Significance
The role of Lewy pathology (LP) in synucleinopathies is incompletely understood. Here we provide a protocol to characterize molecular interactions of LP in primary human disease tissues. Using this protocol, we identified core molecular features of LP. This approach has broad potential to provide consensus for the molecular processes underlying synucleinopathies as well as other proteinopathies.
Keywords: prion, neurodegeneration, iron, immune, mitochondria
Abstract
The intracellular misfolding and accumulation of alpha-synuclein into structures collectively called Lewy pathology (LP) is a central phenomenon for the pathogenesis of synucleinopathies, including Parkinson’s disease (PD) and dementia with Lewy bodies (DLB). Understanding the molecular architecture of LP is crucial for understanding synucleinopathy disease origins and progression. Here we used a technique called biotinylation by antibody recognition (BAR) to label total (BAR-SYN1) and pathological alpha-synuclein (BAR-PSER129) in situ for subsequent mass spectrometry analysis. Results showed superior immunohistochemical detection of LP following the BAR-PSER129 protocol, particularly for fibers and punctate pathology within the striatum and cortex. Mass spectrometry analysis of BAR-PSER129–labeled LP identified 261 significantly enriched proteins in the synucleinopathy brain when compared to nonsynucleinopathy brains. In contrast, BAR-SYN1 did not differentiate between disease and nonsynucleinopathy brains. Pathway analysis of BAR-PSER129–enriched proteins revealed enrichment for 718 pathways; notably, the most significant KEGG pathway was PD, and Gene Ontology (GO) cellular compartments were the vesicle, extracellular vesicle, extracellular exosome, and extracellular organelle. Pathway clustering revealed several superpathways, including metabolism, mitochondria, lysosome, and intracellular vesicle transport. Validation of the BAR-PSER129–identified protein hemoglobin beta (HBB) by immunohistochemistry confirmed the interaction of HBB with PSER129 Lewy neurites and Lewy bodies. In summary, BAR can be used to enrich for LP from formalin-fixed human primary tissues, which allowed the determination of molecular signatures of LP. This technique has broad potential to help understand the phenomenon of LP in primary human tissue and animal models.
Parkinson’s disease (PD) and dementia with Lewy bodies (DLB) are progressive neurodegenerative diseases characterized by intracellular inclusions containing aggregated alpha-synuclein referred to as Lewy pathology (LP). Although strong evidence suggests alpha-synuclein is involved in the disease process, central questions remain regarding the molecular origins and progression of LP. A diverse range of cellular pathways have been implicated in alpha-synuclein–mediated disease processes including dysfunctional synaptic neurotransmission and vesicular trafficking, mitochondrial dysfunction, oxidative stress, neuroinflammation, calcium signaling, glycolysis, autophagy lysosomal dysfunction, and Mitogen-activated protein kinase (MAPK) signaling pathways (1). However, much of the evidence implicating these cellular processes has been derived from cell and animal models, which may or may not closely recapitulate the human disease processes. Defining the molecular details of disease process is important for progress toward the development of disease-modifying therapies.
Alpha-synuclein phosphorylated at serine 129 (PSER129) is a marker of LP. PSER129 is enriched in LP with only trace quantities (<5% of total alpha-synuclein pool) of endogenous alpha-synuclein being phosphorylated (2). Evidence suggests that PSER129 may aggregate more rapidly, and PSER129 may have a role in regulating alpha-synuclein turnover (3). Evidence also suggests that PSER129 accumulates in the brain as PD progresses (4, 5) although it is not clear whether PSER129 occurs prior to, during, or following inclusion formation (6). Several antibodies have been developed to detect PSER129, including the commonly used EP1536Y antibody (Abcam), a rabbit monoclonal antibody against PSER129 that has high specificity for LP (7) and has been used extensively for immunohistochemical (IHC) labeling of LP.
Determining the molecular makeup of LP in the human brain is challenging. Difficulties arise principally from the insoluble nature of LP, which precludes common biochemical methods for determining molecular interactions. In past studies, LP from the human brain has been enriched for by biochemical fractionation methods (8) and isolated by microdissection techniques (9). There are important limitations to each approach. Fractionation disturbs cellular structures and, presumably, LP composition. Microdissection exclusively captures Lewy bodies, but not other lesions (neurites, dots, etc.). Microscopy methods provide accurate data characterizing spatial and molecular makeup of LP, but these methods are limited to known molecular targets. Together, current approaches are insufficient to identify molecular details of LP in the human diseased brain.
Recently, a technique referred to as biotinylation by antibody recognition (BAR) was successfully used to label insoluble cellular components within proximity to an antigen in fixed primary tissues for enrichment and downstream mass spectrometry analysis (10–13). The BAR technique (also called selective proteomic proximity labeling using tyramide, or enzyme-mediated activation of radical sources) labels a target antigen by peroxidase-catalyzed deposition of biotin moieties onto proximal molecules. BAR is particularly well suited for determining protein interactions and subcellular localization of insoluble cellular components (10). LPs are insoluble cellular inclusions, and therefore the BAR technique holds promise to study these structures. Recent demonstrations of the BAR technique, and its utility for proteomic analysis, gave us the impetus to attempt to purify LP and LP interactors from fixed primary human tissues.
Here we applied a modified BAR technique to capture total alpha-synuclein (BAR-SYN1) and LP (BAR-PSER129) interactions in formalin-fixed human brain samples from individuals diagnosed with PD and DLB (SYN), as well as neurologically intact individuals (HC).
Results
Strategy for BAR Labeling LP.
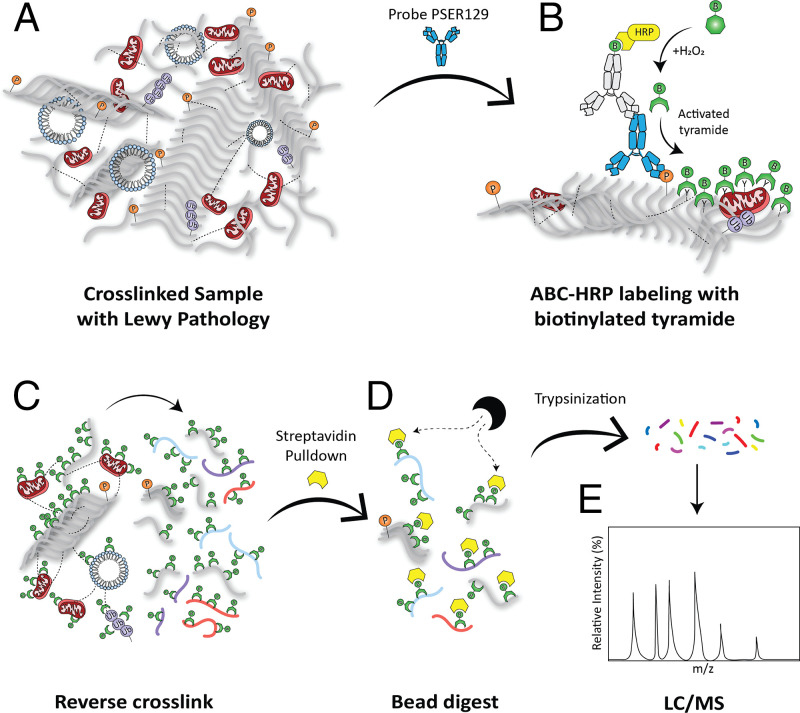
A pictorial summary highlights key considerations for the BAR-PSER129 technique (Fig. 1). First, the PSER129 specific antibody EP1536Y is bound to LP in formalin-fixed tissue samples. Horseradish peroxidase is complexed with EP1536Y using a common avidin biotin complex (ABC) method. HRP-mediated hydrogen peroxide decomposition catalyzes the oxidation of biotinyl tyramide, which subsequently deposits biotin moieties onto proteins proximal to the EP1536Y–ABC complex. Cross-links were reversed by heating the samples in the presence of excess Tris⋅HCl and sodium dodecyl sulfate (SDS). Biotin-labeled proteins captured with magnetic streptavidin beads were then digested and analyzed with LC-MS.
Fig. 1.
Strategy for BAR labeling of LP. (A) Formalin-fixed floating human brain sections were probed with the anti-PSER129 antibody EP1536Y. (B) Using a biotinylated secondary antibody and ABC, horseradish peroxidase (HRP) was concentrated at the EP1536Y antigenic site. Biotin moieties were deposited near the antibody complex via peroxidase catalyzed oxidation of biotinyl tyramine. (C) Once LP was labeled with biotin, formalin cross-links were reversed in the presence of excess Tris and heat. (D) Liberated biotin-labeled LP-associated proteins could then be purified with streptavidin magnetic beads (E) and digested with trypsin for analysis via liquid chromatography–mass spectrometry (LC/MS).
Tissue Distribution of BAR-PSER129 Labeling.
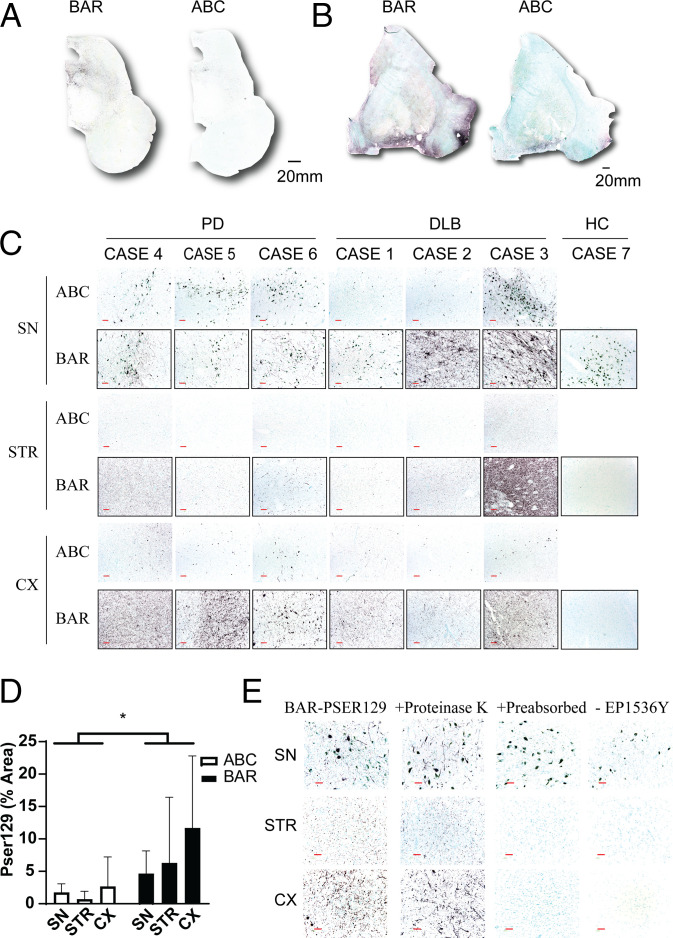
To assess the efficacy of biotin labeling using the BAR-PSER129 technique, we first performed the technique on a striatum containing section and midbrain section of an individuals with PD or DLB using the PSER129 specific antibody EP1536Y. As expected (14), BAR-PSER129 amplification resulted in a substantial signal enhancement in tissues when compared to standard IHC protocol (Fig. 2). In agreement with what has been reported previously with other low abundance antigens (15, 16), DAB staining was readily visible with the naked eye with BAR-PSER129, while the standard IHC protocol often required a microscope to verify staining (Fig. 2 A and B). The visibility appeared to be a result of both the increase signal intensity and the low background associated with high dilutions used with the technique (15). BAR-PSER129 amplification heavily labeled cell bodies of the substantia nigra (SN; Fig. 2A) and punctate structures throughout the striatum (Fig. 2B) and some of adjacent cortical regions.
Fig. 2.
BAR labeling of LP in the synucleinopathy brain. Formalin-fixed floating sections from midbrain and sections adjacent to the anterior commissure of individuals with PD (n = 3), DLB (n = 3), and nonsynucleinopathy controls (HC, n = 3) were labeled using the BAR-PSER129 technique or a standard ABC detection protocol. The distribution of labeling was detected using nickel DAB chromogen. All slides were counterstained with the nuclear stain methyl green. Whole slide scans at 10× magnification of (A) midbrain and (B) putamen sections showing distribution and intensity of BAR labeling and standard ABC detection. (C) The 20× images of regions of interest including the SN, cortex (CX), and striatum (STR). (D) Quantification of PSER129 staining in brain regions of interest. *Two-way ANOVA F(1,30) = 7.008, P = 0.0128. Sections with either BAR-PSER129 or standard ABC were quantified and compared. (E) Assay controls for the BAR-PSER129 labeling. Tissue sections were processed under varying conditions including 1) pretreating tissue with proteinase K, 2) using EP1536Y anti-PSER129 antibody that was preabsorbed with excess immunogen, or 3) omitting the EP1536Y anti-PSER129 antibody from the protocol. Representative 20× images from regions of interest show resulting labeling under each condition. (Red scale bars, 100 μm.)
Next, we applied the BAR-PSER129 technique to tissues from both SYN and HC. We found that in all samples tested BAR-PSER129 resulted in enhanced pathology detection, particularly small punctate structures in the cortex and striatum were easily observed when compared to standard ABC labeling (Fig. 2C). Positionally match images from the same individuals with DLB and PD, show readily apparent small punctate structures throughout the tissue with BAR-PSER129 compared to standard ABC when viewed under high magnification. BAR-PSER129 staining was absent from the SN, cortex, and striatum of all the HC tissues tested, except for rare (<3 instances observed) sparsely labeled unidentified structures morphologically resembling blood vessels (SI Appendix, Fig. S1). Subsequent densitometry analysis of the BAR-PSER129 stained sections showed that the technique labeled a greater area of the PD and DLB tissues than the standard ABC protocol (Fig. 2D). To ensure that the labeling with BAR-PSER129 technique was indeed specific to LP we then performed several control studies testing the specificity of the BAR-PSER129 labeling (Fig. 2E). Brain sections from DLB and PD digested with proteinase K showed similar BAR-PSER129 labeling as undigested sections. In contrast, brain sections incubated with the preabsorbed EP1536Y antibody or processed in the absence of EP1536Y did not show BAR-PSER129 labeling. Under all conditions tested pigmented cells in the SN were observed, which appeared blue/green following the methyl green counterstain. Together these results demonstrate sensitive and specific labeling for PSER129 positive LP using the BAR protocol.
Intracellular Distribution of BAR-PSER129 Labeling.
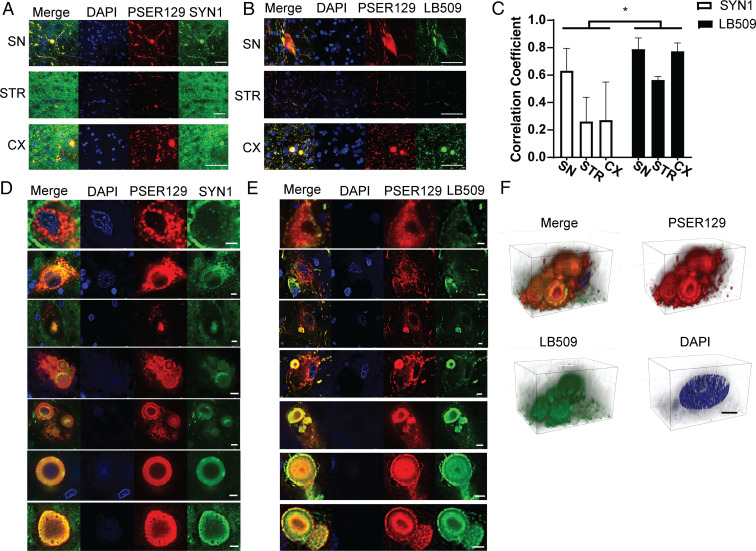
LP are molecularly heterogenous structures with varying morphology and cellular distribution (6, 17). To determine the extent that BAR-PSER129 labeled LP, we performed double labeling using an antibody for total alpha-synuclein (SYN1) and a well characterized antibody used to detect LP (LB509) (18). Double labeling was performed by substituting fluorescent tyramide conjugates for biotinyl tyramide in the BAR protocol, thus providing a direct measure of BAR-labeled intracellular structures. Results show that in SYN brain total alpha-synuclein was ubiquitously expressed with fibers and punctate structures being detected in all samples (Fig. 3A). In contrast, PSER129 signal was restricted to a select few fibers, puncta, and some cell bodies. LB509 labeling largely overlapped with PSER129 which was confirmed by colocalization analysis (Fig. 3 B and C). We did observe, particularly fibers, throughout the afflicted brain regions that labeled exclusively for LB509 and not PSER129. PSER129 and LB509 labeled structures that were granular, diffuse, and compact consistent with spectrum of LP morphologies (19) (Fig. 3 D and E). Interestingly, SYN1 did not label some granular PSER129 positive pathology suggesting that this epitope (amino acids 91 to 99) is concealed in granular structures, possible due to alpha-synuclein membrane interactions (20). Faint DAPI labeling was seen at the core and precipice of many but not all observed LBs. Three-dimensional projections of multiple LBs labeled with PSER129 and LB509 shows the compact spheroid structures with “onion-like” layers characteristic of LBs (3F) (17, 21). In summary, BAR-PSER129 labeling was restricted to intracellular LP structures in the human brain.
Fig. 3.
Intracellular distribution of BAR-PSER129–labeled pathology in the synucleinopathy brain. Formalin-fixed floating brain sections from human synucleinopathy cases were fluorescently labeled by substituting fluorescent tyramides for biotinyl tyramine in the standard BAR protocol. (A) Sections were dual labeled for total alpha-synuclein (SYN1) and phosphoserine 129 alpha-synuclein (PSER129). Representative confocal images are from brain regions of interest. (Scale bars, 50 μm.) (B) Sections dual labeled for the LP marker (LB509) and PSER129. Representative confocal images are from brain regions of interest. (Scale bars, 50 μm.) (C) Colocalization analysis of dual-labeled brain sections. Correlation coefficient was calculated for confocal images acquired under each experimental condition. Subcellular distribution of PSER129 and total (D) alpha-synuclein (SYN1) and (E) LB509 in LP-bearing perikarya. Images depict diversity of LP morphology, including granular morphology to compact circular type. (Scale bars, 5 μm.) (F) Three-dimensional projection images of multiple Lewy bodies in the cytoplasm of a cell labeled with PSER129 and LB509. (Scale bars, 10 μm.) Nuclei were stained in all sections using DAPI. Two-way ANOVA *P < 0.05, n = 3.
Purification and Mass Spectrometry of BAR-Labeled Proteins.
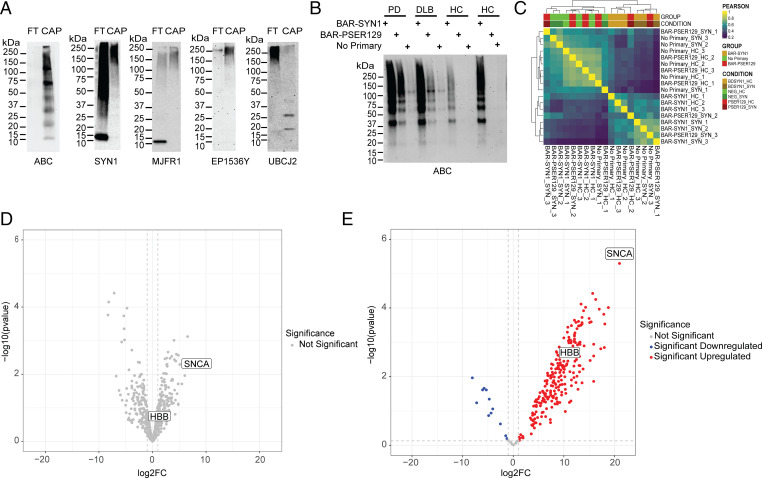
To determine whether we could isolate the BAR-PSER129–labeled pathology from tissue, we then reversed crosslinking and solubilized cross-linked samples using a previous protocol with modifications (22). Prior to cross-link reversal tissue was washed with SDS and 2-mercapitoethanol to remove contaminating primary antibody. Following heat-mediated cross-link reversal, little pellet was observed after centrifugation (i.e., 22,000 × g for 30 min), suggesting the tissue was effectively solubilized. Following streptavidin pulldown of the labeled midbrain and striatal sample, the lysate contained little to no detectable biotin, while the pulldown sample contained abundant biotinylated proteins, at a diverse range of molecular weights (Fig. 4A). We then probed the samples with two alpha-synuclein antibodies reactive toward different epitopes, SYN1 (epitope = amino acids 91 to 99; BD Biosciences) and MJFR1 (epitope = amino acids 118 to 123; Abcam). With both antibodies, we observed predominately high molecular weight species over 75 kDa in the captured sample. Alpha-synuclein that was not captured (i.e., flow-through) consists of a mixture of apparent monomer at 15 kDa and high molecular weight species greater than 37 kDa. Next, we probed for PSER129 using the EP1536Y antibody and found high molecular weight species predominantly in the captured sample. Next, we probed for ubiquitin conjugates and observed several bands approximately at 20 kDa to 25 kDa, consistent with ubiquitinated alpha-synuclein isolated from LP (23). Together, these results demonstrated the BAR-PSER129–captured fractions were immunoreactive for known LP markers.
Fig. 4.
Isolation and mass spectrometry of BAR-labeled LP. BAR-PSER129 was conducted on a midbrain and striatal section from three synucleinopathy (i.e., PD and DLB) and three nonsynucleinopathy (HC) brains (Table 1). These tissues were selected because they showed a similar distribution and amount of LP. Following BAR-PSER129, residual antibody–ABC complexes were stripped from the tissue. Cross-links were then reversed, and proteins were extracted at high temperatures. Extracted biotin-labeled proteins were captured using magnetic streptavidin beads. (A) Western blot analysis of captured proteins (CAP) and flow-through from bead purification (FT). Blots were then probed with ABC to detect biotinylated proteins in the samples (ABC). Blots were also probed with two antibodies against alpha-synuclein (SYN1 and MJFR1). Blots were also probed with anti-pser129 alpha-synuclein antibody (EP1536Y) and the anti-ubiquitin conjugate antibody (UBCJ2). The molecular weight marker has been denoted for each blot. (B) Blot depicting proteins captured for each experimental condition used for mass spectrometry. Three BAR conditions were conducted for each brain, including total alpha-synuclein capture (BAR-SYN1), phosphorylated alpha-synuclein capture (BAR-PSER129), and a no primary capture. Resulting captured proteins were resolved on SDS/PAGE and blotted, and biotin was detected using ABC. (C) Heat map depicting hierarchical clustering of Pearson correlation coefficients for all samples. (D) Volcano plot of BAR-SYN1 identified proteins. No proteins were found to be statistically significant between HC and SYN. (E) Volcano plot of BAR-PSER129–identified proteins; 261 proteins were found to be statistically significant (adj. P value < 0.05). Alpha-synuclein (SNCA) and HBB are denoted on each volcano plot.
Next, we analyzed the BAR-captured LP by mass spectrometry. To do this, the midbrain and striatum of three SYN (two PD and one DLB) and three HC were processed with the BAR protocol, and the captured proteins were digested and identified by mass spectrometry. For each sample, three capture conditions were conducted: 1) BAR-SYN1 (i.e., total alpha-synuclein), 2) BAR-PSER129 (i.e., LP), and 3) primary antibody omission control. Enrichment was confirmed prior to mass spectrometry by blotting and probing with ABC (Fig. 4B). Biotin reactivity was absent from the captured BAR-PSER129 fraction in HC and mostly absent from all samples when the primary antibody was omitted from the BAR protocol. A heat map shows hierarchical clustering of the Pearson correlation between all samples tested (Fig. 4C). BAR-SYN1 in SYN and HC tissue showed similar patterns of protein abundance. In contrast, the BAR-PSER129 abundance profile differed markedly between SYN and HC, with HC having low protein abundance. BAR-PSER129 of HC tissues resembled no primary controls in both SYN and HC tissues. Blocking of endogenous biotin prior to BAR-PSER129 in SYN or HC did not affect the overall distribution of BAR-PSER129 labeling and protein detection (SI Appendix, Fig. S5). Considered with the lack of labeling in primary only condition, these data suggest endogenous biotin was not driving BAR enrichment.
In total, we identified 757 proteins across all capture conditions and samples (Dataset S5). BAR-SYN1 showed enrichment of many proteins over background (i.e., primary omission control), including alpha-synuclein (16.88 log2(FC) ± 3.49), but abundance did not differ between SYN and HC (Fig. 4D). In contrast, BAR-PSER129 capture revealed 261 proteins significantly enriched for SYN when compared to HC (adjusted [adj.] P value < 0.05; Fig. 4E and Dataset S2). The most significantly enriched protein in synucleinopathy samples following BAR-PSER129 was alpha-synuclein (21 log2(FC) > HC), and several well-known PD associated proteins were also enriched, including UCHL1, MAPT, and DJ-1. A heat map of normalized protein abundance for BAR-PSER129 capture samples revealed distinct clusters for HC and SYN (SI Appendix, Fig. S4).
Pathways Analysis of BAR-PSER129–Identified Proteins.
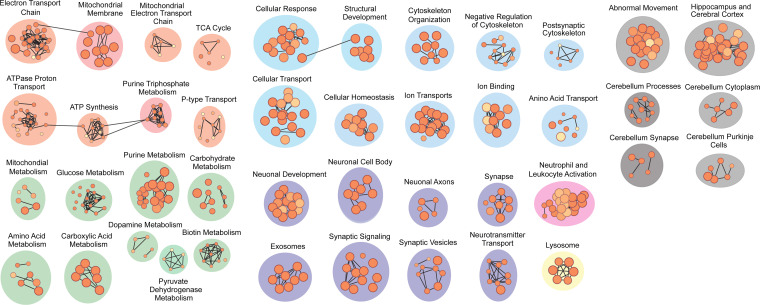
Next, we conducted pathway analysis on the 261 statistically significant BAR-PSER129 proteins. Results showed proteins identified were enriched for 718 annotated processes and pathways (Fig. 5; see Dataset S3 for complete output). Of particular interest, the most enriched KEGG pathway was PD (−log10 adj. P value = 14.75), and the most enriched for GO cellular compartment was vesicles (−log10 adj. P value = 29.73). Indeed, the top six enriched GO cellular compartments involved extracellular vesicles, including extracellular exosomes (−log10 adj. P value = 28.588). Several other neurodegenerative disorders appeared in the top 10 KEGG pathways, including prion disease (−log10 adj. P value = 10.13), Huntington disease (−log10 adj. P value = 10.48), Alzheimer’s disease (−log10 adj. P value = 8.3866), and amyotrophic lateral sclerosis (−log10 adj. P value = 7.16).
Fig. 5.
Pathway enrichment analysis of BAR-PSER129–identified proteins from synucleinopathy brain tissue. Pathway enrichment was performed using gProfiler2 on proteins identified from the BAR-PSER129–labeled PD and DLB brain tissues. Significantly enriched pathways (adj. P value < 0.05) clustered into nodes representing similar pathways were visualized using Cytoscape and Enrichmentmap. Annotations of these molecular nodes were defined using Autoannotate with manual curation.
Subsequent clustering of pathway enrichment analysis was used to provide a visual overview of cellular pathways and processes associated with the BAR-PSER129 captured LP. The clustering analysis allows major themes to be elucidated, as there is much overlap between annotated pathways (24). Results showed several apparent major clusters, including vesicle-mediated synaptic neurotransmission (Fig. 5, purple, pink, and yellow nodes). A single cluster of immune functionalities (Fig. 5, pink node) included neutrophil and leukocyte activation. Lysosomes were also a significant component of LP-associated vesicles. Mitochondrial functionality was a major component of LP, with various metabolic processes and the electron transport chain functions making up the bulk of the enrichment map (Fig. 5, green and orange nodes).
Validation of BAR-PSER129–identified Protein HBB.
Next, we performed several experiments to validate the BAR-PSER129–identified protein hemoglobin beta (HBB). HBB is hypothesized to have functions in neurons such as mitochondrial homeostasis, antioxidant functions, and maintenance of iron metabolism (25, 26). HBB is expressed in many neurons, including midbrain dopaminergic neurons, and HBB down-regulation has been implicated in the pathogenesis of PD (27). However, a physical association of HBB with LP is unclear, and thus we wanted to determine whether BAR-PSER129 was accurately identifying HBB as an LP interacting protein.
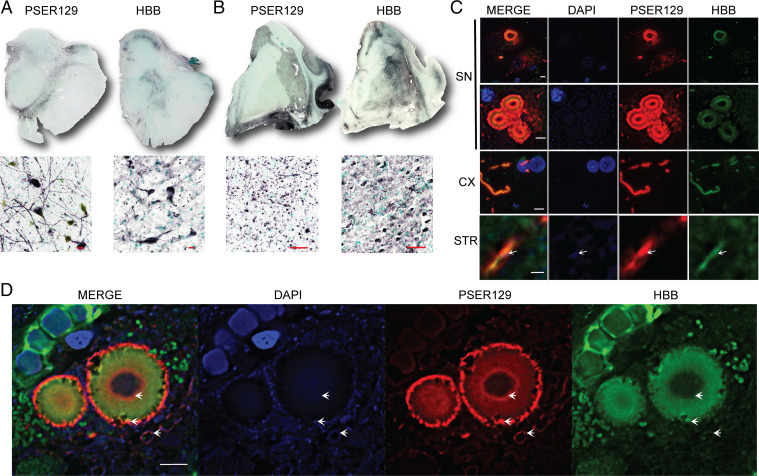
First, we looked at the tissue distribution of HBB in SYN brain tissue sections using a monoclonal antibody raised against HBB (Santa Cruz). Surprisingly, HBB was found in relatively high abundance throughout the brain sections tested. HBB was detected throughout the SN and striatal tissue sections (Fig. 6 A and B). In the striatal sections, intense staining was observed throughout the striatum. Strikingly, the overall tissue distribution of HBB closely resembled PSER129 distribution in the matched tissues. HBB distribution was more limited in the midbrain section, except for strong labeling in the SN. HBB labeling was likely not restricted to dopaminergic processes, as many cells morphologically resembling glia were heavily labeled throughout the midbrain and striatum.
Fig. 6.
BAR-PSER129–identified protein hemoglobin beta (HBB) interaction with LP. Experiments were performed to confirm BAR-PSER129 enrichment of LP. Formalin-fixed human brain sections were immunolabeled for PSER129 and HBB. Nickle DAB detection of immunolabeled PSER129 and HBB in both (A) the human SN and (B) the section containing the STR. (SN scale bars, 20 μm; STR scale bars, 50 μm.) Top of A and B depicts whole section scans obtained using a 4× objective. Bottom is a representative image obtained with a 20× objective. PSER129 and HBB were labeled with fluorescent tyramines and imaged using confocal microscopy. (C) Single z-plane images acquired with a 60× objective showing labeling in the SN, STR, and adjacent cortical regions (CX). SN images depict labeling of a cell containing a single Lewy body, granular-type pathology, and multiple Lewy bodies. (SN scale bars, 5 μm; CX and STR scale bars, 1 μm.) White arrows depict DAPI signal within the PSER129/HBB-labeled Lewy neurite. (D) Single z-plane confocal image showing distribution of HBB and PSER129 in the Lewy body. White arrows highlight DAPI signal in proximity and within the Lewy body (n = 2, case 3 and case 4). Scale bar, 5 μm.
Next, we wanted to determine to what extent PSER129 and HBB colocalized in the tissue. To do this, we double labeled both PSER129 and HBB. Results show that, although HBB can be found throughout the brain tissue, it was detected near and within LP (Fig. 6C). HBB was detected in neurons bearing a single inclusion, granular-type pathology, or even multiple inclusions (Fig. 6C). Although granular PSER129 pathology strongly associated (i.e., adjacent to) with HBB-labeled structures, we observed no instance of overlap between the two signals. In contrast, HBB and PSER129 signals colocalized well in both circular inclusions and Lewy neurites. We also stained double-stranded DNA using the dye DAPI, which has been reported to label the peripheral tubular structures and the core of Lewy bodies (28), but the cellular origin (i.e., nuclear or mitochondrial) of the labeled DNA is unclear. HBB structures adjacent to and at the perimeter of PSER129-positive Lewy bodies weakly labeled for DAPI (Fig. 6D), consistent with HBBs’ localization to mitochondria (29). As observed previously here, and by others (21), the core of many Lewy bodies also weakly labeled for DAPI.
Discussion
LP consists of alpha-synuclein along with numerous other molecules (6, 17, 28). Determining the molecular makeup of LP is critical for understanding synucleinopathies. Here we established BAR-PSER129 as a powerful approach to map LP and LP interactions in the human synucleinopathy brain. There are several apparent advantages of the BAR technique. First, this protocol allows the molecular characterization of LP in primary human disease tissues, which could lead to a deeper understanding of how models of LP represent or do not represent human diseases. Second, this protocol requires a single tissue section and typical laboratory reagents, making the routine processing of many samples feasible. The last major advantage of the BAR technique is that it provides information about the core pathological process for synucleinopathies, while other approaches, for example, bulk tissue RNA sequencing or proteomics, largely measure ancillary cellular processes.
Pathway enrichment analysis was particularly useful for analyzing data produced from BAR-PSER129. The most significant KEGG pathway was “Parkinson’s Disease,” with 12% of identified proteins from the KEGG pathway “Parkinson’s Disease.” Intriguingly, exosomes were the most significantly enriched for GO cellular compartment, and vesicles/extracellular vesicles were also top hits, providing good evidence that LP is found in exosomes in the human diseased brain. This finding has important implications for the hypothesized prion-like spread of LP, as the mechanism of intercellular spread remains unclear, and our data suggest LP may be primarily spreading via exosomes (29). Superpathways, including mitochondrial oxidative phosphorylation, metabolism, and vesicle trafficking, appeared to be the major component of LP, which agrees with the observation of mitochondria and vesicles observed within LB (17). Other common themes in synucleinopathies, such as dopamine metabolism and lysosome, seem to be a minor component of LP structures. Cytoskeletal organization and arrangement were another prominent component to LP, which may be evidence of a structurally regulated cellular process proposed by others (21) as opposed to autonomous alpha-synuclein aggregation. Together, the data support a mode of LP formation where organelles such as mitochondria, vesicles, and lysosomes are in a dynamic process (6). BAR-PSER129–identified proteins significantly overlap with endogenous alpha-synuclein interacting proteins identified in primary rat cortical neurons (30) and disease-implicated proteins identified by differential proteomics of SN tissues (31) (SI Appendix, Fig. S7), suggesting both normal alpha-synuclein interactions and disease interactions are involved with LP.
We confirmed the BAR-PSER129–identified protein HBB localized to LP. HBB is abundant in erythrocytes but is also expressed in many other cell types (32), including neurons of the SN (33). In neurons, HBB localizes to mitochondria, and several studies suggest HBB may play a role in PD (34, 35). HBB may play important roles in oxygen homeostasis, iron utilization, and oxidative phosphorylation, all of which have central roles in synucleinopathy. Several observations made here support the interpretation that HBB within LP was of mitochondrial origin, including 1) mitochondria was the prominent component identified by enrichment analysis and 2) HBB colocalized with DAPI. Previous studies show mitochondria within and surrounding Lewy bodies as well as the DAPI signal at the perimeter and at the core of Lewy bodies (17, 36). Interestingly, we found that PSER129 and HBB were often absent from the core of Lewy bodies, but DAPI remained, suggesting residual mitochondrial DNA or nuclear DNA (36) was being trafficked to the core of LP. Our validation of HBB supports the hypothesis that mitochondria are sequestered into LP (6), but we cannot conclude that HBB has any functional role or significance for LP.
In addition to PSER129, we conducted BAR labeling for total alpha-synuclein using the antibody SYN1. For BAR-SYN1, although many proteins were highly enriched over background (i.e., primary negative condition), no proteins survived multiple corrections when comparing HC and SYN (Dataset S1). This suggests the total alpha-synuclein interactions cannot be used to differentiate disease, and endogenous alpha-synuclein interactions likely dominate the signal for BAR-SYN1, which reduced statistical power, possibly preventing the identification of disease interactions. BAR-SYN1 data provided here are likely a good estimate of endogenous alpha-synuclein interactions in the human brain (Dataset S1). Alpha-synuclein was the 24th most significantly enriched protein for BAR-SYN1 and the most significantly enriched protein for BAR-PSER129, which may result from differences in local target (i.e., alpha-synuclein) concentration between the two BAR captures (e.g., endogenous alpha-synuclein is spread out, while LP alpha-synuclein is concentrated within inclusions). Future studies might differentiate disease from healthy tissues using BAR-SYN1 by increasing the number of cases and thus the statistical power.
Several limitations of the current study include the relatively small sample size for mass spectrometry. Also, data describe LP interactions, irrespective of cellular compartment, cell type, or brain structure in which the LP is found. Furthermore, we do not attempt to differentiate different clinically distinct synucleinopathies (i.e., DLB and PD), which may be a particularly powerful future application of this technique. Lastly, BAR is a powerful technique to determine interactions (i.e., proximity), but it does not determine functionality, and, therefore, further investigation is required to understand how each identified LP interactor fits into the context of disease (37). The pathological process is dynamic, and the data presented here are conceptually a molecular “snapshot”; applying BAR to synucleinopathy models will enable an understanding of how the interactions develop as a function of time.
In conclusion, BAR-PSER129 can be used for sensitive detection and enrichment of LP interactors from formalin-fixed brain tissues. Conceptually, we have provided a framework to explore the molecular details of LP in primary human tissues, and this could instigate a systems-based approach for understanding the molecular architecture of LP (38). Although we explored BAR labeling using two antibodies, this assay could easily be adopted to other high-affinity antibodies, targeting other insoluble pathological structures of interest (e.g., tau tangles or amyloid plaques). Future studies using this technique could explore crucial questions about the molecular nature of LP in the primary disease tissues that could deepen our understanding of synucleinopathies.
Materials and Methods
Brain Tissue Preparation.
Brains were processed as previously described (39). Briefly, 2-cm coronal slabs were immersion fixed in phosphate-buffered 4% paraformaldehyde for 5 d at 4 °C. Slabs were gradually equilibrated in cryoprotectant solution (phosphate-buffered saline [PBS], 2% dimethyl sulfoxide, and 20% glycerol), and 40-μm frozen sections were collected on a sliding knife microtome. Sections were stored in cryoprotectant solution at −20 °C until processed. Tissue sections were in storage for variable times before processing. All tissues utilized for mass spectrometry had a postmortem interval (PMI) of 12 h or less. See Table 1 for details about specimens selected for immunostaining and BAR.
Table 1.
Disease and demographic data for tissue used in study
| Case ID | Diagnosis | Age (yrs.) | Gender | PMI (h) | Disease duration (yrs.) | UPDRS | H&Y | MMSE | Assay |
| 1 | DLB | 87 | M | Unknown | 10 | 31 | 3 | Unknown | IHC |
| 2 | DLB | 94 | F | 9 | Unknown | Unknown | Unknown | 25 | IHC |
| 3 | DLB | 79 | M | 3 | 7 | 49 | 4 | Unknown | IHC, MS |
| 4 | PD | 83 | M | 5 | 7 | 54 | 4 | Unknown | IHC, MS |
| 5 | PD | 72 | M | 10 | Unknown | 70 | 5 | Unknown | IHC |
| 6 | PD | 81 | M | 5 | 13 | 45.5 | 4 | Unknown | IHC, MS |
| 7 | NA | 85 | M | 2.25 | NA | NA | NA | NA | IHC, MS |
| 8 | NA | 78 | F | 2.83 | NA | NA | NA | NA | IHC, MS |
| 9 | NA | 80 | F | 2 | NA | NA | NA | NA | IHC, MS |
UPDRS, Unified Parkinson’s Disease Rating Scale; H&Y, Hoehn and Yahr scale; MMSE, Mini-Mental State Examination; M, male; Unknown, data not available; IHC, sample used for immunohistochemistry; F, female; MS, sample used for mass spectrometry analysis; NA, data point not applicable.
BAR Labeling of Formalin-Fixed Floating Sections.
Floating sections were rinsed two times for 10 min with PBS, postfixed in formalin for 30 min at room temperature, and washed an additional two times with PBS. To quench endogenous peroxidase, sections were incubated for 30 min in PBS containing 3% H2O2 (Sigma). Following quenching, sections were briefly rinsed in PBS and then incubated in primary blocking buffer (1× Tris buffered saline [TBS] pH 7.6, 0.5% Triton X-100, 2% bovine serum albumin, 2% normal goat serum, 0.05% sodium azide) for 1 h at room temperature. Primary antibodies were diluted as follows in primary blocking buffer: anti-PSER129 antibody EP1536Y (Abcam) was diluted 1:50,000, alpha-synuclein antibody SYN1 (BD Biosciences) was diluted 1:10,000, anti-Lewy body antibody LB509 (ThermoFisher) was diluted 1:5,000, and anti-HBB antibody 37-8 (Santa Cruz) was diluted 1:10,000. Sections were then incubated with the primary antibody solution overnight (≥16 h) at 4 °C. For preabsorption studies, we incubated 1µL of EP1536Y and 20 µL of synthetic PSER129 peptide (Abcam) in 1 mL of blocking buffer overnight at 4 °C. The preabsorbed mixture was then added to 50 mL of primary blocking buffer and incubated with tissue sections overnight at 4 °C. A primary negative control was conducted for each case, which consisted of omitting the primary antibody from the protocol. For proteinase K digestion, prior to the fixation step, sections were incubated in digestion buffer (1× TBS pH 7.6, 0.5% Triton X-100, 20 µg of proteinase K per mL) for 10 min at 37 °C, then postfixed for 10 min in formalin, and immunostaining protocol commenced. For mass spectrometry to determine possible endogenous biotin interference, additional BAR-PSER129 (n = 2) was conducted following endogenous biotin block using a kit (Vector Laboratories) according to the manufacturer’s protocol. Briefly, we incubated sections in PBS containing excess Avidin (four drops of Avidin solution per 1 mL of PBS) for 30 min followed by the primary antibody solution containing excess biotin (four drops of Biotin solution per 1 mL of primary antibody solution).
The next day, sections were washed six times for 10 min each time with wash buffer (1× TBS, 0.05% Triton X-100) and incubated with biotinylated antibody (Vector Laboratories) diluted 1:200 in secondary blocking buffer (1× TBS pH 7.6, 0.05% Triton X-100, 2% bovine serum albumin) that contained 2% of the appropriate serum for the secondary antibody used. Sections were then washed three times for 10 min each time with wash buffer and incubated with ABC detection reagent (Vector Laboratories) for 1 h at room temperature. Sections were then washed two times for 10 min each time with wash buffer followed by wash three times for 10 min each time in borate buffer (0.1 M sodium borate pH 8.5). To begin the BAR labeling reaction, tissues were incubated with 100 mL of borate buffer containing 20 µL of stock biotinyl tyramide (Sigma, 5 mg/mL dissolved in dimethyl sulfoxide [DMSO]) or 5 µM fluorescent tyramide conjugates (CF568 and CF640, 10 mM stock dissolved in DMSO; Biotium) and 10 µL of 30% H202 (Sigma-Aldrich) for 30 min at room temperature. Sections were then washed 2 × 10 min with TBS and then three times for 10 min each time with wash buffer. Once BAR labeled, tissue sections can be further processed or stored in wash buffer at 4 °C.
Detection of Labeled Molecules and Quantification.
Once biotin-labeled sections were washed, they could then be probed with ABC reagent as before to detect biotinylation of the tissue. Sections were developed using nickel-enhanced 3,3′-iaminobenzidine (DAB). Following DAB development, sections were counterstained with methyl green, dehydrated in ethanol, cleared with xylene, and coverslipped with Cystoseal 60 mounting medium (ThermoFisher Scientific) for viewing under the microscope. Fluorescent-labeled samples sections were counterstained with the nucleic acid binding dye DAPI (Sigma-Aldrich), mounted onto glass slides, and coverslipped with Fluoromount (Sigma-Aldrich). Both bright-field and confocal images were acquired on the Nikon A1R inverted confocal microscope. Deconvolution of confocal images was conducted using Nikon elements software.
Three images from each region of interest (i.e., SN, striatum, and CX) were acquired from case tissues (n = 6) labeled using either the BAR-PSER129 or the ABC method. Image quantification was performed using a random forest machine learning pixel classifier using ilastik (v.1.3.2), trained on representative images. The random forest pixel classifier was used to predict the probability of all pixels representing positive stain. A custom CellProfiler (v.3.1.5) pipeline was then used to quantify pixels representing positive stain (90% confidence score). The significant difference of positive pixel proportions per image per group was performed using a repeated measures ANOVA in Prism (v7). Results of the analysis were plotted as percent area with PSER129 signal. Colocalization analysis was performed by collecting three single-plane images per labeling condition, per case, from three regions of interest. Pearson’s correlation coefficient was then calculated using Nikon elements software (Nikon).
Capture of Biotinylated Proteins.
To remove biotin-labeled antibody from tissues, sections were washed for 1 h in antibody removal buffer (PBS, 2% SDS, 5% 2-mercapitoethanol) similar to what has been previously described (40). Sections were then rinsed with PBS 3 × 10 min. Sections were placed into 1.5-mL conical tube with 0.5 mL of cross-link reversal buffer [500 mM Tris⋅HCl pH 8.0, 150 mM NaCl, 5% SDS, 2 mM (ethylenedinitrilo)tetraacetic acid pH 8.0, and 2 mM phenylmethylsulfonyl fluoride] and heated to 98 °C for 30 min and 90 °C for 2 h on a heat block (Thermofisher). Samples were mixed well and centrifuged at 22,000 × g for 30 min at room temperature. The supernatant was diluted to 3 mL with TBST (1× TBS containing 1% Triton X-100) and used for the capture protocol.
Fifty microliters of streptavidin magnetic beads (Thermofisher) were washed according to manufacturer’s protocol and incubated with 3 mL of labeled tissue extract for 1 h at room temperature. Beads were isolated using a magnetic stand (Millipore) and washed 3 × 10 min with radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris⋅HCl pH 8.0, 150 mM NaCl, 0.1% SDS, 1% Triton X-100) and then overnight with TBST. The lysate was retained following the pulldown for later characterization. Capture proteins were eluted by heating (98 °C) the beads in 100 µL of SDS polyacrylamide gel electrophoresis (SDS/PAGE) sample buffer (ThermoFisher) for 10 min. Beads were collected using the magnetic stand, and the eluent was retained for Western blot. For mass spectrometry, beads were washed additionally 3 × 10 min with TBS to remove Triton X-100. Beads were collected and stored at −80 °C until digestion for mass spectrometry.
Western Blot.
Five microliters of lysate and 25 µL of bead eluent were resolved on 4 to 20% gradient Bis-Tris gels (ThermoFisher) and blotted onto activated polyvinylidene difluoride using wet transfer with settings at 120 V for 1 h. Following transfer, membranes were rinsed with PBS and then fixed with 4% paraformaldehyde as previously described (41). Membranes were then dried completely and reactivated with methanol just prior to use. Membranes were blocked in TBST containing 5% bovine serum albumin for 2 h at room temperature. Membranes were then incubated overnight at 4 °C with blocking buffer containing one of the following antibodies: 1) anti–alpha-synuclein “SYN1” (BD Lifesciences) and 2) “MJFR1” (Abcam), 3) anti-ubiquitin conjugates “UBCJ2” (Enzo Life Sciences), and 4) anti-PSER129 “EP1536Y” (Abcam). To determine enrichment of biotinylated proteins, one membrane was incubated with prepared ABC reagent, diluted 1:10 in blocking buffer for 1 h at room temperature (Vector Laboratories). The next day, membranes were washed three times for 10 min each time with TBST and incubated for 1 h with either anti-rabbit (1:5,000) or anti-mouse (1:10,000) HRP-conjugated secondary antibodies (Cell Signaling Technologies) diluted in blocking buffer. Membranes were then washed three times for 10 min each time with TBST and detected using chemiluminescence substrate (Bio-Rad).
Mass Spectrometry.
For each case tested, BAR labeling was conducted on a total of two pooled sections, one containing the putamen and one containing the midbrain. In total, we conducted BAR on brain sections from two individuals diagnosed with PD, one diagnosed with DLB, and three individuals that do not have LP (see Table 1 for summary). For every case, we conducted BAR on two antigens, phosphorylated alpha-synuclein and total alpha-synuclein, using two separate primary antibodies, EP1536Y and SYN1, respectively. For each case, we conducted a mock capture, where we omitted the primary antibody from the protocol, which was used as a measure of assay background for the respective case.
Streptavidin beads containing the captured biotinylated proteins were washed in 100 mM Tris⋅HCl pH 7.8. and then resuspended in 100 µL of 100 mM Tris pH7.8 containing 6 M urea. These were then reduced with dithiothreitol, alkylated with iodoacetamide, brought up to 1 mL with 100 mM Tris⋅HCl pH 7.8, and digested with trypsin at 37 °C overnight. This solution was subjected to solid-phase extraction to concentrate the peptides and remove unwanted reagents, followed by injection onto a Waters NanoAcquity HPLC equipped with a self-packed Aeris 3.6-µm C18 analytical column 0.075 mm by 20 cm (Phenomenex). Peptides were eluted using standard reverse-phase gradients. The effluent from the column was analyzed using a Thermo Orbitrap Elite mass spectrometer (nanospray configuration) operated in a data-dependent manner for 54 min. The resulting fragmentation spectra were correlated against the Refseq entries for homosapien using PEAKS Studio 10.5 (Bioinformatic Solutions). Proteins that had at least one unique peptide identified were retained for analysis, and several assay contaminant proteins (i.e., streptavidin, bovine serum albumin, and trypsin) were removed from data prior to analysis.
Data Analysis.
Mass spectrometry analysis was performed in R (v4.0.3) using the package DEP (v1.12.0). In brief, any protein not expressed in all samples in any group was removed, protein expression was normalized using the default vsn method, and missing data were imputed using the quantile regression imputation of left-censored data (QRILC) missing not at random method. Differential expression was performed using a linear regression model with empirical Bayes and Benjamini-Hochberg multiple testing corrected. Pathway analysis was performed using gprofiler2 (v0.2.0). Cytoscape (v.3.1.7), with EnrichmentMap (v.3.1.0) and AutoAnnotate (v.1.3) modules, was used to cluster and visualize the enriched pathways. Heat maps of Pearson correlation coefficients were generated using the R package complexheatmap. Power analysis was not conducted prior to designing experimental groups, and researchers were not blinded to experimental conditions. Mass spectrometry data were uploaded to Proteomics Identifications Database (PRIDE) archive (Project accession: PXD028830 and Project DOI: 10.6019/PXD028830) and are publicly available.
Supplementary Material
Acknowledgments
We thank Eric Spooner at the Whitehead Institute Proteomics Facility for conducting liquid chromatography–mass spectrometry. Brain tissue specimens were provided by generous donations made through the Rush Movement Disorders Brain Bank. Funding was provided by National Institute of Neurological Disorders and Stroke Grant 5R21NS109871-02.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114405119/-/DCSupplemental.
Data Availability
Mass spectrometry data have been deposited in PRIDE (Project accession: PXD028830; Project DOI: 10.6019/PXD028830).
References
- 1.Fujita K. A., et al. , Integrating pathways of Parkinson’s disease in a molecular interaction map. Mol. Neurobiol. 49, 88–102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kellie J. F., et al. , Quantitative measurement of intact alpha-synuclein proteoforms from post-mortem control and Parkinson’s disease brain tissue by intact protein mass spectrometry. Sci. Rep. 4, 5797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Machiya Y., et al. , Phosphorylated alpha-synuclein at Ser-129 is targeted to the proteasome pathway in a ubiquitin-independent manner. J. Biol. Chem. 285, 40732–40744 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker D. G., et al. , Arizona Parkinson Disease Consortium, Changes in properties of serine 129 phosphorylated α-synuclein with progression of Lewy-type histopathology in human brains. Exp. Neurol. 240, 190–204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lue L. F., et al. , Biochemical increase in phosphorylated alpha-synuclein precedes histopathology of Lewy-type synucleinopathies. Brain Pathol. 22, 745–756 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahul-Mellier A. L., et al. , The process of Lewy body formation, rather than simply α-synuclein fibrillization, is one of the major drivers of neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 117, 4971–4982 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delic V., et al. , Sensitivity and specificity of phospho-Ser129 α-synuclein monoclonal antibodies. J. Comp. Neurol. 526, 1978–1990 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia Q., et al. , Proteomic identification of novel proteins associated with Lewy bodies. Front. Biosci. 13, 3850–3856 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leverenz J. B., et al. , Proteomic identification of novel proteins in cortical Lewy bodies. Brain Pathol. 17, 139–145 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bar D. Z., et al. , Biotinylation by antibody recognition—A method for proximity labeling. Nat. Methods 15, 127–133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotani N., et al. , Biochemical visualization of cell surface molecular clustering in living cells. Proc. Natl. Acad. Sci. U.S.A. 105, 7405–7409 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashimoto N., et al. , Proteomic analysis of ganglioside-associated membrane molecules: Substantial basis for molecular clustering. Proteomics 12, 3154–3163 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Li X. W., et al. , New insights into the DT40 B cell receptor cluster using a proteomic proximity labeling assay. J. Biol. Chem. 289, 14434–14447 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toda Y., et al. , Application of tyramide signal amplification system to immunohistochemistry: A potent method to localize antigens that are not detectable by ordinary method. Pathol. Int. 49, 479–483 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Adams J. C., Biotin amplification of biotin and horseradish peroxidase signals in histochemical stains. J. Histochem. Cytochem. 40, 1457–1463 (1992). [DOI] [PubMed] [Google Scholar]
- 16.Krabichler Q., et al. , The centrifugal visual system of a palaeognathous bird, the Chilean Tinamou (Nothoprocta perdicaria). J. Comp. Neurol. 525, 2514–2534 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Shahmoradian S. H., et al. , Lewy pathology in Parkinson’s disease consists of crowded organelles and lipid membranes. Nat. Neurosci. 22, 1099–1109 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Baba M., et al. , Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am. J. Pathol. 152, 879–884 (1998). [PMC free article] [PubMed] [Google Scholar]
- 19.Fares M. B., Jagannath S., Lashuel H. A., Reverse engineering Lewy bodies: How far have we come and how far can we go? Nat. Rev. Neurosci. 22, 111–131 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Fusco G., et al. , Direct observation of the three regions in α-synuclein that determine its membrane-bound behaviour. Nat. Commun. 5, 3827 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moors T. E., et al. , Detailed structural orchestration of Lewy pathology in Parkinson’s disease as revealed by 3D multicolor STED microscopy. bioRxiv [Preprint] (2018). 10.1101/470476. Accessed 25 November 2020. [DOI]
- 22.Kawashima Y., Kodera Y., Singh A., Matsumoto M., Matsumoto H., Efficient extraction of proteins from formalin-fixed paraffin-embedded tissues requires higher concentration of tris(hydroxymethyl)aminomethane. Clin. Proteomics 11, 4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tofaris G. K., Razzaq A., Ghetti B., Lilley K. S., Spillantini M. G., Ubiquitination of alpha-synuclein in Lewy bodies is a pathological event not associated with impairment of proteasome function. J. Biol. Chem. 278, 44405–44411 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Reimand J., et al. , Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 14, 482–517 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shephard F., Greville-Heygate O., Marsh O., Anderson S., Chakrabarti L., A mitochondrial location for haemoglobins—Dynamic distribution in ageing and Parkinson’s disease. Mitochondrion 14, 64–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown N., et al. , Neuronal hemoglobin expression and its relevance to multiple sclerosis neuropathology. J. Mol. Neurosci. 59, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrer I., et al. , Neuronal hemoglobin is reduced in Alzheimer’s disease, argyrophilic grain disease, Parkinson’s disease, and dementia with Lewy bodies. J. Alzheimers Dis. 23, 537–550 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Lashuel H. A., Do Lewy bodies contain alpha-synuclein fibrils? And does it matter? A brief history and critical analysis of recent reports. Neurobiol. Dis. 141, 104876 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Alvarez-Erviti L., et al. , Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol. Dis. 42, 360–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung C. Y., et al. , In situ peroxidase labeling and mass-spectrometry connects alpha-synuclein directly to endocytic trafficking and mRNA metabolism in neurons. Cell Syst. 4, 242–250.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petyuk V. A., et al. , Proteomic profiling of the substantia nigra to identify determinants of Lewy body pathology and dopaminergic neuronal loss. J. Proteome Res. 20, 2266–2282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richter F., Meurers B. H., Zhu C., Medvedeva V. P., Chesselet M. F., Neurons express hemoglobin alpha- and beta-chains in rat and human brains. J. Comp. Neurol. 515, 538–547 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biagioli M., et al. , Unexpected expression of α- and β-globin in mesencephalic dopaminergic neurons and glial cells. Proc. Natl. Acad. Sci. U.S.A. 106, 15454–15459 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shephard F., Greville-Heygate O., Liddell S., Emes R., Chakrabarti L., Analysis of mitochondrial haemoglobin in Parkinson’s disease brain. Mitochondrion 29, 45–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santiago J. A., Potashkin J. A., Blood transcriptomic meta-analysis identifies dysregulation of hemoglobin and iron metabolism in Parkinson’ disease. Front. Aging Neurosci. 9, 73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Power J. H., Barnes O. L., Chegini F., Lewy bodies and the mechanisms of neuronal cell death in Parkinson’s disease and dementia with Lewy bodies. Brain Pathol. 27, 3–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lashuel H. A., Novello S., Lewy body-associated proteins: Victims, instigators, or innocent bystanders? The case of AIMP2 and alpha-synuclein. Neurobiol. Dis. 156, 105417 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Rayaprolu S., et al. , Systems-based proteomics to resolve the biology of Alzheimer's disease beyond amyloid and tau. Neuropsychopharmacology 46, 98–115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu Y., et al. , Nurr1 in Parkinson’s disease and related disorders. J. Comp. Neurol. 494, 495–514 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolognesi M. M., et al. , Multiplex staining by sequential immunostaining and antibody removal on routine tissue sections. J. Histochem. Cytochem. 65, 431–444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee B. R., Kamitani T., Improved immunodetection of endogenous α-synuclein. PLoS One 6, e23939 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data have been deposited in PRIDE (Project accession: PXD028830; Project DOI: 10.6019/PXD028830).