Significance
RAF inhibitors unexpectedly induce ERK activation in normal and oncogenic RAS tumor cells, making them unsuitable for treating RAS-driven cancers. The precise mechanism of this paradox is not fully understood but is believed to be RAS dependent. In this study, we discovered that classical RAS proteins are not essential for RAF inhibitor-induced ERK activation in H/N/KRAS-less mouse embryonic fibroblasts. We further showed that the MRAS/SHOC2 complex is required for the classical RAS-independent paradoxical ERK activation. Our findings provide new insights into the mechanism of paradoxical ERK activation by RAF inhibitors, and they have important therapeutic implications for developing effective RAF inhibitors.
Keywords: MRAS, paradoxical ERK activation, RAF inhibitors, RAS GTPase, RAS-related GTPase
Abstract
RAF inhibitors unexpectedly induce ERK signaling in normal and tumor cells with elevated RAS activity. Paradoxical activation is believed to be RAS dependent. In this study, we showed that LY3009120, a pan-RAF inhibitor, can unexpectedly cause paradoxical ERK activation in KRASG12C-dependent lung cancer cell lines, when KRAS is inhibited by ARS1620, a KRASG12C inhibitor. Using H/N/KRAS-less mouse embryonic fibroblasts, we discovered that classical RAS proteins are not essential for RAF inhibitor-induced paradoxical ERK signaling. In their absence, RAF inhibitors can induce ERK phosphorylation, ERK target gene transcription, and cell proliferation. We further showed that the MRAS/SHOC2 complex is required for this process. This study highlights the complexity of the allosteric RAF regulation by RAF inhibitors, and the importance of other RAS-related proteins in this process.
The classical RAS GTPase family comprises HRAS, NRAS, KRAS-4A, and KRAS-4B. They function as molecular switches at the plasma membrane, cycling between the inactive GDP-bound state (RAS-GDP) and the active GTP-bound state (RAS-GTP). RAS activation from RAS-GDP to RAS-GTP is mediated by guanine nucleotide exchange factors, while GTPase-activating proteins facilitate the reverse. The highly regulated RAS activity links external stimuli to downstream signaling pathways, regulating key cellular functions. Mutations and dysregulation of the RAS-RAF-MEK-ERK pathway are among the most common causes of cancer. HRAS was the first identified oncogene from human tumor DNA, and RAS mutations are found in more than 30% of human cancers (1). Inhibitors that target RAS effector pathways have been developed, but so far these have not provided therapeutic benefit to patients with RAS-driven cancers.
RAF proteins (ARAF, BRAF, and RAF1/CRAF) are key downstream RAS effectors. RAF proteins adopt a closed, monomeric, and inactive conformation in the cytosol. This closed conformation is maintained by the intramolecular interaction between the amino-terminal regulatory domain and the carboxyl-terminal kinase domain. Upon activation, RAS-GTP binds to the RAS binding domain (RBD) of RAF, and recruits RAF to the plasma membrane, where a sequence of conformational changes, phosphorylation, and dephosphorylation events, as well as RAF kinase domain dimerization occur, leading to full RAF activation (2).
Within the RAF carboxyl-terminal kinase domain, the N-terminal lobe (N-lobe) and the C-terminal lobe (C-lobe) are linked through a short flexible hinge. To maintain the active conformation, these two lobes form a closed conformation, with the αC-helix of the N-lobe fixed in the IN position. This allows all the catalytic residues to be close enough and attain the correct orientation to interact. This active conformation is further facilitated by RAF dimerization. The side-to-side dimerization interface is maintained by the interaction between the R509 residues of each αC-helix. This interaction promotes the IN positioning of the αC-helix, and thereby the active conformation (3, 4).
The second-generation ATP-competitive BRAF kinase inhibitors, vemurafenib and dabrafenib, target oncogenic BRAFV600E. They showed clinical efficacy in patients with BRAFV600E/K-driven melanoma, and were approved for the treatment of this disease (5–9). Unlike WT RAF, BRAFV600E/K efficiently activates the ERK pathway as a monomer (10, 11). Binding of the second-generation inhibitor stabilizes the αC-helix in the OUT position and inactivates monomeric BRAFV600E/K kinase activity. However, in RAFWT cells, these inhibitors bind to one RAF protomer with the αC-helix in the OUT position but stabilize the other protomer in the αC-helix IN position. This decreases the affinity of a second inhibitor for the second protomer, subsequently allowing the second inhibitor-free protomer to be activated at nonsaturating inhibitor doses. This results in paradoxical ERK activation in normal cells (3, 4, 12, 13). Patients treated with these inhibitors commonly develop secondary keratoacanthomas and squamous-cell carcinomas. In addition, these inhibitors are also contraindicated for the treatment of tumors with oncogenic RAS (14, 15). To overcome this inhibitor-induced paradoxical activation, new RAF inhibitors with different allosteric mechanisms have been developed.
The third-generation pan-RAF inhibitors (e.g., LY3009120) can bind to CRAFWT, BRAFWT, and BRAFV600E with similar high affinity (16). They were developed to minimize paradoxical ERK activation by lowering the inhibitor concentration required to saturate all RAF protomers; however, they still induce detectable paradoxical ERK activation (17, 18). The pan-RAF inhibitors stabilize the αC-helix in the IN position, promoting an active RAF conformation as well as RAF dimerization (13). Recent studies have also demonstrated that these inhibitors promote RAS–RAF interaction (19, 20). As a result, paradoxical ERK activation is still observed at nonsaturating doses in cells with elevated RAS activity. Notably, structural studies have recently shown that ATP binding to the RAF kinase is required for maintaining the autoinhibited state, and consequently, RAF inhibitors that displace ATP are expected to disrupt the RAF autoinhibited state (21, 22). Together, these new observations indicate that RAF inhibitors that induce the αC-helix in either the OUT or IN position, can cause paradoxical activation through multiple mechanisms.
The RAS-related GTPase (RRAS) subfamily (RRAS, TC21/RRAS2, and MRAS/RRAS3) of the Ras GTPase superfamily is the closest relative to the classical RAS GTPase (23). RRAS GTPases have structural domains that are characteristic of small GTPases: guanine nucleotide binding domain, effector binding domain, switch I, switch II, and a CAAX box. The sequences of the effector binding domain are virtually identical to the classical RAS proteins. Not surprisingly, both the classical RAS and RRAS can be regulated by the same GTPase-activating proteins, as well as bind to the same effectors: RAF, PI3K, and Ral-GDS (24–27). Similar to classical RAS, RRAS possesses transforming activities, albeit weakly (28–33). Mutations of the RRAS subfamily genes occur in human cancer at low frequency, and gain-of-function mutations of RRAS and MRAS were identified in patients with the Noonan syndrome, part of a spectrum of diseases with hyperactive ERK signaling called RASopathies (34, 35). Among the RRAS subfamily, MRAS is believed to play a unique role in regulating the ERK pathway. MRAS dephosphorylates and positively regulates RAF by forming a complex with SHOC2 and protein phosphatase 1 (PP1) at the plasma membrane (36, 37). In addition, like KRAS-4B, the MRAS hypervariable region is farnesylated at the C-terminal cysteine and interacts with the membrane via a polybasic region. Both MRAS and KRAS-4B are found in disordered membrane regions rather than organized lipid rafts, suggesting that they interact with common regulators and effectors in the same signaling cascade (23, 38).
Paradoxical ERK activation is induced by RAF inhibitors with diverse structures and biochemical properties. While we do not fully understand the mechanism, the consensus is that it is RAS-dependent. To our surprise, we found that LY3009120, a pan-RAF inhibitor, induced ERK activation in KRASG12C-dependent lung cancer cells when treated in combination with the KRASG12C inhibitor, ARS1620, prompting us to further investigate this paradox. In this study, we present evidence that ATP competitive RAF inhibitors can induce paradoxical ERK activation independent of classical RAS in mouse embryonic fibroblasts (MEFs). We show that RAF inhibitors induce ERK phosphorylation, and target gene transcription and cellular proliferation in MEFs devoid of classical RAS proteins. We also identify the MRAS/SHOC2 complex to be required for this classical RAS-independent activation.
Results
LY3009120 Induces Paradoxical ERK Activation in ARS1620- or AMG510-Treated Human Cancer Cell Lines.
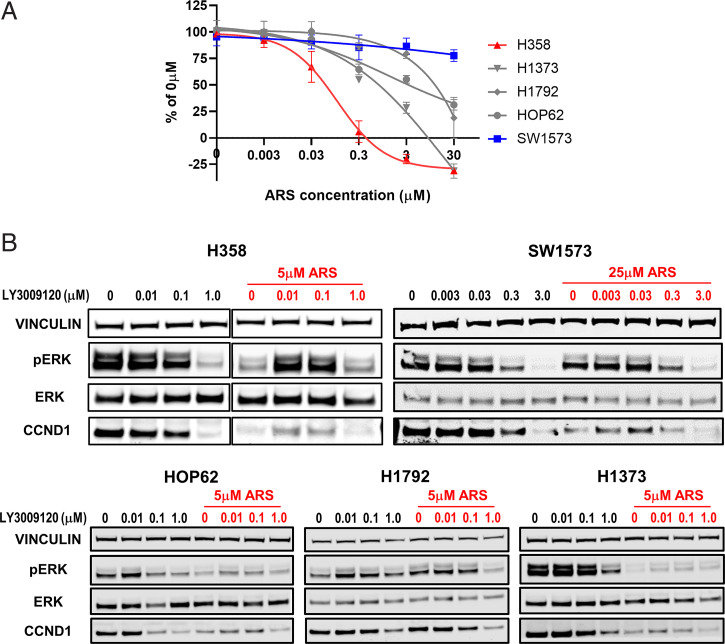
While investigating the combined treatment of the KRASG12C inhibitor, ARS1620, and the pan-RAF inhibitor, LY3009120, we were surprised to find that LY3009120 induced paradoxical ERK activation in some KRASG12C-expressing human cancer cell lines also treated with ARS1620. We treated five human lung cancer cell lines: H358, H1373, H1792, HOP62, and SW1573 with ARS1620 and increasing doses of LY3009120. While all these cell lines express KRASG12C, they showed different sensitivities to ARS1620 in the cell proliferation assay, with H358 being the most sensitive and SW1573 being the least sensitive (Fig. 1). LY3009120 by itself induced either minimal or no paradoxical ERK activation in these cell lines. Surprisingly, of the five cell lines tested, LY3009120-induced biphasic ERK activation was most noticeable in ARS1620-treated H358, as measured by the levels of ERK phosphorylation and CCND1 expression. The nature of activation appears to be biphasic with lower doses of LY3009120-induced ERK activation, while higher doses of LY3009120 did not. The sensitivities to ARS1620 appeared to correlate with the level of biphasic ERK activation induced by LY3009120 in the presence of ARS1620. A smaller degree of LY3009120-induced activation was also observed in ARS1620-treated H1373, which was second most sensitive to ARS1620 among the five cell lines tested (Fig. 1). Similar results were observed in AMG510-treated H358 and H1373 (SI Appendix, Fig. S1). No paradoxical ERK activation was observed in SW1573 even at a higher ARS1620 concentration, although there was a slight increase in CCND1 expression. Collectively, our data demonstrate that LY3009120-induced biphasic ERK activation can occur in KRAS-dependent human cell lines even when the KRAS function is inhibited.
Fig. 1.
LY3009120 induces paradoxical ERK activation in ARS1620-treated human cancer cell lines. (A) Cell viability assay was performed with the indicated cell lines treated with ARS1620 for 96 h. Data are presented as percentages of the untreated cells. Error bars represent SDs from a representative experiment of three experiments. (B) Protein lysates from H358, SW1573, HOP62, H1792, and H1373 treated with LY3009120 alone or in combination with ARS1620 were immunoblotted with indicated antibodies. Representative blots from two experiments that show similar results are shown.
RAF Inhibitors Increase ERK Phosphorylation and Cyclin D1 Expression in H/N/KRAS-less MEFs.
Pan-RAF inhibitors, such as LY3009120, induce minimal paradoxical ERK activation and show antitumor activity in diverse cellular contexts carrying KRAS, NRAS, and BRAF mutations (16). In light of our observation of the KRASG12C-expressing human cancer cell lines (Fig. 1 and SI Appendix, Fig. S1), we hypothesized that classical RAS proteins are not essential for RAF inhibitor-induced ERK activation. We used “RAS-less” MEFs as our cellular model system (39). The parental MEF cell line has both Hras and Nras ablated and expresses only a conditional Kras knockout allele. These single Kras allele parental MEFs can be treated with tamoxifen to yield cells lacking the classical RASs (lacking HRAS, NRAS, and KRAS, referred to as H/N/KRAS-less). Parental and H/N/KRAS-less MEFs were treated for 24 h with increasing doses (0, 0.003, 0.03, 0.3, 3.0 μM) of AZ628 and LY3009120, both pan-RAF inhibitors, dabrafenib, a BRAFV600E inhibitor, and PLX8394, a potent BRAF inhibitor and paradox breaker, in the presence or absence of EGF stimulation (Fig. 2). Consistent with previous findings, dabrafenib induced biphasic paradoxical ERK activation in parental MEFs, while PLX8394 and both pan-RAF inhibitors induced minimal paradoxical ERK activation. Surprisingly, dabrafenib and both pan-RAF inhibitors activated ERK signaling in RAS-less MEFs, while PLX8394 did not change the levels of ERK activation. This paradoxical activation of ERK also produced downstream up-regulation of CCND1, a known target of RAF/MEK/ERK signaling.
Fig. 2.
RAF inhibitors increase ERK phosphorylation and Cyclin D1 expression in H/N/KRAS-less MEFs. Parental MEFs (-4OH-tamoxifen) and H/N/KRAS-less MEFs (+4OH-tamoxifen) were treated with 0, 0.003, 0.03, 0.3, and 3 μM of the indicated inhibitors for 24 h. Cells were serum-starved at time of inhibitor treatment and stimulated for 5 min with EGF (100 ng/mL) prior to harvest. Protein lysates were immunoblotted with indicated antibodies. Representative blots from three experiments that show similar results are shown.
Next, we investigated if RAF and MEK are required for RAF inhibitor-induced paradoxical activation of ERK by using small-interfering RNAs (siRNAs) and small-molecule inhibitors. Araf, Braf, and Craf were simultaneously knocked down with siRNAs in parental and H/N/KRAS-less MEFs and subsequently treated with LY3009120 for 24 h. Simultaneously knocking down all Rafs with siRNAs in parental MEFs proved to be challenging, and it made interpreting the data difficult. Nevertheless, we did observe a small decrease in ERK phosphorylation (SI Appendix, Fig. S2A). Using the same siRNAs and transfection protocol, Araf, Braf, and Craf were more successfully knocked down in H/N/KRAS-less MEFs. When compared to the scrambled siRNA-treated control, knocking down all three RAF isoforms blunted the RAF inhibitor-induced paradoxical activation of ERK, as well as CCND1 and SPRY2 up-regulation in H/N/KRAS-less MEFs (SI Appendix, Fig. S2A). In addition, simultaneous treatment with MEK inhibitors, selumetinib or trametinib, along with LY3009120 completely abolished the effect of the RAF inhibitor (SI Appendix, Fig. S2B). Taken together, these data suggested that RAF inhibitors can induce paradoxical ERK activation in a H/N/KRAS-less cellular system, and that both RAF and MEK are essential. Our data cannot conclude that paradoxical ERK activation is dependent on RAF dimerization, however.
RAF Inhibitors Induce ERK Target Gene Transcription and Cell Proliferation in H/N/KRAS-less MEFs.
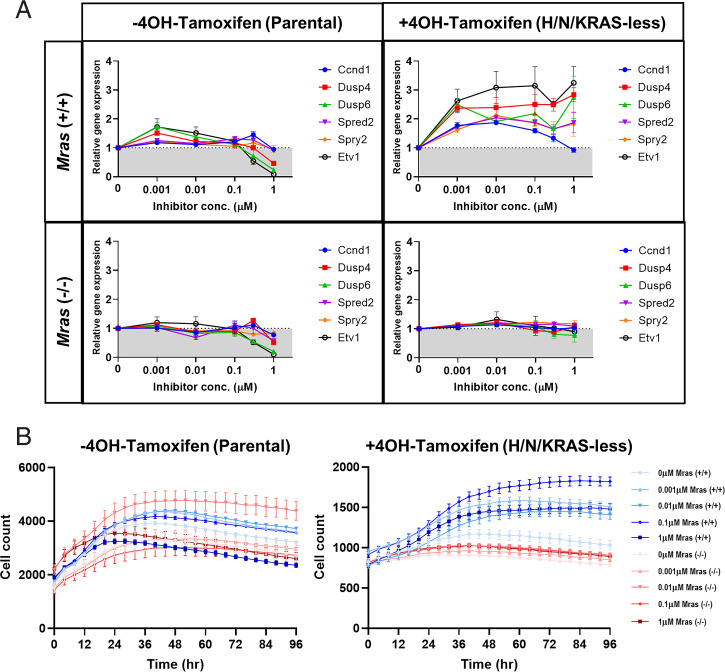
To test whether RAF inhibitor-induced ERK activation in H/N/KRAS-less MEFs would translate into transcriptional activation, ERK target genes were examined using real-time RT-PCR. Ccnd1, Dusp4, Dusp6, Spred2, Spry2, Etv1, Etv4, and Etv6 expression were all induced in a biphasic manner after 24-h treatment with LY3009120 in H/N/KRAS-less MEFs when compared to the parental line (Fig. 3A). LY3009120 induced dose-dependent gene expression at concentrations up to 0.03 and 0.3 μM, but decreased gene expression at higher concentrations. The same biphasic effect was observed with ERK phosphorylation and CCND1 expression induced by LY3009120 in H/N/KRAS-less MEFs (Fig. 2). These same genes did not exhibit any activation after 24-h treatment at the same doses with PLX8394, a paradox breaker, in either the parental or H/N/KRAS-less MEF lines, consistent with its lack of effect on ERK phosphorylation (SI Appendix, Fig. S3). This biphasic induction of ERK target genes demonstrates the ERK phosphorylation induced by RAF inhibitors is indeed capable of activating downstream signaling in a H/N/KRAS-less context.
Fig. 3.
LY3009120 induces ERK target gene transcription and cell proliferation in H/N/KRAS-less MEFs. (A) Total RNA was extracted from parental and H/N/KRAS-less MEFs treated with 0, 0.003, 0.03, 0.3, 1.0, or 3.0 µM LY3008120 for 24 h. Levels of Ccnd1, Dusp4, Dusp6, Spred2, Spry2, Etv1, Etv4, and Etv5 transcript were examined using quantitative RT-PCR. Results are shown as fold-change relative to DMSO-treated control represented by gray shading at a fold-change of 1.0. Error bars represent SEs from a representative experiment of three experiments. (B) Parental and H/N/KRAS-less MEFs were seeded and treated 24 h later with 0.0, 0.001, 0.01, 0.1, or 1.0 µM LY3009120 in serum-free media. Cell numbers were monitored for an additional 4 d using the IncuCyte Live-Cell Analysis System. Error bars represent SDs from a representative experiment of two experiments.
To further understand the potential downstream effects of the paradoxical ERK activation on cell proliferation, parental and H/N/KRAS-less MEFs were treated with LY3009120 and the cell number was tracked using an IncuCyte Live Cell Analysis System over 4 d (Fig. 3B). Parental MEFs treated with LY3009120 at lower doses demonstrated a modest increase in cell numbers compared to the untreated cells, indicating the potential for paradoxical activation in nonsaturating doses. The highest dose of LY3009120 (1.0 μM) did indeed decrease cell proliferation in parental MEFs. H/N/KRAS-less MEFs remained largely quiescent for the whole period, but surprisingly, H/N/KRAS-less MEFs treated with the same various concentrations of LY3009120 became proliferative, with 0.1 μM of LY3009120 causing more than twofold increase in cell numbers. Taken together, these results indicated that RAF inhibitors induce paradoxical activation of ERK in a H/N/KRAS-less context and generate a functional response by inducing ERK target genes which subsequently allow H/N/KRAS-less MEFs to regain a proliferative phenotype.
MRAS Is Required for RAF Inhibitor-Induced Paradoxical ERK Activation.
H/N/KRAS-less MEFs do not express HRAS, NRAS, and KRAS; however, these cells still express RRAS proteins (MRAS, RRAS, and RRAS2), the closest relatives to the classical RAS proteins. To determine if they contribute to the RAF inhibitor-induced paradoxical ERK activation, we generated Mras, Rras, and Rras2 knockout cell lines with CRISPR from the parental MEFs. These cell lines were further treated with tamoxifen to remove Kras. LY3009120-induced paradoxical ERK activation was not affected in H/N/KRAS-less Rras knockout and H/N/KRAS-less Rras2 knockout MEF cell lines, as indicated by the levels of ERK phosphorylation as well as downstream targets, CCND1 and SPRY2 (Fig. 4 A and B). In contrast, knocking out Mras in the H/N/KRAS-less MEFs eliminated the LY3009120-induced paradoxical activation of ERK (Fig. 4C). This was also evident when examining the ERK transcriptionally regulated genes: Ccnd1, Dusp4, Dusp6, Spred2, Spry2, and Etv1 by real-time RT-PCR (Fig. 5A). Treatment of LY3009120 for 24 h induced biphasic activation of the ERK-regulated genes in H/N/KRAS-less MEFs, whereas H/N/KRAS-less MEFs with Mras knocked out and treated with LY3009120 had a substantial reduction in this biphasic activation. Additionally, we examined the proliferative effect of knocking out Mras in H/N/KRAS-less MEFs. The proliferative recovery of H/N/KRAS-less cells in response to the LY3009120 was abolished when Mras was ablated (Fig. 5B). These data further confirmed that Mras is essential for the functional downstream RAF inhibitor-induced paradoxical response observed in the H/N/KRAS-less cellular environment.
Fig. 4.
MRAS is required for RAF inhibitor-induced paradoxical ERK activation in H/N/KRAS-less MEFs. Protein lysates from parental MEFs and H/N/KRAS-less MEFs with Rras knocked-out (A), Rras2 knocked-out (B), and Mras knocked-out (C) treated with 0, 0.01, 0.1, or 1.0 µM LY3008120 for 24 h were immunoblotted with indicated antibodies.
Fig. 5.
MRAS is required for paradoxical up-regulation of ERK target genes and cell proliferation in H/N/KRAS-less MEFs. (A) Total RNA was extracted from MEFs with and without Mras treated with 0, 0.003, 0.03, 0.3, 1.0, or 3.0 µM LY3009120 for 24 h. Levels of Ccnd1, Dusp4, Dusp6, Spred2, Spry2, and Etv1 transcript were examined using quantitative RT-PCR. Results are shown as fold-change relative to DMSO-treated control represented by gray shading at a fold-change of 1.0. Error bars represent SEs of a representative experiment repeated twice. (B) MEFs with or without Mras were treated with 0, 0.001, 0.01, 0.1, or 1.0 µM LY3009120 in serum-free media. Cell numbers were monitored for an additional 4 d using the IncuCyte Live-Cell Analysis System. Error bars represent SDs of a representative experiment repeated twice.
Next, we expanded our analysis with RAS-less MEFs that have Mras or Rit1 transiently knocked down with siRNA. Transiently knocking down Rit1, another RAS-related GTPase, had no effect on the LY3009120-induced ERK activation (SI Appendix, Fig. S5). In contrast, similar to the Mras knockout H/N/KRAS-less MEF cell line, Mras siRNA largely abolished the LY3009120-induced ERK activation and proliferative effect in H/N/KRAS-less MEFs (SI Appendix, Fig. S4). These data from transient Mras knockdown are consistent with our observation with the Mras knockout MEF cell line, supporting the requirement of MRAS in RAF inhibitor-induced ERK activation.
To gain additional insight of the mechanistic role of MRAS in H/N/KRAS-less paradoxical ERK activation, we performed a transient transfection experiment with WT, gain-of-function mutant and loss-of-function mutant Mras. In Mras knockout RAS-less MEFs, transient transfection of MrasG23V, a gain-of-function mutation identified in patients with Noonan syndrome (33), drastically increased ERK signaling on its own, but did not restore the inhibitor-induced paradoxical ERK activation (Fig. 6A). In contrast, transient transfection of WT Mras in Mras knockout H/N/KRAS-less MEFs partially restored the LY3009120-induced ERK activation to a level closer to that of the H/N/KRAS-less MEFs, while mutant Mras lacking the CAAX box (MrasΔCAAX) failed to do so. These data indicate that MRAS membrane localization is crucial to the inhibitor-induced ERK activation in H/N/KRAS-less MEFs.
Fig. 6.
Mechanistic role of MRAS in H/N/KRAS-less paradoxical ERK activation. (A) H/N/KRAS-less MEFs with or without Mras were transiently transfected with the indicated Flag-tagged Mras constructs. Twenty-four hours later, cells were treated with 0, 0.01, 0.1, or 1.0 µM LY3009120 for an additional 24 h. Lysates were collected and immunoblotted with indicated antibodies. Representative blots from three experiments that show similar results are shown. (B) MEFs were treated with LY3009120 for 24 h, and lysates were collected for coimmunoprecipitation with the MRAS antibody, followed by immunoblotting with the BRAF and CRAF antibody. Input lysates were immunoblotted with indicated antibodies. Representative blots from two experiments that show similar results are shown.
We also investigated the interaction of MRAS and BRAF/CRAF in H/N/KRAS-less MEFs treated with LY3009120. We performed MRAS coimmunoprecipitation and showed that MRAS interacts with both BRAF and CRAF in parental MEFs, and the levels of interaction were not affected by LY3009120 (Fig. 6B and SI Appendix, Fig. S6). In contrast, LY3009120 caused an increase in MRAS interaction with BRAF and CRAF at the concentration of 0.01 μM and 0.1 μM in H/N/KRAS-less MEFs. This was similar to the LY3009120-induced biphasic ERK activation, with 0.01 μM or 0.1 μM causing the biggest increase in ERK signaling and cell proliferation. These data supported the notion that MRAS interaction with BRAF/CRAF plays a critical mechanistic role for the paradoxical ERK activation observed in H/N/KRAS-less MEFs.
SHOC2 Is Required for RAF Inhibitor-Induced Paradoxical ERK Activation.
SHOC2 is a known positive regulator of the ERK pathway and a key component of a heterotrimeric complex consisting of MRAS and PP1, which mediates dephosphorylation of the inhibitory S259 site on CRAF (36). Parental and H/N/KRAS-less MEFs were transiently transfected with siRNA targeting Shoc2 and subsequently treated with LY3009120 (0.0, 0.01, 0.1 and 1.0 μM) for 24 h (SI Appendix, Fig. S7). Knockdown of Shoc2 reduced the RAF inhibitor-induced paradoxical ERK activation in H/N/KRAS-less MEFs, as well as CCND1 and SPRY2 biphasic up-regulation. Shoc2 knockdown in the parental MEFs had no effect on the minimal paradoxical ERK activation observed in the presence of LY3009120. Therefore, it can be concluded that SHOC2 in addition to MRAS is required for the RAF inhibitor-induced paradoxical ERK activation.
Discussion
Until the recent discovery of KRASG12C covalent inhibitors, RAS has been considered an “undruggable” target. This dearth of direct RAS inhibitors has led to the clinical investigation of drugging targets downstream of RAS (e.g., RAF and MEK). Early generations RAF inhibitors unexpectedly induce ERK activation in normal and oncogenic RAS tumor cells, making them unsuitable for treating RAS-driven cancers (14, 15). Subsequently, pan-RAF inhibitors were developed trying to eliminate the paradoxical effect. However, minimal paradoxical ERK activation is still detected in KRAS mutated cell lines (17, 18).
RAF inhibitor-induced ERK signaling is believed to be RAS-dependent, and is the combined effects relieved RAF autoinhibition, induced RAF kinase domain dimerization, and induced RAS–RAF interactions (3, 4, 12, 19, 20). We were surprised to see RAF inhibitors (both second-generation ATP-competitive inhibitors and pan-RAF inhibitors) induced paradoxical ERK activation in cancer cells in which KRAS had been inhibited, and in MEFs that lack all classical RAS proteins. The observed paradoxical activation is biphasic. For example, LY3009120 caused a dose-dependent increase of ERK phosphorylation and cell proliferation up to 0.1 μM, but a weaker stimulation at higher doses (Fig. 3). The biphasic activation is similar to the RAF inhibitor-induced paradoxical activation observed in cell lines with activated RAS. It is consistent with the notion that RAF activity is determined by both the number of RAF dimers and the proportions of dimers that are fully inhibitor-bound and inhibited. At subsaturating doses, RAS can stimulate the RAF protomer that is not occupied by an inhibitor, but at higher doses, when all protomers are occupied by inhibitors, ERK signaling is inhibited. The similar biphasic nature of the paradoxical activation led us to hypothesize that the same mechanisms: RAF autoinhibition relief and induction RAF kinase domain dimerization, both potentially contribute to the paradoxical activation. However, our data demonstrated that alternative mechanisms exist to activate RAF in the absence of any classical RAS.
There are close to 20 members in the mammalian RAS GTPase family (40). Classical RAS members have been the focus of studies from the last several decades due to their roles in human cancer, but other members remain poorly characterized. We first investigated the potential role of the RRAS GTPase subfamily, one of the closest relatives to the classical RAS GTPase. Among the three members in the RRAS subfamily, MRAS has been shown to have the highest potential to substitute classical RAS. Indeed, MRAS is believed to evolve independently from other RRAS members, and in ascidian, which lacks classical RAS, it can compensate for RAS function (41). Rodriguez-Viciana et al. (27) performed a detailed study investigating the effector specificity of some of the RAS GTPases, including the classical RAS and RRAS subfamilies. Among all tested RRAS GTPases, MRAS was shown to have the greatest capacity of binding to ARAF, BRAF, and CRAF, although considerably weaker than classical RAS proteins. Consistently, MRAS also demonstrated the highest ability in activating ARAF and CRAF, and concomitantly ERK activation. In addition, MRAS can uniquely form a phosphatase complex with SHOC2 and PP1, to dephosphorylate CRAF at S259 (BRAF at S365) and positively modulate RAF activity. In H/N/KRAS-less MEFs, using either CRISPR-mediated knockout or siRNA-mediated knockdown strategy, MRAS is shown to be essential for the inhibitor-induced paradoxical activation, while RRAS, RRAS2, and RIT1 (another related GTPase) are dispensable (Figs. 4 and 5 and SI Appendix, Figs. S4 and S5).
To gain further insight of the mechanistic role of MRAS in paradoxical ERK activation in RAS-less MEFs, we focused on two aspects of the MRAS requirement: MRAS plasma membrane localization and the MRAS/SHOC2 phosphatase complex (Fig. 6 and SI Appendix, Figs. S6 and S7). In the transient overexpression experiment, the MRAS mutant missing the CAAX box failed to restore the LY3009120-induced ERK activation, while the WT MRAS partially restored it, suggesting that MRAS membrane localization is critical to the paradoxical activation. Intriguingly, expression of the gain-of-function MRASG23V mutant alone induced ERK activation in H/N/KRAS-less MRAS knockout MEFs, but showed a very small degree of inhibitor-induced paradoxical activation. MRASG23V is believed to be mostly GTP bound and constitutively active (35), with an activity level close to the classical WT RAS. The level of paradoxical activation was minimal and similar to that of the parental MEFs. This led us to propose that overexpression of the MRASG23V mutant potentially compensates not only for MRAS but classical RAS. Secondly, we used siRNA to examine the role of SHOC2 in RAS-less paradoxical activation. Knocking down Shoc2 in H/N/KRAS-less MEFs abolished the inhibitor-induced ERK activation, demonstrating the essential role of SHOC2 and likely the function of MRAS in dephosphorylating RAF. Together, these data suggest that both MRAS membrane localization and RAF dephosphorylation by the MRAS/SHOC2 complex are essential for inhibitor-induced ERK activation in H/N/KRAS-less MEFs.
Our model proposes that RAF inhibitors disrupt the closed autoinhibited RAF conformation, which promotes RAF dimerization and RAS–RAF interaction. In normal cells, most if not all RAF proteins bind to the classical RAS. MRAS has a lower affinity for RAF, and mostly binds to RAF through the phosphatase complex it forms with SHOC2 and PP1 (SI Appendix, Fig. S8A). In the absence of the classical RAS, MRAS likely can bind to RAF directly through the RBD, although weakly. The interaction is further augmented due to the open and active conformation of the inhibitor-bound RAF, potentially allowing for easier MRAS access (SI Appendix, Fig. S8B). It is worth noting that in a previous study, overexpression of MRASG23V in H/N/KRAS-less MEFs cannot restore cell proliferation, strongly suggesting that the inhibitor-induced RAF dimerization and relief of RAF autoinhibition is critical for the potential interaction with MRAS (39). We propose that MRAS can substitute for classical RAS to activate RAF and ERK signaling under certain cellular contexts. We also speculate that MRAS may contribute to resistance to KRAS inhibition in the clinical setting.
The level of MRAS interaction with BRAF/CRAF induced by LY3009120 correlates with the induced biphasic ERK activation, strongly implicating its critical mechanistic role (Fig. 6). However, the coimmunoprecipitation experiment cannot distinguish if MRAS directly binds to the inhibitor-primed RAF or simply through the phosphatase complex. In addition, we do not fully understand why the magnitude of the paradoxical activation induced by pan-RAF inhibitors is greater in H/N/KRAS-less MEFs than KRAS-expressing MEFs, but we postulate this is due to the higher MRAS level in the H/N/KRAS-less MEFs. Future investigation is needed to address these questions, which will also shed some light on our understanding of the complicated process of RAF activation and inhibition.
Finally, our analysis with the H/N/KRAS-less MEFs may explain our observations in human cancer cell lines. In some KRASG12C-dependent cell lines, while LY3009120 does not normally cause significant paradoxical ERK activation, it can induce a higher level of ERK activation when RAS function is inhibited (Fig. 1 and SI Appendix, Fig. S1). We postulate that MRAS may play an essential role in this process in human cancer cells. Moreover, it raises the concern that combining both RAS and RAF inhibitors in treating KRASG12C-driven cancers may not have the expected additive or synergistic effect.
In summary, we discovered that classical RAS proteins are not essential for RAF inhibitor-induced paradoxical ERK activation at least in some cellular contexts. We further showed that MRAS, in addition to being part of the SHOC2/PP1 phosphatase complex, may substitute for classical RAS in directly activating RAF on the plasma membrane. This study highlights the complicated allosteric effect of RAF inhibitors on RAF activity, and exposes our lack of comprehensive understanding of the process of RAF activation. Complete knowledge of both aspects of RAF biology is critical to the development of more effective RAF inhibitors and other therapeutics for either RAF- or RAS-driven cancers.
Materials and Methods
Reagents and Cell Culture.
LY3009120 (cat. no. S7842), AZ628 (cat. no. S2746), and PLX8394 (cat. no. S7965) were purchased from Selleckchem and ARS1620 (cat. no. HY-U00418) from MedChem Express. MEFs (DU1473) null for both Hras and Nras were provided by M. Barbacid’s laboratory (CNIO, Madrid, Spain) and sorted based on ploidy to obtain a diploid population. Cells were then cloned to ensure homogeneity and were treated with 600 nM 4-hydroxytamoxifen (cat. no. H7904; Sigma) for 11 d to eliminate the endogenous floxed Kras gene. NCI-H358, NCI-H1373, NCI-H1792, and SW-1573 were obtained from the American Type Culture Collection (ATCC). HOP-62 was obtained from the Biological Testing Branch, Developmental Therapeutics Program, National Cancer Institute.
Immunoblotting.
Cells were lysed in lysis buffer (25 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton) plus Halt protease inhibitor mixture (cat. no. 87786; Thermo Fisher Scientific). Fifteen to 30 mg of protein for each sample was then separated by SDS/PAGE using Invitrogen’s Bolt system, transferred to PVDF membranes (cat. no. 1620262; Bio-Rad), and blocked using Odyssey blocking buffer (cat. no. 927-60001; LI-COR). Membranes were incubated overnight with primary antibodies (SI Appendix, Table S1) in Odyssey blocking buffer plus 0.2% tween-20 (Thermo Fisher Scientific), incubated with LI-COR IRDye 800CW or IRDye 680RD secondary antibodies at 1:15,000, and analyzed by the Odyssey imaging system (LI-COR).
Coimmunoprecipitation.
Cell lysates were lysed in RIPA buffer (cat. no. 89901; Thermo Fisher Scientific) plus Halt Proteinase inhibitors mixture (Thermo Fisher Scientific). Each sample consisted of 400 μg lysate plus 100 μL of M-280 sheep anti-rabbit IgG Dynabeads (cat. no. 11203D; Thermo Fisher Scientific) and 10 μL MRAS antibody. Samples were then separated by SDS/PAGE using Invitrogen’s NuPAGE system, transferred to PVDF membranes (Bio-Rad), and blocked using Odyssey blocking buffer (LI-COR). Membranes were incubated overnight with primary antibodies (SI Appendix, Table S1) in Odyssey blocking buffer plus 0.2% tween-20 (Thermo Fisher Scientific), incubated with LI-COR IRDye 800CW or IRDye 680RD secondary antibodies at 1:15,000, and analyzed by the Odyssey imaging system (LI-COR).
Quantitative RT-PCR.
Total cellular RNA was extracted using the RNA Mini Kit (cat. no. 74106) from Qiagen. Approximately 250 ng of total RNA for each sample was reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit according to manufacturer’s protocol (cat. no. 4368813; Applied Biosystems/Thermo Fisher Scientific). Typically, 5 ng of reverse-transcribed cDNA per sample was used to conduct real-time PCR (real-time PCR) using a QuantiStudio 3 system and SYBR Green Universal Master Mix (cat. no. 4309155; Applied Biosystems/Thermo Fisher Scientific) in triplicate wells. The following primers predesigned by Integrated DNA Technologies (IDT) were used: Tbp, Ccnd1, Spry2, Dusp4, Dusp6, Spred2, Etv1, Etv4, and Etv5 (SI Appendix, Table S2).
siRNA Transient Transfection.
Specific smartpool siRNAs for mouse Mras (cat. no. L-050270-01-0005), Rit1 (cat. no. L-062337-01-0005), Shoc2 (cat. no. L-059319-01-0005), Araf (cat. no. L-059319-01-0005), Braf (cat. no. L-040325-00-0005), Craf (cat. no. L-040149-00-0005), and nontargeting control (cat. no. D-001810-10-05; Dharmacon) were transfected using Lipofectamine RNAiMAX (cat. no. 13778030; Invitrogen/Thermo Fisher Scientific) according to the manufacturer’s protocol. Cells were transfected 24 to 48 h prior to the addition of RAF inhibitors and additionally incubated another 24 h prior to harvesting.
Plasmid DNA Transient Transfection.
Three-hundred thousand cells per well were plated the day before transfection in six-well plates. Cells were transfected using 7.5 μL Lipofectamine 3000 reagent (cat. no. L3000015; Invitrogen/Thermo Fisher Scientific) in addition to 2.5 μg DNA construct plus 5 μL P3000 reagent per well. Transfection was conducted 24 h prior to the addition of RAF inhibitors and cells were additionally incubated for another 24 h prior to harvesting. The following are the plasmids transfected: FLAG tagged human MRAS (1–208); FLAG tagged human MRAS without CAAX box (1–204); and FLAG tagged human MRASG23V (1–208).
CRISPR-Mediated Knockout Cell Lines.
Specific TrueGuide single guide RNA sequences for mouse Mras (cat no. CRISPR588150_SGM), Rras (cat. no. CRISPR488977_SGM)n and Rras2 (cat. no. CRISPR229952_SGM) were obtained from Invitrogen/Thermo Fisher Scientific and were transfected into MEFs using TrueCut Cas9 protein (cat. no. A36496) and Lipofectamine CRISPRMAX Cas9 Transfection Reagent (cat. no. CMAX00001) according to the manufacturer’s recommendations (Invitrogen/Thermo Fisher Scientific). Knockout of specific genes in clones was confirmed using Sanger sequencing or Mi-Seq. Exome sequencing was also performed on all clonal knockout lines. The guide target sequences are as follows: Mras: GATCCTCGTGGCCAACAAGG; Rras: ACTGCGGGGCAAGAGGAATT; and Rras2: GCAATACATGAGGACAGGCG.
IncuCyte Proliferation Assay.
The MEF cell line expressing the NucLight Red was generated by stable transduction with the IncuCyte NucLight Red Lentivirus Reagent (cat. no. 4625; Essen Bioscience); 1,250 cells were seeded for each well of a 96-well plate. The live-cell image was captured, and the cell number determined every 4 h for the indicated time period with the IncuCyte S3 Live-Cell Analysis System.
Supplementary Material
Acknowledgments
We thank our colleagues from the RAS initiative for their ongoing support, in particular Katie Powell, Scott Eury, and Abigail Neish for their technical support in generating the cell lines used in this study. This project has been funded in whole or in part with federal funds from the National Cancer Institute, NIH, under Contract 75N9109D00024. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
Competing interest statement: F.M. is a consultant for the following companies: Amgen; Daiichi Ltd., Frontiers Med, Exuma Biotech, Ideaya Biosciences, Kura Oncology, Leidos Biomedical Research, Inc., PellePharm, Pfizer Inc., PMV Pharma and Quanta Therapeutics. F.M. is a consultant for and cofounder of the following companies (with ownership interest including stock options): BridgeBio; DNAtrix Inc., Olema Pharmaceuticals, Inc., and Quartz. F.M. is the scientific director of the National Cancer Institute RAS Initiative at the Frederick National Laboratory for Cancer Research/Leidos Biomedical Research, Inc. F.M. has been a recipient of research grants from Daiichi Sankyo and Gilead Sciences and has a current grant from Boehringer-Ingelheim.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113491119/-/DCSupplemental.
Data Availability
All study data are included in the main text and/or SI Appendix.
References
- 1.Simanshu D. K., Nissley D. V., McCormick F., RAS proteins and their regulators in human disease. Cell 170, 17–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terrell E. M., Morrison D. K., Ras-mediated activation of the Raf family kinases. Cold Spring Harb. Perspect. Med. 9, a033746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karoulia Z., et al. , An integrated model of RAF inhibitor action predicts inhibitor activity against oncogenic BRAF signaling. Cancer Cell 30, 485–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajakulendran T., Sahmi M., Lefrançois M., Sicheri F., Therrien M., A dimerization-dependent mechanism drives RAF catalytic activation. Nature 461, 542–545 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Bollag G., et al. , Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467, 596–599 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman P. B., et al. , BRIM-3 Study Group, Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364, 2507–2516 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hauschild A., et al. , Dabrafenib in BRAF-mutated metastatic melanoma: A multicentre, open-label, phase 3 randomised controlled trial. Lancet 380, 358–365 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Flaherty K. T., et al. , Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 363, 809–819 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McArthur G. A., et al. , Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): Extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol. 15, 323–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeman A. K., Ritt D. A., Morrison D. K., Effects of Raf dimerization and its inhibition on normal and disease-associated Raf signaling. Mol. Cell 49, 751–758 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao Z., et al. , BRAF mutants evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell 28, 370–383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J., et al. , Allosteric activation of functionally asymmetric RAF kinase dimers. Cell 154, 1036–1046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Yang P. L., Gray N. S., Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 9, 28–39 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su F., et al. , RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N. Engl. J. Med. 366, 207–215 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nazarian R., et al. , Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468, 973–977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng S. B., et al. , Inhibition of RAF isoforms and active dimers by LY3009120 leads to anti-tumor activities in RAS or BRAF mutant cancers. Cancer Cell 28, 384–398 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Henry J. R., et al. , Discovery of 1-(3,3-dimethylbutyl)-3-(2-fluoro-4-methyl-5-(7-methyl-2-(methylamino)pyrido[2,3-d]pyrimidin-6-yl)phenyl)urea (LY3009120) as a pan-RAF inhibitor with minimal paradoxical activation and activity against BRAF or RAS mutant tumor cells. J. Med. Chem. 58, 4165–4179 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Yao Z., et al. , RAF inhibitor PLX8394 selectively disrupts BRAF dimers and RAS-independent BRAF-mutant-driven signaling. Nat. Med. 25, 284–291 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Röck R., et al. , BRAF inhibitors promote intermediate BRAF(V600E) conformations and binary interactions with activated RAS. Sci. Adv. 5, eaav8463 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin T., et al. , RAF inhibitors promote RAS-RAF interaction by allosterically disrupting RAF autoinhibition. Nat. Commun. 8, 1211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park E., et al. , Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes. Nature 575, 545–550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liau N. P. D., et al. , Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization. Nat. Struct. Mol. Biol. 27, 134–141 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Ehrhardt A., Ehrhardt G. R., Guo X., Schrader J. W., Ras and relatives—Job sharing and networking keep an old family together. Exp. Hematol. 30, 1089–1106 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Ohba Y., et al. , Regulatory proteins of R-Ras, TC21/R-Ras2, and M-Ras/R-Ras3. J. Biol. Chem. 275, 20020–20026 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Quilliam L. A., et al. , M-Ras/R-Ras3, a transforming Ras protein regulated by Sos1, GRF1, and p120 Ras GTPase-activating protein, interacts with the putative Ras effector AF6. J. Biol. Chem. 274, 23850–23857 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Movilla N., Crespo P., Bustelo X. R., Signal transduction elements of TC21, an oncogenic member of the R-Ras subfamily of GTP-binding proteins. Oncogene 18, 5860–5869 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez-Viciana P., Sabatier C., McCormick F., Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol. Cell. Biol. 24, 4943–4954 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox A. D., Brtva T. R., Lowe D. G., Der C. J., R-Ras induces malignant, but not morphologic, transformation of NIH3T3 cells. Oncogene 9, 3281–3288 (1994). [PubMed] [Google Scholar]
- 29.Saez R., Chan A. M., Miki T., Aaronson S. A., Oncogenic activation of human R-ras by point mutations analogous to those of prototype H-ras oncogenes. Oncogene 9, 2977–2982 (1994). [PubMed] [Google Scholar]
- 30.Graham S. M., et al. , TC21 causes transformation by Raf-independent signaling pathways. Mol. Cell. Biol. 16, 6132–6140 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrhardt G. R., Leslie K. B., Lee F., Wieler J. S., Schrader J. W., M-Ras, a widely expressed 29-kD homologue of p21 Ras: Expression of a constitutively active mutant results in factor-independent growth of an interleukin-3-dependent cell line. Blood 94, 2433–2444 (1999). [PubMed] [Google Scholar]
- 32.Kimmelman A., Tolkacheva T., Lorenzi M. V., Osada M., Chan A. M., Identification and characterization of R-ras3: A novel member of the RAS gene family with a non-ubiquitous pattern of tissue distribution. Oncogene 15, 2675–2685 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Ward K. R., Zhang K. X., Somasiri A. M., Roskelley C. D., Schrader J. W., Expression of activated M-Ras in a murine mammary epithelial cell line induces epithelial-mesenchymal transition and tumorigenesis. Oncogene 23, 1187–1196 (2004). Correction in: Oncogene 23, 8858 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Flex E., et al. , Activating mutations in RRAS underlie a phenotype within the RASopathy spectrum and contribute to leukaemogenesis. Hum. Mol. Genet. 23, 4315–4327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higgins E. M., et al. , Elucidation of MRAS-mediated Noonan syndrome with cardiac hypertrophy. JCI Insight 2, e91225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez-Viciana P., Oses-Prieto J., Burlingame A., Fried M., McCormick F., A phosphatase holoenzyme comprised of Shoc2/Sur8 and the catalytic subunit of PP1 functions as an M-Ras effector to modulate Raf activity. Mol. Cell 22, 217–230 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Young L. C., et al. , An MRAS, SHOC2, and SCRIB complex coordinates ERK pathway activation with polarity and tumorigenic growth. Mol. Cell 52, 679–692 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Young L. C., Rodriguez-Viciana P., MRAS: A close but understudied member of the RAS family. Cold Spring Harb. Perspect. Med. 8, a033621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drosten M., et al. , Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J. 29, 1091–1104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takai Y., Sasaki T., Matozaki T., Small GTP-binding proteins. Physiol. Rev. 81, 153–208 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Keduka E., et al. , M-Ras evolved independently of R-Ras and its neural function is conserved between mammalian and ascidian, which lacks classical Ras. Gene 429, 49–58 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and/or SI Appendix.