Significance
Understanding cancer’s biomarker, microenvironment, and heterogeneity at the tissue level promises to improve cancer biology and clinical diagnosis and treatment. Many histological imaging techniques have been developed for this purpose. In this study, benefiting from the development of quantum sensing with nitrogen-vacancy (NV) centers in diamond, we proposed and experimentally realized micrometer-resolution magnetic imaging of tumor tissues in an NV-diamond magnetic microscope, magnetically imaging and quantifying cancer biomarkers. Moreover, we achieved the correlated magnetic and optical imaging in the same tissue section. The histological magnetometry exhibits excellent signal stability, negligible magnetic background, and accurate magnetic quantification. Our method thus opens the door to micrometer-resolution magnetic imaging of biological tissues and has the potential to influence research and histopathology of cancer.
Keywords: tumor tissue, histological magnetometry, nitrogen-vacancy center, micrometer-resolution magnetic imaging, absolute magnetic quantification
Abstract
Histological imaging is essential for the biomedical research and clinical diagnosis of human cancer. Although optical microscopy provides a standard method, it is a persistent goal to develop new imaging methods for more precise histological examination. Here, we use nitrogen-vacancy centers in diamond as quantum sensors and demonstrate micrometer-resolution immunomagnetic microscopy (IMM) for human tumor tissues. We immunomagnetically labeled cancer biomarkers in tumor tissues with magnetic nanoparticles and imaged them in a 400-nm resolution diamond-based magnetic microscope. There is barely magnetic background in tissues, and the IMM can resist the impact of a light background. The distribution of biomarkers in the high-contrast magnetic images was reconstructed as that of the magnetic moment of magnetic nanoparticles by employing deep-learning algorithms. In the reconstructed magnetic images, the expression intensity of the biomarkers was quantified with the absolute magnetic signal. The IMM has excellent signal stability, and the magnetic signal in our samples had not changed after more than 1.5 y under ambient conditions. Furthermore, we realized multimodal imaging of tumor tissues by combining IMM with hematoxylin-eosin staining, immunohistochemistry, or immunofluorescence microscopy in the same tissue section. Overall, our study provides a different histological method for both molecular mechanism research and accurate diagnosis of human cancer.
Cancer is one of the most common and most serious human diseases (1). Various medical imaging technologies have been widely used in the clinical examination of tumors (2), while histopathological examination is the gold standard for diagnosing human cancers (3). In addition, for targeted therapy, the screening of sensitive populations relies on quantitative histopathological analysis of tumor local biomarker levels (4). Optical microscopy is the most common method for histological examination. For instance, hematoxylin-eosin (HE) staining, immunohistochemistry (IHC), and immunofluorescence microscopy (IFM) have been developed for many years to obtain the morphological features and the distribution and expression level of biomarkers in tumor tissues (3, 5), respectively. There are many other tissue imaging technologies emerging, such as multispectral imaging based on tyramide signal amplification (6), highly multiplexed imaging based on mass cytometry (7), and label-free spectroscopic imaging (8–10). However, commonly used optical microscopic imaging cannot absolutely quantify the signal intensity and usually suffers from background signals in tissues (11, 12). Furthermore, it is difficult to correlate different optical imaging in the same tissue section, such as HE staining cannot be combined with other optical imaging. Each unique tissue section in imaging mass cytometry is a single-use sample due to its destructive nature. Therefore, developing a tissue-imaging method with outstanding properties remains a persistent pursuit and challenge for biologists and pathologists.
MRI provides a powerful technique for tumor imaging. Conventional MRI has been widely used for in situ imaging from biological research to clinical diagnosis of cancer (2). However, its low spatial resolution, e.g., merely 60 μm resolution of anatomical imaging at 9.4 T (13), limits the applications in tissue-level imaging. Recently developed microscale magnetic-imaging technology based on nitrogen-vacancy (NV) centers in diamond (14) provides a way for breaking the spatial-resolution limitation. The NV center is a nanoscale point defect in diamond and has been proposed as an ultrasensitive quantum sensor to realize nanoscale magnetometry (15–17). Since then, magnetic resonance techniques based on NV centers have been greatly developed and applied in nanoscale magnetic resonance spectroscopy and imaging of biological samples (14, 18–23). Meanwhile, wide-field magnetic imaging using an NV ensemble with submicrometer resolution is more suitable for the detection of large samples (24–30). These previous works are the cornerstones for magnetic resonance researches of microscale biological samples, such as single molecules and cells. However, micrometer-resolution magnetic imaging (or MRI) in biological tissues has not been achieved because of many kinds of technical barriers, which buries the potentially powerful capabilities of NV center-based quantum sensing in histopathology. Combining NV-based wide-field magnetic imaging with an immunomagnetic labeling technique modified from conventional immunocytochemistry (ICC) and immunofluorescence (IF) techniques, here we establish immunomagnetic microscopy (IMM), an approach that is capable of imaging cancer biomarkers in tumor tissues at a cellular resolution of ∼1 μm and quantifying them with absolute magnetic signals.
We here report four technical advancements to achieve an easy-to-use robust IMM of tumor tissues, including 1) quantifying the expression intensity of biomarkers as the absolute magnetic intensity, 2) reconstructing magnetic images using deep-learning algorithms, 3) immunomagnetically labeling tissue samples with magnetic nanoparticles (MNPs), and 4) attaching tissue sections over a large area close to the diamond to meet the limited detection range of NV centers. For the immunomagnetic examination, we developed a diamond magnetic microscope based on NV centers, combining magnetic imaging and multiplexed optical imaging. The wide-field magnetic imaging uses a layer of shallow NV centers below the diamond to detect weak magnetic fields in the biological sample close to the diamond surface.
Applying this approach, we imaged magnetic-labeled human tumor tissues and magnetically quantified the expression of biomarkers, which exhibited unparalleled signal stability and clean magnetic background. Meanwhile, the NV center-based IMM can be combined with conventional optical imaging techniques, such as HE staining, IHC, and IFM, to realize multimodal imaging in the same tissue section and has the potential to influence both fundamental research and clinical diagnosis of human cancer.
Results
Diamond Magnetic Microscope and Sample Preparation.
Principle and setup.
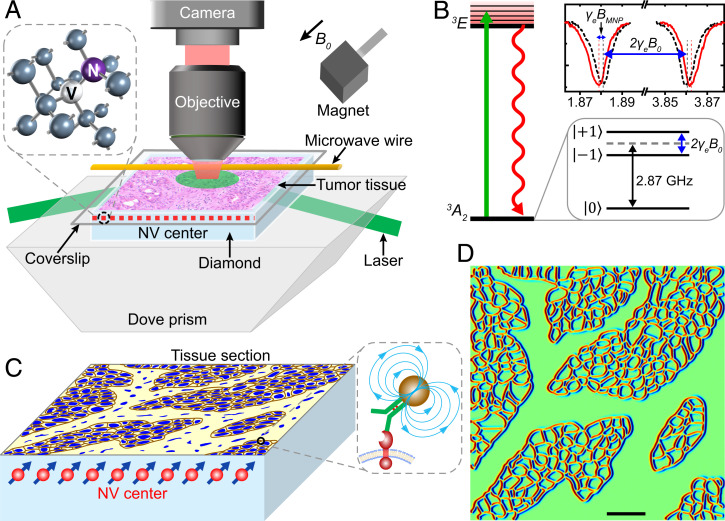
We have built an optically detected magnetic resonance (ODMR) wide-field microscope (Fig. 1A) to achieve tumor tissue magnetic imaging via detecting the continuous-wave (CW) spectrum of NV centers in diamond (Fig. 1B). The magnetic-field detection sensitivity is ∼10 μT/ from an NV sensor volume of about 1 × 1 × 0.1 μm3 in the top surface of diamond with a density of 2 × 1012/cm2 (Materials and Methods and SI Appendix, Fig. S1). The spatial resolution is about 1 μm with a field of view (FOV) of 0.5 × 0.5 mm. We label the target membrane protein of tissue with 20-nm size superparamagnetic nanoparticles and attach the labeled tissue to the diamond surface (Fig. 1C). The MNPs are magnetized by an externally applied magnetic field B0 along an NV axis, and the local magnetic field BMNP from MNPs shifts the peak position of the CW spectrum by a magnitude of γeBMNP (Fig. 1B). The magnitude of BMNP on NV centers depends on the density of MNPs and the distance from NVs. The typical magnetic signal of MNPs in cells and tissues is about 20 μT (SI Appendix, Fig. S2 A and D), which can be detected by our setup. By detecting the magnitude of the frequency shift, we can deduce the signal intensity and the distribution of target proteins in the tissue. We have simulated the magnetic-field signal of randomly distributed MNPs on the cell surface and the magnetic pattern of the MNP-labeled tissue (Fig. 1D and SI Appendix, Magnetic Signal Simulation). The original expression of MNP-labeled proteins can be reconstructed by a deep-learning model (SI Appendix, Deep Learning for Magnetic Signal Reconstruction and SI Appendix, Magnetic Quantification Error Analysis), and the expression intensity of biomarkers can be quantified with the absolute MNP density.
Fig. 1.
Schematic of diamond magnetic microscope and principle of tissue magnetic imaging. (A) Diamond magnetic microscope. The homebuilt wide-field ODMR setup is combined with a commercial optical microscope to achieve both optical and magnetic imaging of tumor tissues. An assumptive tumor tissue on the glass coverslip is located on the diamond surface. The green laser beam (532 nm) illuminates the NV centers layer, and the fluorescence is collected through an objective to an sCMOS camera. A permanent magnet creates a magnetic field B0, and the microwave is delivered by a copper wire. Inset shows the structure of an NV center. The NV centers along the B0 are used to sense the magnetic signal in the tissue. (B) Energy-level diagram of the NV center. Zero-field splitting degenerates the and with 2.87 GHz. Under magnetic-field B0 (here, 354 gauss), the energy level of splits due to the Zeeman effect, which is proportional to B0 (Δ = 2γeB0, for NV gyromagnetic ratio γe = 2.80 MHz/gauss). The peak appears in the CW spectrum when the microwave frequency is resonant with the allowed transition → or → The local magnetic field of MNP shifts the peak positions by a magnitude of γeBMNP. (C) A tissue section is detected by the NV centers. A 100-nm-thick layer of dense NVs is used to image an MNP-labeled tissue section. The distribution of target proteins in the tissue is deduced from the frequency shift caused by MNPs. Brown lines represent MNP-labeled membrane proteins, and blue structures represent cell nuclei. (D) Simulated magnetic image of the tissue in C. We assume that MNP-labeled proteins uniformly distribute on the cell membrane. The protein map is then illustrated here, and the red and blue lines mark the magnetic signal as two poles. (Scale bar, 50 μm.)
Sample preparation and evaluation with magnetic imaging of adherent cells.
As illustrated in SI Appendix, Fig. S6A, tissues and adherent cells were prepared for magnetic labeling using a modified ICC/IF protocol. Primary antibodies were selected to specifically target proteins relevant to cancer, and the biotinylated secondary antibody IgG bound the primary antibody, while the recognition of streptavidin and biotin was used to connect MNPs and secondary antibodies (26, 31). To select MNPs with appropriate particle size, we compared the performance of 20- and 50-nm magnetic particles in immunomagnetic labeling of tumor tissues (SI Appendix, Fig. S7). The 20-nm MNP group displayed the desired permeability and target specificity. In all of the following measurements, the size of the MNP is 20 ± 1 nm in diameter. The MNPs bound to the antigens provided characteristic signals for the magnetic-field imaging. To confine an MNP-labeled tissue section within a few micrometers’ detection range from a shallowly embedded NV layer (with a depth range between 10 and 110 nm below the diamond surface) and improve spatial resolution, we need to keep the tissue section and the diamond as close as possible, i.e., with a distance of ∼1 μm in our experiments, over the entire diamond area and during the magnetic detection. To this end, after air drying, the MNP-labeled sample on the glass coverslip was embedded and fixed on the diamond to ensure that the sample was closely attached to the diamond (Fig. 1 A and C and Materials and Methods). In the meantime, the adhesive-embedding practice preserved the underlying tissue architecture.
To establish the validity of the above strategies, IMM was first performed on cultured adherent cells on a coverslip and a diamond, targeting epithelial cell adhesion molecules (EpCAMs) (SI Appendix, Fig. S6). Comparing the ∼1-μm resolution magnetic images, the magnetic-field magnitude and spatial extension of cells on the coverslip were comparable with those of cells on the diamond, in which case the cells adhered tightly to the diamond surface and the MNP-labeled membrane proteins were close to the NV sensors. Furthermore, we obtained a magnetic image with a resolution of ∼400 nm in cells on the coverslip (SI Appendix, Fig. S6E). According to these experimental results and the simulation of magnetic signals (SI Appendix, Magnetic Signal Simulation), the cells on the coverslip were indeed closely attached to the diamond, with a distance of 1 μm or less, which fulfilled our experimental requirement. These results confirmed our strategies for sample preparation and magnetic imaging based on NV centers.
Immunomagnetic Microscopy of Tumor Tissues.
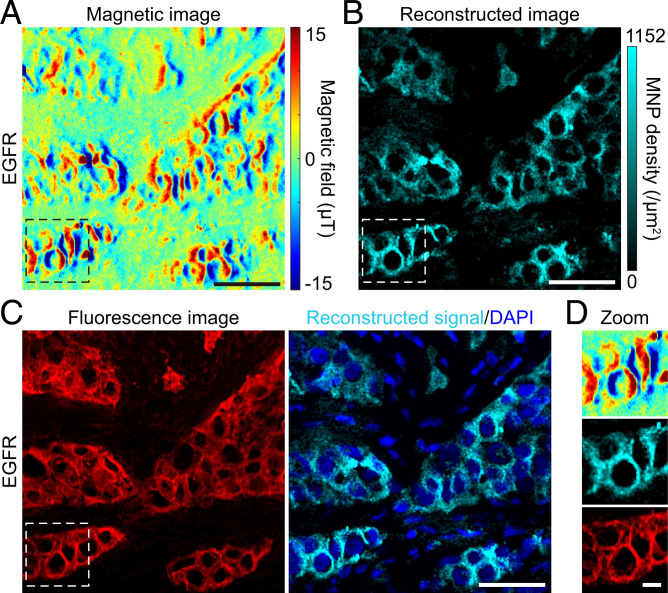
Next, we performed IMM on human tumor tissues. Lung cancer is the most commonly diagnosed cancer and the leading cause of cancer death worldwide (1). Epidermal growth factor receptor (EGFR) is a key regulator protein and therapeutic target of lung cancer. The precise examination of the EGFR expression level and mutations is crucial for its targeted therapy (32, 33). We immunomagnetically labeled and imaged EGFR in a cryosection of a human lung cancer sample. The magnetic image showed a characteristic magnetic-field pattern (Fig. 2A), and the micrometer spatial extension of the magnetic field was consistent with the simulation (Fig. 1D and SI Appendix, Fig. S2). To enable intuitively studying the expression of biomarkers in tissues, we deciphered the relationship between the magnetic-field map and the distribution of corresponding MNP-labeled proteins by using a deep-learning model of conditional generative adversarial networks (cGANs) (34) (SI Appendix, Deep Learning for Magnetic Signal Reconstruction). Benefitting from the superiority of multidimensional structure, the deep-learning network transformed the reconstruction problem into a uniquely resolvable bijection. As a result, we reconstructed the magnetic-field pattern and resolved the distribution of EGFR (Fig. 2B), which matched well with fluorescence micrographs of Cyanine3 (Cy3) labeled on MNPs in the corresponding area (Fig. 2 C and D). The same expression pattern of EGFR locating on the cell membrane and in the cytoplasm in both IMM and IFM (SI Appendix, Fig. S8) confirmed that IMM truly imaged the marker’s expression without apparently changing its distribution in cells of the tissue. Importantly, the magnetic imaging measured the absolute magnetic magnitude, and MNP densities in the reconstructed image suggest the absolute distribution of the marker in the tissue, which can be reproduced in other diamond magnetic microscopes. To evaluate the magnetic quantification accuracy, we analyzed different types of quantification errors, and the typical magnetic quantification errors of IMM are ±5.29% for single pixels and ±1.875% for single cells (SI Appendix, Magnetic Quantification Error Analysis).
Fig. 2.
Immunomagnetic microscopy of lung tumor tissue. (A) A representative magnetic-field image of EGFR proteins in a lung tumor tissue. The biomarker EGFR was immunolabeled with a bimodal magnetic-fluorescent label Cy3-MNP, as shown in SI Appendix, Fig. S6A, and measured in our diamond magnetic microscope. The magnetic-field map displays the protein distribution in the tissue. The typical magnitude of the magnetic signal is between 15 μT and −15 μT. (B) Reconstructed IMM image of the magnetic image in A. The cyan pseudocolor structure represents the reconstructed IMM signal of the marker. The image intuitively shows the distribution of target proteins, which is directly comparable with the fluorescence image. The MNP density represents the expression intensity of the marker. (C) Comparison of fluorescence and IMM images. Cy3 in the red channel represents the original location of MNPs in the same area shown in A and B. DAPI in the blue channel stained cell nuclei. (D) Magnification of magnetic, reconstructed, and fluorescence images. Magnified images display a representative subcellular magnetic-field pattern. The magnetic imaging nearly coincides with the fluorescence imaging. The slight difference is due to the mismatched focused planes between Cy3-fluorescence imaging (a longitudinal plane with the best signal-to-noise ratio in the whole tissue section) and magnetic imaging (a ∼1 μm thickness plane of the section surface facing the diamond). These results were confirmed in five samples in independent experiments. (Scale bars: A–C, 50 μm; D, 10 μm.)
Another apparent advantage of IMM is the ultrastable signal of MNP. We found no evidence of photodamage on MNPs in stained samples even after studying them under intense, continuous laser illumination for several hours. Fluorescent probes generally suffer from light-induced instability. Our examinations revealed that the fluorescence of Cy3 in samples decreased seriously under the illumination of a fluorescent lamp, whereas the magnetic signal had excellent stability (even the samples that had been exposed under the lamp for 1.5 y) (SI Appendix, Fig. S9). The outstanding stability of magnetic signals in IMM (at least about the same as in IHC) allows long-term preservation and future analysis of the samples and guarantees the repeatability of the measurement.
IMM Circumvents the Impact of Background in Tissues.
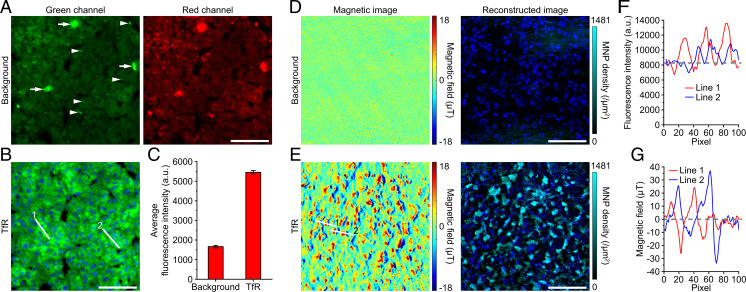
There is barely natural static magnetic-field background in most biological samples, which is the basis of acquiring specific magnetic signals and performing accurate quantification. In comparison, all fluorescence-based methods face a common challenge: The autofluorescence of biological samples affects the detection and quantification of biomarkers (11, 35). For example, liver, kidney, and some other tissues have strong autofluorescence (11, 35, 36), which limits the use of fluorescence imaging. Here, we carried out a set of experiments in human liver cancer samples (Fig. 3). In the liver tumor tissue, we can see obvious uniform autofluorescence in the cytoplasm, as well as scattered punctum and plaque fluorescent structures, especially in the red channel (Fig. 3A). The background fluorescence will seriously affect the subcellular analysis and fluorescence quantification of many biomarkers, whether they are membrane proteins such as transferrin receptor 1 (TfR) (37) (Fig. 3B), proteins in the cytoplasm such as cytokeratins (38), or nuclear proteins such as nuclear proliferative protein Ki67 (39). The results show that the autofluorescence signal accounts for about 30% of the total signal in the immunolabeled sample targeting TfR (Fig. 3 B and C), which will produce a huge statistical deviation in quantification analysis. In contrast, there is no detectable natural magnetic signal in liver tumor tissues (Fig. 3D). Meanwhile, the frequency shift measurement of IMM and the bright fluorescence of the 100-nm NV layer avoid noticeable impact from autofluorescence in tissues (SI Appendix and SI Appendix, Fig. S10). Therefore, IMM detected pure and specific magnetic-field signals from the MNP-labeled biomarker (Fig. 3E). The image contrast of the two methods is also very different, with the signal-to-noise ratio (SNR) mostly less than 2 in fluorescence imaging (Fig. 3F) and more than 10 in IMM (Fig. 3G). Therefore, IMM can image the biomarker more purely and clearly, which also contributes to the magnetic quantification accuracy. These results were confirmed in a paraffin-embedded mouse liver sample (SI Appendix, Fig. S11).
Fig. 3.
IMM of human liver tumor sample. (A) The autofluorescence in a PFA-fixed human liver tumor tissue section was imaged as a background group. Arrowheads and arrows indicate puncta and plaques, respectively. There are more background signals in the red channel. (B) Immunofluorescence image of TfR in the same section as in A. The example images in A and B are in the same imaging area. (C) Quantification of average fluorescence intensity. Data are represented as mean ± SEM, n = 66 (background) and 112 (TfR) areas. (D) Background magnetic-field images in a PFA-fixed liver tissue section. There are only weak measurement noises in the images. The typical magnetic magnitude of the noise is below 1 μT. (E) IMM images of TfR in another section. Shown are DAPI-stained cell nuclei. (F and G) Linear profile analysis of the IFM and IMM images in B and E, respectively. Linear pixels are marked with white lines in B and E. The gray dashed lines represent the baselines. These results were confirmed in two samples in independent experiments. (Scale bars, 100 μm.)
Magnetic Imaging and Quantification of Cancer Biomarkers.
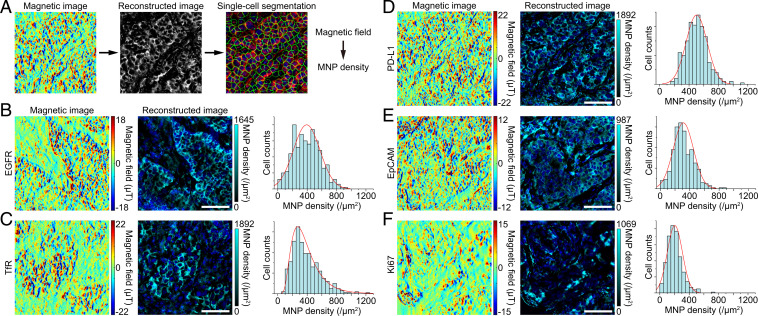
To verify the general applicability of IMM, we carried out a series of histological magnetometry experiments on human lung tumor tissues, labeling and imaging membrane proteins EGFR, TfR, EpCAM, programmed cell death ligand 1 (PD-L1) (40), and nuclear protein Ki67. IMM images from various biomarkers exhibited different staining patterns and expression intensities (Fig. 4). The similar distributions of markers in IFM (SI Appendix, Fig. S8) confirmed the reliability of IMM. We further quantified the single-cell expression distributions of markers from the reconstructed images (Fig. 4). The MNP density-represented expression distributions suggest characteristic absolute expression distributions of the markers.
Fig. 4.
Magnetic images and quantification of a variety of cancer biomarkers. (A) Workflow of single-cell quantification from magnetic images. The magnetic image reconstruction is fulfilled by a deep-learning model, and the reconstructed image is segmented at the single-cell level in a single-cell segmentation software. (B–F) Magnetic images, corresponding reconstructed images, and single-cell expression distributions of five typical biomarkers in lung tumor tissues. Membrane proteins EGFR, TfR, EpCAM, and PD-L1 show obvious cell membrane distribution, while the nuclear localization protein Ki67 mainly locates in the cell nucleus. Color bars represent magnitudes of magnetic signals and MNP densities. Shown are DAPI-stained cell nuclei. The histograms display distinct single-cell expression distributions of different markers. These results were confirmed in five (EGFR), five (TfR), five (PD-L1), three (EpCAM), and three (Ki67) samples in independent experiments, respectively. (Scale bars, 100 μm.)
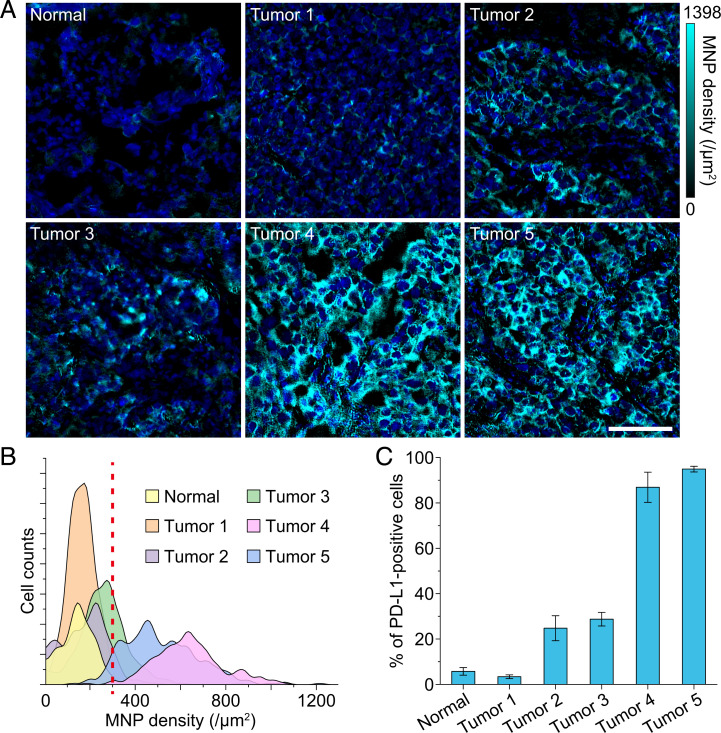
Among the above biomarkers, the immune-inhibitory checkpoint PD-L1, an important target of lung cancer immunotherapy, has been studied extensively (40, 41). However, not all patients are potentially sensitive to PD-L1 inhibitors because of the uncertain and heterogeneous expression of PD-L1 in the tumor microenvironment. Therefore, identification of PD-L1 expression is highly demanded to determine the feasibility of anti–PD-L1 immunotherapy and predict the therapeutic efficacy (41). Here we performed IMM on PD-L1 in five lung tumor samples and a normal case, and there were different distributions and expression levels of PD-L1 (Fig. 5A). In the normal tissue, PD-L1 was almost not expressed, while in the tumor tissues, the expression level was variable. We analyzed the single-cell expression of PD-L1 in each of the representative IMM images in Fig. 5A and delineated the results in the histogram (Fig. 5B). Here, if we set the threshold of PD-L1 positive expression to an MNP density of 300/μm2 (illustrated by the dashed line in Fig. 5B), then the percentages of positive cells were quantified (Fig. 5C). The high-contrast magnetic images and accurate absolute magnetic quantification are helpful to more objectively identify the heterogeneous expression of PD-L1 and more accurately grade the patients. Thus, IMM presents an avenue for the accurate diagnosis related to important targets, which is a prerequisite for individualized targeted therapies.
Fig. 5.
Magnetic quantification of PD-L1 in lung tissues. (A) Reconstructed IMM images of PD-L1 in a normal lung tissue and five lung tumor tissues. We adjusted color bars to the same dynamic range to intuitively show the difference of PD-L1 expression levels among different samples. Shown are DAPI-stained cell nuclei. (Scale bar, 100 μm.) (B) Single-cell expression distributions of PD-L1 in IMM images shown in A. The smoothed histogram shows obvious different expression intensity of PD-L1 in different samples. The red dashed line indicates an assumed positive threshold. (C) Percentages of PD-L1–positive cells were calculated according to the threshold of 300 MNPs/μm2 in B. The normal sample and tumor sample 1 are obviously PD-L1 negative, tumor samples 2 and 3 have low PD-L1 expression, and tumor samples 4 and 5 have high PD-L1 expression. Data are represented as mean ± SEM, n = 7 and 5, 4, 5, 7, and 5 areas for normal sample and tumor samples 1, 2, 3, 4, and 5, respectively.
Taken together, our data demonstrate the utility of IMM for magnetic imaging and absolute magnetic quantification. Under the same magnetic bias field B0, the magnetic-field pattern and signal intensity in a magnetic-labeled sample would not change with the operator, the experimental platform, and the surrounding environment. Therefore, IMM can provide an appropriate standard for the expression of a biomarker of interest in tissues. It is useful for studying a protein’s function and the accurate diagnosis of diseases related to the protein.
Correlated Magnetic and Optical Imaging.
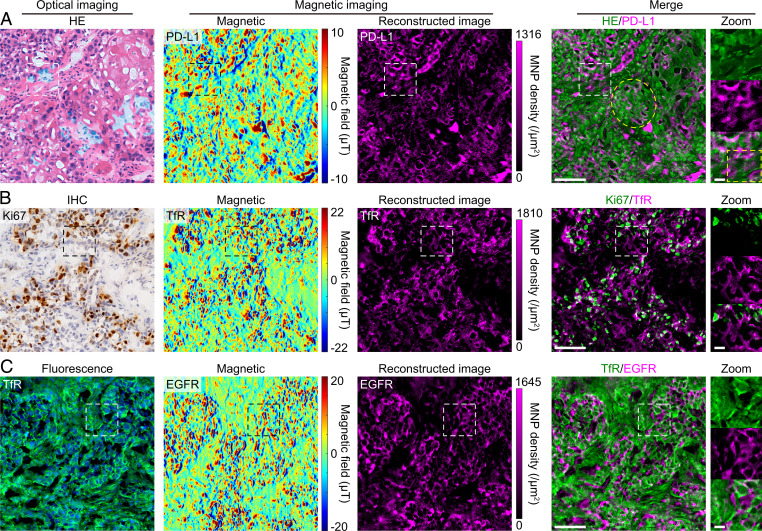
The measurements above have shown the compatibility of magnetic and fluorescence imaging, which benefits from the optical detection manner of ODMR. Here we further investigate multimodal imaging with multiple staining in the same tissue section. Due to the staining and bright fluorescence produced by hematoxylin and eosin (42) (SI Appendix, Fig. S12), it is difficult to correlate the HE, IHC, and fluorescence imaging in the same section. Current multiplexed imaging in adjacent serial sections (43) and sequential imaging in the same section (44) suffer from the registration of multiplexed images. Additionally, sequential imaging can change the antigenicity of the target specimens and is time consuming. In this work, IMM can avoid these problems because the additional magnetism can be detected in parallel with light. Using IMM, although the bright fluorescence from hematoxylin and eosin significantly reduced the contrasts of the NVs’ CW spectrums (SI Appendix, Fig. S13), magnetic fields in the MNP-labeled tissues were effectively mapped by the NV sensors (Fig. 6), which were contributed by the frequency-shift analysis manner of ODMR and the 100-nm NV layer’s strong fluorescence signal. Ultimately, we simultaneously acquired the morphological feature and the biomarker (PD-L1) detail in the same tissue section by correlating HE staining with IMM (Fig. 6A), making it convenient to differentiate PD-L1 expression in tumor cells (indicated by the yellow ellipse in Fig. 6A) and tumor-infiltrating immune cells (yellow box). Correlating IHC and IMM for double labeling of Ki67 and TfR, respectively, we achieved the combination of two approaches with stable signals (Fig. 6B). The IHC imaging generally undergoes endogenous enzymes, chromogenic substrate precipitation, or background chromogenic signals (12), whereas IMM can circumvent many of these issues. In addition, correlated fluorescence imaging and magnetic imaging targeting TfR and EGFR, respectively, were performed in the same section (Fig. 6C). NV-based magnetic imaging hardly interferes with any fluorescence channels.
Fig. 6.
Correlated magnetic and optical imaging in lung tumor tissues. (A) Correlated HE staining and IMM in the same tissue section. Hematoxylin and eosin stained the tissue’s cellular structure, simultaneously producing strong fluorescence signals (SI Appendix, Fig. S12), which significantly reduced the contrast of the NVs’ CW spectrum (SI Appendix, Fig. S13). Nevertheless, the robust IMM resisted the impact of fluorescence from hematoxylin and eosin. Although the magnetic image shows a slightly reduced signal-to-noise ratio, it still clearly resolves the distribution of PD-L1. The yellow ellipse and box in the merged images indicate squamous carcinoma cells and immune cells, respectively. (B) Double labeling of Ki67 and TfR by IHC and immunomagnetism in the same section, respectively. In IHC, DAB immunostained Ki67 proteins and hematoxylin stained cell nuclei. Again, the IMM resisted the impact of DAB and hematoxylin, and we obtained a high-quality magnetic image. (C) Double labeling of TfR and EGFR by IF and immunomagnetism in the same section, respectively. Alexa Fluor 488 in the green channel represents TfR, which was labeled via the routine immunofluorescence procedure. Shown are DAPI-stained cell nuclei. Both imaging methods work well without disturbing each other. The HE and DAB signals were recolored with the green color, and then the optical images were merged with the reconstructed IMM images. These results were confirmed in three (IMM and HE), four (IMM and IHC), and two (IMM and IFM) samples in independent experiments. (Scale bars, 100 μm and 20 μm [zoom].)
Combining NV-based IMM and optical microscopy in the same tissue section realized multimodal imaging for histological examination, thus obtaining additional information from pathological tissues, maximizing pathological data from some limited sample resources, and providing more possibilities for histopathology.
Discussion
In this work, we developed IMM for human tumor tissues utilizing NV centers in diamond as the quantum sensors. In IMM, magnetism was used as an alternative physical quantity besides light and mass for micrometer-resolution tissue imaging. The IMM methodology has accurate absolute magnetic quantification capability with excellent signal stability and negligible magnetic background. Moreover, the correlated IMM and HE staining are significant for studying tumor microenvironment and heterogeneity. The versatile IMM can be extended to histological examinations for diverse cancers and other biological processes and diseases, such as heart disease, inflammation, and neurological disorder (3). This work provides an attractive approach for histological examination of human diseases, which complements existing tissue-imaging approaches and enriches magnetic resonance techniques.
IMM will play a unique role in examining tissues with autofluorescence, endogenous enzymes, or low light transmittance. Background signals in tissue samples hide some information of concern in optical images. Fluorescence imaging provides only relative quantitative information for biomarkers but not absolute quantification, while IHC cannot quantify the signal intensity. For clinical diagnosis, IHC just qualitatively analyzes and grades the pathological sample, which depends on the pathologist’s experience and is subjective. By comparison, magnetic imaging depicts the pure expression of biomarkers without interference from various factors. In principle, we ultimately can achieve absolute quantification of biomarkers based on a definite stoichiometric relationship between the target protein and the specific binding magnetic particles. Based on massive IMM data and other clinical data, standards of quantitative parameters related to biomarkers of interest and general clinical features can be obtained to guide precise clinical diagnosis and treatment. In addition, we propose a combination of IMM with HE staining at the microscale and medical imaging, e.g., MRI, computerized tomography (CT), and positron emission tomography (PET) at the macroscale and in situ levels, to monitor the spatiotemporal changes of the tumor in a cross-scale multimodal manner. Long-term evaluation of responses and tumor progression in patients is needed after clinical treatments (45), in which absolute quantification and signal stabilization of IMM will play a unique role. Further studies will develop three-dimensional IMM and in situ hybridization based on NV centers, which will provide additional opportunities for cancer biology and pathology (46).
Several technical issues also will be addressed in the future. First, IMM using different spin labels or MNPs with different susceptibilities (47) can achieve multichannel magnetic resonance imaging. Besides, high-quality immunomagnetic imaging will benefit from the exploitation of smaller magnetic particles with higher magnetic moment. Biotinylated primary antibodies (26) and antibody-coated MNPs could simplify the immunomagnetic labeling, thereby shortening the processing time. Second, we will try to speed up the examination. At present, to measure an area of 0.5 × 0.5 mm at 1-μm resolution takes ∼10 min. In further works, the closer distance between the sample and the sensor, and the use of an isotope-C12 enriched diamond sensor, will bring higher sensitivity and higher efficiency (48), and then a magnetic image could be acquired in less than 1 min. Third, with a larger bulk diamond, such as 1 × 1 cm, larger tissue specimens can be measured. The increase in data size and complexity should be accompanied by optimized algorithms. The optimization, integration, and modularization of the device will reduce the operating requirements, thereby promoting the popularization of NV-based IMM.
In addition to pathological tissues, our NV-based magnetic microscopy enables imaging, quantification, and analysis of various MRI contrast agents (49), magnetic particles, and magnetic molecules in animal magnetoreception (50) at the tissue level with a submicrometer or subcellular resolution.
Materials and Methods
Diamond Magnetic Microscope.
The diamond magnetic microscope was constructed from an upright microscope (Olympus, BX53), combining a wide-field magnetic imaging system based on NV centers in diamond with a conventional wide-field fluorescence imaging system. The sample stage was carefully designed to allow magnetic imaging via a plug and play mode. A 532-nm laser with adjustable power output (Changchun, OEM-N-532-3W) passed through a dove prism (Thorlabs, PS992-A) to excite the NV ensemble in a bulk diamond with a total internal reflection angle. The prism-assisted excitation raised the laser power upper limit compared with the objective lens-based way. The NV-fluorescence channel comprised a dichroic beam-splitter (Thorlabs, DMLP567R) and a barrier filter (Semrock, BLP01-633R-25). Other fluorescence channels were achieved by mercury lamp excitation, including 4′,6-diamidino-2-phenylindole (DAPI) and other blue channels (Olympus; exciter filter BP340-390, dichroic beam splitter DM410, and barrier filter BA420), Alexa fluor488 and other green channels (BP460-495, DM505, and BA510-550), and Cy3 and other red channels (BP530-550, DM570, and BA575-625). The imaging channels can be expanded by choosing appropriate filters and dichroic beam splitters. A scientific complementary metal-oxide semiconductor (sCMOS) camera (Andor, Neo 5.5) was used in the ODMR experiments and fluorescence image acquisition. Almost all of the fluorescence was collected by a 20× objective with a numerical aperture (N.A.) of 0.75 (Olympus, 60× N.A. 0.7 for the high-resolution imaging) onto the sCMOS camera. A color camera (Thorlabs, CS895CU) was used to acquire color images from HE staining and IHC. The microwave was generated by a radio-frequency generator (SynthNV; WindFreak Technologies, LLC), amplified (Mini-Circuits, ZHL-16W-43-S+), and delivered by a 60-μm copper wire above the glass coverslip. The external magnetic field was produced by a 2-cm cubic permanent magnet (NdFeB, N50).
Fabrication of NV-Diamond Quantum Sensor.
All diamonds used in this work are electronic-grade [100] bulk diamond (2 × 2 × 0.5 mm) grown using chemical vapor deposition (CVD) (Element Six). The diamonds were implanted sequentially with 14N+ ions at energies of 68.9, 49.5, 34.4, 23.0, and 14.4 keV and doses of 5.1 × 1013, 4.2 × 1013, 3.4 × 1013, 2.7 × 1013, and 2 × 1013/cm2, respectively (SI Appendix, Fig. S1). The ion-implanted diamonds were subsequently annealed in a vacuum for 4 h at 1,000 °C to produce NV ensembles with a depth range between 10 and 110 nm below the diamond surface. The estimated final NV density in the shallow layer was 2 × 1012/cm2. Compared with the 10-nm-thick NV layer, the 100-nm layer has a higher fluorescence count, which is more suitable for imaging over a large field of view while maintaining submicrometer spatial resolution for tissue imaging.
Cell Culture.
NCI-H292 cells, a human pulmonary mucoepidermoid carcinoma cell line (ATCC), were grown at 37 °C in a 5% CO2 humidified environment in RPMI-1640 medium (Biological Industries). The medium was supplemented with 10% fetal bovine serum (Biological Industries), penicillin (100 units/mL) (HyClone), and streptomycin (100 mg/mL) (HyClone). Before being used to culture cells, the diamond and the glass coverslip were coated with poly-L-lysine (0.1 mg/mL) for 1 h at room temperature and then washed three times with deionized water. For magnetic labeling and imaging, cells were grown to ∼80% confluency on the diamond and coverslip.
Human Samples.
All tissue specimens from patients with nonsmall cell lung cancer (NSCLC), liver cancer, or benign tumor were collected from the first Affiliated Hospital of Anhui Medical University (Hefei, Anhui, China) and the first Affiliated Hospital of University of Science and Technology of China (Hefei, Anhui, China). The collection of the samples of treatment of naïve patients who underwent surgical resection and the subsequent experiments were approved by the Human Ethics Committee of the University of Science and Technology of China (USTCEC201600004; Hefei, China). Informed consent was obtained from each patient.
Sectioning of Tissues.
Fresh tissues were embedded in OCT (optimal cutting temperature polymer; Leica 14020108926, Leica Biosystems) and snap frozen at −80 °C. Frozen fresh tissues were sectioned at 5 to 10 μm by the cryostat for high-quality sectioning (Leica, CM1950). The paraffin-embedded tissue was sectioned at 5 μm by the rotary microtome (Leica, RM2235). The slide was placed on a poly-L-lysine–treated glass coverslip with a diameter of 25 mm (VMR, 16004-310).
Immunostaining.
The tissue slides were treated as described (3, 12) before staining. The IF staining of tissue-section and cell samples was performed as the routine IF procedure. Briefly, tissue and cell samples were fixed with 4% paraformaldehyde (PFA) for 15 and 10 min at room temperature, respectively. Then, samples were blocked in blocking solution (2% bovine serum albumin [BSA], 0.1% Triton X-100, 1× phosphate-buffered saline [PBS]) for 30 min at room temperature. Subsequently, primary antibodies with different dilutions in incubation buffer (1% BSA, 0.1% Triton X-100, 1× PBS) were added and incubated overnight at 4 °C. Primary antibodies used in this work contain anti-EGFR antibody (EP38Y) (Abcam; ab52894, 1:400 dilution), transferrin receptor monoclonal antibody (H68.4) (Thermo Fisher Scientific; 13-6800, 1:250), EpCAM (VU109) mouse mAb (Cell Signaling Technology; 2929, 1:1,000), anti–PD-L1 antibody (28-8) (Abcam; ab205921, 1:500), anti-Ki67 antibody (SP6) (Abcam; ab16667, 1:250), and anti-Histone H2B antibody (Abcam; ab1790, 1:1,000). After washing five times with PBS at room temperature, each for 5 min, the secondary antibody Alexa fluor 488-AffiniPure goat anti-mouse Immunoglobulin G (IgG) (heavy and light chains, H&L) (Jackson ImmunoResearch, 115-545-003) or Alexa Fluor 488-AffiniPure Goat Anti-Rabbit IgG (H&L) (Jackson ImmunoResearch, 111-545-003) was used at a final concentration of 2 μg/mL and incubated for 1 h at room temperature. After extensive washing and DAPI staining, the labeled samples were examined in the fluorescence microscope.
For the immunomagnetic labeling, all tissue-section and cell samples were processed using the routine ICC/IF protocol almost the same as described above. After the incubation of primary antibodies, biotinylated secondary antibody goat anti-mouse IgG H&L (biotin) (Abcam, ab6788) or goat anti-rabbit IgG H&L (biotin) (Abcam, ab6720) was used at a dilution of 1:1,000 and incubated for 1 h at room temperature. The bound secondary antibodies were labeled by streptavidin-coated iron oxide nanoparticles (20 nm) (Ocean NanoTech, SHS-20) with a concentration of 50 μg/mL (Fe) for 30 min at room temperature. Optionally, Cy3-biotin (0.1 μM) was used to fluorescence label the streptavidin-coated magnetic nanoparticles in the samples for 10 min at room temperature. The Cy3-biotin was derived from the conjugation of Sulfo-Cyanine3 amine (Lumiprobe, 113C0) and EZ-Link Sulfo-NHS-LC-Biotin (Thermo Scientific, 21335), with a mole ratio of 3:1, for 12 h at room temperature. After extensive washing and DAPI staining, the magnetic-labeled samples were examined in the magnetic microscope.
In the magnetic and HE multiplexed staining, the HE staining was carried out according to a standard protocol after the magnetic labeling of PD-L1. Double immunostaining was performed using primary antibodies from different species. For TfR (magnetic) and Ki67 (IHC) double labeling, a mouse TfR (Thermo Fisher Scientific, 13-6800) and rabbit Ki67 (Abcam, ab16667) detection antibody mixture was used, followed by a mixture of goat anti-mouse IgG H&L (biotin) (Abcam, ab6788) and goat anti-rabbit IgG H&L (horseradish peroxidase) (Abcam, ab6721). Di-azo-aminobenzene (DAB) and hematoxylin (Sangon Biotech) were added for immunohistochemical staining. For EGFR (magnetic) and TfR (fluorescence) double labeling, a rabbit EGFR (Abcam, ab52894) and mouse TfR (Thermo Fisher Scientific, 13-6800) detection antibody mixture was used, followed by a mixture of goat anti-rabbit IgG H&L (biotin) (Abcam, ab6720) and Alexa fluor 488-AffiniPure goat anti-mouse IgG (H&L) (Jackson ImmunoResearch, 115-545-003).
Attaching Tissue Sections to Diamonds.
The magnetic-labeled tissue section (or adherent cells) on the coverslip was briefly rinsed with deionized water. After air drying, the section was soaked by a small drop of ultraviolet (UV) curing adhesive (Norland, NBA107). A diamond was then covered on the adhesive, with the side containing NV centers facing the cell layer, and pressed moderately with a customized clamp to bring the sample and the diamond close together. The sandwich structure was carefully transferred to an UV lamp device (Thorlabs, SOLIS-365C) and polymerized for 20 min with an optical power density of ∼15 mW/cm2 to fix the connection. These processes ensured the distance between the sample and the sensor was as close as possible. The fixed sandwich structure was fused on a dove prism by UV curing adhesive with the diamond back facing the dove prism. The glass beyond the diamond region was cut off by a diamond scribing pen (Ted Pella, 54468). The refractive index of the polymer derived from the UV curing adhesive was almost the same as that of dove prism and coverslip, maximally reducing the optical distortion in the light path. Besides, the adhesive-embedding practice avoided the magnetic signal artifacts related to the light scattering and maintained the natural morphological features of tissues.
Magnetic Imaging.
Magnetic imaging was carried out on the homebuilt diamond-based magnetic microscope. The 532-nm excitation laser beam had an average power density of about 300 W/cm2 and an elliptic spot larger than 500 × 500 μm. With the optical polarization and excitation, the state of NV centers was pumped to with a spin state-dependent fluorescence emission, while the resonant microwave induced the transitions between and states. The magnetic-field B0 from the external magnet was applied along an NV axis and magnetized MNPs in the tissue. The projection component of the MNP-generated magnetic-field vector to the NV axis, as well as B0, shifted the peak position of the NV’s CW spectrum due to the Zeeman effect. The Zeeman-shifted NV spin transition can be detected by measuring the fluorescence decrease of NV centers via continuously sweeping the microwave frequency. The fluorescence maps of NVs were acquired with a typical exposure time of 50 ms, and all fluorescence was projected onto and captured by the sCMOS camera. We swept the microwave frequency over the and resonances interleaved point by point with a step size of 0.5 MHz, and the sweeping was repeated 100 times. The switching detection mode reduced possible systematic effects due to thermal drift and internal strain in the diamond. All hardware was controlled by a customized image-acquiring system in LABVIEW, which synchronized the camera and the microwave generator. Collected fluorescence data were binned (bin = 2), preprocessed, and Lorentz fitted using the acceleration package GPUfit (51) in MATLAB 2019b. Subsequently, using a quadratic function, we subtracted the baseline mainly related to the external magnetic field B0 and obtained the magnetic image of MNP-labeled proteins.
Magnetic Image Analysis.
The magnetic-field signals in the magnetic image depend on the distribution and intensity of the magnetic moment in the MNP-labeled tissue sample and the distance from NV sensors (SI Appendix, Magnetic Signal Simulation). We deciphered the relationship between the magnetic-field map and the distribution of corresponding MNP-labeled proteins by using a deep-learning framework (SI Appendix, Deep Learning for Magnetic Signal Reconstruction). Experimental data were slightly denoised using the discrete cosine transform (DCT) filter in MATLAB and then put into the trained network. Magnetic moment images were inferred and the distances were resolved simultaneously. The rationality of the magnetic reconstruction was verified by checking the magnetic reconstruction error and the existence of the error curve minimum (SI Appendix, Fig. S4). Reconstructed IMM images were processed in ImageJ v1.52v for adjusting contrast, cropping, pseudocolor coding, and merging with fluorescence images. The biomarker distributions in the reconstructed IMM images were provided as MNP densities, which were transformed from magnetic moment images with a single MNP moment of 6 × 10−16 emu (electromagnetic unit) (52).
Single-Cell Segmentation and Quantification of Magnetic Images.
For the single-cell expression quantification, we performed the single-cell segmentation on the reconstructed IMM images in the TissueQuest v4.0.1.0151 software (TissueGnostics Gmbh). DAPI was used as a master marker to identify the cell nuclei, assisting the single-cell segmentation. Signals of biomarkers of interest were recognized using the ring mask and the identified cell mask, outsides mode. After automated identification, some errors were corrected by manual operation. Finally, single-cell mean values were collected and analyzed. Histograms of single-cell expression distributions were delineated in Origin 2020. Data in Figs. 3C and 5C were presented as the mean value ± SEM.
Supplementary Material
Acknowledgments
We thank Xuejin Chen, Binxin Yang (University of Science and Technology of China), and Xiaodong Wu (University of Iowa) for helpful discussions on deep learning. The deep learning in this work was performed on the supercomputing system in the Supercomputing Center of the University of Science and Technology of China. This work was supported by the National Natural Science Foundation of China (Grants 81788101, 91636217, 32071450, and T2125011), the National Key R&D Program of China (Grants 2018YFA0306600, 2021YFB3202800, 2016YFA0502400, and 2019YFA0709300), the Chinese Academy of Sciences (CAS) (Grants XDC07000000, GJJSTD20200001, and QYZDY-SSW-SLH004), the Anhui Initiative in Quantum Information Technologies (Grant AHY050000), and the Fundamental Research Funds for the Central Universities.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118876119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Bray F., et al. , Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Bushberg J. T., The Essential Physics of Medical Imaging (Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA, ed. 3, 2012). [Google Scholar]
- 3.Kumar V., Abbas A. K., Aster J. C., Perkins J. A., Robbins Basic Pathology (Elsevier, Philadelphia, PA, ed. 10, 2018). [Google Scholar]
- 4.Hui R., et al. , Pembrolizumab as first-line therapy for patients with PD-L1-positive advanced non-small cell lung cancer: A phase 1 trial. Ann. Oncol. 28, 874–881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzgibbons P. L., et al. , Prognostic factors in breast cancer. College of American Pathologists Consensus Statement 1999. Arch. Pathol. Lab. Med. 124, 966–978 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Stack E. C., Wang C., Roman K. A., Hoyt C. C., Multiplexed immunohistochemistry, imaging, and quantitation: A review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 70, 46–58 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Giesen C., et al. , Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Fernandez D. C., Bhargava R., Hewitt S. M., Levin I. W., Infrared spectroscopic imaging for histopathologic recognition. Nat. Biotechnol. 23, 469–474 (2005). [DOI] [PubMed] [Google Scholar]
- 9.You S., et al. , Intravital imaging by simultaneous label-free autofluorescence-multiharmonic microscopy. Nat. Commun. 9, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camp C. H. Jr., et al. , High-speed coherent Raman fingerprint imaging of biological tissues. Nat. Photonics 8, 627–634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis A. S., et al. , Characterizing and diminishing autofluorescence in formalin-fixed paraffin-embedded human respiratory tissue. J. Histochem. Cytochem. 62, 405–423 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S. W., Roh J., Park C. S., Immunohistochemistry for pathologists: Protocols, pitfalls, and tips. J. Pathol. Transl. Med. 50, 411–418 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sengupta S., et al. , High resolution anatomical and quantitative MRI of the entire human occipital lobe ex vivo at 9.4T. Neuroimage 168, 162–171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wrachtrup J., Finkler A., Single spin magnetic resonance. J. Magn. Reson. 269, 225–236 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Balasubramanian G., et al. , Nanoscale imaging magnetometry with diamond spins under ambient conditions. Nature 455, 648–651 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Maze J. R., et al. , Nanoscale magnetic sensing with an individual electronic spin in diamond. Nature 455, 644–647 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Taylor J. M., et al. , High-sensitivity diamond magnetometer with nanoscale resolution. Nat. Phys. 4, 810–816 (2008). [Google Scholar]
- 18.Schirhagl R., Chang K., Loretz M., Degen C. L., Nitrogen-vacancy centers in diamond: Nanoscale sensors for physics and biology. Annu. Rev. Phys. Chem. 65, 83–105 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Shi F., et al. , Protein imaging. Single-protein spin resonance spectroscopy under ambient conditions. Science 347, 1135–1138 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Lovchinsky I., et al. , Nuclear magnetic resonance detection and spectroscopy of single proteins using quantum logic. Science 351, 836–841 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Schlipf L., et al. , A molecular quantum spin network controlled by a single qubit. Sci. Adv. 3, e1701116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi F., et al. , Single-DNA electron spin resonance spectroscopy in aqueous solutions. Nat. Methods 15, 697–699 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Wang P., et al. , Nanoscale magnetic imaging of ferritins in a single cell. Sci. Adv. 5, eaau8038 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Sage D., et al. , Optical magnetic imaging of living cells. Nature 496, 486–489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinert S., et al. , Magnetic spin imaging under ambient conditions with sub-cellular resolution. Nat. Commun. 4, 1–6 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Glenn D. R., et al. , Single-cell magnetic imaging using a quantum diamond microscope. Nat. Methods 12, 736–738 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barry J. F., et al. , Optical magnetic detection of single-neuron action potentials using quantum defects in diamond. Proc. Natl. Acad. Sci. U.S.A. 113, 14133–14138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis H. C., et al. , Mapping the microscale origins of magnetic resonance image contrast with subcellular diamond magnetometry. Nat. Commun. 9, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fescenko I., et al. , Diamond magnetic microscopy of malarial hemozoin nanocrystals. Phys. Rev. Appl. 11, 034029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCoey J. M., et al. , Quantum magnetic imaging of iron biomineralization in teeth of the chiton Acanthopleura hirtosa. Small Methods 4, 1900754 (2020). [Google Scholar]
- 31.Issadore D., et al. , Magnetic sensing technology for molecular analyses. Lab Chip 14, 2385–2397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pirker R., et al. , EGFR expression as a predictor of survival for first-line chemotherapy plus cetuximab in patients with advanced non-small-cell lung cancer: Analysis of data from the phase 3 FLEX study. Lancet Oncol. 13, 33–42 (2012). [DOI] [PubMed] [Google Scholar]
- 33.da Cunha Santos G., Shepherd F. A., Tsao M. S., EGFR mutations and lung cancer. Annu. Rev. Pathol. 6, 49–69 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Wang T.-C., et al. , “High-resolution image synthesis and semantic manipulation with conditional GANs” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 8798–8807 (2018).
- 35.Robertson D., Savage K., Reis-Filho J. S., Isacke C. M., Multiple immunofluorescence labelling of formalin-fixed paraffin-embedded (FFPE) tissue. BMC Cell Biol. 9, 1–10 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jun Y. W., Kim H. R., Reo Y. J., Dai M., Ahn K. H., Addressing the autofluorescence issue in deep tissue imaging by two-photon microscopy: The significance of far-red emitting dyes. Chem. Sci. (Camb.) 8, 7696–7704 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniels T. R., Delgado T., Rodriguez J. A., Helguera G., Penichet M. L., The transferrin receptor part I: Biology and targeting with cytotoxic antibodies for the treatment of cancer. Clin. Immunol. 121, 144–158 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Barak V., Goike H., Panaretakis K. W., Einarsson R., Clinical utility of cytokeratins as tumor markers. Clin. Biochem. 37, 529–540 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Li L. T., Jiang G., Chen Q., Zheng J. N., Ki67 is a promising molecular target in the diagnosis of cancer (review). Mol. Med. Rep. 11, 1566–1572 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Iwai Y., et al. , Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. U.S.A. 99, 12293–12297 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alsaab H. O., et al. , PD-1 and PD-L1 checkpoint signaling inhibition for cancer immunotherapy: Mechanism, combinations, and clinical outcome. Front. Pharmacol. 8, 561 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Carvalho H. F., Taboga S. R., Fluorescence and confocal laser scanning microscopy imaging of elastic fibers in hematoxylin-eosin stained sections. Histochem. Cell Biol. 106, 587–592 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Cruz-Monserrate Z., Qiu S., Evers B. M., O’Connor K. L., Upregulation and redistribution of integrin alpha6beta4 expression occurs at an early stage in pancreatic adenocarcinoma progression. Mod. Pathol. 20, 656–667 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerdes M. J., et al. , Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl. Acad. Sci. U.S.A. 110, 11982–11987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herbst R. S., et al. , Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis S. M., et al. , Spatial omics and multiplexed imaging to explore cancer biology. Nat. Methods 18, 997–1012 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Issadore D., et al. , Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-Hall detector. Sci. Transl. Med. 4, 141ra92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levine E. V., et al. , Principles and techniques of the quantum diamond microscope. Nanophotonics-Berlin 8, 1945–1973 (2019). [Google Scholar]
- 49.Corot C., Robert P., Idée J. M., Port M., Recent advances in iron oxide nanocrystal technology for medical imaging. Adv. Drug Deliv. Rev. 58, 1471–1504 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Shaw J., et al. , Magnetic particle-mediated magnetoreception. J. R. Soc. Interface 12, 0499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Przybylski A., Thiel B., Keller-Findeisen J., Stock B., Bates M., Gpufit: An open-source toolkit for GPU-accelerated curve fitting. Sci. Rep. 7, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang W., et al. , Magnetoresistive performance and comparison of supermagnetic nanoparticles on giant magnetoresistive sensor-based detection system. Sci. Rep. 4, 1–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.