Abstract
The reemergence of gentamicin-susceptible (Gens) methicillin-resistant Staphylococcus aureus (MRSA) isolates in France between 1992 and 1996 was investigated using a phylogenetic approach (multiprimer randomly amplified polymorphic DNA typing). Eighty-six percent (65 of 85) of the French strains were grouped into one phylogenetic cluster within which all but one Gens strain were grouped into a subcluster. Thus, the reemergence of Gens MRSA strains in France was likely due to the spread of one specific clone which belonged to a cluster comprising most French gentamicin-resistant (Genr) strains. This suggests that the Gens clone has emerged from a Genr strain of this cluster.
In France, the proportion of methicillin-resistant Staphylococcus aureus (MRSA) among S. aureus strains, ranging from 30 to 40%, is one of the highest in Europe (13). In addition, the resistance of MRSA to multiple aminoglycosides, including resistance to gentamicin (Genr), was over 90% in the beginning of the 1990s. Since 1992, several hospitals have observed the reemergence of strains which were susceptible to gentamicin (Gens) (1, 10, 11; L. Gazagne, P. Guedet, and E. Lecaillon, Abstr. 5th Int. Conf. Prev. Infect., abstr. MID/AMR 02, 1998; P. Guedet, L. Gazagne, E. Lecaillon, A. Le Coustumier, R. Bismuth, and “Le College de Bacteriologie, Virologie et Hygiene des Hopitaux de France,” Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. E-122, 1997). These strains were also characterized by the unexpected reappearance of heterogeneous resistance to oxacillin and variable resistance to other antibiotics. It was speculated that changes in the use of antibiotics in French hospitals, especially the decreased consumption of aminoglycosides, could be responsible for the emergence and spread of Gens MRSA strains.
Unlike the situation in Belgium, where the emergence of Gens strains was due to a new MRSA clone (5, 6), it was speculated that French Gens strains emerged from Genr MRSA by means of the loss of the aac6′-aph2" gene, which confers resistance to all aminoglycosides, with conservation of the ant4′ gene conferring resistance to kanamycin, tobramycin, and amikacin (10). Since the aac6′-aph2" gene is frequently carried by transposon Tn4001 (4, 8), it is likely that certain Gens MRSA strains could have emerged from Genr MRSA strains by means of transposon excision or deletion. This hypothesis is supported by the fact that (i) both Genr and Gens strains have closely related pulsed-field gel electrophoresis (PFGE) types, which suggests a recent common ancestor, and (ii) in vitro long-term storage of Genr MRSA at room temperature may result in spontaneously occuring Gens MRSA derivatives (10).
Whereas one report using molecular typing data (PFGE) indicated that one major clone type was responsible for most Gens isolates (1), other reports identified two to three major types (7, 10, 11). As PFGE typing of MRSA is a highly discriminatory method, markers might evolve very rapidly and the evaluation of older genetic relationships is therefore difficult.
The aim of the present study was to further investigate this issue by a population genetics analysis of both Genr and Gens strains isolated in France during the last decade in order to (i) determine if Gens strains in France were derived from a common or different ancestor(s) and (ii) confirm that this ancestor(s) was phylogenetically related or not related to Genr strains. We used a typing method, multiprimer randomly amplified polymorphic DNA (RAPD), which was validated to produce information on the phylogenetic relationships between isolates (2).
Seventy-five French MRSA strains were randomly selected from three sources: (i) 11 Genr and 11 Gens strains of MRSA from a study which included 895 isolates collected from 96 nonuniversity French hospitals in 1995 (Guedet et al., 37th ICAAC), (ii) 9 Genr and 13 Gens strains of MRSA isolated in 1997 to 1998 from the hospital of Perpignan, France (Gazagne et al., Abstr. 5th Int. Conf. Prev. Infect.), and (iii) 12 Genr and 8 Gens strains isolated in 1996 and 11 Genr strains isolated in 1990 from the National Reference Center for Staphylococcal Toxemia in Lyon, France. In addition, 24 strains isolated from other European countries were selected from a previous study and are representative of the diversity of MRSA observed (2).
Multiprimer RAPD typing consists of using the RAPD technique with 10 consecutively selected primers, as already described (2). In brief, DNA was extracted, purified, and quantified before its use for the PCR. Twenty nanograms of template DNA was used in each PCR. The following primers were used separately: A-03 (AGTCAGCCAC), A-13 (CAGCACCCAC), A-17 (GACCGCTTGT), B-05 (TGCGCCCTTC), B-15 (GGAGGGTGTT), F-04 (GGTGATCAGG), N-10 (ACAACTGGGG), N-15 (CAGCGACTGT), R-07 (ACTGGCCTGA), R-20 (ACGGCAAGGA), U-08 (GGCGAAGGTT), and U-19 (GTCAGTGCGG). The amplified products were separated by electrophoresis in agarose gel and stained with ethidium bromide. The Jaccard's distance was used to estimate the genetic difference between each pair of isolates. The unweighted pair-group method with arithmetic averages was used as a clustering method.
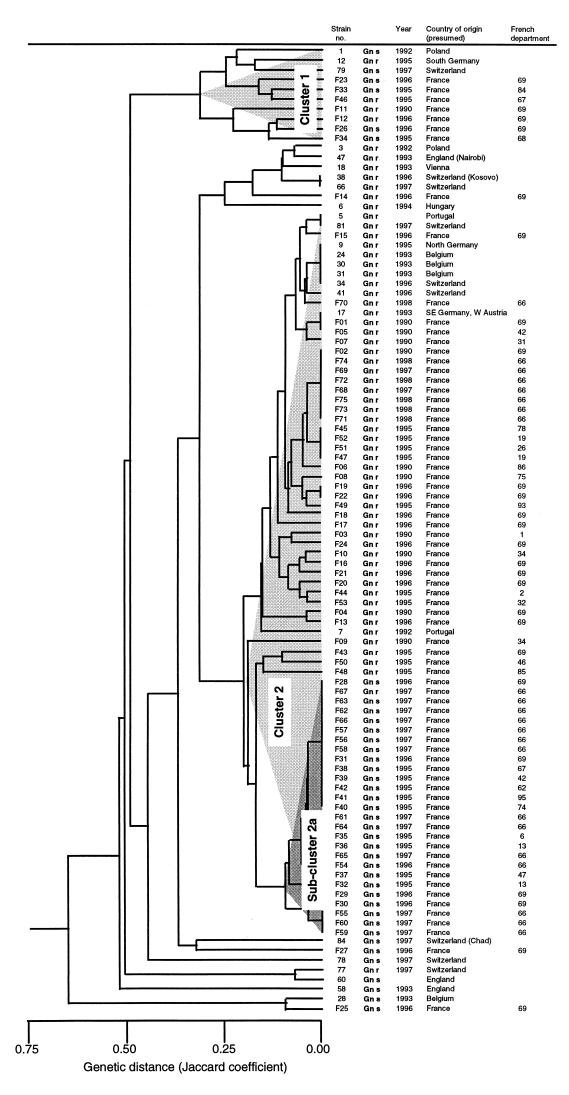
RAPD results from the 75 French MRSA strains and the 24 strains from other European countries with the 10 primers were analyzed, and the phylogenetic relatedness between these strains is shown in the dendrogram of Fig. 1. Most of the French strains were distributed into two clusters of 65 and 7 strains, respectively. Both clusters comprised Gens and Genr strains. The smaller cluster, cluster 1, included three Genr and four Gens strains as well as some other European Genr and Gens strains. The larger cluster, cluster 2, included epidemic clones of Genr MRSA from all over Europe, as well as 39 Genr and 26 Gens strains from France. However, within this cluster, a subcluster showing a relative genetic homogeneity compared to other strains of the cluster could be clearly identified (Fig. 1). This subcluster, labeled 2a, includes all but one French Gens strain. Among the 27 French hospitals from which MRSA of this study was recovered, strains of cluster 2 Genr and Gens were found in 25 different hospitals distributed all over the territory of France; Genr strains were found in 16 hospitals, and Gens strains were found in 10 hospitals. Other Gens strains were found in only three hospitals.
FIG. 1.
Dendrogram (derived using the unweighted pair-group method with arithmetic averages) based on the Jaccard's distance matrix of RAPD results from 100 MRSA strains (75 from France and 25 from other European countries). The susceptibilities to gentamicin (Gens and Genr are represented as Gn s and Gn r) and the year, country, and department of France in which each MRSA strain was isolated are indicated.
Results of multiprimer RAPD typing showed that most of the French MRSA belonged to the same phylogenetic cluster, which comprised both Genr and Gens strains. The fact that the Gens strains were closely related suggests that their emergence in France was mainly due to the spread of one specific clone. Moreover, the fact that this Gens clone belongs to a phylogenetic cluster originally comprising only Genr strains strongly supports the hypothesis that it emerged from a Genr strain, a situation which is different from the one reported in Belgium, where Gens strains are due to the emergence of a new clone represented by strain 28 shown in Fig. 1 (5, 6).
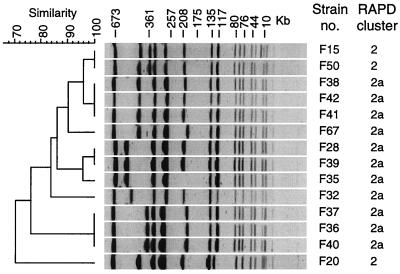
PFGE typing has been performed on 11 strains of cluster 2a (1 strain per French institution) and 3 Genr strains of cluster 2 (Fig. 2). A one-band difference was observed between some Gens and Genr strains (e.g., strains F38 and F50), confirming previously published results (10). However, the close genetic relationship between all strains of cluster 2a, as demonstrated by RAPD, was more difficult to assess with PFGE, since up to seven-band differences were observed between some strains (e.g., F32 and F37) (Fig. 2). For strains belonging to cluster 2, according to RAPD, up to a nine-band difference was observed with PFGE (between strains F20 and F40) (Fig. 2). If standard interpretation criteria for PFGE (12) were used, these strains would be considered unrelated. These results suggest that the markers of PFGE probably evolved more rapidly than those of multiprimer RAPD and thus are less suitable for evaluation of older genetic relationships.
FIG. 2.
PFGE typing of Gens and Genr strains from RAPD clusters 2 and 2a. The Dice coefficient was used to calculate the similarity between each pair of strains.
The decrease in aminoglycoside use in French hospitals was suggested as the cause of the emergence and spread of Gens MRSA strains (1, 7). However, there are three pieces of evidence that this reduction might not be the only factor: (i) the diminution of aminoglycoside use would favor an increase of other Gens strains (e.g., strains of cluster 1) instead of only one clone, (ii) this decrease in use has not been reported in all hospitals, and (iii) it does not explain the increase in susceptibility to other antibiotics (10). The rapid spread of this clone does not seem to be attributable to poor infection control in French hospitals because, in most of them, policies have been reinforced during the last decade. It is likelier that this clone has acquired a fitness advantage during its evolution. In vitro experiments showed that the growth rate of Gens isolates of this clone was higher than that of the Genr strains (9), a finding consistent with a fitness advantage of Gens isolates. In addition, this new Gens clone was characterized by the reappearance of heterogeneous resistance to oxacillin, a characteristic similar to that found in the epidemic Belgium clone 2, which was also characterized by rapid spreading (3). Thus, the emergence and rapid spread of this new Gens clone support the hypothesis that epidemic clones emerge after the acquisition through mutations of some fitness advantage over the other clones. The loss of this fitness advantage during the microevolution of the clone would predict its fading.
In conclusion, the reemergence of Gens MRSA strains in France is likeliest due to the spread of one epidemic clone. This clone belongs to the same phylogenetic cluster that includes most Genr MRSA strains recovered in France, which adds to the evidence that it is derived from a Genr strain.
Acknowledgments
We are grateful to Michel Bernard for technical assistance.
This work was supported by a grant from the Swiss National Foundation for Research (no. 32-56035.98).
REFERENCES
- 1.Aubry-Damon H, Legrand P, Brun-Buisson C, Astier A, Soussy C J, Leclercq R. Reemergence of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus: roles of an infection control program and changes in aminoglycoside use. Clin Infect Dis. 1997;25:647–653. doi: 10.1086/513749. [DOI] [PubMed] [Google Scholar]
- 2.Blanc D S, Banuls A L, Hauser P M, Moreillon P, Francioli P, Tibayrenc M the Swiss MRSA group. Methicillin resistant Staphylococcus aureus: phylogenetic relatedness between European epidemic clones and Swiss sporadic strains. Microb Drug Resist. 2000;6:231–238. doi: 10.1089/mdr.2000.6.231. [DOI] [PubMed] [Google Scholar]
- 3.Blanc D S, Petignat C, Moreillon P, Entenza J, Eisenring M C, Kleiber H, Wenger A, Troillet N, Blanc C H, Francioli P. Unusual spread of a penicillin-susceptible methicillin-resistant Staphylococcus aureus clone in a geographic area of low incidence. Clin Infect Dis. 1999;29:1512–1518. doi: 10.1086/313522. [DOI] [PubMed] [Google Scholar]
- 4.Byrne M E, Gillespie M T, Skurray R A. Molecular analysis of a gentamicin resistance transposonlike element on plasmids isolated from North American Staphylococcus aureus strains. Antimicrob Agents Chemother. 1990;34:2106–2113. doi: 10.1128/aac.34.11.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deplano A, Witte W, van Leeuwen W J, Brun Y, Struelens M J. Clonal dissemination of epidemic methicillin-resistant Staphylococcus aureus in Belgium and neighboring countries. Clin Microbiol Infect. 2000;6:239–245. doi: 10.1046/j.1469-0691.2000.00064.x. [DOI] [PubMed] [Google Scholar]
- 6.De Ryck R, Deplano A, Nonhoff C, Jans B, Suetens C, Struelens M J. Molecular epidemiology and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus (MRSA) in Belgian hospitals: 1992–97. Abstr Clin Microbiol Infect. 1999;5(Suppl. 3):117–118. [Google Scholar]
- 7.Galdbart J O, Morvan A, El Solh N. Phenotypic and molecular typing of nosocomial methicillin-resistant Staphylococcus aureus strains susceptible to gentamicin isolated in France from 1995 to 1997. J Clin Microbiol. 2000;38:185–190. doi: 10.1128/jcm.38.1.185-190.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillespie M T, Lyon B R, Messerotti L J, Skurray R A. Chromosome- and plasmid-mediated gentamicin resistance in Staphylococcus aureus encoded by Tn4001. J Med Microbiol. 1987;24:139–144. doi: 10.1099/00222615-24-2-139. [DOI] [PubMed] [Google Scholar]
- 9.Laurent F, Lelièvre H, Cornu M, Vandenesch F, Carret G, Etienne J, Flandrois J P. Fitness and competitive growth advantage of new gentamicin-susceptible MRSA clones spreading in French hospitals. J Antimicrob Chemother. 2001;47:277–283. doi: 10.1093/jac/47.3.277. [DOI] [PubMed] [Google Scholar]
- 10.Lelièvre H, Lina G, Jones M E, Olive C, Forey F, Roussel-Delvallez M, Nicolas-Chanoine M-H, Bébéar C M, Jarlier V, Andremont A, Vandenesch F, Etienne J. Emergence and spread in French hospitals of methicillin-resistant Staphylococcus aureus with increasing susceptibility to gentamicin and other antibiotics. J Clin Microbiol. 1999;37:3452–3457. doi: 10.1128/jcm.37.11.3452-3457.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemaître N, Sougakoff W, Masmoudi A, Fievet M H, Bismuth R, Jarlier V. Characterization of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus involved in nosocomial spread. J Clin Microbiol. 1998;36:81–85. doi: 10.1128/jcm.36.1.81-85.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voss A, Milatovic D, Wallrauch-Schwarz C, Rosdahl V T, Braveny I. Methicillin-resistant Staphylococcus aureus in Europe. Eur J Clin Microbiol Infect Dis. 1994;13:50–55. doi: 10.1007/BF02026127. [DOI] [PubMed] [Google Scholar]