Significance
The endoplasmic reticulum (ER) is the site at which secreted proteins (such as the hormone insulin) and membrane-bound proteins are folded. ATP-dependent chaperones within the ER help proteins fold. This study describes how two key ER chaperones, BiP and Grp94, work together at a molecular level. BiP binds to Grp94, which enables Grp94 to change conformation and hydrolyze ATP. In short, BiP provides a signal to switch on Grp94 conformational changes that are required to help other proteins fold. This finding helps explain how two chaperones can work together collaboratively in protein folding. Because BiP and Grp94 are members of highly conserved chaperone families, these findings may provide insight into chaperone-assisted protein folding beyond the ER.
Keywords: Grp94, BiP, chaperone, cochaperone
Abstract
Hsp70 and Hsp90 chaperones provide protein quality control to the cytoplasm, endoplasmic reticulum (ER), and mitochondria. Hsp90 activity is often enhanced by cochaperones that drive conformational changes needed for ATP-dependent closure and capture of client proteins. Hsp90 activity is also enhanced when working with Hsp70, but, in this case, the underlying mechanistic explanation is poorly understood. Here we examine the ER-specific Hsp70/Hsp90 paralogs (BiP/Grp94) and discover that BiP itself acts as a cochaperone that accelerates Grp94 closure. The BiP nucleotide binding domain, which interacts with the Grp94 middle domain, is responsible for Grp94 closure acceleration. A client protein initiates a coordinated progression of steps for the BiP/Grp94 system, in which client binding to BiP causes a conformational change that enables BiP to bind to Grp94 and accelerate its ATP-dependent closure. Single-molecule fluorescence resonance energy transfer measurements show that BiP accelerates Grp94 closure by stabilizing a high-energy conformational intermediate that otherwise acts as an energetic barrier to closure. These findings provide an explanation for enhanced activity of BiP and Grp94 when working as a pair, and demonstrate the importance of a high-energy conformational state in controlling the timing of the Grp94 conformational cycle. Given the high conservation of the Hsp70/Hsp90 system, other Hsp70s may also serve dual roles as both chaperones and closure-accelerating cochaperones to their Hsp90 counterparts.
Heat shock proteins maintain protein folding quality control under nonstress conditions, and their function is essential for survival under stress conditions (1). Hsp70 and Hsp90 family chaperones are conserved components of folding quality control from bacteria to humans, and serve critical roles in the endoplasmic reticulum (ER) and mitochondria. Hsp90 often exhibits a minimal capacity to assist client folding when working alone, while becoming functional when operating with Hsp70 (2–4), and the underlying mechanistic explanation is an active area of research (5, 6).
BiP and Grp94 (Fig. 1 A and B) aid in the folding of ER-specific clients ranging from immunoglobulins (7, 8) to insulin-like growth factors (9–11). BiP and Grp94 associate only when BiP adopts the “ADP conformation” (Fig. 1C) (12), a state in which BiP can trap client proteins (13–15). The BiP nucleotide binding domain (NBD) interacts with the Grp94 middle domain (MD) through stabilizing salt bridges that are conserved from bacteria to humans (12, 16, 17). Despite core similarities, Hsp70/Hsp90 systems differ in their regulation. For example, eukaryotic Hsp70/Hsp90 systems in the cytosol can be further regulated via Hop (a cochaperone that bridges Hsp70 and Hsp90). Mechanistic dissection of the Hsp70/Hop/Hsp90 system is complicated by conformational and compositional heterogeneity (18, 19). No Hop equivalent is expressed in the ER, which makes the BiP/Grp94 system a relatively simple system for mechanistic analysis.
Fig. 1.
Overview of BiP and Grp94 conformational cycles. (A) Conformations of BiP in the presence of ATP (Left, PDB ID code 5E84) and ADP (Right, modeled from PDB ID code 2KHO). (B) Conformations of Grp94 in the presence of ADP (Left, open state, PDB ID code 2O1V) and ATP (Right, closed state, PDB ID code 5ULS). (C) Conformation-specific binding between BiP and Grp94.
Grp94, as with other Hsp90 family members, is a homodimer with three domains on each arm: N-terminal domain (NTD), MD, and C-terminal domain (CTD) (Fig. 1B). When ADP is bound, Grp94 adopts an open conformation with an exposed client binding pocket. Upon ATP binding, Grp94 can close, and potentially trap a client between the dimer arms (20). However, Grp94 closure occurs slowly [on the order of 0.1 to 1 per minute (21–23)], suggesting that regulated closure acceleration is necessary. Extensive cochaperone regulation is a hallmark of the cytosol-specific Hsp90 (24), although, in many cases, these cochaperones appear to suppress Hsp90 client folding activity rather than enhance it (25). In the case of Grp94, much less is known about cochaperones; only two have been identified (26, 27), and these appear to specifically recruit certain clients to Grp94 rather than act generally on the Grp94 conformational cycle. A key feature of such cochaperones is their targeted influence on accelerating or decelerating specific steps of the Hsp90 conformational cycle (28–30).
Hsp90 family conformational dynamics are more complex than a simple cycling between one closed and one open state. For example, single-molecule fluorescence resonance energy transfer (smFRET) measurements have identified a Grp94 conformational intermediate (termed C′) populated upon Grp94 opening from the closed state (21). The C′ state is observed in the closed→open half-cycle but is not significantly populated during the open→closed half-cycle. Similarly, two closed conformations have been detected for cytosolic Hsp90 from bulk FRET, smFRET, and electron paramagnetic resonance measurements (31–33). These alternative closed structures are not known with certainty, but one has been proposed from smFRET measurements (34), and a recent report shows cytosolic Hsp90 in a semiclosed conformation when in complex with Hsp70, Hop, and the glucocorticoid receptor (16). Structural studies of Trap1 (the mitochondria-specific Hsp90 paralog) have also identified different closed conformations that are distinct from the well-established closed conformations of cytosolic Hsp90 and Grp94. Trap1 can adopt an asymmetric closed conformation (35) in which the two arms adopt different conformations at their MD/CTD interface region, while their NTD region resembles the closed conformation of cytosolic Hsp90 and Grp94. Trap1 can also adopt a “coiled-coil” conformation in which the N-terminal α-helix adopts a cross-monomer configuration that differs from both the open and closed states (36). The functional roles of these alternative Hsp90 closed conformations are unclear at present. Here we show that BiP is a powerful closure-accelerating cochaperone of Grp94. BiP accelerates Grp94 closure by stabilizing a high-energy conformational intermediate that, our data suggest, resembles the coiled-coil conformation of Trap1.
Results
BiP Accelerates ATP-Driven Closure of Grp94.
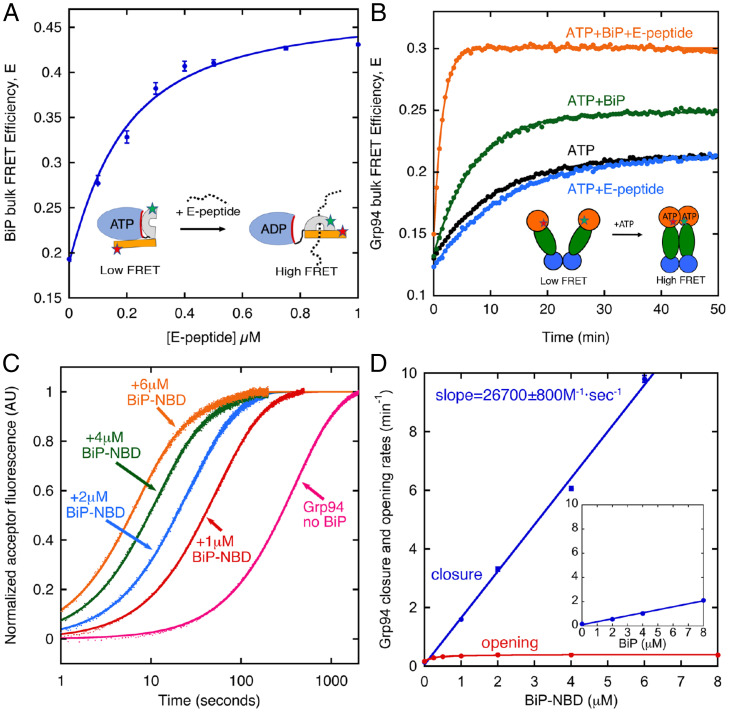
Insulin-like growth factors are a well-established family of Grp94 clients (9–11), and a recent analysis (37) identified a BiP binding site on the E-peptide region of proIGF2 (the proprotein form of IGF2). As in previous work, a FRET assay monitors the conformation of BiP in which the ATP conformation has low FRET efficiency and the ADP conformation has high FRET efficiency (37). Under ATP conditions, E-peptide binding causes BiP to adopt the high FRET ADP conformation (Fig. 2A) in which BiP is capable of binding Grp94 (Fig. 1C). This shift in BiP conformation is likely due to ATP hydrolysis by BiP that is stimulated by the E-peptide (37). We selected a saturating concentration of E peptide and BiP (1 μM for both) to examine their influence on Grp94. Here we used a previously established Grp94 bulk FRET assay in which the donor and acceptor fluorophores are located at residue 91 on the two NTDs (21). In contrast to the characteristically slow ATP-dependent closure of Grp94, BiP and E-peptide collectively cause Grp94 to close quickly (Fig. 2B, orange data). The E-peptide alone has no notable influence (compare black and blue data), whereas the addition of BiP alone results in a modest acceleration of Grp94 closure (green data). These results indicate that, once client binding has shifted BiP into the ADP conformation, BiP can then bind to Grp94 and accelerate Grp94 closure.
Fig. 2.
(A) BiP bulk FRET efficiency measuring lid closure with variable concentration of E-peptide under ATP conditions. Solid line is a fit to Eq. 1 (KD,obs = 0.13 ± 0.01 μM). (Inset) Cartoon shows schematic of BiP conformations, with fluorophore positions shown as stars. (B) Grp94 bulk FRET efficiency measuring ATP-dependent closure. (Inset) Cartoon shows schematic of Grp94 conformations, with fluorophore positions shown as stars. Solid lines are a fit to a single exponential (closure rate with ATP: 0.092 ± 0.002 min−1; ATP+E-peptide: 0.079 ± 0.001 min−1; ATP+BiP: 0.14 ± 0.01 min−1; ATP+BiP+E-peptide: 0.64 ± 0.04 min−1). (C) Stopped-flow bulk FRET acceptor fluorescence of Grp94. Closure is initiated by rapid mixing of 1 mM ATP and variable concentrations of the BiP-NBD. Solid lines are biexponential fits. (D) Fast-phase closure rates (blue circles; the fit values from experiments in C) and opening rates (red circles) measured with variable concentration of the BiP-NBD and full-length BiP (Inset; slope = 4,100 ± 200 M−1⋅s−1). Blue lines are linear fits with slope uncertainty from the fitting error. Grp94 opening is initiated by the addition of 50 µM inhibitor NVP-HSP990. Red line is a fit to a single-site binding model (kopen, no BiP = 0.17± 0.01 min−1, kopen,BiP = 0.40 ± 0.01 min−1, KD,app = 0.25 ± 0.04 μM). Error bars on data points are the SEM for at least three measurements.
The relatively modest closure enhancement from BiP in the absence of E-peptide is expected, because, under ATP conditions, BiP is predominantly in the ATP conformation that cannot bind to Grp94 (Fig. 1C). In contrast, because the BiP-NBD has no such restriction in binding Grp94 under ATP conditions (12), greater Grp94 closure acceleration is expected for the NBD versus the full-length BiP. Indeed, the BiP-NBD causes such rapid closure of Grp94 that stopped-flow experiments are needed. The kinetic curves are biexponential (solid lines, Fig. 2C), and the fast phase rates are shown in Fig. 2D. The mechanistic origin of the slow phase is not evident from these bulk experiments. Both the BiP-NBD and full-length BiP (Fig. 2D, Inset) accelerate ATP-driven closure of Grp94, while having only a modest influence on the opening rate. The BiP-NBD has a melting temperature of 44.7 ± 0.1 °C (52.6 ± 0.3 °C with 1 mM ATP) and an ATP hydrolysis rate that is comparable to full-length BiP (12), demonstrating the NBD is folded and active in these experiments.
The binding of BiP and Grp94 under ATP conditions is saturable (SI Appendix, Fig. 1A), while Grp94 closure rate increases linearly with BiP concentration (Fig. 2D). This behavior can be understood with a model in which the diffusion-limited binding of BiP causes Grp94 to close rapidly (Eqs. 2–4 in Methods). The linearity in Fig. 2D indicates that Grp94 closure is much faster than the unbinding of BiP from Grp94. In this kinetic regime, the slope in Fig. 2D (26,700 ± 800 M−1⋅s−1) represents the association rate of BiP to Grp94, which is in the range of typical diffusion-limited second-order rate constants for the binding of large macromolecules (38).
BiP accelerates Grp94 closure but has a minimal influence on the opening rate (Fig. 2D). Given that ATP hydrolysis is required for opening, we expect that BiP should increase the Grp94 ATPase rate only to the point at which the opening becomes rate limiting, and this is, indeed, what is observed (SI Appendix, Fig. 1B). The increased Grp94 ATPase from BiP quantitively agrees with ATPase predictions using the closure and opening rates of Grp94 in the presence of BiP (solid line in SI Appendix, Fig. 1B is the predicted ATPase based on Eq. 12). These results demonstrate that BiP accelerates Grp94 closure but has a minimal influence on subsequent ATP hydrolysis after Grp94 has closed, although the timing of ATP hydrolysis and phosphate release relative to arm opening is not possible to discern from these data.
BiP Stabilizes a Grp94 Closure Intermediate.
While the influence of BiP on Grp94 closure is dramatic (for example, Grp94 closure is accelerated ∼50-fold at 6 μM BiP-NBD), the causal mechanism is difficult to decipher from bulk experiments alone. We used smFRET measurements with the above FRET pair to determine the mechanism by which BiP accelerates Grp94 closure. All experiments were performed under low-salt conditions (50 mM KCl) in which BiP and Grp94 have a stronger binding affinity (12), whereas previously published smFRET measurements of Grp94 were performed at 150 mM KCl (21). This difference in salt conditions has only a modest effect on the closed state population (47 ± 1% at 150 mM KCl versus 62 ± 1% at 50 mM KCl; see SI Appendix, Table 1) in ATP conditions.
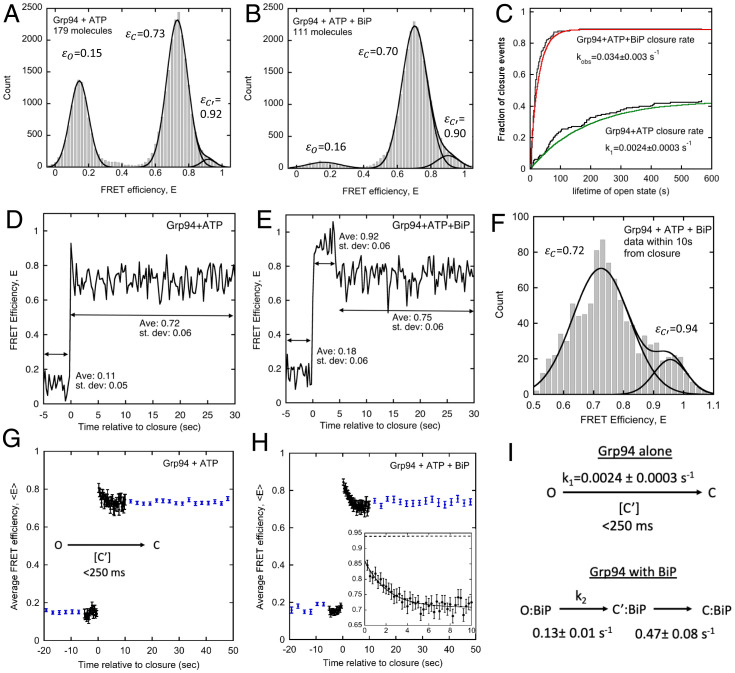
In the absence of BiP under ATP conditions (Fig. 3A), Grp94 adopts a substantial population of the open state (O: FRET efficiency εO = 0.15) and closed state (C: εC = 0.73), and a small population of the C′ state (εC′ = 0.92). In the presence of BiP, Grp94 predominantly populates the closed state (Fig. 3B). Similar to full-length BiP, the BiP-NBD drives Grp94 to the closed conformation (SI Appendix, Fig. 2A). Even in mixed ADP/ATP nucleotide conditions that disfavor closure (21), BiP shifts Grp94 to the closed conformation (SI Appendix, Fig. 2B). The BiP-induced shift toward the Grp94 closed conformation is not an artifact of the particular FRET pair position being used, as we find similar results with donor and acceptor fluorophores at a different pair position (SI Appendix, Fig. 3). SI Appendix, Table 1 provides a summary of Gaussian fitting of smFRET efficiency histograms under ATP conditions.
Fig. 3.
Single-molecule FRET efficiency histograms for Grp94 with (A) ATP and (B) ATP and 4 μM BiP. Solid lines are fits with Eq. 7 (fitting parameters in SI Appendix, Table 1). Fit values of the mean FRET efficiencies for open, closed, and C′ states are reported next to their respective Gaussian curves. Data were collected with alternating excitation (650 μW at 532 nm). (C) Cumulative lifetime distribution of Grp94 open state in presence and absence of BiP. Open states terminate in either closure or photobleaching. Black steps indicate Grp94 open state lifetimes that terminate in closure. Lines are fits to the open state and photobleaching lifetimes (see Methods; open-state photobleach rate: 0.0032 ± 0.0002 s−1; open-state photobleach rate with BiP: 0.0042 ± 0.0010 s−1). (D) Example smFRET record showing Grp94 closure with ATP. (E) Example smFRET record showing an O→C′→C closure event for Grp94 with ATP and 4 μM BiP. Data in D and E were collected with continuous donor excitation (2.6 mW at 532 nm, 250-ms sampling). (F) FRET efficiency histogram for data within 10 s after Grp94 closure in the presence of 4 μM BiP. Solid lines are fits to a two-Gaussian model with the mean FRET efficiencies for the closed and C′ states reported (SI Appendix, Table 1). (G) Average smFRET efficiencies for traces aligned at the time of Grp94 closure. Experiments were performed with both alternating donor/acceptor excitation (blue circles, 2.4-s sampling) and continuous donor excitation (black circles, 250-ms sampling). Error bars are the SEM (from 85 closure events at 2.4-s sampling and 20 closure events at 250-ms sampling). (H) Average smFRET efficiencies for traces aligned at the time of Grp94 closure measured as in G but in the presence of 4 μM BiP. (Inset) Magnified view of continuous excitation data and fit. Error bars are the SEM (from 68 closure events at 2.4-s sampling and 107 closure events at 250-ms sampling). Solid line is an exponential decay fit (decay rate = 0.47 ± 0.08 s−1; baseline = 0.72 ± 0.01; intercept = 0.86 ± 0.01, uncertainties are the SE). The FRET efficiency of C′ state (E = 0.94, from F) is shown as a dashed line. (I) Summary of kinetic influence of BiP on Grp94.
We selected 4 μM full-length BiP as a condition to analyze the kinetic influence of BiP on ATP-dependent closure of Grp94. At this concentration, BiP can shift Grp94 to the closed conformation (Fig. 3B) while having a relatively modest enhancement on closure kinetics compared to the BiP-NBD (Fig. 2D). The modest time constant associated with closure at 4 μM full-length BiP (∼30 s, discussed later) means that single-molecule Grp94 closure transitions are expected to be separated by time intervals long enough to allow detections while using alternating excitation, which is performed with a sampling interval of 2.4 s.
Because the open and closed states of Grp94 have highly distinct FRET efficiencies, closure transitions can be identified with a threshold criterion (see Methods), resulting in 85 closure events for Grp94 without BiP (from 302 molecules) and 145 closure events with 4 μM BiP (from 239 molecules). We also identified all open states that terminate due to dye photobleaching, to correct for this kinetic influence (Methods). The cumulative lifetime distribution shows that BiP dramatically reduces the Grp94 open state lifetime (Fig. 3C), consistent with the faster closure rate measured in bulk experiments (Fig. 2 C and D).
The above experiments were performed with alternating excitation of the donor and acceptor fluorophores and a sampling interval of 2.4 s. We also performed measurements with continuous donor excitation and a 250-ms sampling interval to examine closure with greater time resolution. At this faster sampling rate, an additional influence of BiP becomes apparent. For reference, in the absence of BiP, when Grp94 undergoes ATP-dependent closure, the transition appears to proceed without any appreciable intermediate (Fig. 3D), whereas, when BiP is present, Grp94 closes via a well-populated intermediate with FRET efficiency higher than the closed conformation (Fig. 3E).
Closure transitions measured with a 250-ms sampling interval were identified via the same threshold criteria mentioned earlier. Aligning all identified closure traces at the time of closure yields information about the extent to which a closure intermediate is populated and its lifetime. For Grp94 in absence of BiP, rare closure events show a progression of states consistent with O→C′→C, but the majority appear as a direct O→C transition. As a consequence, the average FRET from these transitions (<E>) exhibits a transient decay from a value only slightly higher than that of the closed state (Fig. 3G). We conclude that, if C′ is an obligate closure intermediate, it must have a lifetime shorter than the 250-ms sampling rate. In contrast, in the presence of BiP, a transient FRET increase is observed after Grp94 leaves the open state but before entering the closed conformation. This transient high FRET state decays with a rate of 0.47 ± 0.08 s−1 (Fig. 3H, Inset). Compiling FRET efficiencies measured within 10 s of the time at which Grp94 leaves the open state results in a histogram with two distinct FRET efficiencies (Fig. 3F). The highest FRET efficiency (ε = 0.94) is comparable to the FRET efficiency observed in individual traces such as that shown in Fig. 3E. We conclude that BiP enables Grp94 to close via a well-populated conformational intermediate with ε ∼0.94 that is on the pathway to the closed state.
Previous work with Grp94 identified a conformational intermediate (termed C′) that is populated prior to opening, which also has a FRET efficiency higher than that of the closed conformation [εC′ = 0.93; see figure 3C in Huang et al. (21)]. For simplicity, we will also refer to the BiP-stabilized closure intermediate as C′, rather than defining a new state for the BiP-stabilized high FRET configuration of Grp94. However, our FRET data cannot rule out the possibility these are structurally distinct states and that BiP causes additional conformational changes to Grp94 that are undetected with our FRET pair.
One notable feature of the data in Fig. 3H is that the amplitude of the transient FRET increase does not match the FRET efficiency of the C′ state (ε = 0.94 from Fig. 3F). This is consistent with a model in which some of the Grp94 closure events are assisted by BiP and proceed via an O→C′→C pathway, while other Grp94 closure events occur without BiP being bound and proceed with an appreciably lower population of this intermediate (as in Fig. 3G). The presence of both types of closure events is an expected outcome of working at 4 μM BiP, which has a modest enhancement on closure kinetics (Fig. 2D). Consequently, we analyze the kinetic data in Fig. 3C with a model in which a given Grp94 molecule undergoes either normal closure or BiP-accelerated closure (Eq. 10). The kinetic consequences of BiP on Grp94 are summarized in Fig. 3I. BiP accelerates the O→C transition 50-fold and extends the lifetime of the C′ state, thus enabling this state to become evident in smFRET traces. As discussed next, these dramatic kinetic changes to Grp94 closure originate from a strong thermodynamic driving force that favors BiP binding to the C′ state of Grp94.
BiP Induces a Dramatic Conformational Change of Grp94 even in the Absence of ATP.
One possible explanation for the BiP-induced acceleration of Grp94 closure is that the C′ state is a high-energy closure intermediate, and BiP lowers an energetic barrier to closure by stabilizing the C′ state. This model predicts that BiP should shift Grp94 to the C′ state in the absence of ATP, and this is, indeed, what we observe (Fig. 4 A and B). At low BiP-NBD concentration (500 nM), rapid transitions between the open and C′ states are evident (see the record in Fig. 4B). Distinct peak FRET efficiencies are consistent with the open and the C′ state values (Fig. 4B: εO = 0.15, εC′ = 0.93). These data, which were collected at a data integration time of 65 ms, show signs of signal averaging in which conformational changes are occurring on a similar timescale as the measurement integration time. Indeed, similar measurements taken at 250-ms and 1.0-s integration times show progressive merging of the open and C′ FRET efficiency peaks (Fig. 4 B–D). Despite the change in shape of the FRET efficiency histograms, the average FRET values remain comparable from datasets taken at different integration times (see <E> values in Fig. 4 A–E).
Fig. 4.
(A) The smFRET efficiency histogram for Grp94 in the absence of ATP with data integration time of 65 ms (10.4 mW at 532 nm). An example smFRET record is shown below the histogram. (B) The smFRET efficiency histogram for Grp94 with 500 nM NBD measured with data integration time of 65 ms. An example smFRET record is shown below the histogram. Rate constants for conformational changes in A and B were determined from Eq. 9. (C and D) The smFRET efficiency histograms for Grp94 with 500 nM NBD measured with data integration time of 250 ms (2.6 mW at 532 nm) and 1 s (650 μW at 532 nm). (E) The smFRET efficiency histogram for Grp94 with 12 μM NBD measured with 1-s integration time. For A–E, red lines are fits with a three-Gaussian model for two states that includes the effect of signal averaging due to rapid conformational changes (see Methods), and blue lines show the individual contribution of each Gaussian component. Mean FRET efficiencies for the open and C′ states are reported next to their respective Gaussian curves. Double asterisks in A and D indicate FRET efficiencies that were set to fixed values due to low state population. Histogram fitting parameters are listed in SI Appendix, Table 2. <E> is the average FRET efficiency for entire histogram. (F) Grp94 FRET in the presence of variable concentrations of BiP-NBD and the absence of ATP. Bulk FRET efficiencies are smaller than smFRET efficiencies because bulk data include dimers with donor but no acceptor, whereas smFRET includes only dimers labeled with both donor and acceptor. Bulk FRET (black circles) and smFRET (blue diamonds, all at 1-s integration time) FRET data were fit with Eq. 5 (black line). Fit values for the BiP affinity for the Grp94 open state (Kd,O) and C′ state (Kd,C′) are reported with the SE. Error bars on data points are the SEM for at least three measurements.
Following the work of Szabo and Eaton, we fit the FRET efficiency histograms in Fig. 4 A–E with a model that accounts for the influence of signal averaging on histogram shape (39, 40). Briefly, two-state conformational transitions that occur on a timescale similar to signal integration lead to FRET efficiency histograms that can be described by a sum of three Gaussian curves, and this fitting can determine the mean FRET efficiencies of the two states as well as their interconversion rates (see Methods). This fitting procedure provides a good fit for the data with Grp94 and BiP in the absence of ATP (solid lines, Fig. 4 A–E), yielding open and C′ populations as well as interconversion rates that are comparable between datasets taken with different integration times (SI Appendix, Table 2). In the absence of BiP, Grp94 only transiently accesses the C′ state (see an example trace in Fig. 4A), resulting in a tail in its FRET efficiency histogram. While these data can be fit with the methods described above, the fitting parameters have large uncertainties due to the low C′ population. The fits indicate that BiP stabilizes the C′ state primarily by slowing the C′→O transition rather than accelerating the O→C′ transition (compare rates in Fig. 4 A and B), a result that is expected if BiP is binding favorably to the C′ state.
We next measured the thermodynamic consequences of BiP binding to Grp94. Higher concentrations of BiP progressively increase the C′ population; for example, at 12 μM BiP-NBD, the majority of Grp94 is in the C′ state (Fig. 4E). We used a concentration series to quantify the degree to which BiP thermodynamically stabilizes the Grp94 C′ state. These experiments were performed with both bulk FRET and smFRET (SI Appendix, Fig. 4 A–E). The average FRET efficiency (<E>) measured via smFRET has a linear relationship with the bulk FRET efficiency (SI Appendix, Fig. 4F), indicating consistency between the methods. Both methods yield a similar concentration-dependent increase in FRET efficiency (Fig. 4F). These data can be fit with a linked equilibrium model in which BiP binds to the open and C′ conformations of Grp94 but binds the C′ state with greater affinity (Kd,O and Kd,C′, Eq. 5). The fit value for Kd,O (1.0 ± 0.2 μM) is similar to the measured value (0.95 ± 0.15 μM; SI Appendix, Fig. 1), whereas the model predicts that BiP will bind the C′ state of Grp94 with much greater affinity (Kd,C′ = 27 ± 4 nM). Overall, we conclude that the binding of BiP thermodynamically stabilizes the C′ state of Grp94 in a preexisting conformational equilibrium, consequently enabling O→C′ transitions to become evident in smFRET traces.
BiP Has a High Affinity for Grp94 When in a “Coiled-Coil” Configuration of the N-Terminal α-Helix.
The C′ conformational intermediate of Grp94 has a FRET efficiency (εC′ = 0.94; Fig. 3F) that is larger than the well-known closed state of Grp94 (εC = 0.73; Fig. 3A), and we sought to understand the structural differences between these two closed conformations. One structure of a Trap1 NTD dimer from the Tsai laboratory [Protein Data Bank (PDB) ID code 5F3K, termed the “coiled-coil” conformation (36)] stands out as a candidate for the C′ state because the fluorophore locations in this conformation are predicted to be closer than in the closed structure of Grp94 (SI Appendix, Fig. 5). Importantly, while the coiled-coil structure was determined for the isolated NTD, full-length Trap1 can adopt a similar conformation. Specifically, the authors identified a cysteine variant at residue L98 in Trap1 (M86 in Grp94) that would only be expected to form a disulfide bond in the coiled-coil conformation but not in the open or closed states. Under highly oxidizing conditions (Cu–phenanthroline as the oxidizing catalyst), Trap1 L98C forms disulfide-linked dimers suggesting that the coiled-coil conformation can be adopted by the full-length dimer. A model of the full-length coiled-coil Trap1 is in supplemental figure 6 of Sung et al. (36), highlighting a fundamentally arrangement of the N-terminal α-helix compared the closed conformation.
If the BiP-stabilized C′ conformation of Grp94 resembles the coiled-coil conformation of Trap1, then BiP should drive the formation of the Grp94 M86C disulfide bond. We constructed M86C as well as two other controls: N91C, which is not expected to oxidize because the cysteines are far apart (23Å, Fig. 5A), and V82C, which is not expected to oxidize based on the coiled-coil structure, but is expected to oxidize based on a plausible alternative arrangement of the Grp94 N-terminal α-helix [PDB ID code 1TC0 (41); SI Appendix, Fig. 5D]. We used oxidized glutathione, rather than Cu–phenanthroline, in our cross-linking experiments, to provide greater control over the kinetics of disulfide formation.
Fig. 5.
(A) Grp94 cross-linking mutations based on the Trap1 “coiled-coil” NTD structure (PDB ID code 5F3K). Trap1 residues corresponding to Grp94 M86, N91, and M178 are highlighted as sticks. Proposed Grp94 coiled-coil conformation is shown as a cartoon model. (B) Fraction of Grp94 M86C cross-linking in the presence of the BiP-NBD under ADP conditions (blue) and ATP conditions (green) versus cross-linking under ADP conditions without BiP (red). Cross-linking fraction was determined by SDS/PAGE gel band quantification. An example gel is shown in SI Appendix, Fig. 6. Cross-linking of 1 μM Grp94 dimer was induced by 10 mM oxidized glutathione (GSSG) with and without 5 μM BiP-NBD. (C) Binding of cross-linked Grp94 M86C and wild-type Grp94 to FITC-labeled BiP-NBD; solid line is a fit with single-site binding model. Error bars are the SEM for at least three measurements.
Grp94 M86C shows minimal disulfide bonding in the absence of BiP and robust oxidation in the presence of BiP (Fig. 5B and SI Appendix, Fig. 6). Greater oxidation is observed under ADP conditions versus ATP conditions (compare blue and green data in Fig. 5B), which is expected because ATP enables Grp94 closure, and the closed state does not cross-link (36). The N91C and V82C negative controls (SI Appendix, Fig. 6B) both exhibit minimal disulfide bonding. We conclude that the BiP-stabilized C′ conformation of Grp94 has a configuration of the N-terminal α-helix that resembles the Trap1 coiled-coil state.
In addition to the cross-monomer arrangement of the N-terminal α-helix, the Trap1 coiled-coil structure also has cross-monomer lid contacts. We constructed two lid-specific cross-linking variants, M178C and Q182C, which do not show enhanced oxidation from BiP (SI Appendix, Fig. 6B). Thus, the coiled-coil conformation provides an insight into the N-terminal α-helix of Grp94 in the C′ state, but other aspects of this conformation remain unclear.
The robust cross-linking of Grp94 M86C from BiP enables testing a key prediction discussed previously. Specifically, Fig. 4F predicts that BiP should bind the Grp94 conformation with a much higher affinity than the open conformation in the absence of ATP. We tested this prediction with a large-scale purification of M86C cross-linked Grp94, as a mimic of the C′ conformation. If the prediction is correct, BiP will have an enhanced affinity for the M86C cross-linked Grp94 construct. Fig. 5C confirms that the M86C cross-linked construct of Grp94 binds BiP with a roughly 50-fold affinity enhancement versus wild-type Grp94.
Discussion
Coordinated chaperoning by Hsp70/Hsp90 chaperones is conserved across prokaryotes (4) and eukaryotes (42), in the ER (12) and mitochondria. However, the mechanism that enables Hsp70/Hsp90 chaperones to work as a coordinated unit has been enigmatic (5, 6). A previous study revealed that Grp94 selectively binds the ADP conformation of BiP (12). Because BiP can trap clients in its ADP conformation, Grp94 should exhibit preferential binding to BiP when it is bound to a client. This is, indeed, what is observed with the E-peptide region of the proIGF2 client, whose binding shifts BiP to the ADP conformation (Fig. 2A), which, in turn, enables BiP to accelerate ATP-dependent closure of Grp94 (Fig. 2B). This mechanism provides an explanation for previous observations of synergy between other Hsp70/Hsp90 systems. For example, just as the E-peptide client enables BiP to enhance Grp94 closure and ATPase (Fig. 2B), the L2 client enables bacterial Hsp70 to enhance the bacterial Hsp90 ATPase (43).
The driving force behind Grp94 closure acceleration is from the NBD of BiP, which has the intrinsic capacity to act as a closure-accelerating cochaperone of Grp94. The enhancement of Grp94 closure by BiP is ∼50-fold (Fig. 3C), which makes it on par with or more effective than the activating cochaperone of cytosolic Hsp90 aha1 (44–46). Given that BiP and Grp94 are two of the most abundant ER proteins, their coordination likely plays an important role in maintaining protein quality control. Previous work identified mutations that disrupt binding between BiP and Grp94 (12). These mutants enable studies of the cellular consequences of impaired coordination between BiP and Grp94.
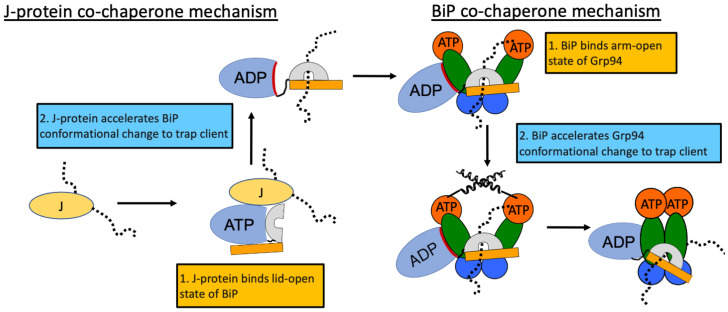
The BiP influence on Grp94 is mechanistically similar to the well-understood J-protein cochaperone influence on BiP, which provides a useful conceptual framework (Fig. 6). In the J-protein/BiP system, a client-bound J-protein can associate with BiP in the lid-open ATP conformation. The J-protein accelerates lid closure via stimulating ATP hydrolysis. Thus, J-proteins bring clients to BiP and also provide a signal for BiP to undergo the client-trapping conformational change. Similarly, a client-bound BiP can associate with Grp94 in the arm-open conformation. BiP accelerates Grp94 arm closure via stabilizing the C′ conformational intermediate. Thus, BiP brings clients to Grp94 and also provides a signal for Grp94 to undergo the client-trapping conformational change.
Fig. 6.
Comparison of J-protein cochaperone mechanism and BiP cochaperone mechanism. An unfolded client is shown by the dotted lines.
Dissecting the mechanism by which BiP accelerates Grp94 closure reveals a high-energy conformational intermediate that plays an important role in determining the timing of closure (Fig. 3). Because BiP stabilizes this conformational intermediate in the absence of ATP (Fig. 4), thermodynamic linkage alone, without any additional effects of kinetic coupling to ATP hydrolysis, is sufficient to explain the closure-enhancing effect of BiP on Grp94. Our data suggest that BiP preferentially binds a Grp94 conformation in which the N-terminal α-helix resembles the Trap1 “coiled-coil” state (Fig. 5). The coiled-coil structure has characteristics expected for a conformational intermediate, such as an open nucleotide pocket like the open conformation (36) and cross-monomer contacts like the closed conformation. A cryoelectron microscopy structure of Hsp90:Hsp70:Hop:GR shows Hsp90 in a semiclosed conformation (16), which is consistent with the high FRET C′ state of Grp94; however, the semiclosed conformation would not be expected to form the Grp94 M86C disulfide cross-link (Fig. 5). Future structural studies are needed to address questions about the BiP/Grp94 complex, such as whether the BiP-stabilized C′ state is the same conformation as the previously identified C′ state populated upon opening.
In the absence of ATP, BiP causes Grp94 to undergo transitions between the open and C′ conformations on the timescale of hundreds of milliseconds (Fig. 4). These rapid conformational changes are in stark contrast to the slow rate of ATP-dependent closure that occurs on the timescale of minutes in the absence of BiP. Fast conformational kinetics observed by smFRET and slow closure measured in bulk have also been observed for the yeast cytosolic Hsp90 (32, 47). Additional studies are needed to determine whether the cytosol-specific Hsp90 also adopts a C′-like state, and how cytosolic Hsp70 influences the structure of Hsp90.
Methods
Protein Expression and Purification.
Wild-type mouse Grp94 (72 to 765), its variants (V82C, M86C, N91C, E131C, M178C, and Q182C), full-length BiP (27 to 655), its variants (G518C/Y636C, D27C), and the BiP-NBD (27 to 411) were all expressed in Escherichia coli BL21* cells. Grp94 and BiP proteins were purified via Ni-NTA, ion exchange, and gel filtration (48). Proteins were stored in 25 mM Tris pH 7.5, 50 mM KCl, 1 mM 2-Mercaptoethanol (BME), and 5% glycerol prior to flash freezing. Fluorophore labeling of Grp94 variants with Alexa Fluor 555 (donor) and Alexa Fluor 647 (acceptor) was performed as in previous work (21).
Mouse E-peptide containing an N-terminal six-histidine tag and S95C mutation was expressed in E. coli BL21* cells. E-peptide was purified from the insoluble fraction of cells. Briefly, inclusion bodies were washed, and insoluble protein was denatured in an 8-M urea, 25 mM Tris buffer containing reducing agent tris(2-carboxyethyl)phosphine (TCEP). Protein was purified by ion exchange chromatography and stored in denaturing conditions (37).
BiP Bulk FRET.
A previously described BiP double-mutant G518C and Y636C was simultaneously labeled with donor and acceptor fluorophores, AlexaFluor 555 C2 maleimide and AlexaFluor 647 C2 maleimide, respectively (37). BiPSBD FRET-labeled protein was diluted to 0.1 μM in 25 mM Tris pH 7.5, 50 mM KCl, 1 mM MgCl2, 1 mM ATP, and 1 mM dithiothreitol and incubated for 20 min at 25 °C. Purified E-peptide was added from denaturing conditions. FRET Efficiency (E) data were collected after 30 min. Fluorometer setup included a donor excitation wavelength at 532 nm, donor emission wavelength at 567 nm, and an acceptor emission wavelength at 668 nm, with 5-nm slit widths. KD value between BiP and E-peptide was calculated using a quadratic single-site binding equation,
| [1] |
where x is the concentration of E-peptide, c is the FRET efficiency value in the absence of E-peptide, a is the FRET E-amplitude, and KD is the dissociation constant between BiP and E-peptide. B is the concentration of BiPSBD FRET-labeled protein.
Grp94 Bulk FRET.
Bulk FRET experiments were started by mixing 125 nM donor-labeled and 125 nM acceptor-labeled Grp94 in 25 mM Tris pH 7.5, 50 mM KCl, 1 mM BME, and 1 mg/mL bovine serum albumin (BSA) at 25 °C for 1 h to allow monomer exchange, then the temperature was adjusted to 30 °C. Data were collected with a FluoroMax-4 spectrofluorometer (Horiba Scientific). Closure kinetics were initiated with 1 mM ATP and 1 mM MgCl2 in the presence or absence of BiP and E-peptide. Reopening kinetics were initiated with 50 μM of the Hsp90 inhibitor NVP-HSP990 (49) after ATP-dependent closure of Grp94 reached a steady state. Excitation of donor was set at 530 nm, while the emission of donor and acceptor were monitored at 565 and 670 nm. Slit widths were set at 1.5 nm for excitation and 4.5 nm for emission. Bulk FRET efficiency was calculated from emission as acceptor/(donor + acceptor).
Stopped-flow FRET experiments were performed using an SX20 stopped-flow spectrometer (Applied Photophysics). Donor-labeled and acceptor-labeled Grp94 monomers were exchanged at room temperature for 90 min. The acceptor fluorescence change from Grp94 closure was monitored by a rapid mixing of various concentrations of BiP-NBD and 1 mM final concentration of ATP/MgCl2. Excitation was set to 532 nm, while the emission was measured by integrating the signals collected beyond 645 nm. The data were fit by a biexponential function, and the rate constants from the fast phase in Fig. 2C were fit by a linear regression based on the following scheme:
| [2] |
where O, C, are the Grp94 open and closed states, and OB is the BiP-bound Grp94 open state. Reopening from the closed state is not included in the above scheme because reopening is slow compared to BiP-accelerated closure (Fig. 2D) and thus does not influence the observed closure rate. The total closure rate of Grp94, ktot, can be calculated as
| [3] |
When k2 is much faster than kon[BiP] and koff, the equation simplifies to a linear concentration dependence,
| [4] |
To determine the BiP affinity to the open and C′ states of Grp94 in Fig. 4F, the bulk FRET efficiency was normalized to the smFRET efficiency by the relationship in SI Appendix, Fig. 4F. This normalized FRET was fit to an equation which relates the fraction of open and C′ states to FRET efficiency. This model assumes that, in the absence of ATP, each Grp94 dimer has two equivalent BiP binding sites, that Grp94 adopts only the open and C′ conformations, and that BiP binds these conformations with different affinities. The measured FRET is determined by the population-weighted FRET efficiency of Grp94 in the open and C′ conformations,
| [5] |
where K is the equilibrium constant between the open and C′ states, Ka,O and Ka,C′ are the association constants for the NBD binding to the open and C′ states, and εO and εC′ are the FRET efficiencies of the open and C′ states, which are fixed at values of εO = 0.16 and εC′ = 0.93. One notable feature of this fitting is that the fit value of Kd,C′ is correlated to the fit value of the equilibrium constant (K) between the open and C′ states (SI Appendix, Fig. 4G), indicating that the exact fit value of Kd,C′ should be interpreted cautiously. However, over a wide range of values for the equilibrium constant, the fit value of Kd,C′ remains in the high-affinity range of 10 nM to 100 nM.
Single-Molecule FRET.
Grp94 with a C-terminal SNAP tag was selectively biotinylated via a benzyl-guanine derivative, as described previously (21). Acceptor-labeled SNAP-Grp94 (N91C) and donor-labeled Grp94 (Grp94-N91C or Grp94-E131C) were monomer exchanged in 50 mM Hepes, pH 8.0, 50 mM KCl, 0.6 mM MgCl2, 2 mM BME, and 0.5 mg/mL BSA at 30 °C for 2 h. Closure was initiated with 600 μM ATP for at least 30 min at 30 °C. The preparation of glass slides and coverslips has been described previously (21). Grp94 was applied to the slide after being diluted to 1 nM with an oxygen scavenging system (0.4% glucose, 1.5 units per µL catalase, 0.04 units per µL glucose oxidase) and triplet-state quencher mixture (2 mM propyl galate, 4 mM 4-nitrobenzyl alcohol, and 4 mM Trolox). Then BiP-NBD or full-length BiP was applied to the slide. Single-molecule total internal reflection fluorescence (TIRF) imaging was performed on a custom microscope as previously described (21). FRET analysis was performed only on molecules with a recorded donor photobleach event, enabling background fluorescence to be subtracted for both the donor and acceptor signals. FRET efficiency values were only calculated for time points where when both the donor and acceptor fluorophores were not photobleached. Donor fluorophore excitation is at 532 nm, using laser powers dictated by the data integration time. Alternating excitation experiments include acceptor excitation with a laser power of 150 μW at 633 nm. All alternating excitation experiments have a sampling interval of 2.4 s with a signal integration time of 1 s for the donor excitation FRET signal.
The smFRET efficiency E is calculated as acceptor/(γ*donor + acceptor) from donor excitation. The γ value is 1.75 as in previous analysis of Grp94 (21). Closure events were identified by a threshold criterion in which at least two frames below a FRET efficiency of 0.4 are followed by at least two frames above a FRET efficiency of 0.4 and having a FRET efficiency change greater than 0.3.
The N observed intervals for Grp94 open-state configurations could terminate due to either a transition to the high FRET closed state (Fig. 3C, black lines) or could alternatively end due to censoring by a photobleach event for one of the dyes. Data were fit using single exponential maximum likelihood routines written in Matlab that account for both the observed transitions and censored events. Such fits produce a rate for the Grp94 open-to-closed transition and a rate for dye photobleaching. The green and red modeled cumulative curves in Fig. 3C incorporate effects from those two rates so that, in each instance, the curves asymptote at the fraction of Grp94 open-state intervals predicted to terminate in an observed transition to the closed state.
We model a single exponential process subject to censoring by maximizing its likelihood function. The likelihood function for a dataset containing nc censored intervals and (N − nc) noncensored intervals may be written as
| [6] |
where each exp(−k Tp) factor is a probability for observing one censored event of duration Tp, and each k exp(−k τi) factor is proportional to the probability for observing one noncensored event of duration τi. Maximizing the likelihood G(k) then yields our estimate for the single exponential rate k.
We may analyze a dataset treating the open-to-closed transitions (low to high FRET transitions) as observed events τI that are sometimes censored by dye photobleaching that contributes to a list of censored Tp intervals. In that instance, the k in Eq. 5 represents the kobs rate of Grp94 closure that we estimate by maximizing G(kobs). We can likewise reverse roles and use the list of dye photobleach intervals as observed events τI that are sometimes censored by an open-to-closed transition that now contributes to our list of Tp censored intervals. In that instance, k in Eq. 5 represents a photobleach rate kb that is estimated by maximizing G(kb). The two calculations use the same sets of open-to-closed intervals and photobleach intervals, merely switching their roles in Eq. 5. Our dataset thus enables an estimate of both the kobs closure and kb dye photobleach rates.
Using both the kobs closure rate and the kb photobleach rate, we can repetitively simulate the time course for an N-interval dataset, thereby producing the model green and red curves in Fig. 3C for the cumulative distribution of noncensored open-to-closed transitions by Grp94.
FRET efficiency histograms were constructed by compiling the efficiency values for each frame for each molecule. FRET efficiency histograms of Grp94 with ATP (except the data in Fig. 3F and SI Appendix, Fig. 3) were fit to a three-state model (open, closed, C′),
| [7] |
where Ai is the histogram area associated with each state, εi is the mean FRET efficiency, and σi is the SD. These three states account for at least 95% of the population in all histograms (see fraction values in SI Appendix, Table 1). States are defined by the mean FRET efficiency: 0.1< εO <0.2; 0.7< εC <0.8; 0.9< εC′ <1.0. All three states are identified by fitting the FRET efficiency histogram data, with the exception of SI Appendix, Fig. 2A where the open-state population is too small to be identified from the fit. In this case, the open-state parameters are fixed at εO = 0.15 and σO = 0.06. For the FRET pair using donor and acceptor labels at residues 131 and 91 (SI Appendix, Fig. 3), the data were fit to a two-Gaussian model because the FRET efficiency of the C′ state is not known for this FRET pair. The FRET efficiency histogram in Fig. 3F was also fit to a two-Gaussian model because only the open and C′ states apply to that analysis, and, for this data, the σC and σC′ were constrained to have the same value to reduce the fitting error.
Histograms of Grp94 with varying concentrations of BiP-NBD with no ATP were fit to a three-Gaussian model for two states, taking into consideration the effects of conformational exchange on the timescale of the data integration time.
The derivation and application of these fitting equations are described in Gopich and Szabo (39) and Chung et al. (40), respectively. The subscripts i = 1, 2 correspond to the C′ and open states, respectively, and i = 0 corresponds to the time-averaged data between the two states. We made one modification to the fitting equations in allowing the variance of the open and C′ Gaussians (σ2) to be treated as a fitting parameter instead of assuming the variance is determined via shot noise. The second half of equation 7 from Chung et al. is therefore modified such that
| [8] |
where T is the data integration time. The fitting parameters used were εO (FRET efficiency of the open state), εC (FRET efficiency of the C′ state), (fraction in the C′ state), k (relaxation rate; ), A (area of the histogram; from equation 4 in Chung et al.) and σ2 (variance of the open and C′ Gaussians). Rate constants, such as those shown in Fig. 4 A and B, can be calculated from the relaxation rate and ,
| [9] |
The O:BiP→C′:BiP rate in Fig. 3I (k2) was determined by an equation relating the observed closure in the presence of BiP (kobs = 0.034 ± 0.003 s−1, from Fig. 3C), the association constant for the binding of full-length BiP to Grp94 [Ka = 82,000 ± 3,000, from Sun et al. (12)], the closure rate in the absence of BiP (k1 = 0.0024 ± 0.0003 s−1, from Fig. 3C), and assuming a rapid preequilibrium of bound and unbound BiP,
| [10] |
The fraction of closure events that proceed through the O:B→C′:B pathway (f) versus the O→C′ pathway is given by
| [11] |
This model provides a prediction of the fraction of closure events expected to occur via the BiP-assisted O→C′→C (94 ± 13%) versus Grp94 closure without BiP, which is close to the measured fraction determined from Fig. 3H (73 ± 11%).
Fluorescence Depolarization Binding Measurements.
The D27C variant of full length and NBD of BiP were labeled in the same way as described previously (12). The binding between wild-type Grp94 and BiP constructs was measured using 50 nM fluorescein isothiocyanate (FITC)-labeled BiP in 25 mM Tris pH 7.5, 50 mM KCl, 1 mM ADP or ATP, 1 mM MgCl2, 1 mM BME, and 1 mg/mL BSA. After incubating at 30 °C for 30 min, Grp94 samples were titrated into the cuvette, and measured using a FluoroMax-4 spectrofluorometer. Due to the high binding affinity of cross-linked Grp94 M86C and BiP, the concentration of FITC-labeled BiP was reduced to 10 nM for a more accurate determination of the binding constant.
Grp94 Cross-Linking.
The M86C, N91C, M178C, and Q182C variants of Grp94 were purified in a similar way except the storage solution contains 0.1 mM TCEP instead of 1 mM BME. A concentration of 1 μM Grp94 dimer was used in all cross-linking assays. Prior to cross-linking, Grp94 was diluted into 25 mM Tris pH 8.0 at 25 °C, 50 mM KCl, 1 mM ADP or ATP, 1 mM MgCl2. Samples with the BiP-NBD were at 5 μM concentration. The cross-linking was initiated with 10 mM oxidized glutathione. The cross-linking kinetics were collected by quenching at different time points using a fresh-made N-ethylmaleimide with a final concentration of 7.5 mM. The quenched Grp94 samples were later boiled and loaded onto a 4 to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) gel. The amounts of cross-linked and non-cross-linked Grp94 were quantified using ImageJ (50).
ATPase Assay.
The ATPase measurements utilize the T229G variant of the BiP-NBD, which itself is hydrolytically inactive (51). The ATPase of Grp94 in the presence and absence of BiP-NBD T229G were measured on a plate reader (BioTeK) using an established enzyme-coupled assay with 2 μM Grp94 monomer (52). The ATPase rate is reported per monomer of Grp94. Buffer conditions were 25 mM Tris pH 7.5, 50 mM KCl, 1 mM MgCl2, 1 mM ATP, and 1 mg/mL BSA at 30 °C. The measured ATPase was compared to the ATPase predicted from Grp94 closure (kc) and opening rates (ko) as
| [12] |
The influence of the BiP-NBD on closure and opening rates is given by the fits in Fig. 2D. The concentration of free [NBD] is determined via
| [13] |
where Kd is the measured affinity between BiP and Grp94 (SI Appendix, Fig. 1A).
Grp94 Fluorophore Dye Position Prediction.
The crystal structures of Grp94 PDB ID codes 2O1V and 5ULS were the input as the O and C state models for FRET positioning and screening (53). The labeling site was defined as the Grp94 N91. The Alexa 488 L linker configuration was applied to the donor, while the Cy5 L linker was applied to the acceptor. After simulating using the simple AV mode, the three-dimensional coordinates of donor and acceptor were randomly picked, and 10,000 pair-wise distances were averaged and reported in SI Appendix, Fig. 5.
Supplementary Material
Acknowledgments
We thank the Dorothee Kern lab for assistance with the stopped-flow measurements, and T.O.S. and J.G. lab members, as well as Thorsten Hugel and Sonja Schmid, for providing helpful feedback on the results. We thank David Agard and Ray Wang for sharing the PDB coordinates of the Hsp90:Hsp70:Hop:GR structure. Research for this project was supported by NIH grants R01 GM115356 (T.O.S.) and R01 GM121384 and R01 GM81648 (J.G.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118793119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Macario A. J., Conway de Macario E., Molecular chaperones: Multiple functions, pathologies, and potential applications. Front. Biosci. 12, 2588–2600 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Morán Luengo T., Kityk R., Mayer M. P., Rüdiger S. G. D., Hsp90 breaks the deadlock of the Hsp70 chaperone system. Mol. Cell 70, 545–552.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Kirschke E., Goswami D., Southworth D., Griffin P. R., Agard D. A., Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 157, 1685–1697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genest O., Hoskins J. R., Camberg J. L., Doyle S. M., Wickner S., Heat shock protein 90 from Escherichia coli collaborates with the DnaK chaperone system in client protein remodeling. Proc. Natl. Acad. Sci. U.S.A. 108, 8206–8211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genest O., Wickner S., Doyle S. M., Hsp90 and Hsp70 chaperones: Collaborators in protein remodeling. J. Biol. Chem. 294, 2109–2120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morán Luengo T., Mayer M. P., Rudiger S. G. D., The Hsp70-Hsp90 chaperone cascade in protein folding. Trends Cell Biol. 29, 164–177 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Melnick J., Aviel S., Argon Y., The endoplasmic reticulum stress protein GRP94, in addition to BiP, associates with unassembled immunoglobulin chains. J. Biol. Chem. 267, 21303–21306 (1992). [PubMed] [Google Scholar]
- 8.Melnick J., Dul J. L., Argon Y., Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature 370, 373–375 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Ostrovsky O., Eletto D., Makarewich C., Barton E. R., Argon Y., Glucose regulated protein 94 is required for muscle differentiation through its control of the autocrine production of insulin-like growth factors. Biochim. Biophys. Acta 1803, 333–341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostrovsky O., Makarewich C. A., Snapp E. L., Argon Y., An essential role for ATP binding and hydrolysis in the chaperone activity of GRP94 in cells. Proc. Natl. Acad. Sci. U.S.A. 106, 11600–11605 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ostrovsky O., Ahmed N. T., Argon Y., The chaperone activity of GRP94 toward insulin-like growth factor II is necessary for the stress response to serum deprivation. Mol. Biol. Cell 20, 1855–1864 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun M., Kotler J. L. M., Liu S., Street T. O., The endoplasmic reticulum (ER) chaperones BiP and Grp94 selectively associate when BiP is in the ADP conformation. J. Biol. Chem. 294, 6387–6396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcinowski M., et al. , Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat. Struct. Mol. Biol. 18, 150–158 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Yang J., Nune M., Zong Y., Zhou L., Liu Q., Close and allosteric opening of the polypeptide-binding site in a human Hsp70 chaperone BiP. Structure 23, 2191–2203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munro S., Pelham H. R., An Hsp70-like protein in the ER: Identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell 46, 291–300 (1986). [DOI] [PubMed] [Google Scholar]
- 16.Wang R. Y.-R., et al. , Structure of Hsp90-Hsp70-Hop-GR reveals the Hsp90 client-loading mechanism. Nature (2021). 10.1038/s41586-021-04252-1. [DOI] [PMC free article] [PubMed]
- 17.Kravats A. N., et al. , Interaction of E. coli Hsp90 with DnaK involves the DnaJ binding region of DnaK. J. Mol. Biol. 429, 858–872 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt C., Beilsten-Edmands V., Robinson C. V., The joining of the Hsp90 and Hsp70 chaperone cycles yields transient interactions and stable intermediates: Insights from mass spectrometry. Oncotarget 6, 18276–18281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgner N., et al. , Hsp70 forms antiparallel dimers stabilized by post-translational modifications to position clients for transfer to Hsp90. Cell Rep. 11, 759–769 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verba K. A., et al. , Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science 352, 1542–1547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang B., Friedman L. J., Sun M., Gelles J., Street T. O., Conformational cycling within the closed state of Grp94, an Hsp90-family chaperone. J. Mol. Biol. 431, 3312–3323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frey S., Leskovar A., Reinstein J., Buchner J., The ATPase cycle of the endoplasmic chaperone Grp94. J. Biol. Chem. 282, 35612–35620 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Dollins D. E., Warren J. J., Immormino R. M., Gewirth D. T., Structures of GRP94-nucleotide complexes reveal mechanistic differences between the hsp90 chaperones. Mol. Cell 28, 41–56 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schopf F. H., Biebl M. M., Buchner J., The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 18, 345–360 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Sahasrabudhe P., Rohrberg J., Biebl M. M., Rutz D. A., Buchner J., The plasticity of the Hsp90 co-chaperone system. Mol. Cell 67, 947–961.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Rosenbaum M., et al. , MZB1 is a GRP94 cochaperone that enables proper immunoglobulin heavy chain biosynthesis upon ER stress. Genes Dev. 28, 1165–1178 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B., et al. , Folding of Toll-like receptors by the HSP90 paralogue gp96 requires a substrate-specific cochaperone. Nat. Commun. 1, 79 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Retzlaff M., et al. , Asymmetric activation of the Hsp90 dimer by its cochaperone Aha1. Mol. Cell 37, 344–354 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Rehn A. B., Buchner J., p23 and Aha1. Subcell. Biochem. 78, 113–131 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Southworth D. R., Agard D. A., Client-loading conformation of the Hsp90 molecular chaperone revealed in the cryo-EM structure of the human Hsp90:Hop complex. Mol. Cell 42, 771–781 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giannoulis A., et al. , Two closed ATP- and ADP-dependent conformations in yeast Hsp90 chaperone detected by Mn(II) EPR spectroscopic techniques. Proc. Natl. Acad. Sci. U.S.A. 117, 395–404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hessling M., Richter K., Buchner J., Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat. Struct. Mol. Biol. 16, 287–293 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Schmid S., Hugel T., Controlling protein function by fine-tuning conformational flexibility. eLife 9, e57180 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolf S., et al. , Hierarchical dynamics in allostery following ATP hydrolysis monitored by single molecule FRET measurements and MD simulations. Chem. Sci. 12, 3350–3359 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavery L. A., et al. , Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism. Mol. Cell 53, 330–343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sung N., et al. , Mitochondrial Hsp90 is a ligand-activated molecular chaperone coupling ATP binding to dimer closure through a coiled-coil intermediate. Proc. Natl. Acad. Sci. U.S.A. 113, 2952–2957 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotler J. L. M., Wei W.-S., Deans E. E., Street T. O., Electrostatics cause the molecular chaperone BiP to preferentially bind oligomerized states of a client protein. bioRxiv [Preprint] (2021). . Accessed 11 January 2022. [DOI] [PMC free article] [PubMed]
- 38.Schreiber G., Haran G., Zhou H. X., Fundamental aspects of protein-protein association kinetics. Chem. Rev. 109, 839–860 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung H. S., et al. , Extracting rate coefficients from single-molecule photon trajectories and FRET efficiency histograms for a fast-folding protein. J. Phys. Chem. A 115, 3642–3656 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gopich I. V., Szabo A., FRET efficiency distributions of multistate single molecules. J. Phys. Chem. B 114, 15221–15226 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Immormino R. M., et al. , Ligand-induced conformational shift in the N-terminal domain of GRP94, an Hsp90 chaperone. J. Biol. Chem. 279, 46162–46171 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Kravats A. N., et al. , Functional and physical interaction between yeast Hsp90 and Hsp70. Proc. Natl. Acad. Sci. U.S.A. 115, E2210–E2219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halpin J. C., Street T. O., Hsp90 sensitivity to ADP reveals hidden regulation mechanisms. J. Mol. Biol. 429, 2918–2930 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LaPointe P., Mercier R., Wolmarans A., Aha-type co-chaperones: The alpha or the omega of the Hsp90 ATPase cycle? Biol. Chem. 401, 423–434 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Wortmann P., Götz M., Hugel T., Cooperative nucleotide binding in Hsp90 and its regulation by Aha1. Biophys. J. 113, 1711–1718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J., Richter K., Reinstein J., Buchner J., Integration of the accelerator Aha1 in the Hsp90 co-chaperone cycle. Nat. Struct. Mol. Biol. 20, 326–331 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Mickler M., Hessling M., Ratzke C., Buchner J., Hugel T., The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nat. Struct. Mol. Biol. 16, 281–286 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Halpin J. C., Huang B., Sun M., Street T. O., Crowding activates heat shock protein 90. J. Biol. Chem. 291, 6447–6455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eccles S. A., et al. , NVP-AUY922: A novel heat shock protein 90 inhibitor active against xenograft tumor growth, angiogenesis, and metastasis. Cancer Res. 68, 2850–2860 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei J., Gaut J. R., Hendershot L. M., In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J. Biol. Chem. 270, 26677–26682 (1995). [DOI] [PubMed] [Google Scholar]
- 52.Leskovar A., Wegele H., Werbeck N. D., Buchner J., Reinstein J., The ATPase cycle of the mitochondrial Hsp90 analog Trap1. J. Biol. Chem. 283, 11677–11688 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Kalinin S., et al. , A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nat. Methods 9, 1218–1225 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.