaryotes. A prototypical example of dynamic polymeric protein structures are micron-scale arrays of microtubules, which assemble into essential cellular machines and tracks such as the mitotic spindle in dividing cells, axonal arrays in neurons, and axonemes in cilia and flagella. The microtubule itself is a complex cylindrical macromolecular assembly of, most commonly, 13 to 15 protofilaments that are composed of repeating α,β-tubulin heterodimers. The intrinsic dynamic instability of microtubules and its regulation by a host of different microtubule-associated proteins (MAPs) are critical for the assembly and disassembly of microtubule arrays (1). How nanometer-scale dynamics of protofilaments (∼4 nm) and microtubules (∼25 nm) result in the organization and remodeling of micron-scale multimicrotubule arrays remains poorly understood.
In vitro reconstitution and visualization by optical microscopy have provided tremendous insights into microtubule dynamic instability and its regulation by MAPs. However, these studies have been limited to single or pairs of microtubules, as light microscopy does not have the resolution to identify individual microtubules within a complex array of multiple microtubules. In addition, it is challenging to image individual protofilaments within each microtubule by this method. Structural intermediates of microtubule-remodeling reactions have been inferred from electron microscopy studies, but the single-snapshot nature of the technique lacks temporal resolution to follow reaction dynamics in real time. To address these technical limitations and offer insights into microtubule array remodeling at single-microtubule and protofilament resolution in real time, we employed atomic force microscopy (AFM) imaging (2–8). This technique allowed the direct visualization of microtubule depolymerization in two different arrays, antiparallel microtubule bundles as found in the mitotic spindle and doublet microtubule arrays that form axonemes in cilia and flagella (9–11).
A critical reaction that governs the size and stability of microtubule arrays is microtubule depolymerization, which is catalyzed by a class of enzymes known as microtubule depolymerases. This reaction is required for rapid large-scale reorganization of the cytoplasm. For example, the mitotic spindle is built and disassembled every time a cell divides, and the cilium is constructed and deconstructed each cell cycle (12–14). In addition to large-scale reorganization of microtubule networks and arrays, microtubule dynamics and its regulation are important for fine-tuning the size of microtubule arrays (15–17). A fundamental conundrum is how the same reaction, the removal of tubulin from microtubules, results in different outcomes, ranging from large-scale remodeling to fine length regulation of microtubule arrays.
Two prototypical depolymerases are the vertebrate kinesin-13 protein MCAK, and the budding yeast kinesin-8 protein Kip3p (18–21). While the nonmotile MCAK and processive Kip3p proteins have different mechanisms for arriving at the microtubule ends, enzymatically, MCAK and Kip3p are both catastrophe factors and catalyze the removal of tubulin from microtubule ends (22–25). Structural studies suggest that, at the microtubule end, both enzymes recognize the curved conformation of tubulin in a similar manner (26–28, 29, 30). Despite these similarities, these proteins differently regulate dynamic instability such that Kip3p limits the distribution of maximum microtubule lengths, whereas MCAK promotes rapid filament shortening (31). These differences are reflected in their distinct functions; kinesin-8 proteins are largely involved in length control of structures, such as the spindle and the cilium, while kinesin-13s are additionally implicated in large-scale cytoskeleton remodeling, such as the depolymerization of interphase microtubules during entry into mitosis and suppression of cilium biogenesis (32–36). However, what underlies the differences in activity of these prototypical kinesin-family depolymerases and how differences in depolymerase activity at the single-microtubule level translate to distinct remodeling of complex microtubule arrays remain unknown.
The AFM imaging reported here reveals the structural dynamics that underlie microtubule array destabilization and provides a framework for linking the action of enzymes on the nanometer-scale protofilaments to the remodeling of micron-scale arrays. The study sheds light on the long-standing question of how different depolymerases are tuned for distinct cellular activities such as rapid remodeling or length control of microtubule arrays. Our findings highlight differences in enzyme activity on the protofilament scale as a critical parameter that governs the fate of microtubules within complex structures, thereby dictating how such arrays are remodeled.
Imaging the Depolymerization of PRC1–Cross-linked Microtubule Arrays by AFM
As a first step toward imaging the depolymerization of microtubule arrays by MCAK, we reconstituted microtubule bundles using the antiparallel cross-linking protein PRC1 (Protein Regulator of Cytokinesis-1) on a mica surface (Fig. 1A) (see Methods and SI Appendix, Methods and Materials and Extended Methods). AFM images of the bundles reveal that PRC1 cross-linking results in two-dimensional (2D) microtubule arrays (Fig. 1 B and C). This is advantageous, as every microtubule in the bundle can be clearly distinguished visually (see Methods and SI Appendix, Methods and Materials and Extended Methods). This is reflected in a height plot as a series of peaks with an average height of ∼30 nm (Fig. 1D). The three-dimensional (3D) rendition of the images, obtained from the surface topography data of Fig. 1B, shows dense linkages connecting the overlapping microtubules in an array, consistent with the cooperative binding of PRC1 to overlapping microtubules (Fig. 1C) (37, 38).
Fig. 1.
MCAK depolymerizes microtubule protofilaments asynchronously. (A) Schematic of an antiparallel microtubule array cross-linked by PRC1 (dotted lines). (B) AFM image of a microtubule bundle cross-linked by 100 nM PRC1. Each microtubule within the flat 2D array can be clearly distinguished. (x-y scale bar, 70 nm.) The z-scale is 0 to 40 nm (dark to light brown). The AFM image is colored according to height from the surface. (C) The 3D rendition of a zoomed-in region from B. (D) The corresponding height profile from dotted line in B. (E) Successive AFM images show depolymerization of individual microtubules within a PRC1-cross-linked bundle by MCAK. The image at 0 min represents the first frame taken after adding MCAK (GMPCPP + taxol microtubules; PRC1: 100 nM; MCAK: 70 nM). (x-y scale bar, 100 nm.) The z-scale is 0 to 40 nm (dark to light brown). (F–K) Two examples of depolymerization from the experiment in E showing stripe-like appearance of depolymerizing protofilaments (boxes 1 and 2). (F and I) The height and (G and J) the corresponding 3D rendition of the AFM time-lapse images. (H and K) Corresponding height profiles from the black dotted line in F and I show that the stripiness in the images corresponds to protofilaments at different heights relative to the surface. (L) Schematic of a microtubule undergoing asynchronous protofilament depolymerization. The scanning rate is 4 min/frame in B and ∼3 min/frame in E with 256 × 256 pixels. The arrow in B and E indicates the scanning direction in the fast axis (SI Appendix, Figs. S1–S3).
We used two main criteria to set the imaging conditions for investigating microtubule dynamics within PRC1-cross-linked bundles (3 to 4 min/frame with 256 × 256 pixels): 1) the spatial and temporal resolution is suitable for imaging the entire array as well as individual microtubules within the bundle and 2) no sample damage is visible in the time frame of the experiment (15 to 30 min). Experiments were performed with both microtubules polymerized with the GTP-analog guanosine 5'-[α,β-methylene]triphosphate (abbreviated as "GMPCPP" hereafter) and microtubules polymerized with GMPCPP and stabilized with taxol (indicated as "GMPCPP + taxol" hereafter). After locating a microtubule bundle by AFM, MCAK was added into the sample chamber at the indicated concentrations, and a time-lapse image series was acquired (note: solution concentrations are reported throughout the manuscript; local concentrations on the mica surface may be different). The rate of microtubule depolymerization by MCAK in the absence of PRC1 detected by AFM is similar to that observed by total internal reflection fluorescence (TIRF) microscopy (SI Appendix, Fig. S1 A–C), which shows that the activity of the enzyme is intact in this assay (SI Appendix, Fig. S1 D and E; as mentioned earlier, the local enzyme concentration in the TIRF and AFM experiments is not directly comparable) (24, 31, 39). The time-lapse AFM imaging of PRC1-cross-linked bundles reveals that depolymerization of individual microtubules within an array by MCAK can be visualized in real time (Fig. 1E, SI Appendix, Fig. S2, and Videos 1 and 2). Depolymerization begins shortly after MCAK addition, as we observed depolymerization of microtubules within the arrays in the first image taken after MCAK addition. This is visualized in sections of microtubule that lack complete tubules (Fig. 1E, time = 0 min). In control experiments without MCAK, PRC1-cross-linked microtubules remain stable and do not depolymerize significantly during the experiment (SI Appendix, Fig. S3).
These observations reveal AFM imaging as a powerful method to examine dynamic changes in a multimicrotubule array with nanometer-scale spatial resolution, thus offering a view of structural changes that are extremely difficult to detect by other imaging methods (40).
MCAK Depolymerizes Microtubule Protofilaments Asynchronously and Propagates Defects within Cross-linked Bundles.
Close examination of the time-lapse AFM images revealed two striking features of the reaction intermediates. First, the loss of microtubules was associated with the appearance of stripe-like features in the AFM images, which correspond to protofilaments. For example, in the magnified time-lapse montages, partial and bidirectional depolymerization is observed at the microtubule ends (Fig. 1F–K and SI Appendix, Fig. S2A). The stripes indicate protofilaments at different heights from the surface, which are resolved because of the high resolution of AFM in the axis perpendicular to the mica surface (SI Appendix, Extended Methods). These observations show that, in the presence of MCAK, the depolymerization of protofilaments within a microtubule is asynchronous (Fig. 1F and I and SI Appendix, Fig. S2A). The stripiness is observed on non-cross-linked as well as cross-linked microtubules with one or two neighbors. These observations suggest that microtubule protofilaments depolymerize at different rates in the presence of MCAK, resulting in an asynchronous loss of protofilaments from the ends (Fig. 1L). To ensure that the features observed do not arise from AFM scanning, we increased the time interval between scans to 5 to 10 min after MCAK addition and found that the features were unaltered and independent of time interval (SI Appendix, Fig. S2 D–E). Similar results were observed with either GMPCPP or GMPCPP + taxol microtubules (Fig. 1F and I and SI Appendix, Fig. S2). Electron microscopy (EM) studies have described the appearance of curved protofilaments during microtubule depolymerization (25, 41). In AFM experiments, these transient structures can only be captured if they lie in the plane of the mica surface, as shown in SI Appendix, Fig. S2B.
Second, in addition to depolymerization from the ends, we observed that breaks can appear in the middle of microtubule arrays. These are likely to be defects formed by the loss of tubulin from the microtubule lattice. We find that defects can be propagated by MCAK, again accompanied by the protofilament-associated stripiness (Fig. 2 A–C and SI Appendix, Fig. S2C). Defects propagate both along the diameter and length of microtubules with different rates (Fig. 2 D and E), suggesting that, in the presence of MCAK, destabilization of interprotofilament interactions is slower than depolymerization along the length of individual protofilaments. The observed depolymerization rates in the two opposite directions along the microtubule length are consistent with the unequal rates of plus- and minus-end microtubule depolymerization by MCAK (Fig. 2D and SI Appendix, Fig. S1) (24). More defects were observed in our taxol-stabilized samples compared to GMPCPP, suggesting that polymerization conditions influence microtubule defects (42). Defect propagation was also observed in TIRF microscopy–based assays with GMPCPP microtubules in the presence of MCAK (300 to 500 nM) (SI Appendix, Fig. S1 A–C). Thus, while the number of defects observed in AFM may be a combination of preexisting lattice defects and any additional ones induced by the AFM tip, the TIRF and AFM data show that MCAK can propagate these defects to induce destabilization of arrays.
Fig. 2.
MCAK propagates defects within cross-linked bundles. (A) Successive AFM images showing depolymerization of individual microtubules within a microtubule bundle by MCAK at the indicated times. The image at 0 min represents the first frame taken after adding MCAK (GMPCPP + taxol microtubules; PRC1: 100 nM; MCAK: 70 nM). (x-y scale bar, 50 nm.) (B) Successive time-lapse montages of a zoomed-in region of a section of microtubule from the experiment in A (box 1) showing defect propagation on three microtubules within the array at the indicated times (arrows). “S” indicates the edge with slow depolymerization, and “F” indicates the edge with fast depolymerization. (x-y scale bar, 30 nm.) (C) The corresponding average height profiles along the length of the microtubule from the dotted line in B. (D). Box plots of the depolymerization rates of “slow (S)” and “fast (F)” events from defect propagation along microtubule length (median: PRC1: 100 nM; MCAK: 70 nM; GMPCPP + taxol microtubules: slow = 1 nm/min, n = 19; fast = 2 nm/min, n = 17; GMPCPP microtubules: slow = 3 nm/min, n = 18; fast = 6 nm/min, n = 18). (E) Box plots of the depolymerization rates from defect propagation events, in “diameter (D)” and “length (L)” directions (median: PRC1: 100 nM; MCAK: 70 nM; GMPCPP + taxol microtubules: diameter = 0.6 nm/min, n = 20; length = 3 nm/min, n = 20; GMPCPP microtubules: diameter = 2 nm/min, n = 22; length = 10 nm/min, n = 22). The scanning rate is ∼3 min/frame with 256 × 256 pixels. The arrow in A indicates the scanning direction in the fast axis. For D and E, the box plots show the median, the inner quartiles, and maximum and minimum values. Statistical calculations used an unpaired t test with Kolmogorov–Smirnov correction for non-Gaussian distribution. * indicates a P value of <0.05. **** indicates a P value of <0.0001 (SI Appendix, Fig. S2).
How does PRC1-mediated bundling of microtubules affect array destabilization by MCAK? To address this, we quantitatively examined the effect of neighboring microtubules in bundles on the depolymerization reaction. Quantitative measurement of the depolymerization rates of microtubules that have zero, one, or two neighbors (SI Appendix, Methods and Materials) showed that bundling has a protective effect on microtubules against MCAK-mediated depolymerization (SI Appendix, Fig. S1E). We find that the depolymerization rates of microtubules with two neighbors are threefold lower than microtubules with zero or one neighbor. Second, microtubule depolymerization rates depend on PRC1 concentration (SI Appendix, Fig. S1E). For instance, the depolymerization rate decreased when the solution concentration of PRC1 was increased 10-fold from 10 nM to 100 nM. This reduction could be alleviated by increasing the MCAK concentration by ∼10-fold. Under all conditions, the presence of two neighboring microtubules has a significant effect on the protection of bundles. This is likely to arise from the highly dense pattern of PRC1 occupancy in overlap regions. Consequently, microtubules with two neighbors, one on either side, are likely to have the least number of exposed plus/minus protofilament ends (hereafter referred to as “exposed protofilament ends”).
The features observed during AFM imaging of microtubule array depolymerization by MCAK suggest that a single or few protofilaments with exposed ends can be effective substrates for MCAK and that these protofilaments are asynchronously removed by the enzyme. Altogether, we demonstrate that microtubule depolymerization can be visualized by AFM in real time at single-microtubule and protofilament resolutions within arrays. By using this imaging modality, we provide a view of protofilament-level depolymerization by a microtubule depolymerase nearly two decades after such a possibility was hypothesized (43, 44). Therefore, we formally show that protofilament-level depolymerization occurs on microtubules within a larger array, in the presence of MCAK. The observed asynchronous loss of protofilaments and defect propagation suggest that MCAK can use exposed protofilament ends as substrates, as would be advantageous for large-scale destabilization of the microtubule arrays.
Structural Dynamics of Microtubule Depolymerization by Kip3p Are Distinct from MCAK.
To investigate whether different depolymerases exhibit distinct structural dynamics, we examined the depolymerization of PRC1-cross-linked microtubule bundles by Kip3p. We mainly focused on GMPCPP-microtubules, since depolymerization of GMPCPP + taxol microtubules by Kip3p is extremely slow (23). We first examined Kip3p activity on single microtubules in the absence of PRC1 (SI Appendix, Fig. S4A). The depolymerization rate increases with Kip3p concentration (SI Appendix, Fig. S4B) and is similar to the rates observed by TIRF microscopy (SI Appendix, Fig. S4 C and D). Depolymerization is primarily seen at one end of the microtubules, although we also observe some slow presumed minus-end depolymerization (SI Appendix, Fig. S4B, Inset). Low levels of minus-end depolymerization are also observed in TIRF assays at concentrations >50 nM (SI Appendix, Fig. S4 C and D). Enhanced local concentration of Kip3p (on the mica substrate or on the cantilever tip) may contribute to Kip3p-induced depolymerization activity from the minus-ends of the microtubules, as has been observed with a nonmotile Kip3p mutant (26). Control experiments confirm that the depolymerization in this experiment is enzyme-mediated (SI Appendix, Fig. S4E). We next examined Kip3p activity on PRC1-cross-linked microtubules. We selected a field in which the ends of several microtubules within the PRC1 bundle were in view, added Kip3p, and collected time-lapse AFM images (Fig. 3A, SI Appendix, Fig. S5 A and B, and Video 3). We observed distinct structural features of PRC1-bound cross-linked and individual microtubules depolymerized by Kip3p in comparison to those depolymerized by MCAK. First, in contrast to MCAK, no stripes were observed at the ends of depolymerizing microtubules, and the protofilaments were lost in unison, leaving only one or a few protofilament remnants (<5 nm in height) that were adhered on the mica surface (Fig. 3 B–D). This is reflected in the shifting of the entire edge of the corresponding height profiles during depolymerization without features of intermediate heights. Second, unlike with MCAK, we rarely observed defect propagation events using the same batch of microtubules. In the rare instances in which we saw depolymerization from the middle of a doubly stabilized GMPCPP–taxol microtubule, we again observed no stripes, suggesting that defects are propagated by Kip3p in unison (i.e., synchronously) (SI Appendix, Fig. S5B). Third, in contrast to MCAK, we did not see a significant effect of PRC1 concentration or neighbors on depolymerization rate (SI Appendix, Fig. S5C). Control experiments in the presence of adenosine triphosphate (ATP) alone confirm that the observed depolymerization of PRC1-cross-linked bundles in the presence of Kip3 is specific to the presence of depolymerases in the assay (SI Appendix, Fig. S3).
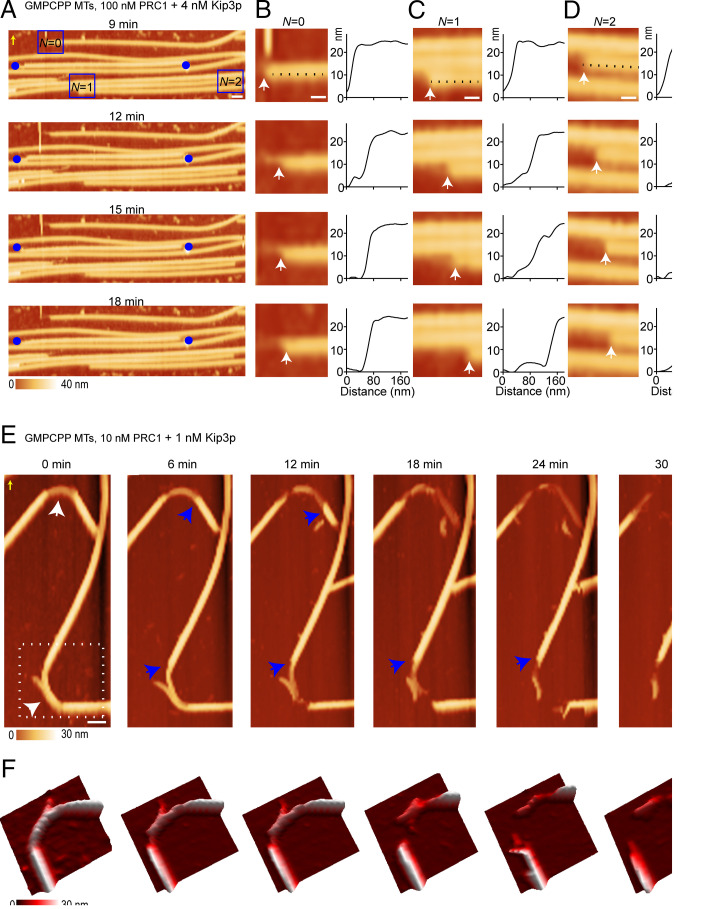
Fig. 3.
Structural dynamics of microtubule depolymerization by Kip3p are distinct from those of MCAK. (A) Successive AFM time-lapse images of a PRC1-cross-linked microtubule bundle in the presence of Kip3p at the indicated times (GMPCPP microtubules, PRC1: 100 nM; Kip3p: 4 nM). The blue circles are fiduciary marks, which show that the microtubules are depolymerizing and not gliding on the surface. (x-y scale bar, 100 nm.) (B–D) Zoomed-in regions from the experiment in A (boxes), showing bluntness at the depolymerizing microtubule end with number of neighbors N = 0 (B), N = 1 (C), and N = 2 (D). The height profiles corresponding to the dotted lines show that protofilaments at the ends of the microtubules are lost synchronously (white arrows). (x-y scale bar, 40 nm.) For A–D, the z-scale is 0 to 40 nm (dark to light brown). (E) Successive AFM time-lapse images show the destabilization of two highly curved regions (white arrows at 0 min) at the indicated times. Over time, the microtubule starts to depolymerize faster from one end with synchronous loss of protofilaments (blue arrows) (GMPCPP microtubules, PRC1: 10 nM; Kip3p: 1 nM). (x-y scale bar, 100 nm.) The z-scale is 0 to 30 nm (dark to light brown). (F) The 3D rendition shows a magnified view of this depolymerization activity (dotted box at 0 min in E). The scanning rate is ∼3 min/frame with 256 × 256 pixels. The yellow arrows in A and E indicate the scanning direction in the fast axis (SI Appendix, Figs. S4–S6).
We also observed that Kip3p destabilizes highly curved microtubules, consistent with the reported accumulation of Kip3 at these sites (26) (Fig. 3 E and F and SI Appendix, Fig. S6A). Destabilization of the curved region was observed neither in the presence of the motile conventional kinesin (K401) nor in the presence of ATP alone (SI Appendix, Fig. S6 B and C). We used time-resolved AFM to follow the structural intermediates of the process by which the curved segment disintegrates in real time in the presence of Kip3p (Fig. 3 E and F, SI Appendix, Video 4) (26). As seen in the zoomed 3D view, the curved section destabilized and depolymerized faster in one direction (Fig. 3F). Again, we observed that depolymerization of the entire microtubule occurs when most of the protofilaments in the curved region are lost. Overall, the features of microtubule depolymerization, such as synchronous depolymerization of protofilaments, lack of defect propagation, and accelerated breaking at curved microtubule segments by Kip3p in the absence of PRC1 (SI Appendix, Fig. S4A and S6A) are similar to those observed in experiments with PRC1 (Fig. 3 A–F and SI Appendix, Fig. S5 A and B). Together, these data suggest that, unlike MCAK, Kip3p depolymerizes protofilament ends synchronously. This is likely due to the accumulation of Kip3p at microtubule ends and a preference to stall at ends rather than at sections of partially exposed protofilament ends, such as those in defects.
Altogether, this suggests that the two depolymerases, Kip3p and MCAK, exhibit distinct preferences in terms of substrate specificity at microtubule ends and defects at the protofilament level and in the context of an array of bundled microtubules. These findings shed light on how the two depolymerases may be tuned for distinct functions. While the properties of MCAK make it well suited for remodeling of arrays and depolymerization at defects, Kip3p activity seems better aligned with a role as a length regulator at microtubule ends.
Visualizing the Depolymerization of Doublet Microtubules Using AFM.
As a step toward examining how other microtubule arrays are remodeled by depolymerases, we focused on the axoneme, an array of nine outer doublet and two central singlet microtubules, which forms the backbone of cilia (Fig. 4A). The doublets are composed of the A tubule, which contains 13 protofilaments, and the B tubule, which is an incomplete microtubule containing 10 protofilaments (45, 46). At the distal cilium tip, the doublets transition into an array composed of singlets (45). Axonemes are one of the most stable arrays of microtubules in cells, and dissociating them into soluble tubulin requires fairly harsh treatments, like sonication and detergent or specific ionic conditions (47, 48). Depolymerases of the kinesin-13 and kinesin-8 families are proposed to act on axonemes to control cilium length and stability (32–34). However, the activity of depolymerases on doublet microtubules has not been visualized, and it is unknown if and how these enzymes depolymerize doublets and impact axoneme stability.
Fig. 4.
Visualizing the activity of MCAK and Kip3p on doublet-microtubules using AFM. (A) Schematic of the axoneme structure. Axonemes consists of nine outer microtubule doublets (MTD). Each doublet contains an A tubule and a B tubule. Outer dynein arms (ODA), present on the A tubule, form a repeating pattern. (B) AFM height image of an MTD. The A and the B tubules in a MTD were distinguished by the height of the tubules in the joined doublet (25 and 35 nm) and by the periodic striations, which are separated by 30 nm (arrows). (x-y scale bar, 50 nm.) (C) The 3D representation of B and the corresponding height profile of the dotted line in B. (D) Successive AFM images show a 2D MTD sheet with MCAK at the indicated times. The zoomed-in region (boxed region) shows the depolymerization activity of alternate tubules in an array (blue arrows) and corresponding height profiles over time (dotted line). The height profiles show the deepening of the minima and changing of the asymmetric peak into a single sharp peak (dotted line, arrow) (-TED sample, MCAK: 7 nM). (x-y scale bars of the zoomed-out and zoomed-in images, 80 nm and 40 nm, respectively.) The z-scale is 0 to 40 nm (dark to light brown). (E) Successive AFM height and amplitude images show a microtubule doublet (D) and a singlet (S) in the presence Kip3p at the indicated times. The corresponding height profiles from dotted line show that the height of the doublet doesn’t change over time, but the height of the singlet reduces with time (dotted lines) (-TED sample, Kip3p: 5 nM). (x-y scale bar, 100 nm.) The z-scale is 0 to 50 nm (dark to light brown). (F) Box plots of the depolymerization rates of A and B tubules in the -TED sample with MCAK and Kip3p. MCAK: A rate = 2 nm/min, n = 12; B rate = 23 nm/min, n = 10; Kip3p: A rate = 0.2 nm/min, n = 22; B rate = 3 nm/min, n = 22. Data were pooled from experiments with protein concentrations less than 10 nM. Statistical calculations used an unpaired t test with Kolmogorov–Smirnov correction for non-Gaussian distribution. * indicates a P value of <0.05. The scanning rate is ∼4 min/frame in B and ∼3 min/frame in D–E with 256 × 256 pixels. The yellow arrows in B, D, and E indicate the scanning direction in the fast axis. For F, the box plot shows the median, the inner quartiles, and maximum and minimum values (SI Appendix, Figs. S7 and S8).
We purified axonemes from Lytechinus pictus sea urchin sperm. We first focused on individual doublets present in this sample. In high spatial resolution mode, the AFM images clearly show two joined tubules, with heights of 25 and 35 nm, respectively (Fig. 4 B and C). Another feature of these microtubule doublets is the periodic repeats separated by ∼30 nm, which are likely to be the outer dynein arms on the A tubule (Fig. 4 B and C). Both of these structural features of microtubules are consistent with cryo-electron microscopy (cryo-EM) structures of the axoneme and previous AFM images of doublet microtubules (49, 50). We examined whether depolymerases act on doublet microtubules by performing AFM experiments with 2D doublet sheets from partially dissociated axonemes composed of two or more doublets linked together (SI Appendix, Fig. S7 A–D). In the presence of MCAK, we observed that one of the tubules depolymerizes from both ends first, followed by the depolymerization of the second tubule (Fig. 4D, SI Appendix, Fig. S7 E and F, and Video 5). This can be visualized in the height plots as a deepening of the minima between adjacent doublets (dotted lines) and the conversion of an asymmetric broad peak to a single sharp peak (indicated by black arrow) (Fig. 4D). The AFM time-lapse data of a microtubule doublet and a singlet in the same field of view in the presence of Kip3p reveal that the doublet depolymerizes very slowly (rate = 4 nm/min), but a single microtubule in the same field-of-view is depolymerized on the same timescale (rate = 26 nm/min) as in vitro polymerized GMPCPP–microtubules (Fig. 4E and Video 6). This is consistent with previous fluorescent studies with the kinesin-8 protein Kif19A, in which single microtubules, nucleated from an axoneme, were depolymerized, while the axoneme itself appears intact (32). Within the doublet, the average depolymerization rate measured for the A and B tubule in the presence of Kip3p was extremely slow (0.2 to 3 nm/min) (Fig. 4F). In contrast, in the presence of MCAK, B tubules are depolymerized at a significantly faster rate compared to A tubules (Fig. 4F; A rate = 2 nm/min; B rate = 23 nm/min). Control experiments with the doublets without enzyme showed no significant depolymerization over the same timescale (SI Appendix, Fig. S7G). Taken together, these data suggest clear differences in how MCAK and Kip3p act on microtubule doublets.
Axonemes are heavily decorated with several proteins. Prior reports have shown that treatment with a solution of tris(hydroxymethyl)aminomethane, ethylenediaminetetraacetic acid and dithiothreitol (Tris-EDTA-DTT; abbreviated as TED) can dissociate a subset of axonemal proteins, particularly the dyneins (51). Our AFM imaging revealed that these TED-treated doublets have a smoother surface in comparison to untreated doublets (SI Appendix, Fig. S8 A and B). We examined if TED treatment alters the depolymerization of doublets. Similar to the doublet samples without TED treatment (Fig. 4), we observed bidirectional depolymerization of one tubule of an isolated doublet with MCAK (SI Appendix, Fig. S8C). TED treatment resulted in faster depolymerization of both tubules (compared to TED-untreated sample with MCAK), with the B tubule being lost at a higher rate than the A tubule (SI Appendix, Fig. S8D; A rate = 16 nm/min; B rate = 38 nm/min). In the presence of Kip3p, TED treatment also permitted slow depolymerization of the B tubule (SI Appendix, Fig. S8 D and E; A rate = 2 nm/min; B rate = 10 nm/min). To ensure that depolymerization did not arise from frequent AFM scanning, we imaged the doublets every 10 min upon adding a depolymerase (SI Appendix, Fig. S8F). The results show that the depolymerization activity of the doublet with MCAK is independent of AFM scanning. In addition, in control experiments without the enzyme, TED-treated doublets showed no significant depolymerization (SI Appendix, Fig. S8G).
Taken together, our results show that doublet microtubules can be enzymatically depolymerized with MCAK and that the rate of depolymerization of the B tubule in a doublet is faster than that of the A tubule. In contrast, doublet microtubules are poor substrates for Kip3p, even when the doublet is stripped of associated proteins. Thus, proteins that have Kip3p-like properties may be selectively functional at the distal cilium tip, where the axoneme is composed mostly of singlet microtubules, to fine-tune cilium length. In contrast, the properties of depolymerases like MCAK make them better suited to depolymerize both singlets and doublets for processes such as cilia disassembly or inhibition of ciliogenesis.
Destabilization of Axonemal Structures by MCAK.
Our observations with the doublet microtubule depolymerization raise the question of how preferential depolymerization of B tubule by MCAK impacts stability of the entire axoneme. We first imaged isolated axonemes adsorbed onto the mica surface. Axonemes were ∼200 nm in height, which is consistent with the diameter of the axoneme from cryo-EM measurements (Fig. 5 A–D and SI Appendix, Fig. S9 A–F) (49). The AFM amplitude image showed longitudinal striations, which likely arise from the nine outer doublets that are around ∼20 to 30 nm apart (Fig. 5B and SI Appendix, Fig. S9B). In AFM imaging experiments with ATP alone, we observed no substantial change in the overall size and integrity of the axoneme over time (Fig. 5E).
Fig. 5.
Destabilization of axonemal structures by depolymerases. (A–C) AFM height (A), amplitude (B), and 3D (C) images of an intact axoneme from L. pictus sea urchin sperm. The AFM amplitude (B) and the 3D (C) images show longitudinal striations, likely from the nine outer doublets, which are ∼20 to 30 nm apart. (x-y scale bar, 300 nm.) The z-scale is 0 to 200 nm (dark to light brown). (D) The height profile of the selected dotted line from height image in A shows a maximum height of ∼200 nm. (E) Successive AFM images of an axoneme with 1 mM ATP at the indicated times. With ATP alone, no significant change was observed in the axoneme structure. (x-y scale bar, 300 nm.) The z-scale is 0 to 300 nm (dark to light brown). (F) Successive AFM images of an axoneme in the presence of MCAK (1 nM) at the indicated times. At t = 0, the axoneme has been partially frayed. (x-y scale bar, 500 nm.) The z-scale is 0 to 60 nm (dark to light brown). (G) Schematic of proposed intermediate that results in unfurling of the axoneme with MCAK. The scanning rate is ∼4 min/frame in A–C and ∼3 min/frame in E–F with 256 × 256 pixels. The yellow arrows in A, E, and F indicate the scanning direction in the fast axis (SI Appendix, Fig. S9).
Next, we selected an axoneme, added depolymerase (t = 0), and monitored the changes in the axoneme structure over time. With the addition of MCAK, we observed a rapid loss of microtubules from the axoneme. For example, we documented that an intact axoneme (∼200 nm in max height) loses most of its tubules upon MCAK addition (SI Appendix, Fig. S9G). Occasionally, we were able to capture intermediates in this reaction due to adsorption of the dissociated microtubules on mica (Fig. 5F, SI Appendix, Fig. S9H, and Video 7). In the example shown in Fig. 5 F and SI Appendix, Video 7, MCAK addition resulted in the rapid unfurling of the axoneme and the scattering of microtubules on the surface.
These data suggest that the preferential depolymerization of one tubule in a doublet by enzymes such as MCAK may result in disintegration of the structure by breaking the links between doublets in an axoneme (Fig. 5G).
Discussion
Collectively, this study demonstrates the power of AFM imaging in visualizing dynamic processes within dense microtubule arrays, in real time and at spatial resolution that allows the observation of individual protofilaments. AFM imaging of depolymerizing microtubule arrays revealed previously unseen structural dynamics and provided mechanistic insights into how differences in the activities of microtubule-remodeling factors at the level of protofilaments and microtubules can lead to differential fates of complex multimicrotubule arrays.
Our data show that two prototypical depolymerases of the kinesin superfamily, the kinesin-13 MCAK and the kinesin-8 Kip3p, depolymerize complex microtubule arrays in distinct ways. How is this achieved at the mechanistic level? The answer to this question has not been clear despite more than two decades of enzymatic and structural studies. From the perspectives of enzymology and substrate preference, Kip3 and MCAK are similar. First, both enzymes use family-specific loops in the motor domains to target curved microtubule ends (26–28). Second, depolymerization by these motors is not strictly coupled to ATP hydrolysis. Instead, the role of ATP hydrolysis is primarily proposed to dissociate the enzymes from tubulin dimers after depolymerization (25, 26, 44, 52). While an obvious difference lies in the diffusive scanning of MCAK and the processive motility of Kip3 on microtubules, these differences largely reflect the mechanism by which the enzymes arrive at microtubule ends and not their depolymerase activity (22–25). Indeed, it is observed that a nonmotile Kip3 can depolymerize microtubules (26). Our data suggest that the observed differences stem from distinct substrate specificity of the depolymerases at the protofilament level (Fig. 6). We propose that MCAK molecules can effectively depolymerize an exposed protofilament end segment that it encounters at the middle or end of the lattice. This is reflected in the observation that MCAK can propagate defects and protofilaments can be asynchronously depolymerized from the ends. In contrast, a short exposed protofilament end, like that in a defect, is not sufficient for Kip3-mediated depolymerization from that site. Instead, Kip3 molecules synchronously depolymerize from the multiprotofilament ends of microtubules, where they accumulate. This is consistent with the previously observed cooperativity in Kip3 activity (53, 54). This difference in protofilament-level substrate preference of the two enzymes leads to distinct outcomes of their activity on cross-linked microtubules and doublets despite similarities in tubulin binding and ATPase activity at microtubule ends.
Fig. 6.
Summary. The schematic illustrates the distinct structural dynamics and intermediates during microtubule depolymerization with different enzymes and its impact on microtubule arrays. In the context of an individual microtubule, 1) the loss of protofilaments is asynchronous with MCAK and synchronous with Kip3p, and 2) MCAK propagates lattice defects, whereas Kip3p does not. In the context of microtubule arrays, 1) cross-linking by PRC1 protects microtubules against depolymerization by MCAK and does not significantly influence the depolymerization by Kip3p, and 2) MCAK depolymerizes doublet microtubules and results in the destabilization of axonemal arrays. This arises from fast depolymerization of one tubule, which compromises the stability of the cylindrical doublet array.
Our results also provide insights into the structural mechanism by which these depolymerases differentially regulate "microtubule aging." Aging refers to the observation that catastrophe results from a multistep process, and its probability increases with microtubule lifetime (31). MCAK reduces this process to a first-order reaction, while Kip3 simply accelerates the rate of steps leading to catastrophe, and, consequently, MCAK is able to disassemble arrays faster than Kip3 (31). Current models propose that the catastrophe-vulnerable state arises from the accumulation of defects and flayed protofilament ends that have fewer laterally stabilized protofilaments (40, 55, 56). How might MCAK and Kip3p differently affect lateral stabilization? The protofilament-level substrate specificity and efficient depolymerization of exposed protofilament ends by MCAK would prevent or greatly reduce the inherent lateral stabilization of protofilaments, thus effectively eliminating the need for accumulation of laterally unstabilized protofilaments over time or aging. On the other hand, the stabilization of curved tubulin at microtubule ends by Kip3 will serve to increase the rate of aging but not the multistep nature of the process. Together, these observations provide a structural mechanism that can serve as a framework for understanding the molecular underpinnings of the aging process and its regulation by depolymerizing enzymes.
The difference in substrate preference at the protofilament level has multiple implications for structural dynamics and regulation of microtubules and the arrays and the role depolymerases play in these processes. First, lattice defects, which are sites with only a few exposed protofilament ends, are propagated by MCAK but not by Kip3p, suggesting that microtubule arrays with a higher level of lattice defects may be especially sensitive to MCAK activity. This may be advantageous for disassembly of microtubule arrays by MCAK, in conjunction with proteins, such as microtubule severing enzymes (57, 58). On the other hand, the inability to act on defects would be advantageous for depolymerases like Kip3 and other homologs of Kip3, such as Kif18A, that suppress microtubule dynamics for length control (59). Second, the restriction of Kip3p activity to microtubule ends makes it less sensitive to the cross-linking of microtubules by PRC1 compared to MCAK. This may arise from lower density of PRC1 cross-links at microtubule ends, the primary site of Kip3p action, either because of crowding of Kip3p at the ends or due to disruption of microtubule geometry at the ends. This feature of Kip3p would allow it to act as an effective length regulator even in the context of densely cross-linked microtubule bundles. It is noteworthy that despite protection by PRC1 cross-links, MCAK remains an overall faster remodeler due to rapid kinetics of tubulin removal and the ability to access exposed protofilaments at both ends of microtubules and at defects. The properties of MCAK also allow it to enzymatically depolymerize one of the most stable microtubule structures in the cell, the doublet microtubules (47, 50, 60). We find that axonemal doublet microtubules can be depolymerized by MCAK, with the B tubule preferentially depolymerized over the A tubule. The lower stability of the B tubule due to fewer microtubule inner proteins (MIPs) and incomplete tubule structure (49, 61, 62) likely exposes protofilaments that are efficiently depolymerized by MCAK. Consequently, MCAK mediates the rapid destabilization of axonemal arrays. We think that the fast depolymerization of one tubule compromises the stability of the axonemal array by breaking the links between doublets. In contrast, proteins like Kip3p, which preferentially depolymerize singlets over doublets, can act as length regulators at the distal cilium tip, which is predominantly comprised of singlet microtubules (63).
As demonstrated here, previously unknown structural dynamics of individual microtubules and protofilaments within complex arrays can be clearly visualized in the AFM time-lapse images. In the case of two major depolymerases, we find that functional dichotomy in the action of depolymerases, which determines how micron-scale arrays are remodeled, can arise from differences in enzyme activity on nanometer-scale protofilaments. These differences in the observed structural dynamics of depolymerizing microtubule-based structures enable a greater diversity of microtubule array remodeling outcomes. This would be beneficial in different cellular contexts in which either large-scale reorganization or fine-tuning of the architecture of cellular structures is required.
Methods
AFM Experiments.
Microtubule adsorption on mica is achieved by supplementing the buffer with 5 mM MgCl2 as described previously (6). To prepare microtubule bundles, GMPCPP or GMPCPP + taxol microtubules, PRC1, and BRB80 buffer with an additional 5 mM MgCl2 were combined in a tube. The microtubule and protein mixture were spun down for 5 min using a tabletop centrifuge, and ∼20 μL of this mixture was deposited on a mica substrate freshly cleaved by Scotch tape. Single-microtubule, doublet, and axoneme samples were diluted with BRB80 and 5 mM MgCl2 and deposited on mica (SI Appendix, Methods and Materials). After 5 min of incubation, ∼10 μL of additional BRB80 buffer was added to the mica before imaging the sample by AFM. All AFM experiments were carried out by tapping mode in liquid with the Asylum Cypher S and ES with a silicon tip (BL-AC40TS, radius: 8 nm; spring constant: 0.09 N/m; Oxford Instruments).
Supplementary Material
Acknowledgments
We thank S. Jiang for purifying MCAK and R. Ohi and D. Pellman for generously sharing Kip3p plasmids and proteins. R.S. was supported by the Pew Biomedical Foundation, the Smith Foundation, and the NIH Director’s New Innovator Award. This work was performed in part at the Center for Nanoscale Systems (CNS), a member of the National Nanotechnology Coordinated Infrastructure Network, which is supported by the NSF under Award No. 1541959. CNS is part of Harvard University.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115708119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Gudimchuk N. B., McIntosh J. R., Regulation of microtubule dynamics, mechanics and function through the growing tip. Nat. Rev. Mol. Cell Biol. 22, 777–795 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Kodera N., Yamamoto D., Ishikawa R., Ando T., Video imaging of walking myosin V by high-speed atomic force microscopy. Nature 468, 72–76 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Ando T., et al. , A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl. Acad. Sci. U.S.A. 98, 12468–12472 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaap I. A., Carrasco C., de Pablo P. J., Schmidt C. F., Kinesin walks the line: Single motors observed by atomic force microscopy. Biophys. J. 100, 2450–2456 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vinckier A., et al. , Immobilizing and imaging microtubules by atomic force microscopy. Ultramicroscopy 57, 337–343 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Hamon L., Curmi P. A., Pastré D., High-resolution imaging of microtubules and cytoskeleton structures by atomic force microscopy. Methods Cell Biol. 95, 157–174 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Dufrêne Y. F., et al. , Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 12, 295–307 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Elie-Caille C., et al. , Straight GDP-tubulin protofilaments form in the presence of taxol. Curr. Biol. 17, 1765–1770 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Duellberg C., Fourniol F. J., Maurer S. P., Roostalu J., Surrey T., End-binding proteins and Ase1/PRC1 define local functionality of structurally distinct parts of the microtubule cytoskeleton. Trends Cell Biol. 23, 54–63 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Goetz S. C., Anderson K. V., The primary cilium: A signalling centre during vertebrate development. Nat. Rev. Genet. 11, 331–344 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanian R., Kapoor T. M., Building complexity: Insights into self-organized assembly of microtubule-based architectures. Dev. Cell 23, 874–885 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodruff J. B., Drubin D. G., Barnes G., Spindle assembly requires complete disassembly of spindle remnants from the previous cell cycle. Mol. Biol. Cell 23, 258–267 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sánchez I., Dynlacht B. D., Cilium assembly and disassembly. Nat. Cell Biol. 18, 711–717 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mchedlishvili N., Matthews H. K., Corrigan A., Baum B., Two-step interphase microtubule disassembly aids spindle morphogenesis. BMC Biol. 16, 14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goshima G., Scholey J. M., Control of mitotic spindle length. Annu. Rev. Cell Dev. Biol. 26, 21–57 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa H., Marshall W. F., Ciliogenesis: Building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 12, 222–234 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Hu Z., Liang Y., Meng D., Wang L., Pan J., Microtubule-depolymerizing kinesins in the regulation of assembly, disassembly, and length of cilia and flagella. Int. Rev. Cell Mol. Biol. 317, 241–265 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Shrestha S., Hazelbaker M., Yount A. L., Walczak C. E., Emerging insights into the function of kinesin-8 proteins in microtubule length regulation. Biomolecules 9, 1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walczak C. E., Gayek S., Ohi R., Microtubule-depolymerizing kinesins. Annu. Rev. Cell Dev. Biol. 29, 417–441 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Reilly M. L., Benmerah A., Ciliary kinesins beyond IFT: Cilium length, disassembly, cargo transport and signalling. Biol. Cell 111, 79–94 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Howard J., Hyman A. A., Microtubule polymerases and depolymerases. Curr. Opin. Cell Biol. 19, 31–35 (2007). [DOI] [PubMed] [Google Scholar]
- 22.M. L. Gupta, Jr, Carvalho P., Roof D. M., Pellman D., Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 8, 913–923 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Varga V., et al. , Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 8, 957–962 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Helenius J., Brouhard G., Kalaidzidis Y., Diez S., Howard J., The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature 441, 115–119 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Desai A., Verma S., Mitchison T. J., Walczak C. E., Kin I kinesins are microtubule-destabilizing enzymes. Cell 96, 69–78 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Arellano-Santoyo H., et al. , A tubulin binding switch underlies Kip3/kinesin-8 depolymerase activity. Dev. Cell 42, 37–51.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogawa T., Nitta R., Okada Y., Hirokawa N., A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell 116, 591–602 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Asenjo A. B., et al. , Structural model for tubulin recognition and deformation by kinesin-13 microtubule depolymerases. Cell Rep. 3, 759–768 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Wang D., et al. , Motility and microtubule depolymerization mechanisms of the kinesin-8 motor, KIF19A. eLife 5, e18101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulder A. M., et al. , A new model for binding of kinesin 13 to curved microtubule protofilaments. J. Cell Biol. 185, 51–57 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gardner M. K., Zanic M., Gell C., Bormuth V., Howard J., Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell 147, 1092–1103 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Niwa S., et al. , KIF19A is a microtubule-depolymerizing kinesin for ciliary length control. Dev. Cell 23, 1167–1175 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi T., Tsang W. Y., Li J., Lane W., Dynlacht B. D., Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell 145, 914–925 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Piao T., et al. , A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 106, 4713–4718 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walczak C. E., Mitchison T. J., Desai A., XKCM1: A Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell 84, 37–47 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Maney T., Hunter A. W., Wagenbach M., Wordeman L., Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 142, 787–801 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bieling P., Telley I. A., Surrey T., A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell 142, 420–432 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Subramanian R., et al. , Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell 142, 433–443 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cooper J. R., Wagenbach M., Asbury C. L., Wordeman L., Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nat. Struct. Mol. Biol. 17, 77–82 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coombes C. E., Yamamoto A., Kenzie M. R., Odde D. J., Gardner M. K., Evolving tip structures can explain age-dependent microtubule catastrophe. Curr. Biol. 23, 1342–1348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.T. Müller-Reichert, D. Chrétien, F. Severin, A. A. Hyman, Structural changes at microtubule ends accompanying GTP hydrolysis: information from a slowly hydrolyzable analogue of GTP, guanylyl (alpha,beta)methylenediphosphonate. Proc. Natl. Acad. Sci. U.S.A. 95, 3661–3666 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rai A., et al. , Taxanes convert regions of perturbed microtubule growth into rescue sites. Nat. Mater. 19, 355–365 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Niederstrasser H., Salehi-Had H., Gan E. C., Walczak C., Nogales E., XKCM1 acts on a single protofilament and requires the C terminus of tubulin. J. Mol. Biol. 316, 817–828 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Moores C. A., et al. , A mechanism for microtubule depolymerization by KinI kinesins. Mol. Cell 9, 903–909 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Satir P., Studies on cilia. 3. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J. Cell Biol. 39, 77–94 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicastro D., et al. , Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc. Natl. Acad. Sci. U.S.A. 108, E845–E853 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orbach R., Howard J., The dynamic and structural properties of axonemal tubulins support the high length stability of cilia. Nat. Commun. 10, 1838 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Binder L. I., Rosenbaum J. L., The in vitro assembly of flagellar outer doublet tubulin. J. Cell Biol. 79, 500–515 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicastro D., et al. , The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944–948 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Owa M., et al. , Inner lumen proteins stabilize doublet microtubules in cilia and flagella. Nat. Commun. 10, 1143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Linck R. W., Flagellar doublet microtubules: Fractionation of minor components and alpha-tubulin from specific regions of the A-tubule. J. Cell Sci. 20, 405–439 (1976). [DOI] [PubMed] [Google Scholar]
- 52.Wagenbach M., Domnitz S., Wordeman L., Cooper J., A kinesin-13 mutant catalytically depolymerizes microtubules in ADP. J. Cell Biol. 183, 617–623 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bugiel M., Chugh M., Jachowski T. J., Schäffer E., Jannasch A., The kinesin-8 Kip3 depolymerizes microtubules with a collective force-dependent mechanism. Biophys. J. 118, 1958–1967 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varga V., Leduc C., Bormuth V., Diez S., Howard J., Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell 138, 1174–1183 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Duellberg C., Cade N. I., Surrey T., Microtubule aging probed by microfluidics-assisted tubulin washout. Mol. Biol. Cell 27, 3563–3573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zakharov P., et al. , Molecular and mechanical causes of microtubule catastrophe and aging. Biophys. J. 109, 2574–2591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharp D. J., Ross J. L., Microtubule-severing enzymes at the cutting edge. J. Cell Sci. 125, 2561–2569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McNally F. J., Roll-Mecak A., Microtubule-severing enzymes: From cellular functions to molecular mechanism. J. Cell Biol. 217, 4057–4069 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du Y., English C. A., Ohi R., The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr. Biol. 20, 374–380 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Witman G. B., Carlson K., Rosenbaum J. L., Chlamydomonas flagella. II. The distribution of tubulins 1 and 2 in the outer doublet microtubules. J. Cell Biol. 54, 540–555 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma M., et al. , Structure of the decorated ciliary doublet microtubule. Cell 179, 909–922.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ichikawa M., et al. , Tubulin lattice in cilia is in a stressed form regulated by microtubule inner proteins. Proc. Natl. Acad. Sci. U.S.A. 116, 19930–19938 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kiesel P., et al. , The molecular structure of mammalian primary cilia revealed by cryo-electron tomography. Nat. Struct. Mol. Biol. 27, 1115–1124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.