Significance
Are olfactory receptor neurons (ORNs) arranged in a functionally meaningful manner to facilitate information processing? Here, we address this long-standing question by uncovering a valence map in the olfactory periphery of Drosophila. Within sensory hairs, we find that neighboring ORNs antagonistically regulate behaviors: stereotypically compartmentalized large- and small-spike ORNs, recognized by their characteristic spike amplitudes, either promote or inhibit the same type of behavior, respectively. Systematic optogenetic and thermogenetic assays—covering the majority of antennal sensilla—highlight a valence-opponent organization. Critically, odor-mixture behavioral experiments show that lateral inhibition between antagonistic ORNs mediates robust behavioral decisions in response to countervailing cues. Computational modeling predicts that the robustness of behavioral output depends on odor mixture ratios.
Keywords: olfactory receptor neurons (ORNs), sensory map, valence opponency, countervailing cues, olfactory sensilla
Abstract
A hallmark of complex sensory systems is the organization of neurons into functionally meaningful maps, which allow for comparison and contrast of parallel inputs via lateral inhibition. However, it is unclear whether such a map exists in olfaction. Here, we address this question by determining the organizing principle underlying the stereotyped pairing of olfactory receptor neurons (ORNs) in Drosophila sensory hairs, wherein compartmentalized neurons inhibit each other via ephaptic coupling. Systematic behavioral assays reveal that most paired ORNs antagonistically regulate the same type of behavior. Such valence opponency is relevant in critical behavioral contexts including place preference, egg laying, and courtship. Odor-mixture experiments show that ephaptic inhibition provides a peripheral means for evaluating and shaping countervailing cues relayed to higher brain centers. Furthermore, computational modeling suggests that this organization likely contributes to processing ratio information in odor mixtures. This olfactory valence map may have evolved to swiftly process ethologically meaningful odor blends without involving costly synaptic computation.
In complex sensory systems, neurons are typically organized into functionally meaningful maps. This arrangement allows specific stimulus attributes, such as color or spatial contrast, to be computed via lateral inhibition (1). In olfaction, however, it is unclear whether such a functional sensory map exists. In both rodents and flies, sensory neurons which project to nearby glomeruli—processing units in the first olfactory relay center—do not necessarily respond to structurally similar odorants (2, 3), suggesting an absence of chemotopic organization at this circuit level.
Might a functional olfactory map instead exist in the periphery? Drosophila melanogaster provides a unique opportunity to address this question, as the receptors, ligands, and behavioral outputs have been characterized for many olfactory receptor neurons [ORNs, (3–8)] (SI Appendix, Table S1). Each sensillum typically houses two and up to four ORNs, which are named “A,” “B,” “C,” or “D” in descending order of their stereotypical extracellular spike amplitudes (9). ORN pairing in a sensillum is also stereotyped—whereby a neuron expressing a particular receptor always neighbors an ORN expressing another specific receptor (4, 6, 8)—implying functional importance for such organization. Indeed, lateral inhibition broadly occurs between compartmentalized ORNs across sensillum types (10).
Interestingly, the sensillum is the only such place in the olfactory circuit where short-range lateral inhibition is commonly observed between specific input channels (10–12). Previous work has revealed that direct ephaptic interaction is sufficient to mediate lateral inhibition between electrically coupled ORNs (11). Furthermore, systematic morphological analysis of Drosophila antennal sensilla through serial block-face scanning electron microscopy shows that the basic anatomical features that support ephaptic coupling between ORNs—the close apposition of neuronal processes in a confined compartment (13, 14)—are conserved across sensillum types (15). Taken together, these studies consistently support the notion that ephaptic coupling occurs broadly across olfactory sensilla. Each sensillum can thus be considered a processing unit for olfactory computation, and understanding the organizing principle of ORN pairing will elucidate whether a functional olfactory map exists and how it is arranged.
Results
ORNs Are Likely Paired Based on Valence.
We began by analyzing responses from ORN pairs to a large odor panel in a published dataset (3). The Euclidean distance between neighboring neurons or any two randomly selected ORNs was not significantly different (SI Appendix, Fig. S1), arguing against the presence of a peripheral chemotopic map. If sensilla are not processing units for odorant identity, what stimulus attribute might be processed instead? Behavioral valence is a likely possibility. While an odor’s valence can be determined by the combinatorial ORN activation and neuromodulatory mechanisms, fly ORNs themselves appear to carry intrinsic valence. A striking pattern emerged from surveying the function of Drosophila ORNs: activation of large-spike neurons tends to positively regulate odor-guided behaviors, such as mediating attraction, stimulating egg laying, or promoting courtship (16–26), while activation of small-spike ORNs tends to have antagonistic effects, such as avoidance or inhibiting egg laying (SI Appendix, Table S1) (16, 27–29). Moreover, some paired neurons play opposite roles in modulating the same behavior. For example, ab4A and its ab4B neighbor promotes and suppresses oviposition, respectively (17, 28). On the basis of animals’ behavioral output, we defined positive or negative valence of a given ORN as promotion or inhibition of a certain behavior upon activation of the neuron. We therefore hypothesized that ORNs housed in the same sensillum antagonistically regulate the same behavior.
To test this hypothesis, we conducted systematic behavioral assays with fictive odors. Given that individual ORNs may not mediate innate behaviors in all contexts, we focused on sensillum types in which odor-induced behavior of at least one ORN has been characterized. This list of published reports—identifying ethologically relevant odor-guided behaviors for individual ORNs—served as the ground truth to direct our choices of sensilla and behavioral contexts in this study (SI Appendix, Table S1). Based on the receptor driver availability and labeling specificity (SI Appendix, Fig. S2), we examined 23 different ORNs, which represent paired neurons in 10 out of the 17 identified antennal sensillum types that house multiple ORNs (6, 30).
Valence Opponency in Place Preference.
We first examined the sensilla which house ORNs known to mediate attraction or aversion. In a proof-of-principle experiment, we expressed a red-shifted channelrhodopsin (CsChrimson) (31) in ab1C, which underlies flies’ aversion to CO2 (27). When fed with the chromophore all trans-retinal, ab1C::CsChrimson flies showed robust avoidance to light as expected (Fig. 1 A–C). Using this validated assay, we further examined ab1A and ab1B, which both mediate attraction to vinegar (16), and found that their optogenetic activation resulted in the expected preferences (Fig. 1D). For ab1D, whose hedonic value was hitherto uncharacterized, we observed aversion upon its activation (Fig. 1D). These results indicate that in the ab1 sensillum, both large-spike ORNs mediate attraction, whereas both small-spike neurons underlie aversion.
Fig. 1.
Valence opponency in place preference. (A) (Top) Optogenetic place preference assay. Inset: Single-sensillum recording; ab1C responded to a 500-ms pulse of 635-nm light. (Bottom) Collective distribution probability along the arena; the illuminated side was adjusted to be on the right (n = 10). (B) Average place preference over time when ab1C was optogenetically activated. Shaded area, SEM. (C–H) Violin plots showing preference indices to the illuminated side when CsChrimson was expressed in the indicated ORN types. Sample traces of single-sensillum recordings were shown to demonstrate optogenetic activation of target ORNs. Circle: average PI from three trials of each experiment; horizontal line: median preference. Negative controls were age-matched siblings without retinal feeding (gray). Results are from sated flies. n = 10 for each condition, **P < 0.01, ***P < 0.001, Wilcoxon rank-sum test.
We next assayed other preference-related sensillum types (SI Appendix, Table S1 and Fig. S2). Among the four previously characterized large-spike ORNs—ab5A, ai2A, ac3IA, and ab9A (18–21)—all mediated attraction when optogenetically activated (Fig. 1 E–H). Conversely, we observed avoidance caused by activation of their respective small-spike neighbors, except for ab9B (Fig. 1 E–H). Of note, we tested both sated and starved flies and found that satiety state did not switch the hedonic value of target neurons (SI Appendix, Fig. S3 A and B), in contrast to findings in Drosophila larvae (32). Taken together, valence opponency was broadly observed in the context of place preference.
Valence Opponency in Oviposition.
Next, we tested our hypothesis in the context of oviposition. Our optogenetic assay recapitulated the antagonistic roles of ab4A and ab4B in regulating egg laying (17, 28) (Fig. 2 A and B). We then extended our analysis to ab10 and ai3, the other two known oviposition-related sensilla (23, 29). Optogenetic activation of ab10B suppressed egg laying, concordant with the neuron’s reported role (29), whereas activation of ab10A enhanced oviposition as predicted (Fig. 2C). We observed a trend of increased, albeit statistically nonsignificant (P = 0.07), oviposition promotion with activation of ai3A (Fig. 2D), consistent with a previous report (23). Meanwhile, activation of the small-spike neighbor that expresses Or43a reduced oviposition (Fig. 2D). These results support our hypothesis and show that valence opponency also holds for oviposition-related sensilla.
Fig. 2.
Valence opponency in oviposition and courtship. (A) Optogenetic oviposition preference assay. (B–D) Violin plots showing oviposition preference indices (OI) to the illuminated side when CsChrimson was expressed in the indicated ORN types. Sample traces of single-sensillum recordings were shown to demonstrate optogenetic activation of target ORNs. Circle: OI for each trial; horizontal line: median preference. Negative controls were age-matched siblings without retinal feeding (gray). n = 20 trials, *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test. (E) (Top) at4 ORNs and their spikes. (Bottom Left) Courtship competition assay. (Bottom Right) Single-sensillum recording of at4A responses to heat generated by an IR laser. (F–H) Normalized cumulative copulation rates of males whose at4A (F), at4B (G), or at4C (H) was thermogenetically activated, when competing with the respective parental controls (black and gray). Cumulative copulation rates were normalized to the total copulation rate of all tested males in the same experiments. (Right) Each circle denotes the end copulation rate of a given experiment with lines connecting data from the same experiments (n = 4 to 6 experiments, total 68 to 94 matches). (I–K) As in E–H, except that TrpA1 was expressed in ac4A (J) or its small-spike Ir76a+ neighbor (K) (n = 5 experiments; total 83 to 85 matches). P values are indicated; one-sample z-test.
Valence Opponency in Courtship.
Does valence opponency also modulate courtship? In D. melanogaster, male courtship is suppressed by the singly housed at1 (33) but promoted by ac4A and at4A (24–26). We began by examining at4A, which expresses the Or47b receptor (25, 26), in a thermogenetic assay where three males of different genotypes—Or47b-GAL4::UAS-TrpA1 and both parent-line controls—competed to mate with one wild-type female (Fig. 2E). Single-sensillum recordings verified at4A’s thermogenetic activation (Fig. 2E). Concordant with published reports (25, 26), the copulation rate of at4A-activated males was markedly higher than in controls (Fig. 2F). Having validated the assay, we further tested the small-spike at4B and at4C and found that their thermogenetic activation indeed suppressed courtship as predicted (Fig. 2 G and H).
A similar antagonistic relationship was observed between ORNs housed in the ac4 sensillum (Fig. 2I). Thermogenetic activation of ac4A increased its spike activity (Fig. 2I) and conferred a copulation advantage as reported (24) (Fig. 2J). In contrast, courtship suppression was observed upon activation of its small-spike neighbor that expresses the Ir76a receptor (Fig. 2K). We excluded the other small-spike ac4 ORN in our analysis because its receptor, Ir75d, is also expressed in other neurons (8), thus preventing specific manipulation of the neuron. Together, these results indicate that valence opponency also underlies ORN pairing in a courtship context, further supporting our hypothesis that ORNs housed in the same sensillum can mediate related but opposite behaviors.
Valence Opponency Is Context-Dependent.
Does valence opponency hold in all behavioral contexts for a given sensillum? To address this question, the ab1, ab4, and at4 sensilla—which we characterized in place preference, oviposition, and courtship, respectively—were further examined under additional contexts. In the ab1 sensillum, activation of ab1A did not influence oviposition or courtship, while ab1C suppressed oviposition but not courtship (SI Appendix, Fig. S4 A–C), indicating that a given ORN does not necessarily influence all behaviors. Further experiments showed valence opponency between ab4A and ab4B for courtship (SI Appendix, Fig. S4 D–F) and between at4A and at4C for place preference but not for oviposition (SI Appendix, Fig. S4 G–I). Collectively, these results suggest that ORNs differentially regulate distinct behaviors and that valence opponency in a given sensillum likely holds only in ethologically relevant behavioral contexts (SI Appendix, Fig. S4J).
In summary, antagonizing ORN pairs were identified in 9 of the 10 examined sensilla (Table 1). Valence opponency is thus broadly observed between compartmentalized ORNs across sensillum types in multiple behavioral contexts. Our results thereby uncover a behavioral valence map in the Drosophila peripheral olfactory system.
Table 1.
Compartmentalized Drosophila ORNs and their behavioral outputs described in this study
| Context | Sensillum | Large-spike ORNs | Small-spike ORNs | ||
| Place preference | ab1 | A (Or42b) | Attraction to vinegar (16) | C (Gr21a/63a) | Aversion to CO2 (27) |
| B (Or92a) | Attraction to vinegar (16) | D (Or10a) | Aversion (this study) | ||
| ab5 | A (Or82a) | Attraction to geranyl acetate (18) | B (Or47a) | Aversion (this study) | |
| ab9 | A (Or69aA/B) | Attraction to a female odor component (19) | B (Or67b) | Attraction (this study) | |
| ai2 | A (Or83c) | Attraction to fruit rind odors (21) | B (Or23a) | Aversion (this study) | |
| ac3 | A (Ir75b) | Attraction to short-chain acids (20) | B (Or35a) | Aversion (this study) | |
| Oviposition | ab4 | A (Or7a) | Promoting egg laying (17) | B (Or56a) | Suppressing egg laying (28) |
| ab10 | A (Or67a) | Promoting egg laying (this study) | B (Or49a) | Suppressing egg laying (29) | |
| ai3 | A (Or19a) | Promoting egg laying by citrus odors (23) | C (Or43a) | Suppressing egg laying (this study) | |
| Courtship | ac4 | A (Ir84a) | Stimulating courtship by food odors (24) | B/C (Ir76a) | Inhibiting courtship (this study) |
| at4 | A (Or47b) | Stimulating courtship by fly pheromones (25, 26) | B (Or65a) | Inhibiting courtship [this and another study (38)] | |
| C (Or88a) | Inhibiting courtship (this study) | ||||
The table summarizes the reported ORN-mediated behaviors that were recapitulated by our optogenetic or thermogenetic assays, as well as the behavioral outputs of hitherto uncharacterized neighboring ORNs. Antennal ORNs are named based on their relative spike size and sensillum identity (b: basiconic; c: coeloconic; i: intermediate; t: trichoid). The receptors are indicated in parentheses.
A Means to Process Countervailing Cues.
What is the functional significance of valence opponency in olfactory processing? We previously showed that lateral inhibition between a pair of ab1 ORNs biases preference to an odor mixture to influence behavior (10). When paired ORNs are concurrently activated by odor mixtures, lateral inhibition enables the dominant cue to be selectively propagated (10, 11). As such, opposing constituents in a mixture are no longer represented in their original proportions; rather, the salient cue is favorably transmitted. Consequently, countervailing cues may no longer be sent as countervailing signals that could mitigate each other at higher olfactory centers (34, 35). Thus, antagonistic odorants are expected to influence behavior more effectively when detected by paired ORNs than by non-neighboring neurons.
To test this prediction, we focused on two pheromone-sensing sensilla. The at1 sensillum houses a single ORN, which expresses Or67d and detects the antiaphrodisiac pheromone cis-vaccenyl acetate (cVA) (33, 36, 37). In the at4 sensillum, the “A” neuron expresses Or47b, responds to palmitoleic acid, and promotes courtship (25), while the small-spike “C” neuron expresses Or88a, responds to methyl palmitate (26), and suppresses courtship (Fig. 2H). Notably in the at4 sensillum, these pheromone ligands only activate the receptors of the target ORNs (25, 26). Using their respective pheromone ligands, paired at4 ORNs were coactivated with a mixture of palmitoleic acid and methyl palmitate, or unpaired at4A and at1 neurons with palmitoleic acid mixed with cVA.
We first verified lateral inhibition of at4A by at4C via single-sensillum recording. We found that strong activation of at4C markedly inhibited at4A only when at4A was also stimulated by palmitoleic acid (Fig. 3A and SI Appendix, Fig. S5A), consistent with previous findings that ephaptic inhibition between grouped ORNs requires odor-induced excitation from both neurons (10). However, no significant inhibition of at4A was observed by the Or67d ligand cVA regardless of palmitoleic acid stimulation (Fig. 3B). Although cVA is reported to also activate at4B (36), we did not observe such responses (Fig. 3B and SI Appendix, Fig. S5B), in agreement with other studies (38, 39). Through lateral inhibition, methyl palmitate–evoked at4C activity, but not cVA–evoked at1 activity, can suppress at4A’s response to palmitoleic acid (Fig. 3 C and D).
Fig. 3.
Ephaptic lateral inhibition processes countervailing olfactory cues. (A) Single-sensillum recording. Representative trace (Top) and raster plot (Bottom) are shown for at4A spikes without (Left) or with its ligand, palmitoleic acid (PA, 10−1 vol/vol dilution), presented in the background (Right). The colored bars indicate odorant stimulations. MP: methyl palmitate (10−2). Parallel experiments, mean ± SEM (n = 12, from 3 to 4 male flies). (B) As with (A), except that recordings were performed with the at1 ligand, cis-vaccenyl acetate (cVA, 10−1), as the transient odorant. Parallel experiments, mean ± SEM (n = 8, from 3 to 4 male flies). (C) Peri-stimulus time histograms of at4A spikes from data shown in (A–B). Line width indicates SEM. (D) Quantification of at4A activity. Each data point represents the basal spike frequency of at4A before methyl palmitate (MP) or cis-vaccenyl acetate (cVA) was presented, as shown in (A and B). (E) Single-pair courtship assays. Cumulative and final copulation rates. Females were perfumed with solvent (acetone) or methyl palmitate (∼87 ng/fly). Lines connect results from parallel experiments. (F) As with (E), except that females were perfumed with cVA (∼38 ng/fly). Numbers of copulated males are indicated in parentheses. (G) Inhibition indices from results in (E and F). (H–J) As with (E–G), except that synaptic transmission was blocked in at4C (H, Or88a::Shits1) or in at1 (I, Or67d::Shits1). (K–M) As with (E–G), except that females were perfumed with palmitoleic acid alone (∼0.1 ng/fly) or with a binary mixture of palmitoleic acid and methyl palmitate (K) or palmitoleic acid and cVA (L). (N–P) As with (K–M), except that synaptic transmission was blocked in at4C (N, Or88a::Shits1) or in at1 (O, Or67d::Shits1). *P < 0.05, **P < 0.01, Wilcoxon rank-sum test. n = 5 to 6, 125 to 150 matches for individual conditions.
Next, we examined pheromone-mediated courtship modulation. When applied directly on females, low doses of palmitoleic acid enhanced copulation in a manner dependent on the male Or47b receptor (SI Appendix, Fig. S6 A and B). Conversely, perfuming females with higher doses of methyl palmitate or cVA resulted in comparable courtship inhibition in a receptor-dependent manner (methyl palmitate: 27%; cVA: 24%) (Fig. 3 E–G, and SI Appendix, Fig. S6 C and D). The modulation was also eliminated when synaptic transmission was blocked by expressing shibirets1 in at4C or at1 neurons (Fig. 3 H–J), thus demonstrating the specificity of these pheromone ligands.
Interestingly, when methyl palmitate or cVA was presented with palmitoleic acid in a mixture, palmitoleic acid–induced courtship was markedly attenuated by methyl palmitate (27%) but to a lesser extent by cVA (13%) (Fig. 3 K–M), despite their similar antiaphrodisiac effects when applied alone (Fig. 3G). As negative controls, we performed similar odor-mixture experiments with receptor mutant flies (Or88a−/− or Or67d−/−), wherein courtship inhibition was eliminated as expected (SI Appendix, Fig. S6 E and F). A parsimonious interpretation of these results is that through ephaptic inhibition, at4C activation attenuates at4A output, thereby selectively propagating at4C signal to suppress courtship. In contrast, at1 activation does not influence at4A peripheral responses, resulting in propagation of countervailing pheromone signals that may mitigate each other at higher brain centers and thus attenuate their efficacy in modulating behavior.
To test whether peripheral lateral inhibition is sufficient to modulate behavior, we blocked synaptic transmission from Or88a ORNs with shibirets1 (Fig. 3H). This manipulation did not affect at4C’s spike responses or its ability to inhibit at4A (SI Appendix, Fig. S7). Strikingly, when palmitoleic acid was coapplied, methyl palmitate reduced courtship by 20% despite the absence of at4C feedforward output (Fig. 3 N and P). These results demonstrate that at4A’s output is indeed attenuated by at4C ephaptic inhibition. In comparison, the antiaphrodisiac effect of cVA was completely abolished in Or67d::shibirets1 males even when palmitoleic acid was coapplied (Fig. 3 O and P).
As negative controls, we performed similar odor-mixture experiments except that palmitoleic acid was replaced by a pheromone ligand for ab4A (17), an ORN type which we found also promotes courtship (SI Appendix, Fig. S4F). As expected, the ab4A ligand 9-tricosene promoted male courtship in a receptor-dependent manner (Fig. 4 A and B). Notably, we no longer observed differential antiaphrodisiac effects between methyl palmitate and cVA when either antiaphrodisiac pheromone was copresented with 9-tricosene, regardless whether the males were wild type, Or67d::shibirets1, or Or88a::shibirets1 (Fig. 4 C–H). These results demonstrate that methyl palmitate in a mixture is not inherently more effective than cVA in suppressing courtship. Taken together, these results highlight a critical role of ephaptic inhibition in processing countervailing olfactory cues detected by compartmentalized neurons.
Fig. 4.
Countervailing cues detected by nonneighboring ORNs. (A) Single-pair courtship assays with a 7-d-old male and a 3-d-old female perfumed with either solvent (acetone) or 9-tricosene (∼7 ng/fly). Cumulative copulation rates of wild-type males; the final copulation rates are shown on the Right. Lines connect results from parallel experiments. The total numbers of copulated males are indicated in the parentheses. (B) As with A, except that Or7a mutant males were tested. (C and D) As with A, except that females were perfumed with 9-tricosene alone (∼7 ng/fly) or with a binary mixture of 9-tricosene and methyl palmitate (C) or 9-tricosene and cVA (D). (E) Inhibition indices from results in (C and D). (F–H) As with C–E, except that synaptic transmission was blocked in at4C (F, Or88a::Shits1) or in at1 (G, Or67d::Shits1). **P < 0.01, Wilcoxon rank-sum test. n = 4 to 6, 100 to 150 matches for individual conditions.
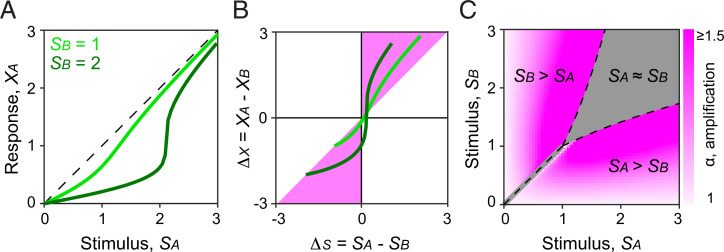
To further investigate the impact of ephaptic coupling on valence contrast, we modeled paired ORNs’ responses to binary mixtures of their respective ligands. In our model, activation of one neuron ephaptically inhibits its neighbor, and the degree of inhibition scales nonlinearly with each neuron’s response, which is determined by the stimulus strength (SA or SB, Fig. 5A ; see Methods for modeling details, and SI Appendix, Figs. S11 and S12). Ephaptic inhibition increases the response difference between paired ORNs (ΔX = XA – XB) relative to the difference between stimuli (ΔS = SA – SB). Given that paired ORNs mediate antagonistic behaviors, the net behavioral output is more robust when valence contrast is enhanced by ephaptic inhibition. The ratio ΔX/ΔS was therefore defined as the degree of valence amplification (α) (Fig. 5B). Our mathematical analysis showed that the magnitude of amplification varies depending on the ratio of respective stimulus strength in a mixture (Fig. 5C). In other words, certain SA to SB ratios are expected to more effectively elicit valence amplification and thus more effectively trigger robust behavioral response. This finding may explain why certain insect pheromone blends, whose constituents typically activate compartmentalized ORNs, can most effectively trigger robust behavior when presented in narrowly defined ratios (40, 41). Therefore, the valence-opponent organization between ephaptically coupled ORNs may have evolved to process ethologically meaningful ratios of odor mixtures.
Fig. 5.
Theoretical modeling of valance enhancement by ephaptic inhibition. (A) Steady-state response of ORNA (XA) as a function of odor stimulus SA. Without ephaptic inhibition, XA is defined to be equal to SA (dashed line, in arbitrary units). When the neighboring ORNB is also activated by SB, XA (green lines) is inhibited ephaptically in a manner that depends on the strength of both SA and SB. (B) Response difference between paired neurons (ΔX) as a function of stimulus difference (ΔS). Ephaptic inhibition amplifies the valence signal if ΔX and ΔS have the same sign, and if |ΔX| > |ΔS|. The simulated responses fall almost entirely within the amplified region (pink-shaded area, see Methods for modeling details). (C) The degree of valence amplification (α) as a function of SA and SB. Ephaptic inhibition amplifies valence in a manner that depends on the ratio of SA and SB. Gray-shaded region: SA ∼ SB, where the response polarity is likely ambiguous and is thus excluded in the analysis (see SI Appendix, Fig. S12 and Methods for modeling details).
Discussion
Here, we identify a valence-opponent olfactory map in D. melanogaster (Table 1). This organization is likely conserved among insects, as other compartmentalized insect ORNs have been reported to detect antagonistic cues (41). However, whereas previous studies reported valence opponency in a small number of select ORN pairs, our study demonstrates that this principle holds broadly across antennal sensilla. Our findings also suggest that the hedonic value of an insect ORN can be predicted based on its relative extracellular spike size. For example, in moths and beetles, attractive sex or aggregation pheromones are typically detected by large-spike ORNs, whose small-spike neighbors instead respond to behavioral antagonists (41). As another example, two subgroups of R. pomonella prefer either hawthorns or apples. The hawthorn flies sense an attractive host plant odorant (3-methyl-1-butanol) with an “A” ORN, which is paired with a small-spike neuron responding to a behavioral antagonist emitted by apples (butyl hexanoate). Conversely for the apple flies, butyl hexanoate is attractive and detected by an “A” ORN, whereas 3-methyl-1-butanol is aversive and detected by a neighboring small-spike neuron (42). In addition, the aversive CO2 cue is detected by a small-spike ORN (ab1C) expressing Gr21a/Gr63a receptors in fruit flies (27, 43, 44); in mosquitoes, however, CO2 is an attractive arousal cue which is in turn sensed by a large-spike ORN (cpA), despite its expression of Gr21a/Gr63a’s orthologous receptors (45).
Our observations also raise many interesting questions for future research. Why do large- and small-spike ORNs signal behaviors of positive and negative valence, respectively? In the context of valence opponency, how does the asymmetric nature of ephaptic interaction between compartmentalized ORNs influence its circuit function given that large-spike ORNs can exert greater ephaptic influence on their small-spike neighbors (11)? Why do some sensory hairs, such as the ab1 and at4 sensilla, house more than one large- or small-spike ORNs? How is the expression of OR genes coordinated for compartmentalized neurons, and what could be the evolutionary driving force for such an arrangement? Moreover, at the circuit level, it will be interesting to determine how paired ORNs are wired to mediate antagonistic behaviors. It is hoped that our study will lay the foundation for future work to address these intriguing and important questions.
Natural odors are notoriously complex, as they often contain countervailing cues. For example, many fruit odors include components that simultaneously activate large- and small-spike ORNs within the same fruit fly sensillum (11). Moreover, in most insect species examined, paired ORNs detect antagonistic components of pheromone blends (41). Furthermore, mixtures of odors with opposing valence elicit strong inhibition in certain attractant-responsive input channels, a phenomenon mediated by GABAergic interneurons in the antennal lobe (34). Such complexity poses a unique challenge to the olfactory system’s ability to evaluate natural odors and guide behaviors, as antagonistic components in an odor mixture may mitigate each other at higher olfactory centers (34, 35). This challenge can be met through the operation of lateral inhibition, which is known to enhance the signal-to-noise ratio of salient cues among concurrent inputs (1). The valence-opponent olfactory map may address this ethologically critical demand by allowing complex olfactory inputs to be filtered for simplification through ephaptic inhibition between grouped ORNs at the first level of the sensory circuit. This arrangement provides a means to both evaluate and shape the countervailing sensory signals relayed to higher brain centers for further processing.
Olfactory coding in both insects and mammals is believed to be distributive, whereby odor identity and hedonic value are determined by unique combinatorial activation patterns of broadly tuned ORNs, which are not held to individually convey valence information (46). In contrast, a small minority of specialist ORNs follows labeled line coding: these neurons respond to a limited number of ethologically salient odors, such as pheromones or predator odors, to mediate robust innate behaviors (47, 48). However, the distinction between generalists and specialists may not apply in insect olfaction. Given that the majority of ORNs examined by others (16–29) and in this study exhibited inherent valence, we hereby propose a hybrid model that combines the valence-labeled line (49) and distributive models. Our hybrid model, which accounts for the inherent hedonic values and valence-opponent organization of ORNs, remains compatible with combinatorial coding for odor identities and context-dependent (50) or learning-mediated modulation of odor valence (51). But importantly, as demonstrated in our synaptic silencing experiments, our model reveals that the entire central activation pattern is not necessarily required to define the overall valence of an odor mixture. Therefore, the peripheral mechanism may have evolved to lessen the computational burden in the central brain by swiftly processing antagonistic cues at the first neurons of the olfactory circuit.
Materials and Methods
Flies.
All flies (D. melanogaster) were housed on standard cornmeal food containing molasses at 25 °C, ∼60% humidity in an incubator with a 12-h light/dark cycle unless noted otherwise. Flies were collected upon eclosion, separated by sex, and used for experiments at indicated ages. The genotypes of fly lines used in this study are listed in SI Appendix, Tables S2 and S3.
Immunohistochemistry and Confocal Imaging.
Female flies expressing mCD8-GFP in the target ORNs were anesthetized on ice. The brains were dissected in phosphate-buffered saline (PBS), fixed in PBS with 4% paraformaldehyde (MPX00553, Fisher Scientific) for 20 min at room temperature (RT). The samples were then washed three times in 0.3% PBT (PBS with 0.3% Triton X-100) and transferred to 0.3% PBT with 10% normal goat serum (NGS) for 2 h at RT. The brains were then stained with rabbit α–green fluorescent protein (GFP) 1:400 (A11122, Life Technologies) and nc82 monoclonal antibody 1:50 (Developmental Studies Hybridoma Bank) in a dilution buffer (PBS with 1% NGS and 0.3% Triton X-100) for 48 h at 4 °C. Samples were washed three times with 0.3% PBT at RT before being stained with secondary antibodies, Alexa 488 goat α-rabbit 1:500 (A31628, Life Technologies), and Alexa 647 goat α-mouse 1:500 (A21236, Life Technologies) in the dilution buffer for 24 h at 4 °C. After three washes in 1% PBT at RT, the brains were mounted in FocusClear (CelExplorer). The samples were imaged on a Zeiss LSM 880 confocal microscope using 40×, N.A.1.2 C-Apochromat water-immersion objective lens. Airyscan images were processed with ZEN (Zeiss) and Fiji (https://imagej.net/Fiji).
Optogenetic Place Preference Assay.
A custom-designed behavioral assay was used to evaluate the place preference of flies when select ORNs were optogenetically activated. The dimensions of the arena were 80 × 35 × 3.5 mm with a sloped edge (52). In this assay, a custom light-emitting diode (LED) board was placed beneath the arena (SI Appendix, Fig. S8). The LED board was composed of interleaving 625-nm red LEDs (VLMR334BACB-GS08CT-ND, Digi-Key) and 850-nm infrared (IR) LEDs (VSMY3850-GS08CT-ND, Digi-Key). The LEDs were powered by programmable direct current (DC) switching power supplies (Model 1697, B&K Precision) and controlled by an Arduino microcontroller with a custom program. An opaque plastic partition (4-cm height) was used to restrict light to one-half of the arena. A diffuser (Rosco) was placed immediately underneath the area for homogeneous illumination.
Female flies expressing the red-shifted channelrhodopsin CsChrimson (31) were collected daily, group housed in 15 per vial, and reared in constant darkness on standard cornmeal medium supplemented with 0.2 mM all trans-retinal (Sigma-Aldrich R2500). Negative controls were age-matched siblings without the retinal supplement. Sated and starved flies were tested separately. For starvation, 24 h prior to experiments, flies were transferred to empty vials containing dampened Kimwipes with either 1 mL 0.2 mM all trans-retinal (experimental groups) or distilled water (control groups). For each experiment, a group of 10 female flies (4-d-old) was tested. A group-fly assay was adopted because collective behavior is known to enhance the robustness of ORN-mediated preference (53). The IR LEDs remained on constantly on both sides of the arena. At the beginning of each trial, flies were acclimated to the arena under darkness (IR only) for 30 s. Subsequently, the red LEDs were turned on to illuminate the right half of the arena for 45 s. After another 30 s of darkness, the red LEDs were switched on at the left side. Each experiment consisted of three trials with a total of six illuminating periods. Red light irradiance was 1.2 μW/mm2. In some experiments (ab1A and ac3A) where less robust preference was observed (0.05 > P > 0.01), higher irradiance (9.1 μW/mm2) was used.
Flies in the arena were recorded at 30 frames/s using a Flea3 camera (Point Gray Research) with fixed lens (Computar) and a long-pass filter (Edmund Optics) from the top of the arena. The location of flies was determined by a custom tracking C++ code. The preference index (PI) of flies to the illuminated side was calculated every half a second based on the positions of flies in the arena. The PI was defined as: PI = (Nlight – Ndark) / Ntotal, where Nlight indicates the number of flies on the illuminated half of the arena, Ndark indicates the number of flies on the dark half of the arena, and Ntotal indicates the total number of flies in the arena (10 flies). For each experiment, the average PI was determined by averaging the preference indices during periods of illumination over the three trials. Data were analyzed offline using custom MATLAB codes.
Although our assay did not involve pulsed light or airflow, their respective effects on fly’s place preference were also examined in preliminary experiments. A previous study employing single-fly optogenetic reference assays paired light stimulation with a constant airflow (54). In pilot experiments, the preference behavior of Gr21a::CsChrimson flies was examined in the absence or presence of airflow (∼25 cm/s). In the absence of airflow, the flies exhibited robust aversion to light (SI Appendix, Fig. S9A), as expected from ab1C’s role in mediating CO2 aversion (27). However, with constant airflow, both the control and experimental flies exhibited a strong tendency to walk upwind, resulting in robust attraction toward the side of air input even when it was illuminated by red LEDs (SI Appendix, Fig. S9B). In another pilot experiment, pulsed red light (1, 5, 10, and 20 Hz, 50% duty cycle) was tested for Gr21a::CsChrimson flies in the absence of airflow. No significant preference difference between pulsed and static light was observed, except in the case of 20-Hz pulsed light where avoidance was attenuated (data not shown). Although varying lighting conditions may influence the degree of preference, it does not alter the polarity of an ORN’s valence (55). Therefore, for simplicity and consistency, static red light was used throughout the optogenetic assays for all ORN types.
Optogenetic Oviposition Assay.
A custom-designed behavioral assay was used to evaluate the oviposition preference of flies when select ORNs were optogenetically activated. This egg-laying assay employed a custom-built array of transparent acrylic that comprised five individual chambers, each measuring 110 × 50 × 5 mm. Placed at the bottom of both sides of each chamber was a 40-mm diameter plastic dish, filled with 1% agarose with 5 mM sucrose (56). A plastic sheet placed on top of the chamber prevented escape by flies, which freely moved between either side of the chamber and laid eggs on the agar dishes. An LED board with 625-nm red LEDs (VLMR334BACB-GS08CT-ND, Digi-Key) was placed beneath the array, while an opaque plastic partition restricted the illuminated field to one side of each chamber (SI Appendix, Fig. S10). Data collected from each chamber constituted one experiment, while results from each array of five chambers constituted one trial.
Flies expressing CsChrimson were collected daily, separated by sex, group housed in 15 per vial, and reared in constant darkness on standard cornmeal medium supplemented with 0.2 mM all trans-retinal (Sigma-Aldrich R2500). Negative controls were age-matched siblings without the retinal supplement. For mating, 15 pairs of 3-d-old females and males were transferred to a new vial containing either the regular (control) or retinal fly food (experimental group) supplemented with diluted yeast paste; mating was allowed to take place over a 24-h period. Subsequently, 15 females were loaded into an individual chamber. The side of illumination was alternated between trials (five experiments per trial and four trials for each condition). Experiments were conducted in a dark incubator at 25 °C for ∼24 h. Red light irradiance was 0.3 μW/mm2 for all ORN types except for ab10A, which was examined with higher irradiance (2.1 μW/mm2). Following each experiment, eggs were counted, and the oviposition preference to the illuminated chamber was calculated as the following: Oviposition index = (Nlight – Ndark) / Ntotal, where Nlight indicates the number of eggs on the illuminated chamber, Ndark is the number of eggs on the other chamber, and Ntotal indicates the total number of eggs. Data were analyzed offline using custom MATLAB codes.
Thermogenetic Courtship Competition Assay.
Male flies expressing TrpA1 in the target ORNs and corresponding parental controls were raised at 21 °C. Naïve males were raised in groups of 10 and transferred to new vials with fresh food every other day as described (57). Prior to courtship assays, flies were acclimated in the behavioral room, which was heated to ∼30 °C, for 30 min. The courtship competition assays were conducted essentially as described (58). In brief, three naïve males of different genotypes and one 3-d-old virgin female (Canton-S) were loaded in a mating chamber (4 cm in diameter and 1 cm in height) positioned atop a Petri dish containing diluted fly food (50% water). The base of the chamber comprises a piece of gauze to allow flies access to food odors. To facilitate fly identification, two males of different genotypes were dusted with fluorescent dyes (UVXPBB and UVXPBR, Llewellyn Data Processing LLC) ∼20 h prior to experiments. Dye application was alternated among genotypes across experiments to minimize dye-induced behavioral bias. Courtship competition assays were conducted at ∼30 °C under 660-nm red light. For each experiment, 30 matches were set up, and only trials in which copulation occurred within 2 h were included in the analysis. Courtship was visually confirmed, and the identity of the mated males and the time of copulation were recorded during the 2-h period. Cumulative copulation rates (CCRs) were normalized to the total copulation rate of all tested males in the same experiments. Data were analyzed using custom MATLAB codes.
Pilot experiments testing males of different ages showed that thermogenetic activation of at4A—known to promote courtship in an age-dependent manner (25)—significantly enhanced courtship in 2-d-old males but not in 7 d old (not shown). The lack of courtship enhancement in the latter condition likely reflects a ceiling effect arising from the high 7-d male copulation around 30 °C (total CCR: 82% from four experiments, total 120 matches; no significant difference was observed between control and experimental groups). Conversely, pilot experiments testing at4C showed significant courtship inhibition only with 7-d-old males, likely reflecting a floor effect arising from the low copulation rate of 2-d-old males (total CCR: 47% from three experiments, total 90 matches; no significant difference between control and experimental groups). Of note, male age was found to influence only the manifestation or degrees of courtship modulation by thermogenetic ORN activation, but never the polarity of the modulation. Therefore, to avoid ceiling or floor effects, 2-d-old males were used in experiments where at4A or ac4A was thermogenetic activated, while 7-d-old males were used for the at4B, at4C, or ac4C experiments.
Pheromone Perfuming Experiments.
Briefly, 0.3 μL either individual pheromones or binary mixtures were directly applied to the abdomen of female flies essentially as described (33). Pheromones were diluted in acetone (vol/vol) prior to experiments (palmitoleic acid: 3 × 10−7, ∼0.1 ng/fly; methyl palmitate: 3 × 10−4, ∼87 ng/fly; cis-vaccenyl acetate: 1.5 × 10−4, ∼38 ng/fly; (Z)-9- tricosene: 3 × 10−6, ∼7 ng/fly). Pilot dosage experiments were conducted to determine appropriate pheromone dilutions such that individual pheromones enhanced or inhibited courtship to similar degrees. After the pheromone dilution (experimental group) or acetone (negative control) was applied to females, the solvent was allowed to evaporate for 1 h prior to experiments. For single-pair courtship assays, one 7-d-old naïve male and one 3-d-old virgin CS female were loaded into a mating chamber (2 cm in diameter and 1 cm in height) placed atop a food patch as described (25). Of note, smaller chambers were used for single-pair courtship assays than the ones for the 3-male–1-female competition assays described above in Thermogenetic Courtship Competition Assay. Courtship assays were conducted at either 25 °C (Fig. 3 E–G and Fig. 4 A and B and SI Appendix, Fig. S6 A–D) or around 30 °C (Fig. 3 H–P and Fig. 4 C–H and SI Appendix, Fig. S6 E and F) under 660-nm red light. Synaptic transmission in select ORNs was blocked around 30 °C when the neurons expressed the temperature-sensitive dynamin mutant shits1 (59, 60). Of note, the UAS-Shibirets1 line used in this study contained a translational enhancer for robust expression (60). Each experiment contained 25 matches. The control and experimental groups were tested in parallel experiments. Copulation was visually confirmed, and the CCR was calculated for the 2-h period. The inhibition index was calculated as follows:
Data were analyzed using custom MATLAB codes.
Single-Sensillum Recordings and Stimuli.
To prepare an antenna for recording, a fly was wedged into the narrow end of a truncated 200-µL pipette tip to expose the antenna, which was subsequently stabilized between a tapered glass microcapillary tube and a coverslip covered with double-sided tape. Extracellular single-sensillum recordings were performed essentially as described (61). Briefly, electrical activity of target ORNs was recorded extracellularly by placing a sharp electrode filled with artificial hemolymph solution (62) in the at4 sensillum (Or47b ORN recordings). For recordings of other sensillum types, 0.6× sensillum lymph Ringer solution (63) was used instead. A reference electrode filled with the same solution was placed in the eye or clypeus. No more than three sensilla from the same antenna were recorded per fly. Alternating current signals (100 to 20,000 Hz) and DC signals were simultaneously recorded on an NPI EXT-02F amplifier (ALA Scientific Instruments) and digitized at 5 kHz with Digidata 1550 (Molecular Devices). ORN spikes were sorted and analyzed offline using Clampfit 10 (Molecular Devices) and Igor Pro (WaveMetrics). Spike responses were averaged, binned at 50 ms, and smoothed using a binomial algorithm to obtain peri-stimulus time histograms. Methyl palmitate (10−2, Sigma P5177) or cis-vaccenyl acetate (Cayman 10010101, 10× dilution from 5% stock) was diluted in ethanol (vol/vol), applied as a 4.5-µL portion to a filter disk and delivered via a 500-ms air pulse at 250 mL/min directly to the antenna from close range as described (61). For recordings with a background odorant, palmitoleic acid (10−1, Cayman 9001798) was applied as a 4.5-µL portion to a filter disk, which was placed in close proximity adjacent to the fly head.
For optogenetic stimulation, newly eclosed female flies expressing CsChrimson in targeted ORNs were reared in constant darkness for 4 d on fly food supplemented with 10 μM all trans-retinal (Sigma R2500). A light stimulus was generated via a LED (635 nm, Universal LED Illumination System, pE-4000, Cool LED). Light pulses (500-ms duration) were controlled by Clampex 10.4 (Molecular Devices). Of note, a wide wavelength range of light (470 to 660 nm) can activate CsChrimson (31). Consequently, in the process of identifying target sensilla for recording, CsChrimson-expressing ORNs were already strongly activated by microscopic light. Such strong and prolonged activation of target ORNs could markedly reduce their spike amplitude to a degree that spikes become too small to be visualized above the level of noise. Therefore, in order to observe 635-nm elicited CsChrimson responses, the ORNs needed to first be dark adapted, and recordings had to be performed with the microscopic light off.
For thermogenetic stimulation, an infrared laser (Roithner, 808 nm, 500 mW, RLTMDL-808-500-5) was directed to the antenna via a 1-mm optic fiber (Roithner, RLTMxL SMA905-1000). The laser power and stimulation duration were controlled by Clampex 10.4 (Molecular Devices). Temperature around the position of the recorded antenna was measured with a K-type thermocouple (Gain Express Holdings 68022), and the stimulus was set at ∼30 °C, which did not elicit any excitatory responses in ORNs without TrpA1 expression (data not shown).
Computational Modeling.
We modeled time-dependent activation of paired ORNs, A and B, as XA(t) and XB(t), respectively. These activation variables obey the following coupled nonlinear ordinary differential equations:
For simplicity, we assumed that the response decays to the baseline linearly with the timescale τ = 1. The parameter wAB describes the strength of ephaptic coupling from ORNB to ORNA, and similarly for wBA. Note that the overall input into ORNA is scaled by the activity level of both neurons (i.e., the second term on the right-hand side of the above equations is proportional to XA and XB), consistent with our previous circuit modeling of ephaptic interactions (11). Furthermore, ephaptic interaction from ORNA to ORNB scales nonlinearly with the activity level of ORNB (scaling exponent, n). We considered the scaling to be nonlinear (n >1) because ephaptic inhibition occurs between paired neurons only when neuronal activity exceeds a certain threshold (10).
Since odor stimuli in our behavioral experiments were sustained, we analyzed the differential equations at the steady state (i.e., ). We reduced the model complexity by assuming that ephaptic interactions are measured in units of wBA (i.e., wBA = 1, wAB = q). By convention, ORNA refers to the larger ORN, so 0 < q ≤ 1. SA and SB indicate the stimulus strength for ORNA and ORNB, respectively (in arbitrary units). In this model, in the absence of ephaptic coupling (wAB = wBA = 0), ORN activity is defined to be equal to the stimulus strength at the steady state: XA = SA, XB = SB. With ephaptic inhibition, ORN responses are expected to be attenuated: XA ≤ SA, XB ≤ SB. To determine the degree of valence amplification by ephaptic inhibition, we considered the response and stimulus differences: ΔX = XA − XB, ΔS = SA − SB. Amplification takes place when ΔX and ΔS have the same sign and when the response difference (ΔX) is larger than stimulus difference (ΔS). We therefore defined the valence amplification index (α) as follows:
Before determining the value of α, we first considered regions in stimulus space where valence amplification is not expected to be meaningful. Note that the stimulus space is defined by SA and SB. We excluded the region where SA ∼ SB (or ΔS ∼ 0) because within this ambiguous region, behavioral output may fluctuate between positive and negative valence, making it difficult to accurately predict the valence polarity of the behavioral response. To mathematically define the excluded region, we began by testing whether ORN spike responses follow a Poisson distribution. If so, the response SD (σ) is expected to be equal to the square-root of the mean (μ) (i.e., ). We therefore analyzed spike responses from 10 different ORN types based on our published data (9) by fitting individual dosage responses to the function, where and m are fitting parameters. Indeed, we found that the average scaling constant ) and exponent (m) were close to 1 and 0.5, respectively, supporting the notion that ORN spike responses follow a Poisson distribution ( = 1.3 ± 0.6; m = 0.58 ± 0.19, SI Appendix, Fig. S11). Having determined that the ORN response distribution is consistent with Poisson, we could then define the meaningful amplification region to be where and where (outside the gray-shaded area in Fig. 5C and SI Appendix, Fig. S12).
Next, we determined α within the meaningful domains of stimulus space by solving the steady-state equations. We considered different degrees of nonlinearity (n = 2, 2.5, or 3) and a range of ephaptic coupling parameter (q = 0.1 to 1). Amplification was broadly observed (α > 1) for all combinations of n and q, suggesting that the validity of our analysis was not affected by the exact value of n or q. However, these two parameters influence the degree of amplification (α) in the stimulus space as a function of SA and SB (SI Appendix, Fig. S12). For certain n and q combinations, there were regions in the stimulus space where there is no amplification (α < 1, white-shaded areas in SI Appendix, Fig. S12). For demonstration purposes, a specific combination of n and q was selected for Fig. 5C (n = 2 and q = 0.9), where amplification was found broadly for a wide range of SA and SB combinations within the meaningful domain of stimulus space.
Quantification and Statistical Analysis.
Place-preference data were analyzed by Wilcoxon rank-sum test because not all ORN datasets passed the Shapiro–Wilk normality test. For oviposition preference, significance was determined by unpaired two-sample Student’s t test, as all datasets passed the Shapiro–Wilk normality test. Courtship competing data were analyzed by one-sample z-test. All pheromone perfuming data collected from parallel experiments were analyzed by Wilcoxon rank-sum test. Courtship inhibition indices between different experiments were analyzed by Wilcoxon rank-sum test. Statistical analysis was performed in MATLAB and RStudio.
Supplementary Material
Acknowledgments
We thank Karen Menuz and Larry Squire for comments on the manuscript, Kalyani Cauwenberghs for assistance with statistical analysis, and Takeo Katsuki for manufacturing the chambers for place preference assays. We also thank the University of California San Diego (UCSD) School of Medicine Microscopy Core (NS047101) and the UCSD Qualcomm Institute Prototyping Lab. This work was supported by a Kavli Institute for Brain and Mind Innovative Research Grant (C.-Y.S.), NIH Grant Nos. R01DC016466, R01DC015519, and R21DC108912 (C.-Y.S.), Air Force Office of Scientific Research Grant Nos. FA-9550-18-1-0051 (R.G.) and FA-9550-19-1-0280 (D.G.), and a Defense Advanced Research Projects Agency Young Faculty Award No. D21AP10162 (J.A.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120134119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.van Hateren J. H., A theory of maximizing sensory information. Biol. Cybern. 68, 23–29 (1992). [DOI] [PubMed] [Google Scholar]
- 2.Soucy E. R., Albeanu D. F., Fantana A. L., Murthy V. N., Meister M., Precision and diversity in an odor map on the olfactory bulb. Nat. Neurosci. 12, 210–220 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Hallem E. A., Carlson J. R., Coding of odors by a receptor repertoire. Cell 125, 143–160 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Hallem E. A., Ho M. G., Carlson J. R., The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Fishilevich E., Vosshall L. B., Genetic and functional subdivision of the Drosophila antennal lobe. Curr. Biol. 15, 1548–1553 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Couto A., Alenius M., Dickson B. J., Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol. 15, 1535–1547 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Silbering A. F., et al. , Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci. 31, 13357–13375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benton R., Vannice K. S., Gomez-Diaz C., Vosshall L. B., Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Bruyne M., Foster K., Carlson J. R., Odor coding in the Drosophila antenna. Neuron 30, 537–552 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Su C.-Y., Menuz K., Reisert J., Carlson J. R., Non-synaptic inhibition between grouped neurons in an olfactory circuit. Nature 492, 66–71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., et al. , Asymmetric ephaptic inhibition between compartmentalized olfactory receptor neurons. Nat. Commun. 10, 1560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson R. I., Early olfactory processing in Drosophila: Mechanisms and principles. Annu. Rev. Neurosci. 36, 217–241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faber D. S., Korn H., Electrical field effects: Their relevance in central neural networks. Physiol. Rev. 69, 821–863 (1989). [DOI] [PubMed] [Google Scholar]
- 14.Jefferys J. G. R., Nonsynaptic modulation of neuronal activity in the brain: Electric currents and extracellular ions. Physiol. Rev. 75, 689–723 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Nava Gonzales C., et al. , Systematic morphological and morphometric analysis of identified olfactory receptor neurons in Drosophila melanogaster. eLife 10, 2021.04.28.441861 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Semmelhack J. L., Wang J. W., Select Drosophila glomeruli mediate innate olfactory attraction and aversion. Nature 459, 218–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin C.-C., Prokop-Prigge K. A., Preti G., Potter C. J., Food odors trigger Drosophila males to deposit a pheromone that guides aggregation and female oviposition decisions. eLife 4, e08688 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlief M. L., Wilson R. I., Olfactory processing and behavior downstream from highly selective receptor neurons. Nat. Neurosci. 10, 623–630 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lebreton S., et al. , A Drosophila female pheromone elicits species-specific long-range attraction via an olfactory channel with dual specificity for sex and food. BMC Biol. 15, 88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prieto-Godino L. L., et al. , Evolution of acid-sensing olfactory circuits in Drosophilids. Neuron 93, 661–676.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Ronderos D. S., Lin C. C., Potter C. J., Smith D. P., Farnesol-detecting olfactory neurons in Drosophila. J. Neurosci. 34, 3959–3968 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Min S., Ai M., Shin S. A., Suh G. S. B., Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 110, 1321-1329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dweck H. K. M., et al. , Olfactory preference for egg laying on citrus substrates in Drosophila. Curr. Biol. 23, 2472–2480 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Grosjean Y., et al. , An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature 478, 236–240 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Lin H.-H., et al. , Hormonal modulation of pheromone detection enhances male courtship success. Neuron 90, 1272–1285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dweck H. K. M., et al. , Pheromones mediating copulation and attraction in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 112, E2829–E2835 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suh G. S. B., et al. , A single population of olfactory sensory neurons mediates an innate avoidance behaviour in Drosophila. Nature 431, 854–859 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Stensmyr M. C., et al. , A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell 151, 1345–1357 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Ebrahim S. A. M., et al. , Drosophila avoids parasitoids by sensing their semiochemicals via a dedicated olfactory circuit. PLoS Biol. 13, e1002318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin C.-C., Potter C. J., Re-classification of Drosophila melanogaster trichoid and intermediate sensilla using fluorescence-guided single sensillum recording. PLoS One 10, e0139675 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klapoetke N. C., et al. , Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogt K., et al. , Internal state configures olfactory behavior and early sensory processing in Drosophila larvae. Sci. Adv. 7, eabd6900 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurtovic A., Widmer A., Dickson B. J., A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature 446, 542–546 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Mohamed A. A. M., et al. , Odor mixtures of opposing valence unveil inter-glomerular crosstalk in the Drosophila antennal lobe. Nat. Commun. 10, 1201 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu Q., Wu Y., Ma L., Yu C. R., Encoding innately recognized odors via a generalized population code. Curr. Biol. 31, 1813–1825 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Goes van Naters W., Carlson J. R., Receptors and neurons for fly odors in Drosophila. Curr. Biol. 17, 606–612 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ha T. S., Smith D. P., A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J. Neurosci. 26, 8727–8733 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pitts S., Pelser E., Meeks J., Smith D., Odorant responses and courtship behaviors influenced by at4 neurons in Drosophila. PLoS One 11, e0162761 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomez-Diaz C., Reina J. H., Cambillau C., Benton R., Ligands for pheromone-sensing neurons are not conformationally activated odorant binding proteins. PLoS Biol. 11, e1001546 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Löfstedt C., Population variation and genetic control of pheromone communication systems in moths. Entomol. Exp. Appl. 54, 199–218 (1990). [Google Scholar]
- 41.Ng R., Wu S. T., Su C. Y., Neuronal compartmentalization: A means to integrate sensory input at the earliest stage of information processing? BioEssays 42, e2000026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tait C., Batra S., Ramaswamy S. S., Feder J. L., Olsson S. B., Sensory specificity and speciation: A potential neuronal pathway for host fruit odour discrimination in Rhagoletis pomonella. Proc. Royal Soc. B: Biol. Sci. 283, 2016210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones W. D., Cayirlioglu P., Kadow I. G., Vosshall L. B., Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445, 86–90 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Kwon J. Y., Dahanukar A., Weiss L. A., Carlson J. R., The molecular basis of CO2 reception in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 104, 3574–3578 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu T., et al. , Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr. Biol. 17, 1533–1544 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramdya P., Benton R., Evolving olfactory systems on the fly. Trends Genet. 26, 307–316 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Haverkamp A., Hansson B. S., Knaden M., Combinatorial codes and labeled lines: How insects use olfactory cues to find and judge food, mates, and oviposition sites in complex environments. Front. Physiol. 9, 49 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dewan A., Pacifico R., Zhan R., Rinberg D., Bozza T., Non-redundant coding of aversive odours in the main olfactory pathway. Nature 497, 486–489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liman E. R., Zhang Y. V., Montell C., Peripheral coding of taste. Neuron 81, 984–1000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Badel L., Ohta K., Tsuchimoto Y., Kazama H., Decoding of context-dependent olfactory behavior in Drosophila. Neuron 91, 155–167 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Davis R. L., Olfactory memory formation in Drosophila: From molecular to systems neuroscience. Annu. Rev. Neurosci. 28, 275–302 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Simon J. C., Dickinson M. H., A new chamber for studying the behavior of Drosophila. PLoS One 5, e8793 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramdya P., et al. , Mechanosensory interactions drive collective behaviour in Drosophila. Nature 519, 233–236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bell J. S., Wilson R. I., Behavior reveals selective summation and max pooling among olfactory processing channels. Neuron 91, 425–438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tumkaya T., Stewart J., Burhanudin S., Claridge-Chang A., Optogenetic olfactory behavior depends on illumination characteristics. bioRxiv [Preprint] (2019). https://www.biorxiv.org/content/10.1101/559674v1. Accessed 25 January 2022.
- 56.Schwartz N. U., Zhong L., Bellemer A., Tracey W. D., Egg laying decisions in Drosophila are consistent with foraging costs of larval progeny. PLoS One 7, e37910 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sethi S., et al. , Social context enhances hormonal modulation of pheromone detection in Drosophila. Curr. Biol. 29, 3887–3898 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ng R., et al. , Amplification of Drosophila olfactory responses by a DEG/ENaC channel. Neuron 104, 947–959 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kitamoto T., Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J. Neurobiol. 47, 81–92 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Pfeiffer B. D., Truman J. W., Rubin G. M., Using translational enhancers to increase transgene expression in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 109, 6626–6631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ng R., Lin H.-H., Wang J. W., Su C.-Y., Electrophysiological recording from Drosophila trichoid sensilla in response to odorants of low volatility. J. Vis. Exp. 125, e56147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J. W., Wong A. M., Flores J., Vosshall L. B., Axel R., Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell 112, 271–282 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Kaissling K. E., Thorson J., “Insect olfactory sensilla: Structure, chemical and electrical aspect of the functional organization” in Receptors for Neurotransmitters, Hormones, and Pheromones in Insects: Proceedings of the Workshop on Neurotransmitter and Hormone Receptors in Insects held in Cambridge, 10-12 September 1979, Sattelle D. B., Hall L. M., Hildebrand J. G., Eds. (Elsevier/North-Holland Biomedical Press, 1980), pp. 261–282. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.